An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm (original) (raw)
Abstract
The oomycete Phytophthora infestans causes late blight, the potato disease that precipitated the Irish famines in 1846 and 1847. It represents a reemerging threat to potato production and is one of >70 species that are arguably the most devastating pathogens of dicotyledonous plants. Nevertheless, little is known about the molecular bases of pathogenicity in these algae-like organisms or of avirulence molecules that are perceived by host defenses. Disease resistance alleles, products of which recognize corresponding avirulence molecules in the pathogen, have been introgressed into the cultivated potato from a wild species, Solanum demissum, and R1 and R3a have been identified. We used association genetics to identify Avr3a and show that it encodes a protein that is recognized in the host cytoplasm, where it triggers _R3a_-dependent cell death. Avr3a resides in a region of the P. infestans genome that is colinear with the locus containing avirulence gene ATR1NdWsB in Hyaloperonospora parasitica, an oomycete pathogen of Arabidopsis. Remarkably, distances between conserved genes in these avirulence loci were often similar, despite intervening genomic variation. We suggest that Avr3a has undergone gene duplication and that an allele evading recognition by R3a arose under positive selection.
Keywords: microsynteny, Phytophthora infestans, hypersensitive response, linkage disequilibrium, hemibiotroph
Phytophthora infestans causes late blight, the devastating potato disease responsible for the Irish famines in the mid-1840s (1). For decades, disease control has involved regular applications of agrochemicals, although recent widespread occurrence of new fungicide-resistant strains has led many to consider this pathogen as a reemerging threat to global food security (2, 3). It is one of >70 Phytophthora species that are arguably the most devastating pathogens of dicotyledonous plants. Other economically important phytophthoras include Phytophthora sojae, cause of soybean root rot; Phytophthora palmivora, cause of cocoa black pod; Phytophthora cinnamomi, cause of dieback and root rot in >2,000 plant species; and the recently identified Phytophthora ramorum that is decimating oak trees in the United States and throughout Europe (4). Despite a fungus-like filamentous growth habit, they are related to brown algae in the kingdom Stramenopiles (5). Whereas fungal pathogenicity has been intensively studied, little is known about the molecular genetics of oomycete pathogenicity or about pathogen molecules that are recognized by host defenses (1, 4).
Over the last 75 years, potato breeders have introduced at least 11 late blight resistance (R) alleles from Solanum demissum into the cultivated potato (6). The products of R alleles recognize the products of corresponding avirulence (Avr) alleles in races of P. infestans, triggering disease resistance and a localized programmed cell death called the hypersensitive response (HR). Recently, the potato R3 locus was shown to contain two tightly linked genes, R3a and R3b, with distinct specificities (7). R3a has since been cloned and encodes a presumed cytoplasmic coiled-coil nucleotide binding site leucine-rich repeat protein (8). Although several P. infestans Avr loci have been genetically mapped (1, 9), no Avr genes have yet been reported, mainly due to difficulties encountered with positional cloning, such as high levels of repetitive DNA and aberrant segregation at the target locus. Our knowledge of the molecular basis of race evolution in this pathogen is thus limited.
Many AVR proteins from fungal plant pathogens possess N-terminal type II signal peptides (SPs) for secretion and exhibit significant sequence variation between pathogen races. Thus, Avr alleles in virulent races of the tomato pathogen Cladosporium fulvum (10, 11) and the barley pathogen Rhynchosporium secalis (12) contain SNPs yielding secreted proteins that do not elicit HR on plants containing corresponding R alleles. We therefore predicted (13) that a candidate gene approach of screening ESTs for genes encoding secreted proteins, followed by association genetic [linkage disequilibrium (LD)] studies, would be an alternative route to identify P. infestans Avr genes.
A strategy for predicting Phytophthora extracellular protein (pex) genes was reported that identified 142 such genes from P. infestans (14). One EST, Pex147 (encoding a 147-aa protein; GenBank accession no. BE776395), was similar to a P. sojae gene, avr1b, encoding an elicitor of HR in soybean plants containing Rps1b (15). The PEX147 protein product was found in P. infestans culture filtrate, demonstrating that it was secreted (14). We used an association genetics approach to show that Pex147 was likely to be Avr3a in P. infestans and confirmed that this was the case by transient expression of alleles of the gene in potato genotypes containing a variety of R genes and by coexpression of Avr3a and R3a in Nicotiana benthamiana.
Materials and Methods
P. infestans Isolates and Analyses of Polymorphism. P. infestans isolates were from the Scottish Crop Research Institute culture collection (Table 1, which is published as supporting information on the PNAS web site). DNA was prepared by using the DNeasy plant minikit (Qiagen, Valencia, CA). PCR primers (Pex147F, 5′-CCATGCGTCTGGCA AT TATGCT-3′; Pex147R, 5′-CTGAAAACTAATATCCAGTGA-3′) were used to amplify Pex147 from each isolate. PCR products were purified by using the QIAquick PCR purification kit (Qiagen) and were directly sequenced by using the Applied Biosystems Bigdye v3.1 Terminator sequencing kit or used to generate a radiolabeled probe for hybridization to the P. infestans bacterial artificial chromosome (BAC) library (see below).
