Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration (original) (raw)
Abstract
Spinocerebellar ataxia type 7 (SCA7) is characterized by cone-rod dystrophy retinal degeneration and is caused by a polyglutamine [poly(Q)] expansion within ataxin-7, a protein of previously unknown function. Here, we report that ataxin-7 is an integral component of the mammalian STAGA (SPT3-TAF9-ADA-GCN5 acetyltransferase) transcription coactivator complex, interacts directly with the GCN5 histone acetyltransferase component of STAGA, and mediates a direct interaction of STAGA with the CRX (cone-rod homeobox) transactivator of photoreceptor genes. Consistent with these results, chromatin immunoprecipitation assays document retinal-specific association of CRX, GCN5, and acetylated histone H3 with CRX target genes. RNA interference studies also implicate ataxin-7 and GCN5 in CRX-dependent gene activation, and histone deacetylase inhibitors restore the compromised expression of a CRX target gene in an ataxin-7-deficient background. Significantly, in relation to SCA7, poly(Q)-expanded ataxin-7 gets incorporated into STAGA and, in a dominant-negative manner, inhibits the nucleosomal histone acetylation function of STAGA GCN5 both in vitro and, based on chromatin immunoprecipitation assays, in SCA7 transgenic mice. These results suggest that the normal function of a poly(Q) disease protein may intersect with its pathogenic mechanism, an observation with significant implications for the molecular basis of all poly(Q) disorders and ultimately for their treatment.
Keywords: SCA7, transcription, neurodegeneration, poly(Q), CRX
The poly(Q)-repeat diseases are a group of neurodegenerative disorders sharing the common feature of glutamine tract expansion within unrelated proteins as their mutational basis (1). Blindness due to retinal degeneration (2) distinguishes SCA7 from other neurodegenerative diseases and is caused by a poly(Q) expansion (3) within ataxin-7, but the mechanism of pathogenesis remains unknown. Although it is generally agreed on that misfolding of the expanded glutamine tract is a critical step in the neurotoxicity, the pathways underlying the pathogenic cascade in neurons remain ill defined. Expression of poly(Q) expansions in cells and model organisms can disrupt a wide range of cellular processes that include proteasome-dependent protein degradation, axonal/vesicular transport, mitochondrial function, and transcription (4–6). Determining which cellular pathology is central to poly(Q) disease pathogenesis is especially challenging because the different poly(Q) diseases display selective neuronal vulnerability despite relatively widespread expression of individual mutant gene products throughout the central nervous system. Consequently, poly(Q) neurodegeneration is likely to be protein-context-dependent, such that the pathogenic process in each disease may stem from the protein–protein interactions and/or normal function of each respective disease protein.
The human STAGA complex is a transcriptional coactivator with histone acetyltransferase (HAT) activity, as well as UV-damaged DNA-binding activity (7), and is equivalent to the human PCAF complex (8) and the yeast SAGA (9)/SLIK (10)/SALSA (11) complexes. STAGA and PCAF complexes differ from the related TFTC (TBP-free TAF-containing) complex (12) in that they do not contain TAFs 2, 4, 5, 6, and 7. These complexes contain either GCN5 or PCAF as the HAT that, after activator-dependent recruitment of the complex, specifically acetylates histone H3 tails in adjacent nucleosomes and facilitates transcription initiation (13).
Whereas ataxin-7 has been suggested to be a STAGA component on the basis of a previous bioinformatics analysis (14) and a biochemical analysis in yeast (15), and recently has been identified as a component of the TFTC complex (16), our work documents functions of WT and poly(Q)-expanded ataxin-7 within STAGA both in vitro and in vivo. We further propose the SCA7 pathogenesis mechanism as a transcriptional dysregulation that involves a loss of STAGA-mediated HAT activity as a result of the dominant-negative action of the poly(Q)-expanded ataxin-7. We also document the ability of histone deacetylase (HDAC) inhibitors to help rescue transactivation defects resulting from inhibition of STAGA coactivator function.
Methods
Constructs and Stable Cell Lines. FLAG-tagged ataxin-7 expression constructs F-SCA7–24Q (WT), F-SCA7–92Q, and F-92Q NT (1–229) (NT, amino-terminal) were generated in pTriEx-4-Hygro (Invitrogen) by using a PCR cloning strategy with PrP-SCA7-c24Q/-c92Q (17) as templates. HEK293T cells were transfected with F-ataxin-7–24Q or F-ataxin-7–92Q and selected with hygromycin (0.2 mg/ml, Invitrogen) for 14 days to establish cell lines that stably express the desired proteins. Single hygromycin-resistant colonies expressing F-ataxin-7–24Q and F-ataxin-7–92Q were expanded for nuclear extract (NE) preparations.
