Functional and Antigenic Characterization of Human, Rhesus Macaque, Pigtailed Macaque, and Murine DC-SIGN (original) (raw)
Abstract
DC-SIGN, a type II membrane protein with a C-type lectin binding domain that is highly expressed on mucosal dendritic cells (DCs) and certain macrophages in vivo, binds to ICAM-3, ICAM-2, and human and simian immunodeficiency viruses (HIV and SIV). Virus captured by DC-SIGN can be presented to T cells, resulting in efficient virus infection, perhaps representing a mechanism by which virus can be ferried via normal DC trafficking from mucosal tissues to lymphoid organs in vivo. To develop reagents needed to characterize the expression and in vivo functions of DC-SIGN, we cloned, expressed, and analyzed rhesus macaque, pigtailed macaque, and murine DC-SIGN and made a panel of monoclonal antibodies (MAbs) to human DC-SIGN. Rhesus and pigtailed macaque DC-SIGN proteins were highly similar to human DC-SIGN and bound and transmitted HIV type 1 (HIV-1), HIV-2, and SIV to receptor-positive cells. In contrast, while competent to bind virus, murine DC-SIGN did not transmit virus to receptor-positive cells under the conditions tested. Thus, mere binding of virus to a C-type lectin does not necessarily mean that transmission will occur. The murine and macaque DC-SIGN molecules all bound ICAM-3. We mapped the determinants recognized by a panel of 16 MAbs to the repeat region, the lectin binding domain, and the extreme C terminus of DC-SIGN. One MAb was specific for DC-SIGN, failing to cross-react with DC-SIGNR. Most MAbs cross-reacted with rhesus and pigtailed macaque DC-SIGN, although none recognized murine DC-SIGN. Fifteen of the MAbs recognized DC-SIGN on DCs, with MAbs to the repeat region generally reacting most strongly. We conclude that rhesus and pigtailed macaque DC-SIGN proteins are structurally and functionally similar to human DC-SIGN and that the reagents that we have developed will make it possible to study the expression and function of this molecule in vivo.
Attachment of human immunodeficiency virus (HIV) to the cell surface can occur independently of envelope (Env) protein interactions with CD4, the major HIV type 1 (HIV-1) receptor. A variety of cell surface molecules have been shown to support virus attachment and to increase the efficiency of virus infection (10, 11, 19, 20). Cellular proteins incorporated into virus particles can also impact virus attachment and infection efficiency (3, 7, 9, 15). DC-SIGN is a type II integral membrane protein that avidly binds primary and lab-adapted HIV-1, HIV-2, and simian immunodeficiency virus (SIV) strains but does not, by itself, mediate virus infection (5, 12, 14). Rather, DC-SIGN appears to function as a universal attachment factor for primate lentiviruses. Binding of DC-SIGN to Env is dependent largely if not exclusively on carbohydrate recognition, involving interactions between the lectin binding domain of DC-SIGN and the gp120 subunit of Env (5, 12). It is not known if DC-SIGN also binds to the gp41 transmembrane domain subunit. A closely related homologue of DC-SIGN, termed DC-SIGNR (17), also binds and transmits multiple virus strains (1, 14).
DC-SIGN is of particular interest because its expression is largely restricted to immature dendritic cells (DCs) and certain types of macrophages in vivo (6; E. J. Soilleux, L. S. Morris, G. Leslie, J. Chehimi, J. Trowsdale, L. J. Montaner, R. W. Doms, D. Weissman, N. Coleman and B. Lee, submitted for publication). Natural ligands of DC-SIGN include ICAM-3 and ICAM-2, indicating that DC-SIGN may play an important role in DC trafficking and in interactions with naïve T lymphocytes (4, 6). In addition, DC-SIGN can mediate binding of virus to DCs in vitro and once bound virus can remain infectious for days (5). Interestingly, virus bound to DCs via DC-SIGN can be efficiently presented or transmitted to receptor-positive cell types (5). This finding raises the possibility that DC-SIGN-positive DCs may serve as a conduit for HIV transmission, providing a mechanism by which virus can usurp the normal trafficking pathways of DCs and be delivered from mucosal surfaces to lymphoid organs (5). Thus, it will be important to further characterize the expression patterns of DC-SIGN in vivo and the mechanisms by which DC-SIGN interacts with and transmits virus. In addition, it will be important to study DC-SIGN homologues from species used as animal models for HIV and AIDS and from mice, as this species affords an opportunity to study the normal functions of DC-SIGN in vivo.
In this study, we report the cloning of rhesus macaque, pigtailed macaque, and murine DC-SIGN. Rhesus and pigtailed macaque DC-SIGN proteins were highly similar to human DC-SIGN. By contrast, murine DC-SIGN exhibited significant homology to human DC-SIGN in the lectin binding domain and transmembrane domain of the protein but not in other regions. All three of these proteins bound ICAM-3 as well as HIV and SIV strains. In addition, rhesus and pigtailed macaque DC-SIGN molecules could transmit bound virus to receptor-positive cell types. By contrast, virus bound to murine DC-SIGN was not transmitted to receptor-positive cells, indicating that binding of virus to a C-type lectin protein does not always result in efficient virus transmission. Using a bacterial fusion protein as an immunogen, we produced and characterized a panel of monoclonal antibodies (MAbs) to DC-SIGN, identifying antibodies to at least two determinants in the repeat region, to the lectin binding domain, and to the extreme C terminus of the protein. One of the MAbs was DC-SIGN specific—it did not cross-react with human DC-SIGNR. Nearly all of the MAbs reacted with DC-SIGN on the surface of peripheral blood-derived DCs (PBDCs), and many cross-reacted with pigtailed and/or rhesus macaque DC-SIGN. None reacted with murine DC-SIGN. Our results indicate that DC-SIGN from rhesus and pigtailed macaques is functionally and antigenically similar to human DC-SIGN, while murine DC-SIGN exhibits important sequence and functional differences. The DC-SIGN-specific MAbs described here will be useful for studying DC-SIGN and DC-SIGNR expression patterns in vitro and in vivo in humans and nonhuman primates.
MATERIALS AND METHODS
Cells, antibodies, and reagents.