Analyses of P. infestans BAC Clones, BAC Sequencing, and Annotation. Hybridization of coding sequences (CDSs) (PCR-amplified by using primers shown in Table 2, which is published as supporting information on the PNAS web site) and Pex147 to the P. infestans BAC library and BAC DNA preparations were as described in ref. 16. Pi-BAC-61F2 (GenBank accession no. AC146942) was sequenced at the Broad Institute (Cambridge, MA). Pi-BAC-35J4 (accession no. AJ893356) and Pi-BAC-49P21 (accession no. AJ893357) were sequenced at The Wellcome Trust Sanger Institute. BAC sequencing and assembly were essentially performed as described in ref. 17. Clones were sequenced to 8-fold coverage and finished.
Annotation was performed by using artemis software (18). Functional assignments were based on assessment of blast and fasta searches against public databases, and for domain predictions, searches such as interpro (19), tmhmm 2.0 (20), and signalp 2.0 (www.cbs.dtu.dk/services/SignalP) were carried out. The comparison of the avirulence loci of Hyaloperonospora parasitica and P. infestans was carried out by using blast comparisons viewed with the artemis comparison tool (K. Rutherford, unpublished).
Analyses of Transcript Levels. Maintenance of P. infestans isolate 88069; preparation of nonsporulating mycelium, sporangia, zoospores, germinating cysts, and germinating cysts developing appressoria; and inoculations of potato were carried out as described in refs. 21 and 22. For real-time RT-PCR, material was prepared, RNA extracted, and experiments conducted as described previously by using the actA gene from P. infestans as a constitutively expressed endogenous control (21). Real-time RT-PCR primers for Pex147 were 5′-CGCCATAAACTTTGCAACCA-3′ and 5′-TGCCGGCTGAATCGTGTAT-3′ (amplicon size = 92 bp), primers for Pex147-2 were 5′-GGTGGCAGCACAAGAGGC-3′ and 5′-GCAGCTATTGTAGATCCGATTGTATC-3′ (amplicon size = 104 bp), and primers for Pex147-3 were 5′-TTGGTGGCAGCACAATGGT-3′ and 5′-CCCAGGTGCATCAGGTAGCT-3′ (amplicon size = 120 bp). Repeated amplifications, on independent occasions with different cDNA samples, resulted in similar expression profiles.
Expression Constructs, Particle Cobombardment, and Agrobacterium-Infiltration Experiments. The pex147 ORF was PCR-amplified from heterozygous P. infestans isolate SC95–1.1.2 (Table 1) by using primers 5′-CACA_CCATGG_GTCTGGCAATTA-3′ (introducing an NcoI site at the 5′ end) and 5′-GTTTCAGCA_TCTAGA_ATCGGATTTTCTG-3′ (introducing an XbaI site at the 3′ end) and cloned into pRTL2 (23) for particle cobombardment and primers 5′-GGAA_ATCGAT_TCTCTCAGCTCCCCAGGGTTCCAC-3′ (introducing a ClaI site at the 5′ end) and 5′-GGAAGCGGCCGCCACGAGCGTTTCAGCAGTTAGAATCGG-3′ (introducing a NotI site at the 3′ end) for cloning into potato virus X–Agrobacterium tumefaciens binary vector pGR106 (24). The alleles were cloned without SP-encoding sequences into pRTL2 by using additional primer 5′-GGAA_CCATGG_ACCAAACCAAGGTCCTGG-3′ and into pGR106 by using additional primer 5′-GGAA_ATCGAT_ATGGACCAAACCAAGGTCCTGGTG-3′, introducing methionine in place of isoleucine at position 22 [predicted to be the SP cleavage site by using signalp (www.cbs.dtu.dk/services/SignalP)]. PCR products digested with NcoI and XbaI or ClaI and NotI were ligated into pRTL2 or pGR106 (respectively), electroporated into Electromax DH10B Escherichia coli, and selected on LB ampicillin (pRTL2) or LB kanamycin/tetracycline (pGR106) by using standard protocols. Individual clones were sequenced to ensure that errors had not been introduced.