Protein Purification and Analysis. The STAGA complex was purified from NEs of cell lines expressing F-HA-SPT3, F-SCA7–24Q, F-SCA7–92Q, and F-PCAF, respectively, by anti-FLAG immunoprecipitation using M2-agarose (Sigma) as described in ref. 7. Protein identification by mass spectrometry (MS) was as described in ref. 7. The p110 band of STAGA was identified as ataxin-7 by tandem MS analysis from the following peptide sequences: FDVLLAEHK, IPPVPSTTSPISTR, QVSSSSSSP-STPSGLS, SVPSSPMSR, and GPPTGSPAESIK. For gelfiltration chromatography, NE (0.2 ml) was analyzed on a Sephacryl S-300 (Amersham Pharmacia Biosciences) column (10 ml, 0.4 cm2 × 25 cm) in BC300 (20 mM Tris·HCl, pH 7.9/20% glycerol/0.2 mM EDTA/1 mM DTT/0.5 mM PMSF/0.3 M KCl)/0.05% Nonidet P-40, and the high-molecular-weight fractions were analyzed by immunoblot. Recombinant His6-p300, F-PCAF, and F-SCA7–92Q NT were expressed from baculovirus vectors and purified by using Ni-NTA or anti-FLAG M2 agarose resin (Sigma) as described in ref. 18.
Immunoprecipitation (IP), Immunoblot, and HAT Assays. For IP assays, ataxin-7 antibody (K) (17) or rabbit IgG was immobilized on an ImmunoPure Protein A orientation kit (Pierce) with dimethylpimelimidate according to the manufacturer's instructions. Y-79 NE was nutated with anti-ataxin-7 and anti-IgG beads for 8 h at 4°C. After extensive washes with BC300/0.1% Nonidet P-40, bound proteins were eluted with low-pH buffer (0.1 M glycine, pH 2.5/20% glycerol/0.3 M KCl/0.2 mM EDTA), analyzed by SDS/PAGE, and immunoblotted with antibodies against TRRAP (sc-11411, Santa Cruz Biotechnology); p300 (sc-584, Santa Cruz Biotechnology); ataxin-7 “K” (17); GCN5 (sc-20698, Santa Cruz Biotechnology); TAF5L and ADA3 (gift of Y. Nakatani, Harvard University, Cambridge, MA); and TBP, TAF9, TAF12, SPT3, hADA2b, and STAF36 (antibodies from the Roeder laboratory; antibodies to hADA2b and STAF36 were raised in rabbits immunized against purified full-length His6-tagged proteins). For HAT assays post-IP, beads were eluted with Gentle Elution Buffer (Pierce). HAT assays were performed by using 1.5 μg of core histones or oligonucleosomes purified from HeLa cells (7). For the nucleosomal HAT-inhibition assay, 1 pmol of His6-p300 or PCAF complex containing 2 pmol of PCAF was preincubated with 1 pmol of F-ataxin-7–24Q/-92Q or 10 pmol of F-ataxin-7–92Q NT for 10 min at 30°C before standard HAT assay in a total volume of 25 μl at 30°C for 45 min.
Chromatin Immunoprecipitation (ChIP). ChIP assays were performed on pooled retinas (four for each antibody) or livers (30 mg) from adult WT and PrP-SCA7 92Q mice (17) as described in ref. 19. Further details and primer sequences are provided in Supporting Text, which is published as supporting information on the PNAS web site.
RNA Interference (RNAi) and Transfection Assays. HEK293 cells were transfected with individual siRNAs directed against ataxin-7, GCN5, p300, luciferase (LUC), or nonspecific control. Forty-eight hours after siRNA transfection, the cells were transfected with reporter and activator DNAs. The dual LUC assay (Promega) was performed 48 h after DNA transfection. Where indicated, an HDAC inhibitor mixture of suberoylanilide hydroxamic acid (10 μM; Biomol, Plymouth Meeting, PA) and sodium butyrate (5 mM, Sigma) was added to the cell-culture medium 24 h before the LUC assays. siRNA target sequences and other details are provided in Supporting Text.
Further Details. Details of protein–protein interaction assays, RNAi, ChIP, and immunohistochemistry are provided in Supporting Text.
