Division Planes Alternate in Spherical Cells of Escherichia coli (original) (raw)
Abstract
In the spherical cells of Escherichia coli rodA mutants, division is initiated at a single point, from which a furrow extends progressively around the cell. Using “giant” rodA ftsA cells, we confirmed that each new division furrow is initiated at the midpoint of the previous division plane and runs perpendicular to it.
Normal Escherichia coli cells are rods that double in length and then divide across the shorter axis to give two equal cells. If division is prevented, cells grow to form elongated, multichromosomal cylinders (filaments). When filaments divide, a septum forms between each pair of segregated chromosomes to produce cells of normal length, each with a single chromosome, and the septal planes are parallel to one another along the length of the long rod (4, 8).
The cylindrical shape of E. coli cells depends on the presence of two membrane proteins, RodA and PBP 2 (15, 16, 20, 21). If either is inactivated, E. coli cells become spherical. If the physical separation of round sister cells is prevented by immobilizing the cells on agar, they give the appearance of dividing in alternating planes at 90° to one another (9, 13). Recently it has been suggested that this appearance might be an artifact due to cells rotating in place on the agar after each division (7). In order to test this explanation, we have made very large spherical cells that divide rapidly into smaller progeny cells without moving or bending. We show here that these “giant” coccal E. coli cells do indeed divide along planes alternating at right angles to one another and that the third division is probably in the same plane as the first.
E. coli K-12 strain KJB24 was constructed by cotransducing a rodA(Am) mutation (5) and a linked Tn_5_ into the suppressor-free (Sup0) strain W3110. Kanamycin-resistant transductants were screened individually for the spherical cell phenotype. These transductants showed extensive lysis in rich medium but were stable in minimal medium. KJB24 is a spontaneous variant that grows well in both media (the reason for the increased stability is now known and will be reported elsewhere). In order to produce even larger cells, a temperature-sensitive division mutant, KJB28 [leu::Tn_10 ftsA.12_(Ts) rodA(Am) Sup0], was constructed by transducing KJB24 with P1 prepared on strain TKF12 (leu::Tn_10 ftsA.12_) and selecting tetracycline-resistant, temperature-sensitive transductants.
The division furrow is initiated at a single point and grows as an arc around the cell.
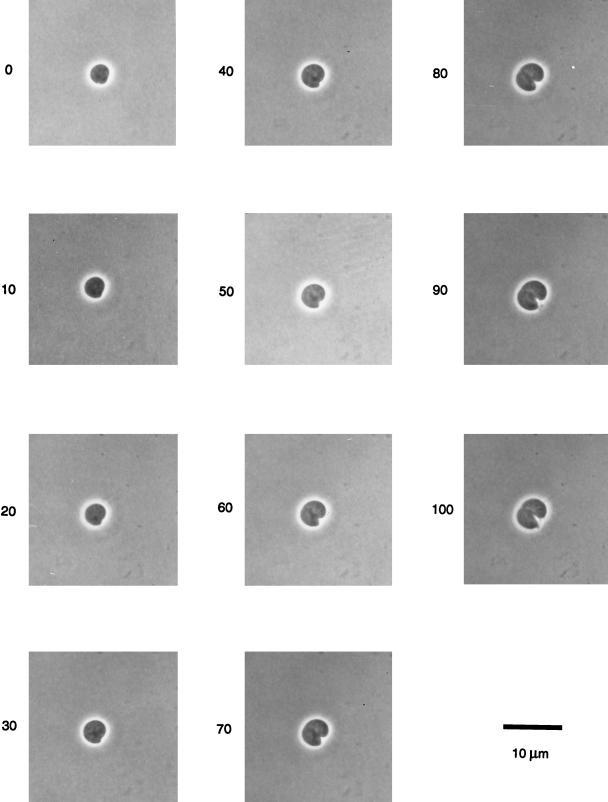
Previous observations (2, 9, 13) have suggested that the division of spherical cells is asymmetrical, beginning on one side of the cell only. Since this fact is important to our argument, we have shown this for a KJB28 cell, grown for 90 min without division at 42°C before being placed on a nutrient broth (NB) agar surface at 30°C (Fig. 1). This cell had a volume (∼27 μm3) equal to that of about 20 average rod-shaped (W3110) or 6 coccal (KJB24) cells growing in this medium. Division begins as a localized deformation of the cell surface at a single point, from which a furrow extends and deepens until the two ends meet. The two surfaces of the division furrow are separate from the outset and the newly formed divided surfaces appear to be almost flat, so that two hemispherical sister cells of equal size are produced.
