Inhibition of Human Immunodeficiency Virus Rev and Human T-Cell Leukemia Virus Rex Function, but Not Mason-Pfizer Monkey Virus Constitutive Transport Element Activity, by a Mutant Human Nucleoporin Targeted to Crm1 (original) (raw)
Abstract
The hypothesis that the cellular protein Crm1 mediates human immunodeficiency virus type 1 (HIV-1) Rev-dependent nuclear export posits that Crm1 can directly interact both with the Rev nuclear export signal (NES) and with cellular nucleoporins. Here, we demonstrate that Crm1 is indeed able to interact with active but not defective forms of the HIV-1 Rev NES and of NESs found in other retroviral nuclear export factors. In addition, we demonstrate that Crm1 can bind the Rev NES when Rev is assembled onto the Rev response element RNA target and that Crm1, like Rev, is a nucleocytoplasmic shuttle protein. Crm1 also specifically binds the Rev NES in vitro, although this latter interaction is detectable only in the presence of added Ran · GTP. Overexpression of a truncated, defective form of the nucleoporin Nup214/CAN, termed ΔCAN, that retains Crm1 binding ability resulted in the effective inhibition of HIV-1 Rev or human T-cell leukemia virus Rex-dependent gene expression. In contrast, ΔCAN had no significant affect on Mason-Pfizer monkey virus constitutive transport element (MPMV CTE)-dependent nuclear RNA export or on the expression of RNAs dependent on the cellular mRNA export pathway. As a result, ΔCAN specifically blocked late, but not early, HIV-1 gene expression in HIV-1-infected cells. These data strongly validate Crm1 as a cellular cofactor for HIV-1 Rev and demonstrate that the MPMV CTE nuclear RNA export pathway uses a distinct, Crm1-independent mechanism. In addition, these data identify a novel and highly potent inhibitor of leucine-rich NES-dependent nuclear export.
Retroviral replication requires the expression of both fully spliced and incompletely spliced forms of the initial, genome-length proviral transcript (7, 25). However, eukaryotic cells encode factors that inhibit the nuclear export of incompletely spliced (i.e., largely immature) mRNAs of both cellular and retroviral origin. To avoid this problem, retroviruses have evolved the ability to access nuclear RNA export pathways that are, at least in part, distinct from the nuclear export pathway used by fully spliced mRNAs (7, 25).
The best-understood retroviral RNA export mechanism is the one utilized by human immunodeficiency virus type 1 (HIV-1) and many other complex retroviruses, including human T-cell leukemia virus type 1 (HTLV-1) (7, 25). HIV-1 encodes a regulatory protein, termed Rev, that induces the specific nuclear export of incompletely spliced HIV-1 mRNA species (12, 14, 30). Rev function is therefore essential for the expression of proteins encoded by these late viral mRNAs, including the Gag structural protein. In contrast, Rev function is entirely dispensable for expression of the fully spliced, early HIV-1 mRNAs that encode Rev as well as the regulatory proteins Tat and Nef. Rev contains an RNA binding motif that directly interacts with an RNA stem-loop structure, termed the Rev response element (RRE), located in all incompletely spliced HIV-1 transcripts (10, 30, 32, 52). Rev also contains a leucine-rich nuclear export signal (NES) that allows the resultant RNA-protein complex to access a nuclear export pathway distinct from that used by fully spliced mRNAs of either HIV-1 or cellular origin (13, 28, 31, 50).
In contrast to complex retroviruses such as HIV-1 and HTLV-1, simple retroviruses such as Mason-Pfizer monkey virus (MPMV) and avian leukemia virus do not encode a nuclear export factor equivalent to Rev or Rex (5, 37, 46). Nonetheless, these retroviruses face the same difficulty of having to express both fully spliced and incompletely spliced forms of one primary transcript. Both MPMV and avian leukemia virus have been shown to contain _cis_-acting RNA stem-loop structures, termed constitutive transport elements (CTEs), that mediate the nuclear export of their genomic RNAs (5, 37). Recent data strongly suggest that MPMV CTE-mediated nuclear RNA export is dependent on a cellular CTE binding protein, termed Tap, that is the human homolog of the yeast nuclear RNA export factor Mex67 (24). Interestingly, although the CTE and RRE/Rev can functionally substitute for each other in mediating nuclear RNA export, they appear to act via different pathways. Thus, in Xenopus oocytes, overexpression of the Rev NES inhibits Rev function and also 5S rRNA and U snRNA export but fails to inhibit CTE or cellular mRNA export. Conversely, overexpression of the MPMV CTE inhibits both CTE RNA and cellular mRNA export but fails to inhibit Rev function or 5S rRNA or U snRNA export (39, 41).
Initial efforts to define the nuclear export pathway utilized by HIV-1 Rev led to the demonstration that the Rev NES could interact specifically with a cellular nucleoporin-like protein termed hRIP/Rab in both yeast and mammalian two-hybrid assays (4, 21, 45). The hRIP/Rab protein proved able to interact with Rev when Rev was bound to the RRE and also specifically bound NESs found in HTLV-1 Rex and in other, distinct Rev proteins, such as visna maedi virus (VMV) Rev. Significantly, overexpression of hRIP/Rab was observed to modestly enhance Rev function when Rev expression was limiting or when a Rev protein bearing an attenuated NES was used (4, 21). Subsequent experiments demonstrated, however, that the Rev NES could also specifically interact with several other human nucleoporins, including Nup98, Nup153, and Nup214/CAN, thus demonstrating that the Rev NES-hRIP/Rab interaction was not unique (20, 44). More disconcerting were reports that the Rev NES-hRIP/Rab interaction, while readily detectable by the two-hybrid assay, could not be observed in vitro in assays using recombinant proteins (44). This latter finding raised the possibility that this interaction was instead mediated by an evolutionarily highly conserved factor expressed in both yeast and human cells. Several recent reports suggest that this factor is the cellular protein Crm1 (15, 22, 35, 38, 43). Crm1 is related in sequence to known protein nuclear import factors, such as importin β, and is known to associate with cellular nucleoporins, including particularly Nup214/CAN (16). Evidence that Crm1 is a direct cellular cofactor for the Rev NES that also bridges the Rev-nucleoporin interaction includes the following: (i) the Rev NES can be shown to directly interact with human Crm1 (hCrm1) in vitro (15); (ii) leptomycin B (LMB), a drug believed to specifically inhibit Crm1 function (36), blocks Rev-dependent nuclear export (15, 22, 38, 51); and (iii) yeast Crm1 (yCrm1) is required for Rev function in yeast cells and is also required for detection of the Rev-nucleoporin interaction in the yeast two-hybrid assay (35, 43).