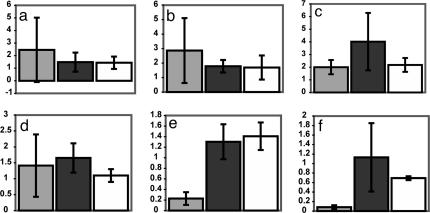
Biolistic cobombardment was performed by using the Bio-Rad PDS-1000/He Biolistic Particle Delivery System. One-micrometer gold particles were coated with 0.83 μg/mg each pRTL2::gfp and pRTL2 expressing test sequences. Leaves from 4- to 6-week-old potato plants were cut down the midrib. Half-leaves from cv. Bintje or cv. Craigs Royal lacking either R gene-mediated (r) or field resistances to P. infestans were cobombarded alongside half-leaves from potato genotypes containing R3a (cv. Pentland Ace), R3b (SW8563-0182), R1 (cv. Craigs Snow White), R2 (1512c[16]), or R10 (3681ad[1]) to receive similar quantities of particles. Transgenic cv. Desireé containing R3a (T68.4-002) was similarly screened with adjacent nontransgenic cv. Desireé, which, again, lacks either R gene-mediated or field resistances to P. infestans. Half-leaf pairs were cobombarded at 1,550 psi and incubated in light for 20–24 h at 20°C followed by 18–24 h in dark at 20°C. The half-leaves were imaged by using a Bio-Rad MRC 1000 confocal laser scanning microscope, and GFP was excited by using blue laser light at 488 nm. Numbers of GFP-fluorescing cells were quantified. For each half-leaf pair, the percentage of GFP fluorescence was determined, and the figure for the R gene-containing cultivar was divided by that observed on Bintje, yielding a GFP fluorescence ratio. For each line, three leaf pairs from independent plants were cobombarded with pRTL2::gfp and pRTL2 expressing each test sequence. A further three leaf pairs were bombarded with particles coated with pRTL2::gfp alone. These experiments were repeated, and the means and standard deviations are presented in Fig. 4.
Fig. 4.
Cobombardment demonstrates recognition of AVR3a by plants containing R3a. Truncated (E80 M103) avr3a and (K80 I103) Avr3a alleles were transiently coexpressed with gfp in potato genotypes containing R1 (a), R2 (b), R3b (c), R10 (d), and R3a (e), each compared with cv. Bintje (r), and in transgenic cv. Desiree expressing R3a compared with cv. Desiree (f). Typically, more GFP-expressing cells were observed on R gene-containing genotypes when gfp was bombarded alone than on adjacent Bintje leaves (a_–_e), resulting in GFP fluorescence indexes >1 (where 1 is the average total GFP fluorescence on Bintje). In each histogram, the average GFP fluorescence index is presented after coexpression with Avr3a (light gray) or avr3a (dark gray) and expression of gfp alone (white).
Agrobacterium infiltration was performed in N. benthamiana by using Agrobacterium tumefaciens strain AGL0 carrying the pBINplus::R3a construct (8) and strain LBA4404 carrying the pGR106::Avr3a_K80 I103 and pGR106::Avr3a_E80 M103 (both without SP) constructs. Recombinant A. tumefaciens cultures were grown and induced before infiltration as described in ref. 25, except that culturing was performed in LB supplemented with 50 μM kanamycin. A. tumefaciens cultures carrying the R3a and Avr3a constructs were mixed in a 2:1 ratio before centrifugation. For transient coexpression, the harvested cells were resuspended in MMA buffer containing 200 μM acetosyringone (25) to a final OD of 0.4. [One liter of MMA contains 5 g of MS salts (Duchefa, Haarlem, The Netherlands), 1.9 g of MES (_N_-morpholinoethanesulfonic acid; Sigma), and 20 g of sucrose, pH adjusted to 5.6 with 1 M NaOH (25).] For transient expression of the Avr3a genes, cells harvested from A. tumefaciens cultures carrying the Avr3a_K80 I103 or Avr3a_E80 M103 constructs were resuspended to a final OD of 0.133 and infiltrated into 5- to 6-week-old leaves with a 3-ml syringe. Symptom development was monitored starting from 3 days after infiltration.
Results and Discussion
Identification of Avr3a Through Association Genetics. Primers were designed to PCR-amplify Pex147 from 55 P. infestans strains isolated from countries in Europe and North and South America that had been virulence-tested on potato genotypes containing 9 of the 11 S. demissum R genes. After sequencing the PCR products, only three SNPs were found, changing amino acids S19C, E80K, and M103I (Fig. 1). These SNPs revealed only two alleles that showed 100% correlation with virulence phenotype on cv. Pentland Ace, containing R3a (Table 1). Presence of the C19 K80 I103 (C-K-I) allele was associated with avirulence, whereas virulent isolates were homozygous for the S19 E80 M103 (S-E-M) allele.
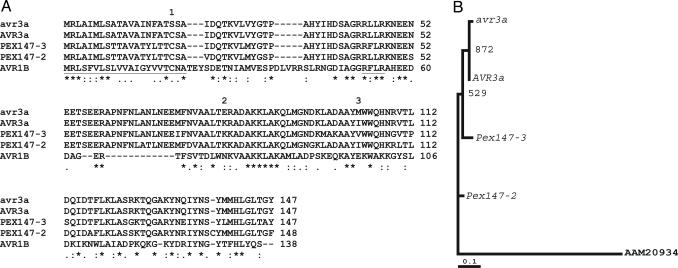
Fig. 1.