293T cells were used for transient-transfection experiments. To obtain DCs from peripheral blood mononuclear cells (PBMC), we used the method originally described by Sallusto et al. with minor modifications (16, 21). Briefly, monocytes were purified from PBMC by discontinuous Percoll gradient centrifugation. The low-density fraction (monocyte enriched) was depleted of B, T, and, in certain experiments, NK cells by using magnetic beads (Dynal, Lake Success, N.Y.) specific for CD2, CD16, CD19, and CD56. This resulted in highly purified monocytes as determined by flow cytometry using anti-CD14 (95%) or anti-CD11c (98%) MAb. To generate immature DCs, purified monocytes were cultured in either RPMI 1640 supplemented with glutamine (2 mM) and HEPES (15 mM) or in RPMI 1640 with granulocyte-macrophage colony-stimulating factor (GM-CSF) (50 ng/ml) and interleukin 4 (100 ng/ml). Phycoerythrin-conjugated goat anti-mouse Fab fragment (Caltag, Burlingame, Calif.) was used as secondary antibody for flow cytometry experiments. DC-SIGN hybridoma supernatants or mouse ascites were used at the indicated concentrations. For enzyme-linked immunosorbent assay (ELISA) screening the following peptides were used: PEKSKLQEIYQELTRLKAA (ND), WTFFQGNCYFMSNSQRNWHD (LD1), TWMGLSDLNQEGTWQWVDG (LD1), CAEFSGNGWNDDKCNLAKFWIC (LD3), and KKSAASCSRDEEQFLSPAPATPNPPPA (C terminus). The isotyping of the MAbs was carried out using a commercially available kit as directed by the manufacturer (Boehringer Mannheim, Indianapolis, Ind.).
Plasmids.
The human DC-SIGN, the human DC-SIGN mutants, and the human DC-SIGNR constructs have been previously described (12, 14). The cloning of pigtailed macaque DC-SIGN cDNA was as follows. Heparinized blood was collected by standard venipuncture from a healthy SIV- and simian retrovirus (SRV)-negative pigtailed macaque (Macaca nemestrina). PBMC were isolated by Ficoll-Hypaque density gradient and were plated at 5 × 107 per 10 ml of RPMI medium supplemented with 10% fetal bovine serum, 10 mM HEPES, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) for 2 h at 37°C. Nonadherent cells were removed with two washes of phosphate-buffered saline (PBS). The adherent cells were further cultured with 800 U of recombinant human GM-CSF/ml and 500 U of recombinant human interleukin 4 (R&D Systems, Minneapolis, Minn.)/ml. The culture media were replaced every 2 to 3 days, and after 7 days, the immature DCs were harvested and lysed and total RNA was prepared using the Qiagen RNeasy kit. The pigtailed macaque DC-SIGN homologue was cloned by reverse transcriptase PCR (RT-PCR) using SIGN-1 (5′-AGA GTG GGG TGA CAT GAG TGA CTC-3′) and SIGN-END (5′-GTG AAG TTC TGC TAC GCA GGA G-3′) primers. Both oligo(dT) priming and random hexamer priming were used for cDNA synthesis. A portion of each cDNA reaction was used for PCR amplification under the following conditions: 94°C for 3 min followed by 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 2 min. This procedure yielded a 1.1-kb fragment from both the oligo(dT)- and random hexamer-primed cDNA reactions. Products were subsequently blunt end cloned into pBluescript KS(+) and were sequenced. One representative clone was used for further study. The sequence of this clone was deposited (see end of Materials and Methods). Finally, the pigtailed macaque DC-SIGN cDNA was excised from pKS+ with _Eco_RI and _Sal_I and cloned into pcDNA3 using the _Eco_RI and _Xho_I sites.
The cloning of rhesus macaque DC-SIGN cDNA was as follows. The rhesus macaque DC-SIGN cDNA was amplified by RT-PCR using total RNA from spleen as the template. Total RNA was obtained by homogenization of snap-frozen rhesus macaque spleen tissue in Trizol (Life Technologies, Rockville, Md.) and by isopropanol precipitation. Reverse transcription was performed using oligo(dT) and avian myeloblastosis virus RT (Promega, Madison, Wis.) at 45°C for 45 min. PCR was subsequently performed using the resulting cDNA as template and primers YKCRHDCSIGNF1 (5′ATGAGTGACTCCAAGGAACCAA-3′) and YKCRHDCSIGNR3 (5′-CTACGCAGGAGGGGGGTTTGGGGT-3′) at 1 μM with 1.5 mM MgCl2 and Taq polymerase for 30 cycles of amplification. Gel-isolated RT-PCR product was agarose gel purified, subcloned into the pGEM-T in vitro transcription vector (Promega), and DNA sequenced using automated and manual strategies. Finally, rhesus macaque DC-SIGN was cloned into pcDNA3 and sequenced. It is to be noted that the primers were designed according to the human coding sequence of DC-SIGN; therefore, the sequence integrity of the first 7 and last 8 amino acids cannot be guaranteed.
Murine DC-SIGN was identified by homology search using the GenBank database sequence for human DC-SIGN, resulting in several mouse expressed sequence tags that could be assembled into a 0.75-kb core sequence. This sequence was subjected to a second GenBank database search that provided information about the genomic organization of the gene (accession number AC073706). RNA (Trizol; Life Technologies) for cDNA synthesis was obtained from murine DCs generated from the bone marrow of C57/BL6 mice cultured for 10 days in vitro in the presence of 200 U of rhesus macaque GM-CSF (Peprotech, Rocky Hill, NJ)/ml. First-strand cDNA was prepared using oligo(dT) and the SuperScript kit (Life Technologies), and murine DC-SIGN was amplified using Expand High Fidelity Polymerase (Roche, Indianapolis Ind.). End primers for PCR amplification based on genomic information were 5′ primer TGA CTC CAC AGA AGC CAA GAT GC; and 3′ primer, AAT GAA ACT ATG ATA AAT GCA GAG GAT GAA. The PCR product was gel purified and cloned into pCR2.1 TOPO vector (Invitrogen, Carlsbad, Calif.). The N-terminally truncated sequence, i.e., the one missing the 8 N-terminal amino acids (MSDSTEAK), is 100% identical to the sequence under the accession number AK007656 (murine pancreas cDNA-enriched library). This murine DC-SIGN construct was subcloned into pcDNA3 and pcDNA4. Subsequently, we added to this construct the eight N-terminal residues from human DC-SIGN and found that this molecule functioned identically to the original murine DC-SIGN construct.
Protein production and purification.