FIG. 1.
Single cell of strain KJB28 [rodA(Am) ftsA.12(Ts) Sup0] dividing on NB agar at 30°C, following 90 min of growth at 42°C without division. Photographs were taken at 10-min intervals as indicated by numbers next to the frames.
[Note: KJB28 cells do not form partial constrictions at 42°C, unlike mutants carrying _ftsA_(Ts) and _rodA_(Ts) alleles (3). This difference appears to be because rodA(Ts) cells are rods at 30°C and retain their poles for several generations at 42°C, whereas rodA(Am) cells never have poles (unpublished observations)].
Cell division takes place in alternating planes in rodA cells.
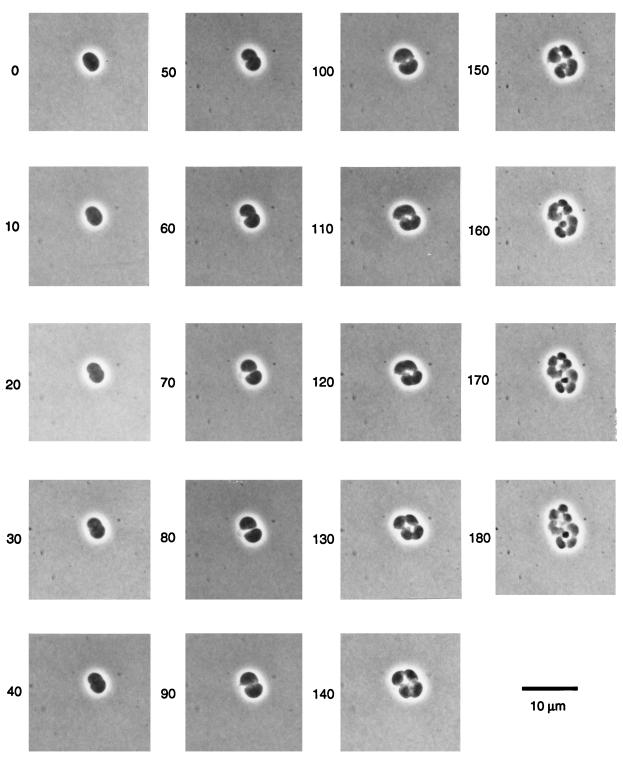
Figure 2 shows successive divisions of a giant KJB28 cell (volume, ∼20 μm3 at time zero) on NB agar at the permissive temperature. The formation of an almost-flat plane on the newly divided side of each sister cell makes it clear that the cells do not bend or rotate during division. The figure also clearly shows that new division furrows start in the middle of the previous division plane and run at 90° to that plane. This pattern was seen in every KJB28 cell examined. Each cell also elongates slightly between divisions, at 90° to the direction of elongation of its mother cell. Similar alternation of the axis of cell elongation of each division has been reported previously for cells of Neisseria gonorrhoeae (24).
FIG. 2.
Successive divisions of a KJB28 cell on NB agar at 30°C, following 90 min of growth at 42°C without division. Photographs were taken at 10-min intervals as indicated by numbers next to the frames.
The joint activities of the RodA and PBP 2 proteins somehow ensure that a cell has a cylindrical shape, grows by elongation, divides after each replication of its single chromosome, and has division planes that run at right angles to its long axis. Therefore, these may properly be considered to be cell cycle proteins because, in their absence, cells switch to a “default” cycle in which cells are spherical, large, and multichromosomal; in which divisions take place by asymmetric growth of division furrows; and in which division planes alternate by 90°. Like “natural” cocci (6, 10–14, 17–19, 22), rodA mutants are large, multichromosomal cells but rapidly segregate mutations into pure clones (1). We think that such obligate homozygosity is likely to stem from a fixed spatial relationship between chromosomes and division planes in the cell (9).
Figure 2 also shows that the third division of a spherical cell (observable at 150 min) starts in the center of the second division plane, i.e., parallel to the plane of the first division. Divisions therefore occur in only two dimensions, as in, for example, Lampropedia and Micrococcus, rather than in three dimensions, as in Sarcina. It is interesting to speculate about what distinguishes these two different patterns of division, and whether yet another morphogene in E. coli restricts the division of rodA cells to two dimensions.