Protein alignments and phylogeny of AVR3a, avr3a, and PEX147-like paralogues. (A) Multiple alignment (using www.ebi.ac.uk/clustalw) of predicted proteins (AVR3a and avr3a) derived from the Avr3a (avirulent) and avr3a (virulent) alleles (initially termed Pex147) with those from the two paralogous P. infestans Pex147-2 and Pex147-3 sequences and the AVR1b protein from P. sojae. The N-terminal type II SP (first 21 aa) and the RXLR motif, found also in the_H. parasitica_ ATR1NdWsB protein (32), are underlined. The locations of the three amino acid polymorphisms are indicated by numbers above the amino acids. (B) Maximum likelihood estimation of the phylogeny of Avr3a, avr3a, and _Pex147_-like paralogues using a nucleotide alignment based on the amino acid alignment in A. The tree was constructed by using dnaml from the phylip package, with the P. sojae Avr1b sequence (AAM20934) as an outgroup. Bootstrap values from 1,000 random resamplings of the data are indicated. The alignment supports the closer evolutionary relationship of the Avr3a allele, rather than the avr3a allele, to the flanking Pex147-2 and Pex147-3 paralogues.
The possibility that the observed association resulted from genetic hitchhiking (26) was considered; Pex147 could simply be closely linked to Avr3a rather than the gene itself. In sexual populations, hitchhiking decreases with physical distance because of recombination. After hybridization of Pex147 to a P. infestans BAC library (16), BAC clones were identified from which SNPs flanking the gene were sought to investigate the region of LD. The sequence of Pi-BAC-49P21 revealed Pex147 and two additional _Pex147_-like sequences (Fig. 1). A further _Pex147_-like sequence contained a frameshift mutation and was annotated as a pseudogene (Fig. 2). The occurrence of SNPs 9.6 kb upstream and 10.9 kb downstream of Pex147 (Fig. 2) in 10 P. infestans isolates showed no association with the Avr3a-R3a phenotype (Table 1), indicating breakdown in LD and ruling out the Pex147-2 and Pex147-3 paralogues as candidate Avr3a genes. Within the 20-kb region defined by these SNPs, only four additional CDSs were identified (results not shown), none of which encoded proteins with SPs, and Pex147 was thus selected as the most likely Avr3a candidate.
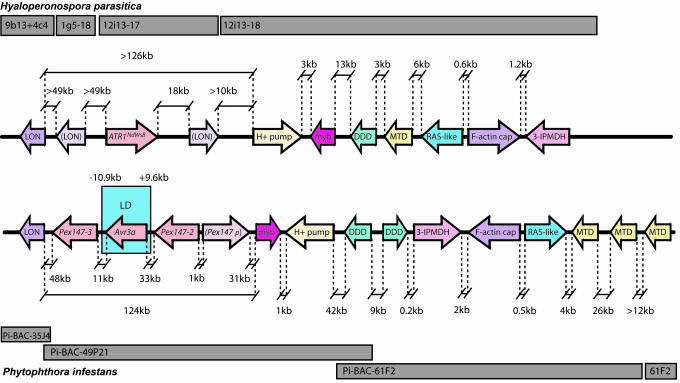
Fig. 2.
Conservation of synteny between H. parasitica ATR1NdWsB (Upper) and P. infestans Avr3a (Lower) loci. Sequenced BACs are indicated as gray blocks. Arrows indicate positions and transcriptional orientation of CDSs encoding Lon protease (LON), partial LON sequences (bracketed), vacuolar proton ATPase (H+ PUMP), Myb transcription factor (myb), dimethyl dihydrodiol dehdrogenase (DDD), methylenetetrahydrofolate dehydrogenase (MTD), a RAB (RAS-like), F-actin capping protein (F-actin cap), and 3-isopropylmalate dehydrogenase (3-IPMDH). Distances (in kb) between sequences (not to scale) are indicated. Probes PCR-amplified (see Table 2 for primers) from each P. infestans sequence hybridized to the P. infestans BAC library (16) did not indicate colocalization of these sequences elsewhere in the genome (data not shown). The block of LD defined by primers 9.6-kb upstream and 10.9-kb downstream of Avr3a (initially termed Pex147) is indicated, as are the flanking Pex147-2 and Pex147-3 paralogues and a _Pex147_-like pseudogene (Pex147p).
Pex147 Is Up-Regulated Before and During Potato Infection. Real-time RT-PCR was used to investigate the expression of Pex147 and the flanking Pex147-2 and Pex147-3 paralogues in P. infestans preinfection cell types (sporangia, zoospores, germinating cysts, and germinating cysts forming appressoria) and infected susceptible potato cv. Bintje 12, 24, 33, 48, 56, and 72 h postinoculation (hpi). Expression of each gene was compared with the ActA gene from P. infestans, which has been used as a constitutively expressed endogenous control (21, 22). Although PCR products of expected sizes were generated for Pex147-2 and Pex147-3 from genomic DNA template, no products were detected from cDNA templates in the case of Pex147-3, and expression was only weakly detected in the case of Pex147-2 and only in cDNA from axenic cultured, nonsporulating mycelium. In contrast, expression of Pex147 was readily detectable in preinfection and infection stages and, in these samples, was compared with the level of its expression in a calibrator sample, cDNA from cultured nonsporulating mycelium, which was assigned the value of 1.0. Pex147 was up-regulated >100-fold in germinating cysts developing appressoria and showed elevated levels of expression throughout infection, with an early peak of expression at 24 hpi, in the biotrophic phase of infection, and >200-fold elevation of expression at 72 hpi, in the necrotrophic phase of the interaction (Fig. 3). Repeated amplifications on independent occasions with different cDNA samples resulted in similar expression profiles. The up-regulation of Pex147 immediately before and during potato infection supports a potential role in pathogenicity. In contrast, the failure to detect such expression of the flanking Pex147-2 and Pex147-3 paralogues supports the LD analysis, ruling them out as Avr3a candidates.