Results
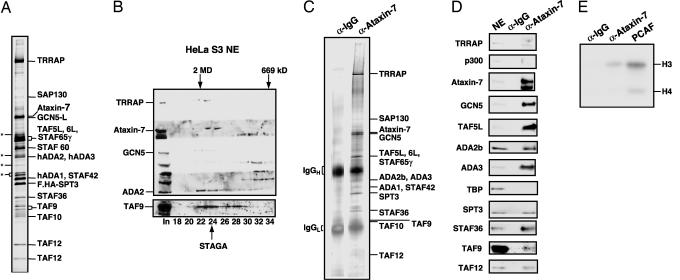
Ataxin-7 Is an Integral Subunit of STAGA. The 110-kDa subunit of STAGA (7) was identified by tandem MS analysis as ataxin-7 (Fig. 1_A_). To confirm a stable association of ataxin-7 with other STAGA components, NE from HeLa S3 cells was subjected to gel-filtration chromatography on Sephacryl S-300. Immunoblot of the 1.8-MDa, high-molecular-weight fraction revealed coelution of ataxin-7 with STAGA core components TRRAP, GCN5, ADA2, and TAF9 (Fig. 1_B_). Silver stain analysis of anti-ataxin-7-purified STAGA from NE of Y-79 retinoblastoma cells revealed a stoichiometric association of ataxin-7 with other STAGA components (Fig. 1_C_). Immunoblot of this anti-ataxin-7 affinity-purified STAGA complex confirmed the presence of most STAGA subunits (namely TRRAP, ataxin-7, GCN5, TAF5L, ADA2b, ADA3, SPT3, STAF36, TAF9, and TAF12) and the absence of p300 and TBP (Fig. 1_D_). This anti-ataxin-7 affinity-purified STAGA complex was demonstrated to be functional based on the presence of histone H3-biased HAT activity on free histones (Fig. 1_E_), a characteristic of GCN5. To establish the presence of GCN5 in a STAGA complex containing a stoichiometric amount of ataxin-7, STAGA was isolated by anti-FLAG affinity chromatography from NE of a 293T line (F-24Q) that stably expresses FLAG-ataxin-7–24Q. The F-24Q-purified complex was shown to contain GCN5 and other STAGA components (TRRAP, ADA2b, SPT3, and TAF12) by immunoblot (Fig. 2_A_, lane 3) and, as well, a histone H3-biased HAT activity on a physiological chromatin substrate (Fig. 2_B_, lane 3).
Fig. 1.
Ataxin-7 is an integral subunit of the STAGA complex. (A) SDS/PAGE/silver stain analysis of affinity-purified STAGA from FLAG.HA-SPT3 HeLa cells. The p110 band was identified as ataxin-7 by tandem MS analysis. Asterisks indicate nonspecific bands. (B) Immunoblot of Sephacryl S-300 fractions after chromatography of HeLa S3 NE. (C) SDS/PAGE/silver stain analysis of control (IgG resin) or anti-ataxin-7 affinity-purified STAGA from Y-79 cell NE. (D) Immunoblot of IPs from C with indicated antibodies. Y-79 NE served as positive control. (E) Fluorogram of HAT assay of IPs from C, using free histones as substrate. Recombinant PCAF served as a positive control.
Fig. 2.
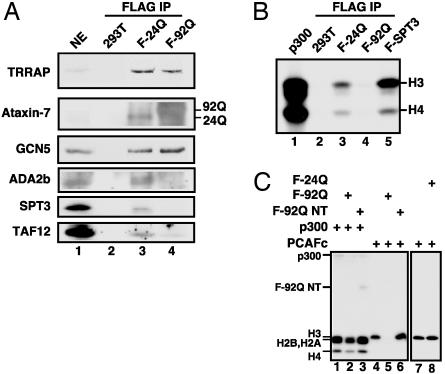
Poly(Q)-expanded ataxin-7 is incorporated into STAGA and inactivates its HAT function in a dominant-negative manner. (A) Immunoblot of anti-FLAG IPs of NE from control 293T cells (lane 2) or from 293T lines stably expressing FLAG-ataxin-7–24Q (lane 3) or FLAG-ataxin-7–92Q (lane 4). 293T NE served as positive control (lane 1). (B) Fluorogram of HAT assays using an oligonucleosome substrate on IPs from A, a highly concentrated STAGA complex (lane 5), and recombinant p300 (lane 1). (C) Fluorogram of solution HAT assays with an oligonucleosome substrate and purified proteins FLAG-ataxin-7–24Q/-92Q, FLAG-ataxin-7–92Q NT, His6-p300, and affinity-purified PCAF complex added as indicated (top).