To express the ecto- and lectin domains of DC-SIGN, we used the bacterial pBAD/TOPO ThioFusion expression system (Invitrogen). Briefly, the ecto- and lectin domains of DC-SIGN were amplified by PCR using the following pairs of primers: 5′-GGTCCCCAGCTCCATAAGTCA-3′ (p5N unique) and 5′-CGC AGG AGG GGG GTT TGG GGT-3′ (p3C lectin) and 5′-TGC CAC CCC TGT CCC TGG GAA TGG-3′ (p5N lectin) and p3C lectin, respectively. Both fragments were cloned into the pBAD/Thio-TOPO expression vector by TA cloning. Insert-containing colonies were used to transform TOP10 Escherichia coli cells and were screened for protein expression. DNA from expressing colonies was extracted and sequenced for sequence integrity. A 1-liter culture for each of the two clones was grown and induced, and the proteins were purified under denaturing conditions as specified by the manufacturer. The purified proteins were dialyzed against PBS and quantified, and their integrity was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on gels stained with Coomassie blue.
DC-SIGN MAb and rabbit polyclonal serum production.
BALB/c mice were immunized weekly four times intradermally with 100 μg of bacterial ectodomain of human DC-SIGN. Four days after the last injection, spleen cells were obtained and fused to the murine myeloma cell line SP2, as previously described (1a). Hybridoma supernatants were screened by ELISA for reactivity with the human DC SIGN ectodomain immunogen but not against a negative control protein (thioredoxin) and were cloned by limiting dilution. A rabbit polyclonal serum was generated against the ectodomain of human DC-SIGN (Cocalico Biologicals Inc., Reamstown, Pa.) by standard immunization procedures using Freund's incomplete adjuvant.
Flow cytometry.
CaPO4-transfected 293T cells or in vitro-derived DCs were stained in fluorescence-activated cell sorter (FACS) buffer (PBS supplemented with 3% fetal calf serum and 0.02% sodium azide) for 30 min on ice with either hybridoma supernatants at a final dilution of 1/20 or ascites at a final concentration of 10 μg/ml. The samples were washed and incubated with phycoerythrin-conjugated goat anti-mouse Fab fragments (Caltag) (1/100) for 30 min on ice and were then washed and resuspended in FACS buffer containing 2% paraformaldehyde. The samples were analyzed with a FACScan (Becton Dickinson, San Jose, Calif.) cell analyzer using the CellQuest software for data evaluation. Dead cells were excluded on the basis of their forward and side scatter characteristics.
Western blots.
Purified bacterial proteins or cleared cell lysates from 293T cells transfected with the indicated construct were analyzed by immunoblotting. Proteins were detected with a 1:20 dilution of DC-SIGN MAb hybridoma supernatant or by DC-SIGN MAb ascites at 0.5 μg/ml.
Assessment of DC-SIGN-mediated virus binding and infection in trans.
The efficiency of DC-SIGN-mediated virus transfer was assessed in a cocultivation assay as previously described (12, 14). Briefly, 293T cells were transfected with the different DC-SIGN clones, and 24 h after transfection, cells were seeded in 96-well dishes. The following day, the transfected cells were incubated with luciferase reporter virus for 3 to 5 h at 37°C. Thereafter, the cells were washed several times with fresh Dulbecco's modified Eagle medium. To assess virus binding, the cells were lysed in 0.5% Triton X-100, and the amount of bound viral antigen was quantified using commercially available p24 or p27 ELISA kits (Coulter Beckman, Miami, Fla.). To determine infection in trans, the cells were cocultivated with C8166 T cells. Two days after cocultivation, the medium was changed, and 24 h later, the cells were lysed. Luciferase activity in 25-μl cell lysate was determined using a commercially available kit (Promega).
ICAM-3 binding assays.
Soluble Fc–ICAM-3 protein (R&D Systems) was iodinated by using Iodogen (Pierce). Specific activities of 500 to 2,000 Ci/mmol were obtained by using 5 μg of protein with 500 μCi of Na125I for 20 min in 5-ml glass tubes precoated with 10 μg of Iodogen by chloroform evaporation. Radiolabeled proteins were purified from free Na125I by separation through a 0.3-ml Dowex column prepared in a 1-ml syringe and were preequilibrated in a mixture containing 50 mM HEPES (pH 7.4), 5 mM MgCl2, 1 mM CaCl2, 1% bovine serum albumin (BSA), and 150 mM NaCl. Protein fractions were eluted in the void volume of the column, and the fractions containing peaks of labeled protein were combined. Human DC-SIGN-, human DC-SIGNR-, pigtailed macaque DC-SIGN-, rhesus macaque DC-SIGN-, and murine DC-SIGN-transfected 293T cells (48 h at 37°C) were washed once with PBS and resuspended in binding buffer (50 mM HEPES (pH 7.4), 2 mM magnesium chloride, 2 mM calcium chloride, and 0.5% BSA). Cells (n = 106) were incubated with 50,000 cpm of Fc–ICAM-3 for 60 min at room temperature. Cells were collected onto Brandel-grade GF/B filters with wash buffer (same as binding buffer plus 150 mM sodium chloride and no BSA) using a cell harvester. Filters were counted using a Wallac Wizard 1470 automatic gamma counter. Percent binding was determined by dividing the counts from the filters by the input radioactivity after deduction of the background counts obtained on pcDNA3-transfected cells.
Nucleotide sequence accession number.
The sequence of the representative clone described above has been deposited in GenBank under the number AF343727. The sequence of the cloned rhesus macaque DC-SIGN has been deposited in GenBank under the number AF369755.
RESULTS
Cloning and sequence analysis of rhesus macaque, pigtailed macaque, and murine DC-SIGN.