In this paper we have characterized the interaction between hCrm1 and NES sequences from HIV-1 Rev, HTLV-1 Rex, and VMV Rev in more detail and report evidence further validating hCrm1 as the cellular target of these retroviral regulatory proteins. We also demonstrate that a Nup214/CAN nucleoporin fragment that retains hCrm1 binding ability is a highly potent inhibitor of Rev-dependent, but not CTE-dependent, nuclear RNA export.
MATERIALS AND METHODS
Molecular clones.
The following expression plasmids have been previously described: the parental mammalian expression plasmid pBC12/CMV (8); plasmids expressing wild-type or mutant forms of HIV-1 Rev, VMV Rev, or HTLV-1 Rex fused to the VP16 activation domain (4); the pSLIIB/Cat indicator plasmid (47); plasmids expressing the wild-type or M10 or M5 mutant form of HIV-1 Rev (28) or wild-type VMV Rev or HTLV-1 Rex (3, 48); prokaryotic expression plasmids encoding glutathione _S_-transferase (GST) fusions to Rev or the Rev M10 mutant (4); the HIV-1 Rev indicator construct pDM128/CMV (26, 31), encoding the chloramphenicol acetyltransferase (cat) gene and the HIV-1 RRE located between two HIV-1-derived splice sites, and equivalent constructs containing the VMV RRE (pDM128/CMV/VRRE) (18), the HTLV-1 Rex response element (RXRE) (pDM128/RXRE) (23), or a polylinker in place of the RRE (pDM128/CMV/PL) (18); the mammalian two-hybrid indicator construct pG6(−31)HIVLTRΔTAR (42); the HIV-1 provirus expression plasmid pNL-Luc-E− (6); and pBC12/CMV-based expression plasmids encoding CD4, CCR-5, or β-galactosidase (β-Gal) (1, 18).
The GAL4-Crm1 expression plasmid, encoding the GAL4 DNA binding site (residues 1 to 147) linked to full-length hCrm1 (a gift of G. Grosveld), was prepared by PCR using primers that introduced a unique _Bgl_II site at the 5′ end and a unique _Xho_I at the 3′ end of the hCrm1 open reading frame. Sequencing of the Crm1 gene revealed a single difference (serine 793 to proline) in the deduced amino acid sequence compared to the published hCrm1 sequence (16). While this change could represent a PCR artifact, we note that this residue is a proline in both the Saccharomyces cerevisiae and Schizosaccharomyces pombe homologs of hCrm1 (16). The in vitro hCrm1 expression plasmid was generated by insertion of this same _Bgl_II/_Xho_I Crm1 DNA fragment into the expression plasmid pGEM3zf (Promega). The prokaryotic GST-Crm1 fusion protein expression plasmid was generated by insertion of hCrm1 into pGEX4T-1 (Pharmacia) digested with _Bam_HI and _Xho_I.
The pDM128/CTE indicator plasmid was generated by insertion of a PCR-generated MPMV DNA sequence containing the full-length CTE (a gift of E. Hunter) (5) into the polylinker _Bgl_II site present in pDM128/CMV/PL. A truncated form of Nup214/CAN (ΔCAN; residues 1864 to 2090 of human Nup214/CAN; a gift of G. Grosveld) was expressed by using pBC12/CMV and was initially isolated by PCR amplification using (i) a 5′ primer that introduced both a unique _Nco_I site and a consensus translation initiation codon and (ii) a 3′ primer that introduced a unique _Xho_I site. A pBC12/CMV-based plasmid expressing a Crm1-Tat fusion protein was generated by mutation of the Crm1 termination codon to a glycine codon, which also inserted a unique _Bgl_II site. The full-length HIV-1 tat gene was then PCR amplified from pcTat (28), using primers that introduced 5′ _Bgl_II and 3′ _Bam_HI sites, and cloned in frame into pBC12/CMV at the introduced Crm1 _Bgl_II site. The resultant plasmid encodes full-length hCrm1 fused to HIV-1 Tat separated by a three-amino-acid linker sequence (Gly-Asp-Leu).
Cell culture and transfection.
COS, HeLa, and 293T cells were maintained as previously described (1, 19, 28) and transfected by using DEAE-dextran, calcium phosphate, or Lipofectamine (9). Levels of DNA used in each transfection experiment are given in the relevant figure legend, with pBC12/CMV/βgal included as an internal control in all transfections. The mammalian two-hybrid assay has been described elsewhere (4), as have methods for quantifying levels of CAT, luciferase, and β-Gal expression in transfected cells (3, 40).
Protein microinjection.
A fusion protein consisting of GST linked to full-length hCrm1 was expressed in bacteria and purified in the absence of detergents as previously described (19). The GST-Crm1 protein was then concentrated to 2 mg/ml and supplemented with 1 mg of rhodamine isothiocyanate-conjugated rabbit immunoglobulin G (IgG). After microinjection of HeLa cells cultured on glass coverslips, the injected cells were incubated at 37°C for a further 30 min prior to fixation using 3% paraformaldehyde in phosphate-buffered saline (PBS). The subcellular localization of the injected protein was then determined by double-label immunofluorescence (19). The GST-Crm1 fusion protein was visualized with an anti-GST monoclonal antibody (Santa Cruz Biotechnology) followed by a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody (Cappel).
In vitro Rev binding.
A GST-Ran fusion protein, containing the Q69L mutation of Ran that inhibits hydrolysis of bound GTP (2), was expressed and purified on glutathione affinity resin as previously described (49). The Ran protein was then cleaved from GST by treatment with factor Xa protease and dialyzed against 100 mM NaCl–20 mM potassium phosphate (pH 7.0). Prior to use, the Ran protein was incubated for 30 min with 10 mM GTP in dialysis buffer supplemented with 1 mM magnesium acetate. Columns containing purified GST-Rev and GST-M10 cross-linked to an Affigel 10 matrix (Bio-Rad) were prepared as previously described (4). The Crm1 protein was expressed by in vitro translation in the presence of [35S]methionine and then diluted with binding buffer as previously described (4). Prior to loading onto the Rev or M10 affinity column, samples were incubated with either Ran · GTP (final concentration, 1 μM) or an equivalent volume of dialysis buffer, supplemented with 10 mM GTP and 1 mM magnesium acetate, for 20 min at 4°C. After extensive washing with binding buffer, bound proteins were eluted with 100 mM glycine (pH 2.8). The eluate was then diluted 1:10 with PBS, concentrated in a Centricon 10, and analyzed by sodium dodecyl sulfate (SDS-PAGE) and fluorography.
HIV-1 infection assay.