Incorporation of Poly(Q)-Expanded Ataxin-7 Into STAGA Disrupts Its Assembly and HAT Activity. The discovery of ataxin-7 as a core component of STAGA led us to consider the possibility that poly(Q)-expanded ataxin-7 may somehow disrupt a STAGA coactivator function in SCA7. An immunoblot of anti-FLAG affinity-purified STAGA from a HEK293T line (F-92Q) that stably expresses FLAG-ataxin-7–92Q demonstrated F-ataxin-7–92Q solubility and incorporation into a STAGA complex that contains stoichiometric amounts of TRRAP and GCN5 but diminished levels of ADA2b, SPT3, and TAF12 (Fig. 2_A_, lane 4). Most importantly, incorporation of the mutant poly(Q) (92Q) form of ataxin-7 into STAGA dramatically reduced its ability to acetylate histone H3 on a chromatin substrate (Fig. 2_B_, lane 4) relative to that seen with equimolar amounts of F-ataxin-7–24Q-purified (lane 3) or highly concentrated F-HA-SPT3-derived STAGA (lane 5) complexes, suggesting that incorporation of mutant ataxin-7 into STAGA abolishes its GCN5-mediated HAT activity.
Poly(Q)-Expanded Ataxin-7 Inhibits the HAT Activity of STAGA in a Dominant-Negative Manner. To further explore the mechanism by which poly(Q)-expanded ataxin-7 inhibits the HAT activity of STAGA on a physiological substrate, we carried out HAT assays using purified p300 and the STAGA-related PCAF complex in the absence and presence of recombinant ataxin-7 (F-24Q), poly(Q)-expanded ataxin-7 (F-92Q), or an NT fragment (F-92Q NT) of poly(Q)-expanded ataxin-7 containing residues 1–229. Significantly, the full-length F-92Q mutant, but not F-24Q or F-92Q NT, dramatically inhibited histone H3-biased nucleosomal acetylation by the PCAF complex but only marginally inhibited acetylation by p300 (Fig. 2_C_).
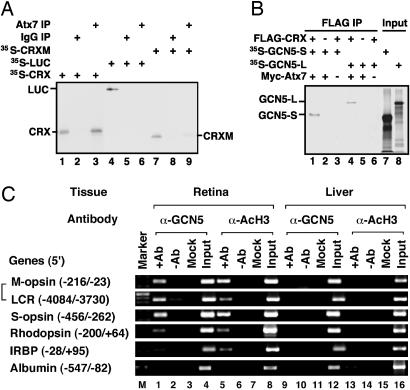
CRX Is a STAGA-Dependent Transcription Factor. Because ataxin-7 is known to interact with a photoreceptor-specific factor (CRX) that regulates the transcription of numerous genes (17), we tested whether CRX can recruit STAGA in an ataxin-7-dependent manner. First, a purified ataxin-7-containing STAGA complex was found to interact with an _in vitro_-translated CRX, but not with luciferase (LUC) and only weakly with a CRX mutant (Q96/97W-ΔQ7) that is incapable of productively interacting with ataxin-7 (19) (Fig. 3_A_). Second, GCN5-S or GCN5-L, a key component of the STAGA complex, bound to FLAG-CRX only in the presence of ataxin-7 (Fig. 3_B_). Immunoblotting confirmed the presence of Myc-ataxin-7 and FLAG-CRX in the anti-FLAG pull-downs (Fig. 7, which is published as supporting information on the PNAS web site). Furthermore, in vitro coimmunoprecipitation assays indicated that ataxin-7 interacts with GCN5-S and that this interaction is facilitated by the presence of CRX (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 3.
CRX interacts with STAGA through ataxin-7. (A) SDS/PAGE/fluorographic analysis of interactions of 35S-labeled _in vitro_-translated proteins (LUC, CRX, and CRX-Mutant) with IPs from Fig. 1_C_. Additions were as indicated (top), and inputs (2%) are shown in lanes 1, 4, and 7. (B) SDS/PAGE/fluorographic analysis of interactions of 35S-labeled _in vitro_-translated proteins (GCN5-S and GCN5-L) with FLAG-CRX in the presence and absence of Myc-ataxin-7–92Q (Myc-Atx7). Similar results were obtained with Myc-ataxin-7–10Q (data not shown). Additions were as indicated (top). Corresponding empty vector controls are represented by “-.” Inputs (2%) are shown in lanes 7 and 8. (C) ChIP analysis of acetylated histone H3 and GCN5 on CRX-target genes in mouse retina vs. liver tissues. PCR primers covered the regulatory regions indicated in parentheses. M-opsin, M-cone opsin; LCR, locus control region of M-cone opsin; S-opsin, S-cone opsin; IRBP, interphotoreceptor-binding protein. Input (pre-IP), + and - antibody (Ab), and mock (no DNA) lanes are marked. Similar results were obtained by using an isotype-matched antibody (rabbit IgG, Santa Cruz Biotechnology) as with the no antibody control.