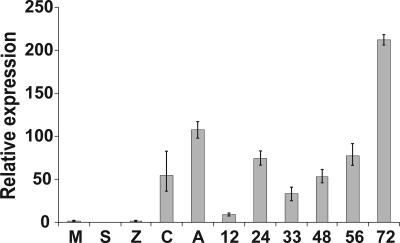
Fig. 3.
Pex147 is up-regulated before and during potato infection. Real-time RT-PCR analysis of the expression of Pex147 (avr3a), using cDNA templates derived from P. infestans 88069 (virulent on R3a genotypes; Table 1) nonsporulating mycelium (M) grown in pea broth, sporangia (S), zoospores (Z), germinating cysts (C), germinating cysts forming appressoria (A), and infected susceptible cv. Bintje 12, 24, 33, 48, 56, and 72 h postinoculation.
Coexpression in Potato of Pex147 Alleles with gfp Demonstrates That Pex147 Is Avr3a. The S-E-M (virulent) and C-K-I (avirulent) alleles of Pex147 and each allele truncated to remove SP-encoding sequences (and thus lacking the S19C polymorphism) were transiently expressed in potato genotypes lacking a late blight R gene (cv. Bintje or cv. Craigs Royal) or containing R3a, R3b, R1, R2, and R10. In each case, transient expression of the alleles involved biolistic cobombardment with a second vector expressing gfp as a marker of cell vitality. Similar approaches were used to indicate and quantify HR triggered by Avr genes from the bacterium Pseudomonas syringae (27), the fungus Magnaporthe grisea (28), and the oomycete H. parasitica (29). There was no reduction in GFP fluorescence on leaves from any plants cobombarded with alleles possessing SP-encoding sequences, relative to that observed when GFP was bombarded alone (data not shown). Furthermore, no difference in GFP fluorescence was seen in leaves from Bintje or Craigs Royal compared with plants containing R3b, R1, R2, or R10 that were cobombarded with alleles lacking SP-encoding sequences (Fig. 4). However, a 5-fold reduction in GFP fluorescence was observed in Pentland Ace (R3a) cobombarded with the truncated K-I sequence, indicating a reduction in vital cells consistent with triggering HR (Fig. 4). This analysis was extended to transgenic Desireé expressing R3a (8). A dramatic reduction in GFP fluorescence was observed only with the truncated K-I sequence in the presence of the R3a transgene (Fig. 4), and Pex147 was renamed Avr3a.
Coexpression of Avr3a and R3a in N. benthamiana. We also assessed whether Avr3a triggers _R3a_-dependent HR by using agrobacterium infiltration in another solanaceous plant, N. benthamiana. Coinfiltration of N. benthamiana leaves with an A. tumefaciens strain carrying a construct expressing R3a and a strain carrying a construct expressing the truncated K-I Avr3a sequence resulted in a confluent cell death response (Fig. 5). In contrast, coinfiltration of the R3a strain with a strain carrying a vector expressing the truncated E-M avr3a sequence, or infiltrations of the individual A. tumefaciens strains and other controls, failed to elicit visible cell death (Fig. 5).
Fig. 5.
Transient expression and coexpression of Avr3a and R3a in N. benthamiana. Leaves of N. benthamiana plants were infiltrated with A. tumefaciens carrying pGR106::Avr3a_K-I or pGR106::avr3a_E-M (expressing mature forms of the proteins lacking SPs) alone or mixed with an A. tumefaciens strain carrying pBINplus::R3a. For transient coexpression of Avr3a and R3a, the A. tumefaciens solutions were mixed in a 1:2 ratio before infiltration. Photographs of symptoms were taken 6 days postinfiltration. Circles indicate the infiltrated area on the leaf panels for each treatment. This is a representative leaf from multiple assays and experiments.