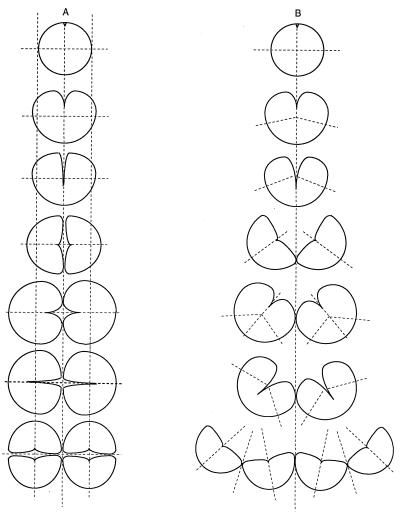
A recent paper has challenged the conclusion that KJB24 cells divide in alternating planes (7). KJB24 cells grown in a viscous solution of Methocel formed chains and irregular clumps of cells, from which it was concluded that division planes had in fact been parallel to one another, as they are in rod-shaped cells. The irregular clumps were dismissed as being due to the movement of cells in situ after division, so that their presumed poles came to face in irregular directions. While we agree that rotation of cells in the viscous liquid may indeed have taken place, we could argue that this might well have caused the formation of the chains of cells, even though divisions actually took place in alternating planes. Figure 3B shows the way in which we think that chains of cells form in Methocel (compared to on agar). As has been shown conclusively (2, 3, 9; also this paper), division furrows in spherical E. coli cells begin at a single point and proceed around the cell in an ever-deepening arc. If the sister cells are free to move during this process then the deepening furrow opens out the pair of cells so that they rotate through 90° until they are attached only at the point where the furrow has completed its traverse (as we see when cells are observed dividing in liquid medium). Initiation of the next set of furrows in each daughter cell therefore takes place approximately parallel to the first division plane, as shown in Fig. 3B. (If the second set of furrows had actually been across the same cell axis, then the cells would not have formed a chain, but rather a sort of daisy.) We think that by ignoring the fact that division furrows are asymmetric in spherical cells, it was not realized that pairs of dividing cells will open out and rotate about their point of attachment, unless they are immobilized on agar.
FIG. 3.
Proposed explanation for the appearance of chains of KJB24 cells after growth in Methocel. (A) On NB agar, cells are prevented from moving as they divide so that alternating planes of cell division give rise to tetrads. (B) In Methocel, cells are held less firmly in place so that as the division furrow progresses from one side, the cells fall apart. New division furrows are initiated in the middle of the completed division planes and appear to be parallel or nearly parallel to the first division plane. The process repeats to give a kinked chain of cells (or, sometimes, an irregular clump).
Our results show that in spherical cells, the division furrow begins from a single point on the cell surface, and it has been shown that this is also the case for FtsZ assembly (2). One of the most interesting questions arising from this work is, therefore, that of what provides the focus for FtsZ assembly at this single site on the cell membrane. A clue to the answer may be given by the observation that the point of origin of each new furrow is the midpoint of the plane of the previous division: perhaps some elements of the previous division machinery remain at this point and provide a nucleus for the assembly of the next division arc. This idea will be the subject of a separate communication.
Finally, it is interesting to compare the behavior of rodA E. coli cells with that of other kinds of spherical-celled bacteria. There are both gram-positive and gram-negative coccal species, e.g., species of the gram-positive genera Streptococcus, Staphylococcus, Micrococcus, Lampropedia, and Sarcina and species of the gram-negative genus Neisseria. Not all of these show the same pattern of division. Streptococcus spp. form long chains of small spherical cells, i.e., successive division planes are parallel to one another as in rod-shaped bacteria, whereas the other cocci divide successively in different planes. Staphylococcus and Sarcina divide in three planes which alternate by 90°, forming cubical packets of eight cells. (The Staphylococcus cells are not attached to one another except by single junctions and therefore do not remain together in an orderly way [23], but the Sarcina cells form very regular packets.) Lampropedia cells divide successively in only two planes, producing flat sheets of regularly arranged cells. Micrococcus radiodurans characteristically forms only pairs of cells that separate before their next division, but the planes of the nascent septa can be seen in stained preparations (17). The first two planes are at right angles (forming a tetrad) but the third set of septa form at an intermediate angle. Our observations indicate that successive septal orientations in giant rodA ftsA cells (Fig. 2) most closely resemble those of Lampropedia.
Acknowledgments
This work was supported by a Programme Grant from the Medical Research Council (United Kingdom).