To determine whether CRX is capable of recruiting a functional STAGA complex to its target photoreceptor genes in vivo, we performed ChIP assays using adult mouse retinas. Using this assay, we previously showed that CRX and ataxin-7 cooccupy the regulatory region of several CRX target genes (19). Here, we performed similar assays with antibodies against GCN5 and its modified substrate, acetylated histone H3 (AcH3). We found that both GCN5 and AcH3 are associated with the regulatory regions of several CRX-dependent genes (rhodopsin, S- and M-cone opsins) in mouse retina but not in liver where CRX-dependent genes are not expressed (Fig. 3_C_). These results indicate that CRX recruits a functional GCN5-containing STAGA complex onto its targets in vivo. Such recruitment appears to be mediated by CRX, because it does not occur either on non-CRX target genes such as albumin (Fig. 3_C_) or on the3′ regions of CRX-target genes in the retina (Fig. 9, which is published as supporting information on the PNAS web site), where CRX does not bind.
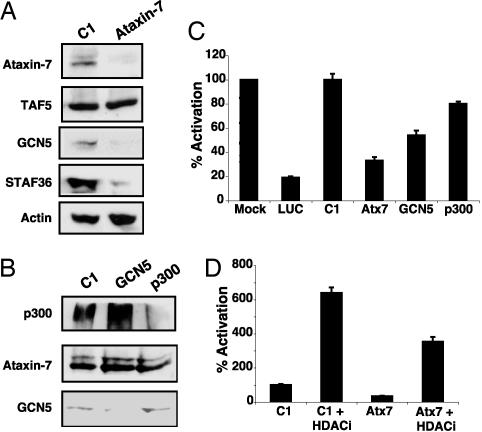
Ataxin-7 Is Required for the Stability and Coactivator Function of STAGA. To determine the physiological relevance of the CRX–STAGA interaction, we examined the role of STAGA in CRX-mediated transactivation of its target photoreceptor genes using RNAi in HEK293 cells transiently transfected with CRX and the neural retina leucine zipper protein (NRL). Previous studies have established that CRX and NRL can synergistically activate a rhodopsin promoter-LUC reporter in this assay (17). Immunoblotting confirmed the specificity and efficacy of the siRNA-mediated knockdowns (Fig. 4 A and B). Interestingly, whereas siRNAs against GCN5 or p300 showed selective knockdowns of the respective proteins (Fig. 4_B_), ataxin-7 siRNAs knocked down the levels not only of ataxin-7 but also STAGA subunits GCN5 and STAF36 (7) [a novel component of STAGA homologous to the yeast Sgf29 (15)] while TAF5 and actin levels were unaltered (Fig. 4_A_). These results suggest that ataxin-7 may play a role in the assembly or maintenance of a stable, functional STAGA complex. Independent additions of ataxin-7 and GCN5 siRNAs reduced activation to 33% and 54%, respectively (Fig. 4_C_). The inhibition was specific, because the unrelated control siRNA had no effect, and siRNA against another HAT (p300) had only a modest inhibitory effect. Furthermore, this inhibition was restricted to CRX/NRL-regulated promoters because levels of cotransfected Renilla LUC reporter, expressed from the CMV promoter, were unaltered (data not shown).
Fig. 4.
CRX/NRL-mediated activation of the rhodopsin promoter depends on a functional STAGA complex. (A and B) Immunoblot of whole-cell extracts from HEK293 cells transfected with siRNAs as indicated (top). (C) Effects of ataxin-7, GCN5, and p300 siRNAs on expression of a transfected rhodopsin promoter-driven LUC reporter in HEK293 cells. LUC and nonspecific control (C1) siRNAs served as positive and negative controls, respectively. The activation observed from mock-transfected HEK293 cells was arbitrarily set as 100%. (D) HDAC inhibitor-mediated recovery of ataxin-7 siRNA-compromised rhodopsin-promoter activity in transfected HEK293 cells. Control siRNA (C1) and ataxin-7 siRNA transfected cells (as in C) were treated with suberoylanilide hydroxamic acid and butyrate (HDAC inhibitors) as indicated. Error bars represent mean plus standard deviation in C and D from three independent experiments.
HDAC Inhibitors Help Rescue Loss of STAGA HAT Function. Transcriptional homeostasis within cells is maintained by the interplay between HATs and the opposing HDACs. We thus performed ataxin-7 RNAi experiments in the presence of the HDAC inhibitors suberoylanilide hydroxamic acid and sodium butyrate. Exposure to HDAC inhibitors reversed ataxin-7 siRNA-mediated inhibition of CRX/NRL-dependent transactivation (Fig. 4_D_), confirming that STAGA coactivation of CRX transcription relies on its intrinsic HAT activity.