Stocks of the NL-Luc-E− HIV-1 provirus were prepared by Lipofectamine transfection of 293T cells (in 35-mm-diameter dishes) with 1 μg of pNL-Luc-E− and 1 μg of a pCMV5-based plasmid that expresses the JR-FL envelope (40). Simultaneously, other 293T cell cultures (in 35-mm-diameter dishes) were transfected with 250 μg of pBC12/CMV, 250 μg of pBC12/CCR-5, and either 500 μg of pcRev, 500 μg of pM10, 500 μg of pBC12/ΔCAN, 250 μg of pBC12/ΔCAN plus 250 μg of pBC12/CMV, or 500 μg of the pBC12/CMV parental plasmid. After 35 h of incubation at 37°C, the virus-containing supernatant media were harvested, pooled, and passed through a 0.2-μm-pore-size filter, and 1 ml of virus supernatant was overlaid on each target cell culture (40). After incubation at 37°C for 22 h, the virus supernatant was aspirated and each target culture was washed four times with medium prior to addition of 2 ml of fresh medium. Where appropriate, LMB was added at this time. After 24 h of further incubation, supernatants were harvested and analyzed for p24_gag_ expression levels by using a commercial enzyme-linked immunosorbent assay (ELISA) (Dupont). The cell monolayer was then washed with PBS and lysed with luciferase lysis buffer (Promega), and luciferase levels were determined with a luminometer (40).
RNA analysis.
Cytoplasmic RNA was isolated from 293T cells at ∼48 h after transfection as previously described (28), and equivalent samples were subjected to electrophoresis on a 1% formaldehyde–agarose gel. The RNA was then transferred to a Hybond-N filter (Amersham) and probed with a 32P-labeled random-primed probe directed against the predicted 3′ noncoding region shared by the spliced and unspliced transcripts encoded by pDM128/CMV. Hybridization, washes, and autoradiography were performed as previously described (28).
RESULTS
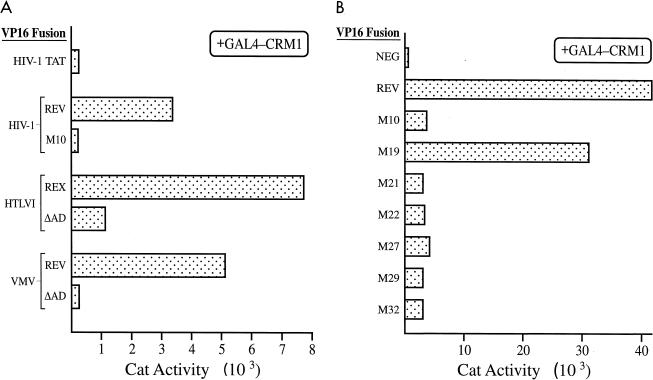
Previously, it has been demonstrated that hCrm1 can interact in vitro with the leucine-rich NESs found in HIV-1 Rev, in IκB, and in mitogen-activated protein kinase kinase but not with the M10 mutant of the Rev NES or with mutants of the IκB or mitogen-activated protein kinase kinase NES bearing alanines in place of leucine (15, 22, 38). It has also been demonstrated that yCrm1 can bind specifically to the protein kinase inhibitor α NES or the Rev NES, as shown by two-hybrid analysis of yeast cells (35, 43). To further confirm that hCrm1 binding indeed fully correlates with NES function, as also previously demonstrated for hRIP/Rab binding (3, 4, 17, 20, 21, 45), we examined the ability of hCrm1 to bind to wild-type and NES mutant forms of HIV-1 Rev, HTLV-I Rex, and VMV Rev in a mammalian two-hybrid assay. As shown in Fig. 1A, hCrm1 could strongly interact with the wild-type forms of all three of these proteins but failed to interact when the M10 missense mutation (28) was introduced into the HIV-1 Rev NES or the M4 mutation (48) was introduced into the VMV Rev NES. Introduction of a mutation into HTLV-1 Rex (Leu 90 and 92 to Ala) that blocks both Rex function and NES activity (3) resulted in an ∼85% reduction in the intensity of, but did not completely inhibit, the mammalian two-hybrid interaction with hCrm1. Similar results were observed with a second Rex NES mutant (residues 90 to 93 all to Gly) that also lacks detectable NES function (3, 11).
FIG. 1.
Specific interaction between hCrm1 and functional leucine-rich NESs detected in the mammalian two-hybrid assay. (A) The hCrm1 protein was expressed fused to the GAL4 DNA binding domain, while the indicated retroviral proteins were expressed as VP16 transcription activation domain (AD) fusions. COS cell cultures (in 100-mm-diameter dishes) were transfected by the DEAE-dextran procedure, using 1 μg of the pG6(−31)HIVLTRΔTAR reporter plasmid, 0.5 μg of the pGAL4-Crm1 expression plasmid, 1 μg of the relevant VP16 fusion protein expression plasmid, and 1 μg of the pBC12/CMV/βgal internal control plasmid. Transfected cells were harvested at ∼48 h posttransfection and analyzed for CAT and β-Gal expression levels as previously described (4, 18). (B) As for panel A except that COS cell cultures (on 35-mm-diameter dishes) were transfected by the calcium phosphate procedure, using 2 μg of GAL4-Crm1, 2 μg of each VP16 fusion expression plasmid, 0.5 μg of the pG6(−31)HIVLTRΔTAR indicator, and 0.1 μg of pBC12/CMV/βgal. The indicated mutants of the Rev NES (75-LPPLERLTLD-84) have been described elsewhere (31) and are as follows: M10, Leu 78 and Glu 79 to Asp and Leu, respectively; M19, Pro 77 to Asp; M21, Leu 81 and Thr 82 to Asp and Leu; M22, Leu 83 and Asp 84 to Asp and Leu; M27, Leu 78 to Ala; M29, Leu 83 to Ala; M32, Leu 78, Leu 81, and Leu 83 all to Ala. The only active Rev mutant is M19. These data are representative of three separate transfection experiments.
This laboratory has previously described (31) an extensive set of point missense mutations within the Rev NES, and it has been demonstrated, by both ourselves and others, that the activity of these NES mutants completely correlates with their ability to bind hRIP/Rab, as well as certain other nucleoporins such as Nup214/CAN, in the yeast two-hybrid assay (4, 20, 21, 44, 45). We examined whether the activity of these NES mutants also correlates with their ability to bind hCrm1, as would be predicted if hCrm1 were required to bridge the Rev-hRIP/Rab interaction; the data obtained demonstrate a complete correlation between Rev NES function and hCrm1 binding in the mammalian two-hybrid assay (Fig. 1B).
hCrm1 can bind HIV-1 Rev when Rev is assembled on the RRE.