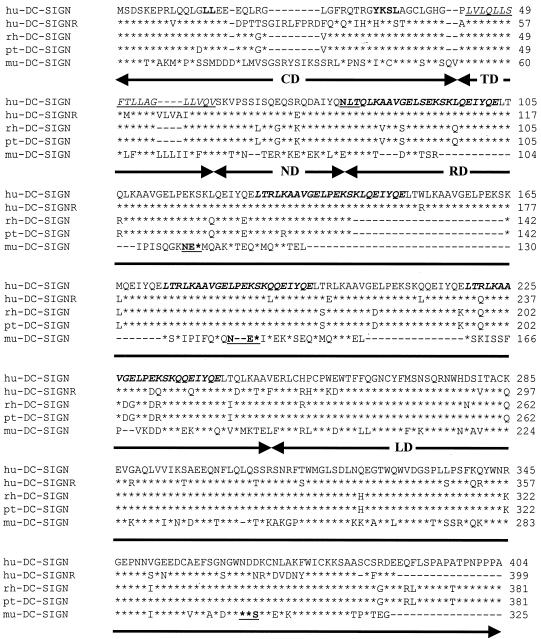
Because the SIV/macaque system should make it possible to directly test the role of DC-SIGN in virus transmission in vivo, we cloned rhesus and pigtailed macaque DC-SIGN. Murine DC-SIGN was also cloned to provide an additional tool for the evaluation of the panel of MAbs described below and to exploit the outstanding opportunity that the murine system affords to study normal DC-SIGN function in vivo. Pigtailed macaque and murine DC-SIGN molecules were cloned from RNA extracted from in vitro-cultured DCs, while rhesus macaque DC-SIGN was cloned from spleen RNA. All three clones were placed into a mammalian expression vector. Rhesus and pigtailed macaque DC-SIGN differed from each other by only 5 amino acids and were highly similar to human DC-SIGN, sharing approximately 87% amino acid identify overall (Fig. 1). The degree of homology was highest in the lectin binding domain, with 93% amino acid identity. Endocytosis signals present in the cytoplasmic domain and the single N-linked glycosylation site were also conserved (Fig. 1). The rhesus and pigtailed macaque DC-SIGN clones that we analyzed, however, had 6.5 repeats of the 23-residue sequence, whereas human DC-SIGN had 7.5 repeats of this sequence. Variability in the number of repeat regions has been noted in DC-SIGNR, a DC-SIGN homologue in which the two most common isoforms contain seven and five repeats (1). However, both isoforms function as virus attachment factors (1, 14). Whether there is variability in the number of repeat sequences in rhesus and pigtailed macaque sequences remains to be determined.
FIG. 1.
Protein sequence comparison of DC-SIGN molecules of human, simian, and mouse origin. Human DC-SIGN (hu-DC-SIGN), human DC-SIGNR (hu-DC-SIGNR), rhesus macaque DC-SIGN (rh-DC-SIGN), pigtailed macaque DC-SIGN (pt-DC-SIGN), and murine DC-SIGN (mu-DC-SIGN) protein sequences were aligned using ClustalW. Asterisks indicate amino acid identity with the human DC-SIGN sequence. Dashes indicate gaps introduced to maximize amino acid identity. The consensus dileucine and YXXL internalization motifs are in boldface. The putative transmembrane domain is underlined and italicized. The repeat motifs are indicated in alternating fashion by boldface and italics or plain text. The N-linked glycosylation motifs are underlined and boldfaced. Arrows below the alignment highlight the different domains of the DC-SIGN molecule: CD, cytoplasmic domain; TD, transmembrane domain; ND, N-terminal domain; RD, repeat domain; LD, lectin binding domain.
Murine DC-SIGN shared 68% identity with human DC-SIGN in the lectin binding domain. However, the remainder of the molecule was quite divergent (Fig. 1). Murine DC-SIGN contained a cytoplasmic domain estimated to be 51 residues long that lacked the endocytosis signals present in the primate DC-SIGN sequences examined in this research. Murine DC-SIGN also lacked the N-linked glycosylation site present in the neck domain of the primate DC-SIGN molecules but contained three N-linked consensus sites not present in these molecules, including one in the lectin binding domain. While there was 81% amino acid identity between murine DC-SIGN and human DC-SIGN in the transmembrane domain, significant differences were present in the repeat region. Murine DC-SIGN contained approximately 2.5 copies of a motif that had some similarity to the human sequence. For example, the murine repeat region was glutamine rich, with a predicted secondary structure displaying significant alpha-helical content that was similar to the human sequence. Thus, the repeat region of murine DC-SIGN is much shorter than that of human and nonhuman primate DC-SIGN and contains significant sequence differences.
Expression and functional analysis of rhesus macaque, pigtailed macaque, and murine DC-SIGN.
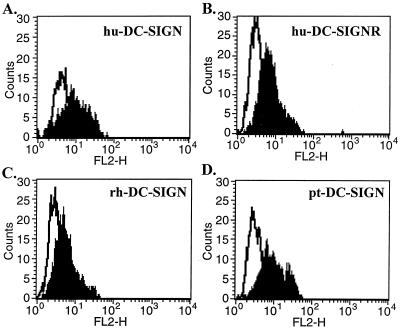
Due to their high degree of similarity, we reasoned that rhesus and pigtailed macaque DC-SIGN molecules would be detected by a rabbit antiserum that we generated against the entire ectodomain of human DC-SIGN. Human 293T cells were transfected with plasmids encoding human, rhesus macaque, pigtailed macaque, or murine DC-SIGN, stained with the antiserum, and analyzed by FACS. We found that antiserum to the ectodomain of human DC-SIGN specifically reacted with cells transiently expressing human DC-SIGNR as well as with those expressing rhesus and pigtailed macaque DC-SIGN (Fig. 2). The antiserum did not recognize murine DC-SIGN (data not shown). To confirm that murine DC-SIGN was in fact expressed, we placed an AU1 antigenic tag at the C terminus of the protein. It has previously been shown that an AU1 tag at this position does not impact the expression or function of human DC-SIGN (13). We found that AU1-tagged murine DC-SIGN was expressed at high levels (data not shown). We consistently observed less intense staining of cells expressing rhesus macaque DC-SIGN but noted that the antiserum recognized pigtailed macaque DC-SIGN, which differs from rhesus DC-SIGN at only four positions in the ectodomain, as efficiently as it recognized human DC-SIGN. This makes it likely that rhesus macaque DC-SIGN was not expressed as well as human and pigtailed macaque DC-SIGN under the conditions examined here.
FIG. 2.
Flow cytometry analysis of DC-SIGN molecules of human, simian, and mouse origin. 293T cells were transfected with human DC-SIGN (hu-DC-SIGN) (A), human DC-SIGNR (hu-DC-SIGNR) (B), rhesus macaque DC-SIGN (rh-DC-SIGN) (C), pigtailed macaque DC-SIGN (pt-DC-SIGN) (D), and murine DC-SIGN (mu-DC-SIGN) (data not shown). Forty-eight hours posttransfection, the cells were analyzed for DC-SIGN expression by flow cytometry using a rabbit polyclonal serum generated against the ectodomain of human DC-SIGN. Transfected cells stained with the preimmune-phase sera were used as negative controls (white profile) for the cells transfected with DC-SIGN constructs (shaded profile).