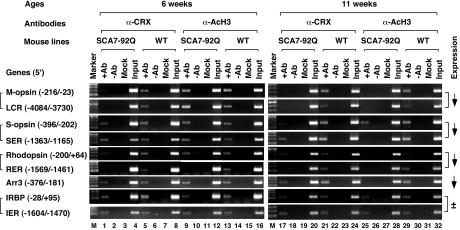
Dominant-Negative Inhibition of STAGA HAT Activity in Retinas of SCA7–92Q Mice. To determine whether suppression of STAGA HAT activity by ataxin-7–92Q occurs in vivo, we performed ChIP assays on retinas from SCA7–92Q transgenic mice that recapitulate SCA7 retinal degeneration (17). ChIP assays were performed with antibodies against CRX and AcH3 on 6-week-old (presymptomatic) and 11-week-old (moderate retinal degeneration) mice. Both conventional and quantitative real-time PCR were then performed for the regulatory regions of four CRX-dependent genes (rhodopsin, S- and M-cone opsins, and cone arrestin) and one CRX-independent gene (IRBP) (20). Standard PCR analyses revealed a marked reduction in the presence of AcH3 on the promoter/enhancer regions of the CRX-dependent photoreceptor genes in the 11-week-old SCA7–92Q mice but not in littermate controls or in 6-week-old mice (Fig. 5). The reduced levels of AcH3 were not seen for the CRX-independent genes IRBP and rhodopsin kinase (Fig. 5 and data not shown), suggesting that reduction of AcH3 is specific for the CRX-dependent genes. Importantly, these results parallel the changes in gene expression profiles previously documented in the retinas of these mice (17) and thus indicate that defects in STAGA-mediated histone H3 acetylation may account for the transcription dysregulation that has been linked to SCA7 retinal degeneration (17, 21).
Fig. 5.
HAT activity alterations in SCA7 result from a dominant-negative effect on the STAGA complex. ChIP assays on retinas from SCA7–92Q transgenic mice and their nontransgenic WT littermates reveal differences in occupancy of CRX target and nontarget gene regulatory regions. Quantitation of the occupancy differences is provided in Table 1. Brackets group together the enhancer/promoter pair regulating each gene. On the far right, previously reported gene expression alterations for this SCA7 mouse model of retinal degeneration are as indicated (17). SER, S-cone opsin enhancer region; RER, rhodopsin enhancer region; Arr3, cone arrestin 3; IER, IRBP enhancer region. See the legend of Fig. 3_C_ for other definitions. A 100-bp DNA ladder was used as a size marker.
To determine whether the reduction of AcH3 at CRX-target genes in SCA7 retina is due to a defect in STAGA recruitment or to a dominant-negative effect of poly(Q)-ataxin-7 on STAGA HAT activity, we quantified the association of CRX and GCN5 with the same set of photoreceptor genes by ChIP. We observed a consistently reduced CRX occupancy at the regulatory regions of the four CRX-dependent genes but not for the CRX-independent gene IRBP in SCA7–92Q retina at both 6 and 11 weeks of age (Fig. 5). This finding is in contrast to the progressive decrease in AcH3 on these genes from 6 to 11 weeks of age. To confirm these observations, we performed quantitative real-time PCR analysis on the immunoprecipitated chromatin, normalized the results with that of the input, and compared the differences between these two ages in SCA7–92Q vs. WT mice. This analysis revealed a much more dramatic reduction in AcH3 levels than in CRX-binding for these two time points (Table 1). For example, the AcH3 level for the cone arrestin promoter (Arr3) is reduced by 67%, whereas CRX occupancy is reduced only by 1%. Thus, a reduction in CRX occupancy cannot fully account for the dramatic transcription dysregulation seen in SCA7 retinal degeneration in these mice. This finding was independently confirmed by ChIP assays of GCN5 occupancy at these photoreceptor genes. To our surprise, the association of GCN5 with the photoreceptor genes in the retina of SCA7–92Q mice remained unchanged up to 11 weeks of age (Fig. 10, which is published as supporting information on the PNAS web site). Possible explanations are provided in Supporting Text.