REFERENCES
- 1.Addinall S G, Begg K J, Donachie W D. Is partitioning of chromosomes in E. coli random (or worse)? J Cell Biochem. 1993;17E:286. [Google Scholar]
- 2.Addinall S G, Lutkenhaus J. FtsZ spirals and arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22:231–237. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- 3.Begg K J, Donachie W D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985;163:615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begg K J, Donachie W D. Experiments on chromosome separation and positioning in Escherichia coli. New Biol. 1991;3:475–486. [PubMed] [Google Scholar]
- 5.Begg K J, Takasuga A, Edwards D H, Dewar S J, Spratt B G, Adachi H, Ohta T, Matsuzawa H, Donachie W D. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J Bacteriol. 1990;172:6697–6703. doi: 10.1128/jb.172.12.6697-6703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Contreras A, Casadesús J. Tn10 mutagenesis in Azotobacter vinelandii. Mol Gen Genet. 1987;209:276–282. doi: 10.1007/BF00329654. [DOI] [PubMed] [Google Scholar]
- 7.Cooper S. Division pattern of a round mutant of Escherichia coli. J Bacteriol. 1997;179:5582–5584. doi: 10.1128/jb.179.17.5582-5584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donachie W D. The cell cycle of Escherichia coli. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 9.Donachie W D, Addinall S, Begg K J. Cell shape and chromosome partition in prokaryotes or, why E. coli is rod-shaped and haploid. Bioessays. 1995;17:1–9. doi: 10.1002/bies.950170616. [DOI] [PubMed] [Google Scholar]
- 10.Donachie W D, Begg K J. Cell length, nucleoid separation, and cell division of rod-shaped and spherical cells of Escherichia coli. J Bacteriol. 1989;171:4633–4639. doi: 10.1128/jb.171.9.4633-4639.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driedger A A. The ordered growth pattern of microcolonies of Micrococcus radiodurans: first generation sectoring of induced lethal mutations. Can J Microbiol. 1970;16:1133–1135. doi: 10.1139/m70-191. [DOI] [PubMed] [Google Scholar]
- 12.Hansen M T. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J Bacteriol. 1978;134:71–75. doi: 10.1128/jb.134.1.71-75.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwaya M, Goldman R, Tipper D J, Feingold B, Strominger J L. Morphology of an Escherichia coli mutant with a temperature-dependent round cell shape. J Bacteriol. 1978;136:1143–1158. doi: 10.1128/jb.136.3.1143-1158.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maldonado R, Garzón A, Dean D R, Casadesús J. Gene dosage analysis in Azotobacter vinelandii. Genetics. 1992;132:869–878. doi: 10.1093/genetics/132.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuhashi S, Kamiryo T, Blumberg P M, Linnett P, Willoughby E, Strominger J L. Mechanism of action and development of resistance to a new amidino penicillin. J Bacteriol. 1974;117:578–587. doi: 10.1128/jb.117.2.578-587.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuzawa H, Hayakawa K, Sato T, Imahori K. Characterization and genetic analysis of a mutant of Escherichia coli K-12 with rounded morphology. J Bacteriol. 1973;115:436–442. doi: 10.1128/jb.115.1.436-442.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray R G E, Hall M, Thompson B G. Cell division in Deinococcus radiodurans and a method for displaying septa. Can J Microbiol. 1983;29:1412–1423. doi: 10.1139/m83-217. [DOI] [PubMed] [Google Scholar]
- 18.Phadnis S H, Dimri G P, Das H K. Segregation characteristics of multiple chromosomes of Azotobacter vinelandii. J Genet. 1988;67:37–42. [Google Scholar]
- 19.Punita S, Jafri, Reddy M A, Das H K. Multiple chromosomes of Azotobacter vinelandii. J Bacteriol. 1989;171:3133–3138. doi: 10.1128/jb.171.6.3133-3138.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spratt B G. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977;131:293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamaki S, Matsuzawa H, Matsuhashi M. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J Bacteriol. 1980;141:52–57. doi: 10.1128/jb.141.1.52-57.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tirgari S, Moseley B E B. Transformation in Micrococcus radiodurans: measurement of various parameters in evidence for multiple, independently segregating genomes per cell. J Gen Microbiol. 1980;119:287–296. [Google Scholar]
- 23.Tzagoloff H, Novick R. Geometry of cell division in Staphylococcus aureus. J Bacteriol. 1977;129:343–350. doi: 10.1128/jb.129.1.343-350.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westling-Häggström B, Elmros T, Normark S, Winblad B. Growth pattern and cell division in Neisseria gonorrhoeae. J Bacteriol. 1977;129:333–342. doi: 10.1128/jb.129.1.333-342.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]