An interesting finding that appeared to strongly support the relevance of hRIP/Rab for Rev function was the demonstration that hRIP/Rab could bind to Rev when Rev was assembled on the RRE in vivo (4). The assay used to demonstrate this result took advantage of the finding that Tat, the RNA sequence-dependent transcriptional transactivator of the HIV-1 long terminal repeat (LTR) promoter element, also functions when recruited to the HIV-1 LTR via a heterologous RNA target site, rather than by its normal transactivation response element (TAR) RNA target (47). In this earlier experiment, we substituted the TAR element with the RRE stem-loop IIB (SLIIB) RNA binding site for Rev and then demonstrated that expression of Rev together with a Tat-hRIP/Rab fusion protein, but not expression of either protein alone, resulted in recruitment of the Tat-hRIP/Rab fusion protein to the SLIIB RNA target and, hence, activation of the HIV-1 LTR promoter (4).
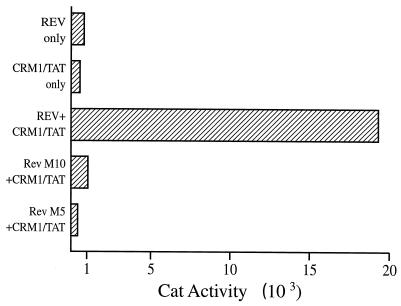
In Fig. 2, we demonstrate that an hCrm1-Tat fusion protein is also effectively recruited to the RRE RNA target by an intact Rev NES. Specifically, coexpression of both Rev and hCrm1-Tat results in a marked activation of transcription directed by an HIV-1 LTR linked to the SLIIB RNA target. Rev alone or hCrm1-Tat alone did not activate transcription, while inactivation of the Rev NES, in the Rev M10 mutant (28), or of the Rev RNA binding motif, in Rev M5 (28), entirely blocks this effect. We therefore conclude that Rev can effectively bind to hCrm1 when assembled onto the RRE, which is a clear prediction of the hypothesis that the hCrm1-Rev interaction is critical for late HIV-1 RNA export from the cell nucleus.
FIG. 2.
hCrm1 binds to Rev when Rev is bound to the RRE. The pSLIIB/CAT indicator construct contains an HIV-1 LTR in which the TAR RNA binding site for Tat has been substituted by the RRE SLIIB RNA binding site for Rev. When a Crm1-Tat fusion protein is coexpressed with Rev, the pSLIIB/CAT indicator is activated by recruitment of Tat to the SLIIB RNA target by the Crm1-Rev protein-protein interaction. This effect is not observed when either protein is expressed alone or if the Rev NES (M10) or the Rev RNA binding site (M5) is mutated. 293T cells (in 35-mm-diameter dishes) were transfected by the calcium phosphate procedure with 500 ng of pSLIIB/CAT, 1,000 ng of pcRev, pM10, or pM5, 500 ng of pBC12/Crm1/Tat, and 100 ng of pBC12/CMV/βgal. The parental pBC12/CMV plasmid was substituted where necessary as a negative control. The data are representative of four separate transfection experiments.
Rev binding to hCrm1 is dependent on Ran · GTP.
While three papers have reported a direct interaction between leucine-rich NESs and hCrm1 in vitro (15, 22, 38), there is a significant discrepancy with regard to the requirement for Ran · GTP. Specifically, while Fornerod et al. (15) have reported that the Crm1-NES interaction is dependent on the presence of Ran · GTP, and indeed involves the formation of an obligate NES-Crm1-Ran · GTP ternary complex, both Ossareh-Nazari et al. (38) and Fukuda et al. (22) have reported a readily detectable hCrm1-NES interaction in the absence of any added Ran · GTP. This issue is of critical importance in that the high-level expression of Ran · GTP in the eukaryotic nucleus, and its effective absence in the cytoplasm, has been proposed to underlie the directionality of nuclear transport pathways (27, 33).
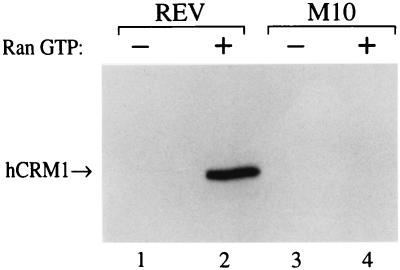
We examined whether hCrm1 protein, synthesized in an in vitro translation reaction in the presence of [35S]methionine, could interact with HIV-1 Rev, or the Rev M10 mutant, on an affinity column in the presence or absence of added Ran · GTP. As shown in Fig. 3, efficient binding of hCrm1 to Rev was detected only in the presence of Ran · GTP. This binding was NES dependent, in that no binding was observed with the Rev M10 mutant. These data therefore strongly support the proposal of Fornerod et al. (15) that the high level of Ran · GTP present in the cell nucleus is critical for the efficient recruitment of hCrm1 to the Rev NES and presumably to all other leucine-rich NESs.
FIG. 3.
The Rev NES forms a ternary complex with hCrm1 and Ran · GTP in vitro. The hCrm1 protein was synthesized in vitro, in the presence of [35S]methionine, using a rabbit reticulocyte coupled transcription-translation system (Promega). The labeled proteins were then divided into two equal aliquots, and one was supplemented with recombinant Ran · GTP. The reaction mixtures were then again divided into two equal parts, and each fraction was loaded onto affinity columns bearing recombinant GST-Rev or the GST-M10 NES mutant. Bound proteins were eluted and resolved by SDS-PAGE. The data are representative of two separate protein interaction experiments.
hCrm1 is a nucleocytoplasmic shuttle protein.