Table 1. Changes in promoter occupancy in the retina of SCA7–92Q WT mice.
| CRX | AcH3 | |||||
|---|---|---|---|---|---|---|
| Genes | 6 weeks | 11 weeks | Δ (6W-11W) | 6 weeks | 11 weeks | Δ (6W-11W) |
| M-opsin | 58.1 ± 3.2* | 48.4 ± 0.7* | 9.7 | 96.8 ± 1.2 | 72.5 ± 0.9* | 24.3 |
| LCR | 23.7 ± 1.4* | 24.1 ± 1.7* | -0.4 | 102.3 ± 2.6 | 79.6 ± 1.0* | 22.7 |
| S-opsin | 37.8 ± 3.9* | 38.6 ± 0.4* | -0.8 | 67.8 ± 0.4* | 42.0 ± 0.3* | 25.8 |
| SER | 80.5 ± 1.0* | 56.1 ± 0.9* | 24.4 | 96.9 ± 1.0 | 9.9 ± 0.5* | 87.0 |
| Rhodopsin | 82.2 ± 0.5* | 75.0 ± 1.6* | 7.2 | 104.6 ± 1.7 | 67.2 ± 0.5* | 37.4 |
| RER | 15.7 ± 0.1* | 11.4 ± 0.4* | 4.3 | 2.2 ± 0.3* | 7.4 ± 1.4* | -5.2 |
| Arr3 | 71.1 ± 1.3* | 69.8 ± 0.3* | 1.3 | 77.9 ± 0.5* | 10.9 ± 0.2* | 67.0 |
| IRBP | 99.0 ± 0.7 | 98.4 ± 0.6 | 0.6 | 101.5 ± 0.7 | 100.2 ± 1.0 | 1.3 |
| IER | 105.2 ± 0.1 | 104.1 ± 0.9 | 1.1 | 104.3 ± 0.8 | 103.9 ± 0.8 | 0.4 |
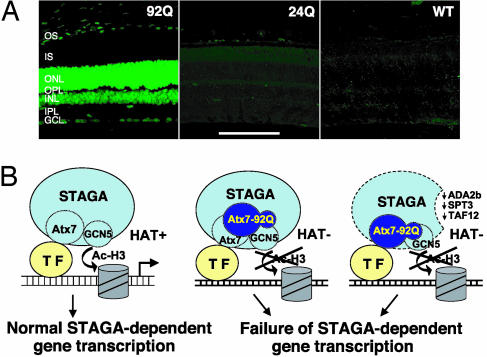
Whereas GCN5 occupancy at CRX-regulated photoreceptor genes was unchanged in the SCA7 mice based on ChIP analysis, GCN5 immunostaining of retinal sections from the SCA7 mice surprisingly revealed a dramatic increase in signal intensity (Fig. 6_A_) that is probably due to compensatory up-regulation of GCN5 expression as mentioned in Supporting Text. The markedly increased GCN5 immunostaining pattern could be detected in presymptomatic SCA7 mice and persisted with onset and progression of the retinal degeneration (Fig. 11, which is published as supporting information on the PNAS web site).
Fig. 6.
Mutant ataxin-7 effects on GCN5 function. (A) Retinal sections from 13-week-old SCA7–92Q transgenic mice (92Q), SCA7–24Q (24Q), and nontransgenic littermates (WT) were immunostained with an anti-GCN5 antibody (green). Although confocal images reveal intense immunostaining of photoreceptor nuclei (ONL) and bipolar/interneuron nuclei (INL) in the 92Q mice, no appreciable GCN5 immunostaining is detectable in the 24Q or nontransgenic mice. OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. (Scale bar, 100 μm.) (B) Models indicating (i) normal function of WT ataxin-7 (Atx7), as an integral GCN5-interacting subunit of STAGA, in facilitating STAGA recruitment and GCN5-mediated histone H3 acetylation through interactions with a promoter-bound transcription factor (TF) such as CRX (Left), and (ii) dominant-negative inhibition of GCN5 function (histone acetylation) within promoter-bound STAGA by poly(Q)-expanded ataxin-7 (Atx7–92Q) after interactions with WT ataxin-7-containing STAGA (Center) or after incorporation into STAGA in place of WT ataxin-7 resulting in reduced incorporation of ADA2b, SPT3, and TAF12 (Right).
Discussion
Earlier studies in yeast have shown that TAF12 is essential for the nucleosomal HAT activity of SAGA and for SPT3 incorporation (9), and that association of recombinant GCN5 with the bridging factor ADA2 enables GCN5 to acetylate nucleosomes (22). Thus, our data indicate that depletion of critical proteins that regulate GCN5-dependent nucleosome recognition and STAGA HAT function help explain the mechanism of transcriptional dysfunction for the poly(Q)-expanded ataxin-7 protein in SCA7 pathogenesis. Because a recent study has indicated that a highly conserved “zinc-binding domain” between residues 311 and 406 of ataxin-7 may mediate its interaction with certain subunits that are shared between the STAGA and TFTC complexes (16), the 92Q NT fragment of ataxin-7 used in the present study may not be capable of exerting any HAT interference because of a failure to interact with the STAGA complex. This observation clearly demonstrates a trans dominant-negative mechanism of HAT inhibition by full-length poly(Q)-expanded ataxin-7 that is also observed in a more pronounced, activator-dependent manner by ChIP studies from retinas of SCA7 mice (Fig. 5 and Table 1).