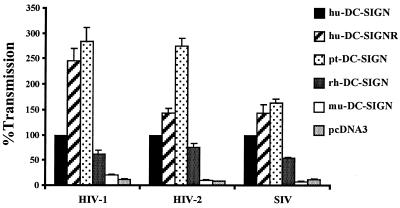
To determine if rhesus, pigtailed macaque, and murine DC-SIGN could bind and transmit HIV and SIV, we expressed the indicated DC-SIGN molecules in 293T cells. The next day, HIV-1, HIV-2, or SIV luciferase reporter virus was added for 3 to 5 h, after which unbound virus was removed by vigorous washing. Receptor-positive C8166 T cells were added, and virus infection was measured 3 days later by quantifying luciferase activity in the cell lysate. Each infection was performed in triplicate in each experiment. We found that pigtailed and rhesus macaque DC-SIGN transmitted all three virus strains to receptor-positive cells (Fig. 3). In a representative experiment, HIV-2 ROD that was bound to DC-SIGN-positive cells infected C8166 cells, giving an average relative light unit value of 5,341. When 293T cells expressing vector alone were used, only 519 relative light units were transmitted. Similar values were obtained with the other viruses. While there was excellent reproducibility within an experiment, differences in absolute signals were evident between experiments due to the fact that different virus stocks were used and due to the fact that the indicated attachment factors were expressed transiently. Therefore, to normalize data between experiments, the amount of virus transmitted to C8166 cells from cells expressing human DC-SIGN was set to 100% for each virus.
FIG. 3.
Rhesus and pigtailed macaque DC-SIGN but not murine DC-SIGN transmits HIV-1, HIV-2, and SIV. 293T cells were transiently transfected with the indicated DC-SIGN expression vectors; incubated with HIV-1 (NL4-3), HIV-2 (ROD10), and SIV (SIVmac239 MER Env) replication-competent luciferase reporter viruses; vigorously washed; and cocultivated with C8166 T cells. The luciferase activity in the cultures was determined 3 days after the start of the coculture. We used pcDNA3 as a negative control. All data are normalized to the transmission obtained, with human DC-SIGN set at 100%. A representative experiment out of two done in triplicate is shown ± standard error of the mean.
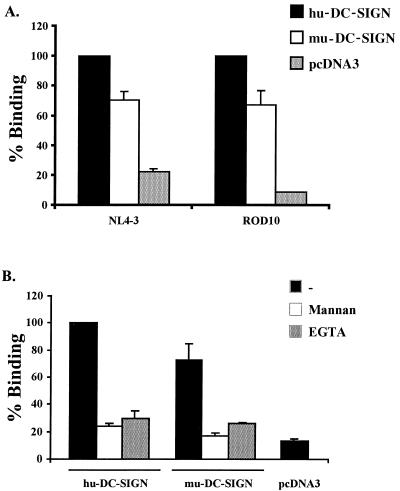
We found that pigtailed macaque DC-SIGN transmitted virus more efficiently than human DC-SIGN, while rhesus macaque DC-SIGN transmitted virus somewhat less efficiently. However, it has previously been shown that DC-SIGN expression levels have a significant impact on virus binding and transmission (12), and rhesus macaque DC-SIGN was expressed less efficiently than human DC-SIGN. Thus, the differences in transmission efficiency seen here could be due to variability in expression levels. In contrast to cells expressing primate DC-SIGN molecules, cells expressing murine DC-SIGN did not transmit virus under the conditions tested (Fig. 3). We therefore tested the ability of cells expressing murine DC-SIGN to bind virus and found that both HIV-1 NL4-3 and HIV-2 ROD10 bound specifically to cells expressing murine DC-SIGN, though at levels slightly below those obtained with human DC-SIGN (Fig. 4A). In one experiment, 1,347 pg of HIV-2 ROD10 bound to DC-SIGN-positive cells, 910 pg bound to cells expressing murine DC-SIGN, and 48 pg bound to cells expressing vector alone. Similar signal-to-noise ratios were obtained in other experiments, but due to variability inherent in transient-expression assays, we normalized the amount of virus bound to human DC-SIGN-expressing cells in each experiment to 100%.
FIG. 4.
Murine DC-SIGN efficiently binds HIV-1 and HIV-2. DC-SIGN of human and murine origin was transiently overexpressed in 293T cells. (A) The cells were pulsed with replication-competent luciferase reporter viruses, vigorously washed, and lysed in 0.5% Triton X-100, and the p24 or p27 content of the lysates was quantified by ELISA. (B) Binding experiment done with HIV-1 NL4-3 in the presence of 20 μg of mannan/ml or 5 mM EGTA prior to washing. The data are normalized to virus binding by human DC-SIGN and show the results of an experiment performed in triplicate. Comparable results were obtained in an independent experiment. −, no treatment.
Binding could be largely eliminated by preincubation of the cells with either mannan or EGTA, consistent with the binding activity of a C-type lectin (Fig. 4B). Thus, the failure of cells expressing murine DC-SIGN to transmit virus is not due to an inability to bind virus. This finding indicates that virus binding to a C-type lectin does not necessarily result in virus transmission. Clearly, more detailed studies will have to be performed in order to investigate the ability of murine DC-SIGN to bind different virus strains and to transmit viruses to receptor-positive cells in different cellular contexts. Finally, soluble, iodinated ICAM-3 bound specifically to cells expressing human, rhesus macaque, pigtailed macaque, or murine DC-SIGN (data not shown).
Production of immunogens and MAb generation.
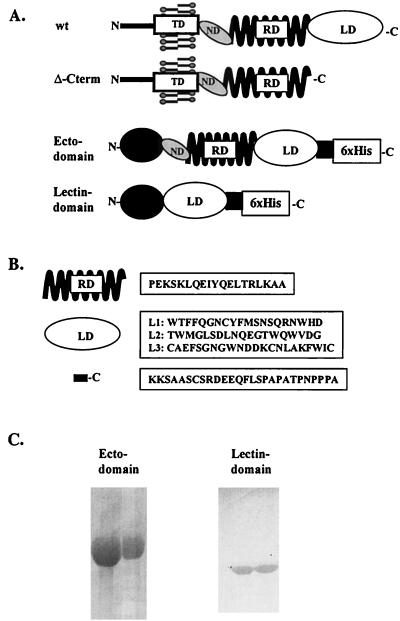
To provide the immunological reagents needed to study the role of DC-SIGN and DC-SIGNR in HIV/SIV pathogenesis, we used the bacterial pBAD/TOPO ThioFusion expression system to express the human DC-SIGN ectodomain and lectin binding domain (Fig. 5A). Both proteins were produced and purified to homogeneity using a nickel affinity column under denaturing conditions and were partially renatured by dialysis against PBS. The resulting proteins were pure, as judged by Coomassie blue staining following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 5C), and the ectodomain protein was used to immunize BALB/c mice. After three immunizations, a fusion between spleen cells and SP2 myeloma cells were performed and a hybridoma was generated. The hybridomas were screened by ELISA for reactivity against the ectodomain and lectin binding domain proteins (Table 1). Thioredoxin was used to identify antibodies that reacted against this portion of the fusion protein. We identified 16 clones that reacted with the ectodomain protein but not with thioredoxin alone (Table 1). Two clones (nos. 6 and 23) recognized both the ectodomain and lectin binding domain proteins.