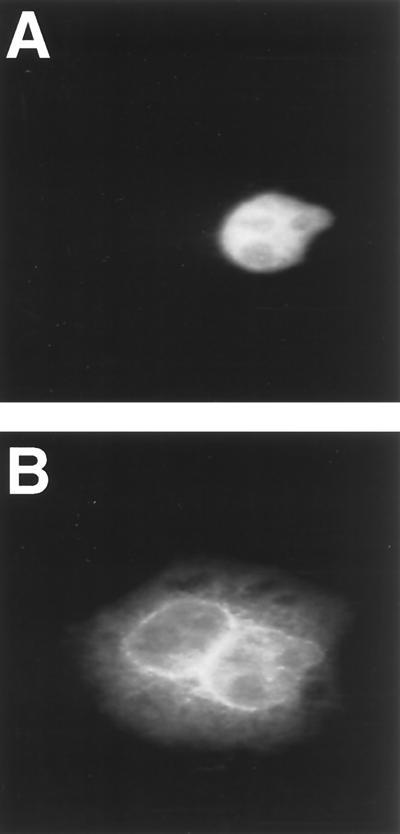
If hCrm1 is indeed required for delivery of Rev, and the bound RRE RNA, to the cell cytoplasm, then hCrm1 should continuously shuttle between nucleus and cytoplasm, as previously shown for HIV-1 Rev (34). To test this hypothesis, we expressed hCrm1 in bacteria in the form of a GST-Crm1 fusion protein and then purified this recombinant protein in the absence of detergent. To test for nucleocytoplasmic shuttling, we mixed the purified GST-Crm1 with a rabbit IgG tracer protein and then microinjected this mixture into one nucleus of a binuclear HeLa cell. As shown in Fig. 4A, the injected IgG tracer remains in the injected nucleus. In contrast, the injected GST-Crm1 rapidly relocalizes such that accumulation at the nuclear membrane and within the nucleoplasm of both the injected and uninjected nucleus is readily detectable. A significant level of cytoplasmic hCrm1 is also observed. These data are consistent with a previous report that localized hCrm1 to nuclear pore complexes within the nuclear membrane and also to the nucleoplasm (16). These data are also consistent with a report demonstrating that yCrm1 is rapidly mislocalized to the cytoplasm upon inhibition of protein nuclear import in yeast cells (43).
FIG. 4.
hCrm1 is a nucleocytoplasmic shuttle protein. Recombinant purified GST-Crm1 was mixed with a rabbit IgG tracer and then injected into one nucleus of a binuclear HeLa cell. While the IgG tracer remains behind (A), the Crm1 fusion protein relocalizes to the other nucleus, and in particular to the nuclear membrane, as well as to the cytoplasm (B).
ΔCAN specifically blocks leucine-rich NES function.
It has previously been demonstrated that yCrm1 can interact with a subset of nucleoporins, including hRIP/Rab, Nup98, Nup153, and Nup214/CAN, as determined by yeast two-hybrid analysis (35), and we have confirmed this result for hCrm1 in assays using hRIP/Rab, Nup153, and Nup214/CAN (11). The interaction between hCrm1 and Nup214/CAN appears particularly likely to be physiologically relevant in that hCrm1 and Nup214/CAN can be recovered from cells in a protein complex together with a third nucleoporin, termed Nup88 (16). The binding site for hCrm1 on Nup214/CAN has been localized to residues 1864 to 2090, i.e., to the extreme carboxy terminus. It has further been shown that overexpression of this fragment of Nup214/CAN, which we term ΔCAN, results in the relocalization of hCrm1 away from the nuclear membrane and into the nucleoplasm (16). This observation suggests that ΔCAN may block the ability of Crm1 to bind to the nuclear pore complex by competing with authentic nucleoporins for binding to Crm1. If this is the case, then ΔCAN should function as a specific inhibitor of leucine-rich NES-dependent nuclear export but not necessarily of other export pathways. We therefore examined the ability of ΔCAN to block the nuclear export, and hence translation, of CAT mRNAs containing the HIV-1 or VMV RRE, the HTLV-1 RXRE, or the MPMV CTE. As described previously (18, 23, 26, 31), in these pDM128-based indicator constructs the cat gene and the relevant retroviral RNA target are located in an intron, flanked by HIV-1-derived splice sites, and efficient CAT expression is therefore dependent on the induced nuclear export of an unspliced CAT mRNA.
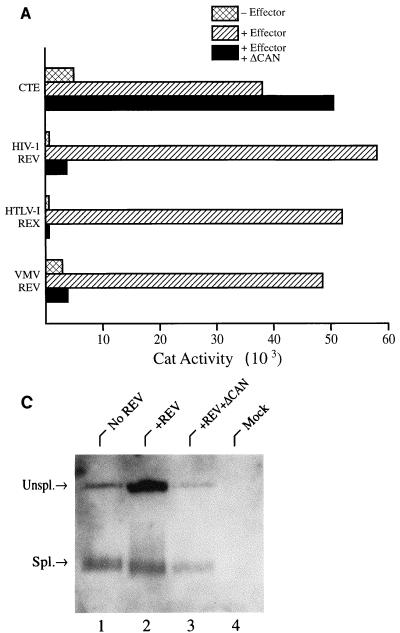
As shown in Fig. 5A and as previously reported (18, 23, 26, 31), expression of unspliced CAT mRNAs containing the HIV-1 RRE, VMV RRE, or HTLV-1 RXRE was strongly induced by expression of the cognate viral RNA export factor. An analogous control is not possible for the CTE, as this depends on an endogenous human RNA export factor (5, 25). However, insertion of the CTE clearly gave rise to a marked increase in the level of CAT expression compared to an equivalent CAT mRNA lacking the CTE (Fig. 5A).
FIG. 5.
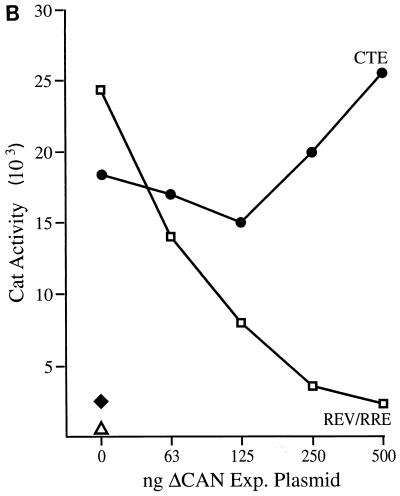
The ΔCAN protein is a specific inhibitor of leucine-rich NES-dependent mRNA expression. (A) The pDM128/CMV Rev indicator construct contains the cat gene and the HIV-1 RRE located in an intron defined by HIV-1 splice sites (26, 31). Efficient CAT expression is therefore dependent on the Rev-induced nuclear export of an unspliced form of this RNA. Similar indicator plasmids containing the VMV RRE, the HTLV-1 RXRE, or the MPMV CTE were also tested. A pDM128/CMV derivative lacking any inserted RNA target site (pDM128/CMV/PL) served as a negative control for pDM128/CTE. 293T cells (in 35-mm-diameter dishes) were transfected with 50 ng of the relevant HIV-1 Rev, VMV Rev, or HTLV-1 Rex reporter plasmid, 50 ng of the relevant effector plasmid, 100 ng of the pBC12/CMV/βgal internal control plasmid, and 500 ng of pBC12/CMV/ΔCAN. The pBC12/CMV negative control plasmid was added to maintain a level of 1,050 ng of DNA per transfection. Plasmid pDM128/CTE or pDM128/CMV/PL was transfected at 500 ng per culture, together with 100 ng of pBC12/CMV/βgal and 500 ng of either pBC12/ΔCAN or pBC12/CMV. The data are representative of three separate transfection experiments. (B) As for panel A except that the pBC12/ΔCAN expression plasmid was introduced at increasing levels, up to 500 ng per transfection. Total transfected DNA was held constant by substitution of pBC12/CMV. ⧫, CAT activity induced by the pDM128/CMV/PL negative control for pDM128/CTE; ▵, CAT activity induced by pDM128/CMV in the absence of HIV-1 Rev. (C) Northern analysis of cytoplasmic unspliced (Unspl.) and spliced (Spl.) RNA expression in 293T cells transfected with the pDM128/CMV indicator construct and plasmids expressing HIV-1 Rev and ΔCAN, as described for panel A. The parental pBC12/CMV plasmid served as a negative control.