Cobombardment of Avr3a alleles and gfp in _R3a_-containing potato genotypes and coexpression of Avr3a alleles and R3a in N. benthamiana each indicated recognition of the C-K-I AVR3a form by R3a. Moreover, AVR3a was recognized after removal of SP-encoding sequences, indicating that recognition occurs in the host cytoplasm and is likely to depend on one or both of the E80K and M103I polymorphisms, because the S19C polymorphism is within the SP. Cytoplasmic recognition of AVR3a is consistent with the presumed cytoplasmic localization of R3a (8). Intracellular recognition of avirulence proteins from plant pathogenic bacteria (27) and fungi (28, 30) has been demonstrated, and here we report that cytoplasmic recognition is also the case for the oomycete AVR3a protein. Bacterial effector proteins, some of which are perceived by R proteins and are thus termed AVR proteins, are often translocated into plant cells by a type III secretion system (T3SS), where they interact with and manipulate host defenses (31). No T3SS-like systems have yet been identified in hemibiotrophic or biotrophic fungi or oomycetes, but it is perhaps likely that specialized systems exist for delivering effector proteins into host cells to manipulate defense pathways. The results for AVR3a contrast with the P. sojae AVR1b protein, which elicited an HR upon extracellular infiltration (15). However, it is possible that infiltration of large quantities of protein could lead to uptake of AVR1b into the host cell through endocytosis.
Analysis of Avr3a and ATR1NdWsB Loci Reveals Conservation of Microsynteny. Analysis of >250 kb spanning Avr3a revealed a surprising finding: Several CDSs were strikingly similar to CDSs flanking ATR1NdWsB (32) in H. parasitica (Table 2), suggesting similar gene content in these avirulence loci. Genes encoding methylenetetrahydrofolate dehydrogenase (MTD), a RAS-like protein, F-actin capping protein, and 3-isopropylmalate dehydratase (3-IPMDH) (Fig. 2) were colinear in the two loci and similar distances apart, although they were apparently inverted relative to other genes. It is interesting to note that MTD- and dimeric dihydrodiol dehydrogenase-encoding sequences are duplicated in P. infestans, and this duplication may have coincided with inversion (Fig. 2). Three intact copies of Pex147 and a _Pex147_-like pseudogene again indicate gene duplication in P. infestans, and, if extended with similar frequency throughout the genome, such duplication would contribute significantly to global gene redundancy. This phenomenon is marked in the yeast Debaryomyces hansenii and distinguishes it from other yeasts (33). Three Lon protease (LON)-encoding sequences in H. parasitica, two of which are partial sequences and, therefore, possibly pseudogenes, indicate that gene duplication is not exclusive to P. infestans in this ancestral locus (Fig. 2).
ATR1NdWsB (32) and Avr3a share little sequence similarity but are in strikingly similar locations in their respective loci and are flanked on one side by one or more LON-encoding sequences and on the other side by vacuolar proton ATPase subunit (H+ PUMP)- and myb-encoding genes (Fig. 2). The H+ PUMP- and myb-encoding genes are again similar distances apart and are also transcriptionally inverted, suggesting numerous structural rearrangements in the Avr3a and ATR1NdWsB loci. Remarkably, the distances between intact LON sequences and the myb- or H+ PUMP-encoding sequences are also similar, suggesting that an unknown mechanism of positional conservation has acted at sites within these oomycete genomes, between which are regions of variation. A similar phenomenon has been noted in studies of conservation of synteny between rice and Arabidopsis thaliana (34). Although recently grouped together in the Peronosporales based on molecular phylogenies (35), P. infestans and H. parasitica are not particularly closely related (36). Considering the high sequence diversity observed in the Peronosporales, the colinearity is therefore notable.
Within these colinear loci, the avirulence genes have apparently evolved differently. In H. parasitica, considerable allelic variation was observed in ATR1NdWsB (32). In P. infestans, gene duplication and divergence has provided a mechanism for generating variation in _Avr3a_-like sequences, and numerous synonymous and nonsynonymous nucleotide polymorphisms distinguish these genes. The avr3a allele is distinguished from the Pex147-2 gene by 10 synonymous and 22 nonsynonymous polymorphisms and is distinguished from Pex147-3 by 8 synonymous and 20 nonsynonymous polymorphisms. The Pex147-2 and Pex147-3 paralogues are distinguished from each other by 9 synonymous and 21 nonsynonymous polymorphisms (Fig. 1). Nevertheless, only two alleles of Avr3a were observed in 55 P. infestans individuals, and the SNPs distinguishing them all result in nonsynonymous amino acid substitutions, suggesting that diversifying selection, perhaps driven by coevolution with host plants, has acted on this gene. Interestingly, diversifying selection has also acted on the R3a gene (8) and its paralogues, suggesting a coevolutionary “arms race” between R3a and Avr3a. The amino acids specific to AVR3a (C19 K80 I103) are shared with PEX147-2, and two are shared with PEX147-3. The absence of other SNPs, or of selectively neutral SNPs between Avr3a and avr3a, suggests that the virulent allele arose from an avirulent progenitor sequence under positive selection (Fig. 1).