FIG. 5.
Human DC-SIGN-derived constructs and reagents used to generate DC-SIGN MAbs. (A) Scheme of the different constructs made in pcDNA3, i.e., wild-type human DC-SIGN and Δ-C-term, and scheme of the two bacterial proteins produced, i.e., ectodomain and lectin binding domain. All were derived from human DC-SIGN. For abbreviations, see legend to Fig. 1. (B) Peptides derived from the human DC-SIGN molecule and used in the screening of the MAbs, with the domains from which they were derived depicted on the left side. RD, repeat domain; LD, lectin binding domain; C, C terminus of DC-SIGN. (C) Coomassie blue-stained protein gels of the purified bacterial ectodomain and lectin binding domain proteins.
TABLE 1.
ELISA reactivity of DC-SIGN MAbsa
| MAb | Isotype | Reactivity for: | ||||
|---|---|---|---|---|---|---|
| Ectodomain | Lectin binding domain | C terminus | L1, L2, and L3 | Repeat domain | ||
| 4 | IgG1 κ | + | − | − | − | − |
| 11 | IgG1 κ | + | − | − | − | − |
| 25 | IgG1 κ | + | − | − | − | − |
| 28 | IgG2a κ | + | − | − | − | − |
| 37 | IgG2a κ | + | − | − | − | − |
| 55 | IgG2a κ | + | − | − | − | − |
| 56 | IgG1 κ | + | − | − | − | − |
| 62 | IgG1 κ | + | − | − | − | − |
| 63 | IgG1 κ | + | − | − | − | − |
| 5 | IgG1 κ | + | − | − | − | + |
| 20 | IgG1 κ | + | − | − | − | + |
| 29 | IgG1 κ | + | − | − | − | + |
| 42 | IgG1 κ | + | − | − | − | + |
| 51 | IgG1 κ | + | − | − | − | + |
| 23 | IgG1 κ | + | + | − | − | − |
| 6 | IgG1 κ | + | + | + | − | − |
Epitope mapping and antibody characterization.
A total of 16 DC-SIGN-specific MAbs were analyzed in greater detail. Of these, 13 were of the immunoglobulin G1 (IgG1) isotype and 3 were IgG2a molecules. Peptides previously used to generate rabbit antisera to DC-SIGN were used for epitope mapping studies (Fig. 5B). Clone DC6, directed against the lectin binding domain, recognized the C-terminal peptide (Table 1), thus mapping its epitope to the extreme C-terminal end of DC-SIGN. Five clones (nos. 5, 20, 29, 42, and 51) reacted against the repeat domain peptide. None of the other peptides was recognized by any of the hybridomas.
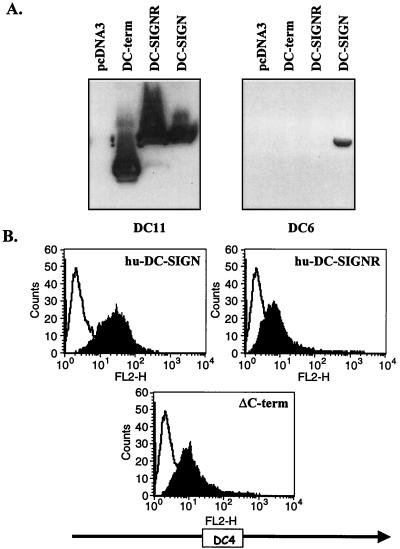
Each hybridoma was also examined for the ability to recognize full-length DC-SIGN and DC-SIGNR, as well as DC-SIGN lacking the lectin binding domain (Δ-C-term), by Western blotting and flow cytometry (Fig. 6). All of the MAbs recognized full-length DC-SIGN by Western blotting, suggesting that they recognized conformation-independent determinants. We confirmed that clones DC6 and DC23 were specific for the lectin binding domain by Western blotting (Fig. 6A and data not shown). FACS analyses in which Δ-C-term was expressed on the surface of 293T cells were consistent with the Western blot analyses (Fig. 6B and data not shown).
FIG. 6.
Characterization of DC-SIGN MAbs. (A) Western blot reactivity of DC11 and DC6 with lysates obtained from cells expressing either pcDNA3, Δ-C-term, human DC-SIGNR, or human DC-SIGN. (B) Flow cytometry analysis of human DC-SIGN-, Δ-C-term-, and human DC-SIGNR-transfected 293T cells with MAb DC4. Overlay histograms are shown where the white profile represents the staining obtained with an IgG1 isotype match control and the black profile shows the staining obtained with DC4. In both cases hybridoma supernatants were used at 10 μg/ml.
Antibody reactivity with pigtailed macaque, rhesus macaque, and murine DC-SIGN.
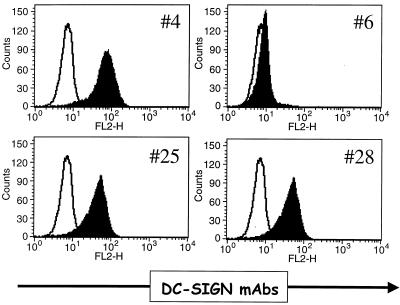
All of the MAbs were tested for the ability to recognize human DC-SIGNR, pigtailed macaque DC-SIGN, rhesus macaque DC-SIGN, and murine DC-SIGN by FACS. MAb DC6, which recognized an epitope in the C-terminal 27 residues of DC-SIGN, did not cross-react with rhesus DC-SIGN or pigtailed macaque DC-SIGN, which differ from the human sequence at four positions within the C-terminal epitope (Table 2). DC6 also failed to bind to human DC-SIGNR, which lacks much of the C-terminal epitope. MAb DC23, which binds to an epitope in the lectin binding domain, and DC56, which binds to the repeat domain, failed to bind to pigtailed macaque DC-SIGN and human DC-SIGNR but did bind to rhesus macaque DC-SIGN. Five additional MAbs which mapped to the repeat domain of human DC-SIGN recognized pigtailed macaque DC-SIGN and human DC-SIGNR but not rhesus macaque DC-SIGN, while seven MAbs recognized all three primate DC-SIGN molecules as well as human DC-SIGNR (Table 2). None of the MAbs recognized murine DC-SIGN. The ability of murine DC-SIGN to bind virus (Fig. 4) indicated that it was expressed at the cell surface, so the failure of any of our MAbs or antisera to recognize murine DC-SIGN reflects lack of cross-reactivity rather than lack of murine DC-SIGN expression. Finally, all of the MAbs that we have tested stained DC-SIGN on the surface of PBDCs. In general, the MAbs to the repeat region stained PBDCs more intensely than did MAbs to the lectin binding domain (Fig. 7 and data not shown). Despite being able to bind to DC-SIGN on the surface of cells, none of the MAbs potently inhibited binding of soluble ICAM-3 to DC-SIGN, nor did any of the MAbs effectively block virus transmission (data not shown). Given the role of carbohydrate recognition in DC-SIGN interactions with Env, it is likely that MAbs to the lectin binding domain, which were underrepresented in our panel of antibodies, will have a greater likelihood of inhibiting DC-SIGN function.