As predicted above, expression of the ΔCAN protein indeed resulted in a dramatic inhibition in the expression of CAT mRNAs dependent on HIV-1 Rev, VMV Rev, or HTLV-I Rex for nuclear export. In contrast, expression of ΔCAN did not inhibit the MPMV CTE-dependent expression of a similar CAT mRNA and also did not inhibit the expression of an internal control β-Gal mRNA (Fig. 5A) (11). To further confirm the specificity of this inhibition, we also performed a dose-response experiment that measured the effect of increasing levels of ΔCAN on the expression of CAT mRNAs that depend on the MPMV CTE or on the Rev/RRE pathway for nuclear export. As shown in Fig. 5B, while ΔCAN produces a clear dose-dependent inhibition of Rev/RRE-dependent CAT expression, it does not inhibit, and may even slightly enhance, CTE-dependent cat gene expression.
It has been previously demonstrated that activation of CAT expression from the pDM128 indicator plasmid by HIV-1 Rev correlates with the increased cytoplasmic expression of the unspliced mRNA that encodes CAT (26). This result is confirmed in Fig. 5C (compare lanes 1 and 2), which also shows that coexpression of ΔCAN prevents this Rev-induced increase in cytoplasmic unspliced CAT mRNA expression without affecting the level of expression of the spliced, noncoding RNA derived from the pDM128 indicator plasmid (Fig. 5C, lane 3).
ΔCAN inhibits late, but not early, HIV-1 gene expression in infected cells.
If ΔCAN is a specific inhibitor of Rev-dependent nuclear RNA export, but not of RNA transport via the normal cellular mRNA export pathway, then ΔCAN should selectively block late, but not early, HIV-1 gene expression. Similarly, one would expect LMB, which is believed to be a specific inhibitor of Crm1 function (36), to also selectively block late HIV-1 gene expression. To test this hypothesis, we used an HIV-1 proviral construct, termed pNL-Luc-E−, that contains the luciferase (luc) indicator gene substituted in place of the nef gene (6). Because Nef is an early HIV-1 gene product that is expressed from a Rev-independent, fully spliced mRNA (7), luc expression here serves as a readily quantifiable measure of the level of early viral gene expression. The Gag protein, which is expressed in a Rev-dependent manner (7), can be accurately quantified to determine the level of late HIV-1 gene expression. The pNL-Luc-E− proviral construct also lacks a functional env gene and must therefore be complemented in trans by using an HIV-1 env expression plasmid (6). Importantly, this means that only a single round of HIV-1 infection can occur.
To test the effects of ΔCAN and LMB on HIV-1 late and early gene expression, we first prepared a stock of infectious NL-Luc-E− provirus by transfection of 293T cells with the pNL-Luc-E− proviral expression plasmid together with a second plasmid that expresses the JR-FL envelope protein (40). At the same time, target cells for infection were generated by transfection of 293T cells with expression plasmids encoding the CD4 and CCR-5 receptors for HIV-1 (40). As a result, only transfected cells are susceptible to infection by HIV-1. In addition, these cells were also cotransfected with expression plasmids encoding either wild-type HIV-1 Rev, the dominant negative Rev M10 NES mutant, or the ΔCAN protein. At 34 h after transfection, the virus-containing supernatant medium was recovered from the pNL-Luc-E−-transfected culture, filtered to remove any cells, and then used to infect the CD4/CCR-5-expressing 293T cell cultures. At 22 h after HIV-1 infection, all cultures were washed with fresh medium to remove residual p24_gag_ protein. In addition, two cultures that were transfected with CD4 and CCR-5 expression plasmids only were supplemented with 0.2 or 0.5 ng of LMB per ml. Finally, at 46 h after infection, the supernatant media were harvested for analysis of Gag protein expression levels by ELISA, while the infected cells were lysed to determine luciferase expression levels (40).
As shown in Table 1, 293T target cells expressing only the CD4 and CCR-5 gene products are readily infected with the JR-FL envelope-pseudotyped NL-Luc-E− virus, as shown by the high-level expression of both the luciferase indicator gene product and p24_gag_ protein. While coexpression of HIV-1 Rev in the target cells had little effect on either luciferase or Gag protein expression, coexpression of the dominant negative Rev M10 mutant inhibited Gag protein expression by ∼98% while having little or no effect on luc expression, as predicted from earlier research (28, 29). Similarly, while cotransfection of intermediate or high levels of a ΔCAN expression plasmid into the CD4/CCR-5-expressing 293T cells had little effect on luc expression, ΔCAN almost entirely blocked Gag protein expression. Finally, treatment of CD4/CCR-5-expressing 293T cells with a moderate (0.2 ng/ml) or high (0.5 ng/ml) level of the Crm1-specific inhibitor LMB resulted in a marked (∼83%) or essentially complete (∼99%) inhibition of HIV-1 Gag protein expression yet again had little effect on luciferase production. The fact that M10, ΔCAN, and LMB each proved able to effectively block expression of the late HIV-1 gag gene product while early HIV-1 gene expression, measured here by substitution of the luc gene in place of nef, was essentially unaffected shows that each of these reagents neither inhibits HIV-1 infection per se nor blocks the expression of mRNAs that exit the nucleus via the standard mRNA export pathway.
TABLE 1.