The allelic diversity in the ATR1NdWsb (32) and Avr3a genes are in stark contrast, and it should be noted that H. parasitica isolates were derived from natural populations coevolving with wild A. thaliana, whereas P. infestans isolates were largely obtained from cultivated potato in countries distant from Mexico, a center of diversity for the pathogen. It is well documented that P. infestans underwent a series of extreme genetic bottlenecks during its panglobal distribution (37), and a lack of allelic diversity in Avr3a may reflect this. It will thus be interesting to compare Avr3a allele frequency and diversity in natural Mexican P. infestans populations with those found in agricultural populations elsewhere.
Conclusions
We report the use of association genetics to identify the P. infestans Avr3a gene and show that its product is recognized in an R3a_-dependent manner in the host cytoplasm. Analysis of the Avr3a locus revealed unexpected conservation of synteny with the locus containing ATR1NdWsB in H. parasitica. This colinearity supports comparative genomics as an approach to investigate the evolution of pathogenicity in oomycetes. The isolation of R3a (8) and Avr3a represents an opportunity for detailed investigation of the earliest recognition events in a potato–_P. infestans R-AVR interaction and of subsequent signaling pathways leading to disease resistance. It also opens a door to studies of molecular mechanisms potentially underlying the biotrophic and necrotrophic phases of the P. infestans infection cycle. The current economic importance of this pathogen and its roles in the history of Ireland and in the establishment of the field of plant pathology means that this investigation is a significant step forward in applied and fundamental late blight research.
Supplementary Material
Supporting Tables
Acknowledgments
We thank Vivianne Vleeshouwers and Richard Visser (both of Wageningen University) for tissue samples (T68.4-002 and SW8563-0182) and Petra Boevink (Scottish Crop Research Institute) for pRTL2::gfp and for critically reading the manuscript. This work was supported by the Scottish Executive Environment and Rural Affairs Department, the Biotechnology and Biological Sciences Research Council, and the National Science Foundation. J.I.B.B. and S.K. were supported by National Science Foundation Plant Genome Research Program Grant DBI-0211659.
Author contributions: S.C.W., A.P.R., S.K., J.L.B., and P.R.J.B. designed research; M.R.A., S.C.W., J.I.B.B., E.V., A.O.A., A.P.R., K.B., I.C., N. Hamlin, B.W., A.F., A.L., M.A.Q., and C.C. performed research; S.H. contributed new reagents/analytic tools; M.R.A., S.C.W., L.P., U.B., N. Hall, M.B., S.K., and P.R.J.B. analyzed data; and M.R.A., S.C.W., L.P., S.K., and P.R.J.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SP, signal peptide; Avr, avirulence; LD, linkage disequilibrium; BAC, bacterial artificial chromosome; CDS, coding sequence; HR, hypersensitive response; R, resistance; pex, Phytophthora extracellular protein.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AC146942 (Pi-BAC-61F2), AJ893356 (Pi-BAC-35J4), and AJ893357 (Pi-BAC-49P21)].
References
- 1.Birch, P. R. J. & Whisson, S. C. (2001) Mol. Plant Pathol. 2**,** 257–263. [DOI] [PubMed] [Google Scholar]
- 2.Schiermeier, Q. (2001) Nature 410**,** 1011. [DOI] [PubMed] [Google Scholar]
- 3.Duncan, J. M. (1999) Microbiol. Today 26**,** 114–116. [Google Scholar]
- 4.Kamoun, S. (2003) Eukaryot. Cell 2**,** 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sogin, M. L. & Silberman, J. D. (1998) Int. J. Parasitol. 28**,** 11–20. [DOI] [PubMed] [Google Scholar]
- 6.Gebhardt, C. & Valkonen, J. P. T. (2001) Annu. Rev. Phytopathol. 39**,** 79–102. [DOI] [PubMed] [Google Scholar]
- 7.Huang, S., Vleeshouwers, V. G. A. A., Werij, J. S., Hutten, R. C. B., van Eck, H. J., Visser, R. G. F. & Jacobsen, E. (2004) Mol. Plant–Microbe Interact. 17**,** 428–435. [DOI] [PubMed] [Google Scholar]
- 8.Huang, S., van der Vossen, E. A. G., Huang, H., Vleeshouwers, V. G. A. A., Zhang, N., Borm, T. J. A., van Eck, H. J., Baker, B., Jacobsen, E. & Visser, R. (2005) Plant J. 42**,** 251–261. [DOI] [PubMed] [Google Scholar]
- 9.van der Lee, T., Robold, A., Testa, A., van't Klooster, J. W. & Govers, F. (2001) Genetics 157**,** 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joosten, M. H. A. J., Vogelsang, R., Cozignsen, T. J., Verberne, M. C. & de Wit, P. J. G. M. (1997) Plant Cell 9**,** 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luderer, R., Takken, F. L., de Wit, P. J. G. M. & Joosten, M. H. (2002) Mol. Microbiol. 45**,** 875–884. [DOI] [PubMed] [Google Scholar]
- 12.Rohe, M., Gierlich, A., Hermann, H., Hahn, M., Schmidt, B., Rosahl, S. & Knogge, W. (1995) EMBO J. 14**,** 4167–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bos, J. I. B., Armstrong, M. Whisson, S. C., Torto, T. A., Ochwo, M., Birch P. R. J. & Kamoun, S. (2003) New Phytol. 159**,** 63–72. [DOI] [PubMed] [Google Scholar]
- 14.Torto, T. A., Li, S., Styer, A., Huitema, E., Testa, A., Gow, N. A. R., van West, P. & Kamoun, S. (2003) Genome Res. 13**,** 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shan, W., Cao, M., Leung, D. & Tyler, B. (2004) Mol. Plant–Microbe Interact. 17**,** 394–403. [DOI] [PubMed] [Google Scholar]
- 16.Whisson, S. C., van der Lee, T., Bryan, G., Waugh, R., Govers, F. & Birch, P. R. J. (2001) Mol. Genet. Genomics 266**,** 289–295. [DOI] [PubMed] [Google Scholar]
- 17.Harris, D. E. & Murphy, L. (2001) in Methods in Molecular Biology, eds. Starkey, M. P. & Elaswarapu, R. (Humana, Totowa, NJ), pp. 217–234.