TABLE 2.
Flow cytometry reactivity of DC-SIGN MAbsa
| Clone | Isotype | Reactivity for: | ||||
|---|---|---|---|---|---|---|
| Human DC-SIGN | Human DC-SIGNR | Pigtailed macaque DC-SIGN | Rhesus macaque DC-SIGN | Murine DC-SIGN | ||
| 4 | IgG1 κ | + | + | + | + | − |
| 20 | IgG1 κ | + | + | + | + | − |
| 25 | IgG1 κ | + | + | + | + | − |
| 28 | IgG2a κ | + | + | + | + | − |
| 37 | IgG2a κ | + | + | + | + | − |
| 42 | IgG1 κ | + | + | + | + | − |
| 51 | IgG1 κ | + | + | + | + | − |
| 55 | IgG2a κ | + | + | + | + | − |
| 5 | IgG1 κ | + | + | + | − | − |
| 11 | IgG1 κ | + | + | + | − | − |
| 29 | IgG1 κ | + | + | + | − | − |
| 62 | IgG1 κ | + | + | + | − | − |
| 63 | IgG1 κ | + | + | + | − | − |
| 23 | IgG1 κ | + | − | − | + | − |
| 56 | IgG1 κ | + | − | − | + | − |
| 6 | IgG1 κ | + | − | − | − | − |
FIG. 7.
PBDC staining with a panel of DC MAbs. PBDCs were stained with a panel of DC MAbs. The white profile represents labeling with an isotype match control antibody and the black profile represents the staining with the corresponding DC MAb. In both cases the hybridoma supernatants were used at 1:20 dilution.
DISCUSSION
The infection of macaques with SIV or simian/human immunodeficiency viruses is the most commonly used animal model for HIV and AIDS. Although the viral receptors of macaque origin show high sequence homology to their human counterparts, several amino acid differences which affect receptor function have been described. For example, a single-amino-acid exchange in rhesus macaque CCR5 compared to the human sequence allows CD4-independent entry of many SIV strains (2), whereas a single-amino-acid substitution in rhesus macaque STRL33 blocks entry of SIV strains, which otherwise can use the human receptor for efficient entry (13). Thus, while we anticipated that the sequence and function of rhesus macaque DC-SIGN would be highly similar to those of its human counterpart, serving as both a virus attachment and transmission factor, it was important to clone and characterize it nonetheless to determine if the SIV/macaque system will make it possible to address the role of DC-SIGN in virus transmission and pathogenesis.
Rhesus DC-SIGN and pigtailed macaque DC-SIGN differed from each other at only five amino acid positions and were highly similar to human DC-SIGN. The only significant difference was in the repeat region, where rhesus and pigtailed macaque DC-SIGN contained 6.5 copies of a 23-amino-acid repeat sequence, compared to 7.5 copies in the human protein. However, this did not appear to have significant functional consequences, since both rhesus DC-SIGN and pigtailed macaque DC-SIGN supported HIV-1, HIV-2, and SIV binding and transmission. DC-SIGNR, a homologue of DC-SIGN expressed on certain types of endothelial cells that also functions as a universal viral attachment factor for HIV-1, HIV-2, and SIV strains (1, 14), exists in several isoforms which differ in the number of repeat sequences that they contain (1). Whether rhesus DC-SIGN and pigtailed macaque DC-SIGN exhibit similar variation or if they always contain 6.5 copies of the 23-amino-acid repeat is not known.
In contrast to the primate DC-SIGN molecules studied here, murine DC-SIGN exhibited considerable sequence variation, especially outside the lectin binding domain. Despite this, murine DC-SIGN was a type II membrane protein that proved capable of binding ICAM-3 as well as HIV-1 and HIV-2. However, while cells expressing murine DC-SIGN bound virus, they proved incapable of transmitting virus efficiently to receptor-positive cells under the conditions tested. The reason for this is not clear at present, but this observation shows that virus binding and transmission are dissociable functions, indicating that DC-SIGN may do something other than simply tether virus to the cell surface. A previous mutagenesis study indicated that the repeat region is important for DC-SIGN function (12). The lectin binding domain in murine DC-SIGN shares 68% amino acid identity with human DC-SIGN but lacks a repeat region. Instead, murine DC-SIGN contains a 116-amino-acid region that separates the transmembrane and lectin binding domain regions, compared to a 170-amino-acid region in human DC-SIGN. Whether this accounts for the failure of murine DC-SIGN to transmit virus could be tested through the construction of chimeric molecules.
The repeat region of DC-SIGN appears to be highly immunogenic, with the bulk of our MAbs mapping to determinants within this domain. Only a subset of the MAbs to this domain recognized a peptide based on the repeat sequence, indicating that the antibodies that we produced to the repeat region recognized at least two antigenic determinants. Given that carbohydrate recognition is important for DC-SIGN interactions with Env, it is perhaps not surprising that antibodies to the repeat region failed to block either ICAM-3 or virus binding to cells expressing DC-SIGN. To make recovery of antibodies to the lectin binding domain likelier, it may be important to immunize animals with the lectin binding domain alone or to produce soluble, native forms of DC-SIGN in the hopes of eliciting antibodies to conformational determinants in the lectin binding domain that will potently block DC-SIGN–ligand interactions.