Specific inhibition of late, but not early, HIV-1 gene expression by Rev M10, ΔCAN, and LMB
| 293T culture infected with NL-Luc-E− virus | Early HIV-1 gene expressiona | Late HIV-1 gene expressionb | Relative late HIV-1 gene expressionc |
|---|---|---|---|
| CD4 + CCR-5 only | 120,847 | 23,000 | 100 |
| CD4 + CCR-5 + Rev | 76,324 | 13,000 | 90 |
| CD4 + CCR-5 + Rev M10 | 55,590 | 175 | 2 |
| CD4 + CCR-5 + ΔCAN (250 μg) | 72,795 | 235 | 2 |
| CD4 + CCR-5 + ΔCAN (500 μg) | 57,766 | 115 | 1 |
| CD4 + CCR-5 + LMB (0.2 ng/ml) | 52,690 | 1,720 | 17 |
| CD4 + CCR-5 + LMB (0.5 ng/ml) | 53,419 | 125 | 1 |
| CD4 only | 375 | 25 |
ΔCAN inhibits Rev NES function.
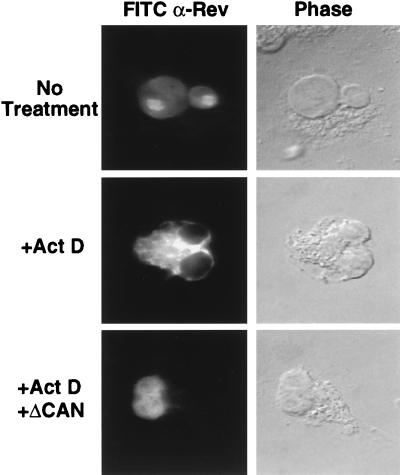
If ΔCAN inhibits Rev function by preventing the targeting of ribonucleoprotein complexes containing Rev and Crm1 to the nuclear pore complex, then ΔCAN should also prevent export of the Rev protein itself. Rev contains both a leucine-rich NES and a basic nuclear localization signal (13, 28) and has been shown to rapidly shuttle between the nucleus and cytoplasm of expressing cells (34). However, because Rev import into the nucleus is normally more efficient than Rev export, Rev demonstrates a largely nuclear subcellular localization at steady state (28, 34). For unknown reasons, inhibition of RNA synthesis by treatment of cells with RNA polymerase inhibitors such as actinomycin D reduces the efficiency of Rev nuclear import and leads to the accumulation of Rev in the cell cytoplasm (34), a result that is reproduced in Fig. 6. Importantly, the redistribution of Rev from the nucleus to the cytoplasm in the presence of actinomycin D has been shown to be dependent on the activity of the Rev NES (34). Therefore, if ΔCAN indeed blocks Rev NES function, then ΔCAN should prevent the cytoplasmic accumulation of wild-type Rev seen in actinomycin D-treated cells. As shown in Fig. 6, this is indeed what is observed.
FIG. 6.
The ΔCAN protein inhibits Rev NES function. COS cells were transfected (9) with the wild-type HIV-1 Rev expression plasmid pcRev together with a second plasmid expressing ΔCAN or with plasmid pBC12/CMV as a negative control. At ∼48 h after transfection, some cultures were treated with actinomycin D (Act D; 5 μg/ml) for 4 h, and all cultures were then fixed by using 3% paraformaldehyde in PBS. The subcellular localization of the Rev protein was determined by immunofluorescence, as previously described (28), using a polyclonal rabbit anti-Rev antiserum and FITC-conjugated goat anti-rabbit IgG.
DISCUSSION
The demonstration of a specific interaction between the Rev NES and hRIP/Rab, as well as several cellular nucleoporins, in both the yeast and mammalian two-hybrid assays (4, 21, 45) appeared to represent an important step forward in understanding how leucine-rich NESs function, given the known importance of nucleoporins in the regulation of nucleocytoplasmic transport (27, 33). The evidence supporting the relevance of this interaction included a strikingly complete correlation between NES function not only for HIV-1 Rev mutants but also for mutants in a range of other retroviral and cellular proteins bearing leucine-rich NESs (3, 4, 17, 20, 21, 44, 45). In addition, the Rev protein could be shown to interact with hRIP/Rab when Rev was bound to the RRE (4), and hRIP/Rab also proved able to modestly enhance Rev function when Rev activity was attenuated by low expression or mutation (4, 20). However, the inability to demonstrate a direct interaction between Rev and hRIP/Rab in vitro (44) clearly raised the possibility that this interaction was being bridged by a conserved factor present in both human and yeast cells. If this interaction is indeed bridged, then the bridging factor should recapitulate many of the activities for demonstrated hRIP/Rab, including a complete correlation between NES function and factor binding and the ability to bind Rev when Rev is bound to the RRE.
As noted above, several groups have recently presented data arguing that Crm1 is the bridging factor between Rev and nucleoporins and, thus, the direct cofactor for Rev NES function (15, 22, 35, 38, 43). Evidence in support of this hypothesis includes the demonstration of a direct interaction between hCrm1 and wild-type but not M10 mutant form of the Rev NES in vitro (15); the demonstration that LMB, a proposed specific inhibitor of hCrm1 function (36), blocks leucine-rich NES function in vivo (15, 22, 38, 51); and, most compellingly, the demonstration that yCrm1 is required both for Rev function and for detection of Rev nucleoporin interactions in yeast cells (35, 43). Nevertheless, the evidence supporting the specificity of the Rev-hCrm1 interaction has thus far remained well short of that reported earlier for the Rev-hRIP/Rab interaction.
In this study we attempted to test whether hCrm1 indeed satisfies the expected properties for the factor that bridges the Rev NES-nucleoporin interaction. We also tested the relevance of the predicted hCrm1-nucleoporin interaction for Rev function by targeting hCrm1, using a dominant negative mutant of the nucleoporin Nup214/CAN, a known target for hCrm1 binding in vivo (16). Using the mammalian two-hybrid assay, we observed that hCrm1 binding is indeed strongly predictive of NES function in not only HIV-1 Rev but also VMV Rev (Fig. 1). The HTLV-1 Rex protein did, however, behave somewhat anomalously in that NES mutations that entirely block Rex NES, and indeed Rex, protein function (3) retained a significant, albeit low, level of hCrm1 binding activity (Fig. 1A). The reason for this residual activity, which was readily demonstrable for two distinct Rex NES mutants, is unclear at present, especially given that these same mutations entirely block detection of the Rex-hRIP/Rab interaction in the yeast two-hybrid assay (3, 4). Nevertheless, the data presented in Fig. 1 do, in sum, demonstrate a strong correlation between leucine-rich NES function and hCrm1 binding.