- 18.Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A. & Barrell, B. (2000) Bioinformatics 16**,** 944–945. [DOI] [PubMed] [Google Scholar]
- 19.Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M. D., et al. (2000) Bioinformatics 16**,** 1145–1150. [DOI] [PubMed] [Google Scholar]
- 20.Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. (2001) J. Mol. Biol. 305**,** 567–580. [DOI] [PubMed] [Google Scholar]
- 21.Avrova, A. O., Venter, E., Birch, P. R. J. & Whisson, S. C. (2003) Fungal Genet. Biol. 40**,** 4–14. [DOI] [PubMed] [Google Scholar]
- 22.Liu, Z., Bos, J. I., Armstrong, M., Whisson, S. C., da Cunha, L., Torto-Alalibo, T., Win, J., Avrova, A. O., Wright, F., Birch, P. R. J. & Kamoun, S. (2005) Mol. Biol. Evol. 22**,** 659–672. [DOI] [PubMed] [Google Scholar]
- 23.Restrepo, M. A., Freed, D. D. & Carrington, J. C. (1990) Plant Cell 2**,** 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, L. Hamilton, A. J., Voinnet, O., Thomas, C. L., Maule, A. J. & Baulcombe, D. C. (1999) Plant Cell 11, 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van der Hoorn, R. A., Laurent, F., Roth, R. & De Wit, P. J. (2000) Mol. Plant–Microbe Interact. 13**,** 439–446. [DOI] [PubMed] [Google Scholar]
- 26.Barton, N. H. (2000) Philos. Trans. R. Soc. London B 355**,** 1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leister, R. T., Ausubel, F. M. & Katagiri, F. (1996) Proc. Natl. Acad. Sci. USA 93**,** 15497–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia, J., McAdams, S. A., Bryan, G. T., Hershey, H. P. & Valent, B. (2000) EMBO J. 19**,** 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen, R. L., Bittner-Eddy, P. D., Grenville-Briggs, L. J., Meitz, J. C., Rehmany, A. P., Rose, L. E. & Beynon, J. L. (2004) Science 306**,** 1957–1960. [DOI] [PubMed] [Google Scholar]
- 30.Dodds, P. N., Lawrence, G. J., Catanzariti, A., Aycliffe, M. A. & Ellis, J. G. (2004) Plant Cell 16**,** 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collmer, A., Lindeberg, M., Petnicki-Ocwieja, T., Schneider, D. J. & Alfano, J. R. (2002) Trends Microbiol. 10**,** 462–469. [DOI] [PubMed] [Google Scholar]
- 32.Rehmany, A. P., Gordon, A., Allen, R. L., Armstrong, M. R., Whisson, S. C., Kamoun, S., Tyler, B. M., Birch P. R. J. & Beynon, J. L. (2005) Plant Cell, in press. [DOI] [PMC free article] [PubMed]
- 33.Dujon, B., Sherman, D., Fischer, G., Durrens, P., Caseregola, S., Lafontaine, I., de Montigny, J. Marck, C., Neuvéglise, C., Talla, E., et al. (2004) Nature 430**,** 35–44.15229592 [Google Scholar]
- 34.Han, F., Kilian, A., Chen, J. P., Kudrna, D., Steffenson, B., Yamamoto, K., Matsumoto, T., Sasaki, T. & Kleinhofs, A. (1999) Genome 42**,** 1071–1076. [DOI] [PubMed] [Google Scholar]
- 35.Riethmüller, A., Voglmayr, H., Göker, M., Weiss, M. & Oberwinkler, F. (2002) Mycologia 94**,** 834–849. [DOI] [PubMed] [Google Scholar]
- 36.Göker, M., Voglmayr, H., Riethmüller, A., Weiss, M. & Oberwinkler, F. (2003) Can. J. Bot. 81**,** 672–683. [Google Scholar]
- 37.Goodwin, S. B., Cohen, B. A. & Fry, W. (1994) Proc. Natl. Acad. Sci. (USA) 91**,** 11591–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Tables