An important question raised by the discovery of DC-SIGN as a specific attachment factor for primate lentiviruses concerns its potential role in virus transmission in vivo. DCs in the submucosa may at times be among the first cells encountered by HIV during sexual transmission (8, 18). If virus binds to these cells and retains infectivity, it could be transported to lymphoid organs as a consequence of normal DC trafficking. The SIV/macaque model affords an opportunity to test this hypothesis, provided that macaque DC-SIGN functions like its human homologue with regards to virus binding and transmission. We have previously shown that human DC-SIGN binds and transmits SIV strains (12). Here we show that both rhesus macaque DC-SIGN and pigtailed macaque DC-SIGN exhibit virus binding and transmission functions and that many of our MAbs recognize these proteins. If expression of DC-SIGN in these nonhuman primates is similar to that in humans, then studies employing antibodies that inhibit virus–DC-SIGN interactions could be used to test the role of DC-SIGN in virus transmission. In addition, the MAbs described here can be used to study DC-SIGN and DC-SIGNR expression and to further define the relationship between expression levels of DC-SIGN and its virus binding and transmission functions.
ACKNOWLEDGMENTS
We thank H. Ni and D. Weissman for supplying in vitro-derived dendritic cells, and we thank B. Lee for useful reagents.
This work was supported by NIH R01 35383 and 40880 grants to R.W.D. and by the Centers for AIDS Research at the University of Pennsylvania. This work was also supported by a Burroughs Wellcome Fund Translational Research Award and an Elizabeth Glaser Scientist Award from the Pediatric AIDS Foundation to R.W.D. J.T.K. is supported by R01 AI47725. F.B. was supported by a fellowship from the Swiss National Science Foundation (grant number 823A-61172). S.P. was supported by a fellowship from the Deutsche Forschungsgemeinschaft (DFG). T.A.R. was supported by NIH grant R01 HL62056.
REFERENCES
- 1.Bashirova A A, Geijtenbeek T B, van Duijnhoven G C, van Vliet S J, Eilering J B, Martin M P, Wu L, Martin T D, Viebig N, Knolle P A, KewalRamani V N, van Kooyk Y, Carrington M. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J Exp Med. 2001;193:671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Brass L F, Pizarro S, Ahuja M, Belmonte E, Blanchard N, Stadel J M, Hoxie J A. Changes in the structure and function of the human thrombin receptor during receptor activation, internalization, and recycling. J Biol Chem. 1994;269:2943–2952. [PubMed] [Google Scholar]
- 2.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fortin J-F, Cantin R, Bergeron M G, Tremblay M J. Interaction between virion-bound host intercellular adhesion molecule-1 and the high-affinity state of lymphocyte function-associated antigen-1 on target cells renders r5 and x4 isolates of human immunodeficiency virus type 1 more refractory to neutralization. Virology. 2000;268:493–503. doi: 10.1006/viro.2000.0190. [DOI] [PubMed] [Google Scholar]
- 4.Geijtenbeek T B, Krooshoop D J, Bleijs D A, van Vliet S J, van Duijnhoven G C, Grabovsky V, Alon R, Figdor C G, van Kooyk Y. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat Immunol. 2000;1:353–357. doi: 10.1038/79815. [DOI] [PubMed] [Google Scholar]
- 5.Geijtenbeek T B H, Kwon D S, Torensma R, van Vliet S J, van Duijnhoven G C F, Middel J, Cornelissen I L M H A, Nottet H S L M, KewalRamani V N, Littman D R, Figdor C G, van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 6.Geijtenbeek T B H, Torensma R, van Vliet S J, van Duijnhoven G C F, Adema G J, van Kooyk Y, Figdor C G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 7.Hioe C E, Bastiani L, Hildreth J E, Zolla-Pazner S. Role of cellular adhesion molecules in HIV type 1 infection and their impact on virus neutralization. AIDS Res Hum Retrovir. 1998;14(Suppl. 3):S247–S254. [PubMed] [Google Scholar]
- 8.Hu J, Gardner M B, Miller C J. Simian immunodeficiency virus rapidly penetrates the cervicovaginal mucosa after intravaginal inoculation and infects intraepithelial dendritic cells. J Virol. 2000;74:6087–6095. doi: 10.1128/jvi.74.13.6087-6095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao Z, Roos J W, Hildreth J E. Increased infectivity of HIV type 1 particles bound to cell surface and solid-phase ICAM-1 and VCAM-1 through acquired adhesion molecules LFA-1 and VLA-4. AIDS Res Hum Retrovir. 2000;16:355–366. doi: 10.1089/088922200309232. [DOI] [PubMed] [Google Scholar]
- 10.Mondor I, Ugolini S, Sattentau Q J. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olinger G G, Saifuddin M, Spear G T. CD4-negative cells bind human immunodeficiency virus type 1 and efficiently transfer virus to T cells. J Virol. 2000;74:8550–8557. doi: 10.1128/jvi.74.18.8550-8557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pöhlmann S, Baribaud F, Lee B, Leslie G J, Sanchez M D, Hiebenthal-Millow K, Münch J, Kirchhoff F, Doms R W. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J Virol. 2001;75:4664–4672. doi: 10.1128/JVI.75.10.4664-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pöhlmann S, Lee B, Meister S, Krumbiegel M, Leslie G, Doms R W, Kirchhoff F. Simian immunodeficiency virus utilizes human and sooty mangabey but not rhesus macaque STRL33 for efficient entry. J Virol. 2000;74:5075–5082. doi: 10.1128/jvi.74.11.5075-5082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pöhlmann S, Soilleux E J, Baribaud F, Leslie G, Morris L S, Trowsdale J, Lee B, Coleman N, Doms R W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc Natl Acad Sci USA. 2001;98:2670–2675. doi: 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizzuto C D, Sodroski J G. Contribution of virion ICAM-1 to human immunodeficiency virus infectivity and sensitivity to neutralization. J Virol. 1997;71:4847–4851. doi: 10.1128/jvi.71.6.4847-4851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sallusto F, Mackay C R, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science. 1997;277:2005–2007. doi: 10.1126/science.277.5334.2005. [DOI] [PubMed] [Google Scholar]
- 17.Soilleux E J, Barten R, Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 18.Spira A I, Marx P A, Patterson B K, Mahoney J, Koup R A, Wolinsky S M, Ho D D. Cellular targets of infection and route of viral dissemination after an intravaginal inoculation of simian immunodeficiency virus into rhesus macaques. J Exp Med. 1996;183:215–225. doi: 10.1084/jem.183.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ugolini S, Mondor I, Parren P, Burton D, Tilley S, Klasse P J, Sattentau Q J. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J Exp Med. 1997;186:1287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ugolini S, Mondor I, Sattentau Q J. HIV-1 attachment: another look. Trends Microbiol. 1999;7:144–149. doi: 10.1016/s0966-842x(99)01474-2. [DOI] [PubMed] [Google Scholar]
- 21.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature. 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]