Additional observations that support the proposed role of hCrm1 in Rev NES function include the demonstration that hCrm1 is able to effectively bind to the Rev NES when Rev is assembled on the RRE (Fig. 2) and the observation that hCrm1 is a nucleocytoplasmic shuttle protein (Fig. 4). These are clearly both properties that would be predicted for the Rev NES cofactor. We also observed that Rev, but not Rev M10, was able to bind to hCrm1 in vitro in a Ran · GTP-dependent manner. This latter finding confirms the earlier, analogous results of Fornerod et al. (15) but appears to contradict the observation by both Fukuda et al. (22) and Ossareh-Nazari et al. (38) of efficient hCrm1 binding to leucine-rich NESs in the absence of added Ran · GTP. This is an important issue given the proposed critical role of the exclusively nuclear pool of Ran · GTP in providing directionality to nucleocytoplasmic transport pathways (27, 33), and it will therefore be important to determine the basis for this discrepancy.
An interesting prediction of the hypothesis that hCrm1 is the cellular cofactor for HIV-1 Rev is that the truncated form of Nup214/CAN, here termed ΔCAN, should function as a selective inhibitor of leucine-rich NES, and hence HIV-1 Rev, function. This prediction is based on the finding that overexpression of ΔCAN blocks the ability of hCrm1 to interact with the nuclear pore complex (16), which is presumably a critical step in nuclear export. In fact, ΔCAN overexpression proved able to effectively and specifically block HIV-1 Rev-, VMV Rev-, and HTLV-1 Rex-dependent nuclear RNA export (Fig. 5) and also inhibited the nuclear export of the Rev protein itself (Fig. 6). In contrast, ΔCAN expression did not affect nuclear RNA export mediated by the MPMV CTE (Fig. 5) or the expression of fully spliced mRNAs exported via the standard, as yet ill-defined, cellular mRNA export pathway (Table 1; Fig. 5C). These data therefore strongly support the proposal, based on cross-competition data, that Rev-dependent and CTE-dependent nuclear RNA export occur via distinct pathways (39, 41) and further suggest that docking of MPMV CTE-dependent RNAs, or of cellular mRNAs, at the nuclear pore complex is likely mediated by protein-protein interactions distinct from those utilized by hCrm1.
As a final test of the relevance of hCrm1 for Rev function, we examined the ability of ΔCAN and of the cytotoxic drug LMB, a reported specific inhibitor of hCrm1 function (36), to block late but not early HIV-1 gene expression in infected cells. The positive control for this experiment was the Rev M10 NES mutant, which is known to exert a dominant negative phenotype in vivo (28, 29). In fact, Rev M10, ΔCAN, and LMB all proved able to potently block HIV-1 late (i.e., Rev-dependent) gene expression in HIV-1-infected cells while exerting at most a minimal effect on early, Rev-independent HIV-1 gene expression. Of interest, while these agents all act as inhibitors of Rev function, they presumably employ very different mechanisms. The Rev M10 mutant, which lacks a functional NES, is believed to inhibit Rev function by competing for binding to the RRE RNA target (28, 29). The Rev M10 mutant is therefore a highly specific inhibitor of HIV-1 Rev function and does not exert any detectable general toxicity (29). In contrast, both LMB and ΔCAN directly target the cellular protein hCrm1, which is likely critical for the nuclear export of a range of cellular proteins bearing leucine-rich NESs, as well as for the nuclear export of 5S rRNA and several U snRNAs (7, 13, 17, 33, 50). As a result, LMB is cytotoxic (36), and we also noted evidence of toxicity in ΔCAN-expressing cells if they were cultured for more than 4 or 5 days after transfection (11). Therefore, although these latter two agents are certainly selective inhibitors of Rev function, only Rev M10, and other equivalent transdominant Rev mutants, is truly specific. However, LMB and ΔCAN should prove to be valuable reagents for demonstrating whether the nuclear export of a particular protein or RNA is indeed hCrm1 dependent.
ACKNOWLEDGMENTS
The first two authors contributed equally to this report.
We thank Gerard Grosveld for the hCrm1 and ΔCAN coding sequences, Eric Hunter for the MPMV CTE, Nat Landau for the pNL-Luc-E− proviral expression vector, and Minoru Yoshida for LMB. We also thank Ted Benson for performing microinjection assays.
This research was funded by the Howard Hughes Medical Institute. A.E. is the recipient of a Gobierno Vasco/Eusko Jaurlaritza postdoctoral fellowship, while T.M.R. is supported by the Duke Interdisciplinary Research Training Program in AIDS grant from the National Institute of Allergy and Infectious Diseases (2T32AI07392).
REFERENCES
- 1.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. HIV-1 induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 5.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjöld M-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 7.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 8.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 9.Cullen B R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 10.Daly T J, Cook K S, Gray G S, Maione T E, Rusche J R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989;342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 11.Echarri, A., H. P. Bogerd, and B. R. Cullen. Unpublished data.
- 12.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 14.Fischer U, Meyer S, Teufel M, Heckel C, Lührmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 16.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fridell R A, Bogerd H P, Cullen B R. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fridell R A, Partin K M, Carpenter S, Cullen B R. Identification of the activation domain of equine infectious anemia virus Rev. J Virol. 1993;67:7317–7323. doi: 10.1128/jvi.67.12.7317-7323.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 20.Fritz C C, Green M R. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- 21.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 23.Gröne M, Hoffmann E, Berchtold S, Cullen B R, Grassmann R. A single stem-loop structure within the HTLV-I Rex response element is sufficient to mediate Rex activity in vivo. Virology. 1994;204:144–152. doi: 10.1006/viro.1994.1518. [DOI] [PubMed] [Google Scholar]
- 24.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 25.Hammarskjöld M-L. Regulation of retroviral RNA export. Semin Cell Dev Biol. 1997;8:83–90. doi: 10.1006/scdb.1996.0127. [DOI] [PubMed] [Google Scholar]
- 26.Hope T J, Huang X, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 28.Malim M H, Böhnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 29.Malim M H, Freimuth W W, Liu J, Boyle T J, Lyerly H K, Cullen B R, Nabel G J. Stable expression of transdominant Rev protein in human T cells inhibits human immunodeficiency virus replication. J Exp Med. 1992;176:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 31.Malim M H, McCarn D F, Tiley L S, Cullen B R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 33.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 34.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 35.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 36.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 37.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 39.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjöld M-L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, utilizes factors required for the export of cellular mRNAs. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 42.Southgate C D, Green M R. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- 43.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 44.Stutz F, Izaurralde E, Mattaj I W, Rosbash M. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 46.Tabernero C, Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 48.Tiley L S, Malim M H, Cullen B R. Conserved functional organization of the human immunodeficiency virus type 1 and visna virus Rev proteins. J Virol. 1991;65:3877–3881. doi: 10.1128/jvi.65.7.3877-3881.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Truant R, Fridell R A, Benson R E, Bogerd H, Cullen B R. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein. Mol Cell Biol. 1998;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 51.Wolff B, Sanglier J-J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 52.Zapp M L, Green M R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]