The importance of ataxin-7 for the integrity and function of STAGA would explain why siRNAs targeting ataxin-7 reduce CRX/NRL-mediated transactivation more than siRNAs targeting GCN5, because knockdown of ataxin-7 would lead to a loss of all STAGA functions. Mechanistically distinct functions, namely TBP interaction via SPT3, that act independently of the GCN5 HAT activity have been demonstrated for the yeast SAGA complex (23). Importantly, however, these findings reveal that STAGA is the major coactivator for CRX-dependent gene transcription.
Our in vitro analyses of HAT activity in normal and poly(Q)-ataxin-7-containing STAGA complexes, together with RNAi, ChIP, and immunostaining assays for STAGA components and corresponding histone modifications, support a model in which poly(Q)-expanded ataxin-7 produces a defect in histone H3 acetylation at CRX-regulated photoreceptor genes by inhibiting GCN5 HAT activity (Fig. 6_B_). Thus, we conclude that the transcription dysregulation in SCA7 retinal degeneration results from a dominant-negative effect of poly(Q)-expanded ataxin-7 on the GCN5 subunit of the STAGA complex in which ataxin-7 normally resides.
Our documentation of intact STAGA interactions with CRX, and of ataxin-7 interactions with CRX and GCN5, suggests CRX transcription coactivation via ataxin-7-dependent recruitment of STAGA, with its GCN5-mediated HAT activity. Our results are strongly supported by independent studies showing that the yeast orthologue of ataxin-7, Sgf73, is a core component of the yeast SAGA complex and is essential for the structural stability of the SAGA complex. Importantly, this work indicates that ΔSgf73 mutants can be rescued by human WT ataxin-7 but not by a human poly(Q)-expanded (60Q) ataxin-7 that gets incorporated into the yeast SAGA complex and inhibits its ability to acetylate a chromatin substrate [see companion article (24)].
The most compelling aspects of our study relate to the molecular basis of transcription dysregulation in SCA7 retinal degeneration. Our data indicate that poly(Q)-expanded ataxin-7 gets incorporated into the STAGA complex and thereby disrupts the assembly of a functional complex. Loss of key components may cause impaired nucleosome recognition and acetylation. Our data also indicate that poly(Q)-expanded ataxin-7 can inhibit the HAT activity of the normal STAGA complex in vitro, but whether this results from displacement of the WT ataxin-7 from STAGA or from a secondary interaction with STAGA (including GCN5) remains to be determined (Fig. 6_B_). Our findings suggest that the normal function of ataxin-7 as a component of STAGA explains how poly(Q) expansion within ataxin-7 produces retinal degeneration in SCA7. Although previous studies of poly(Q) diseases have proposed that normal function may be relevant to poly(Q) disease pathogenesis (25), our results provide evidence for a dominant-negative mechanism in poly(Q) disease pathology.
Supplementary Material
Supporting Information
Acknowledgments
We thank S. Berger (Wistar Institute, Philadelphia) for hADA2 anti-serum and the GCN5-S expression construct; Y. Nakatani for providing the FLAG-PCAF HeLa cell line, F-PCAF baculovirus, and GCN5, ADA3, and TAF5L antibodies; J. Park for help in HeLa oligonucleosome preparation; C. Bhattacharya, J. Huang, D. E. Possin, and J. Liu for technical assistance; D. Zack (The Johns Hopkins University, Baltimore) for the FLAG-CRX construct; S. Malik for critical reading of the manuscript; and members of the Roeder laboratory for helpful discussions. The siRNA Selection Program developed by the Whitehead Institute for Biomedical Research (2003) was used for designing siRNAs in the 3′ UTR of ataxin-7. V.B.P. was supported by a postdoctoral fellowship from the Hereditary Disease Foundation. This work was supported by The Rockefeller University (to R.G.R.); National Institutes of Health Grants EY12543 (to S.C.), EY02687 (to the Department of Ophthalmology and Visual Sciences of Washington University), EY14061 (to A.R.L.S.), and EY01730 (to the University of Washington Medical Center); the Knights Templar Foundation (G.-H.P.); Prevent Blindness America (S.C.); and unrestricted funds from Research to Prevent Blindness (to the Department of Ophthalmology and Visual Sciences of Washington University). S.C. is a Research to Prevent Blindness Sybil B. Harrington Scholar. A.R.L.S. is a Paul Beeson Physician Faculty Scholar in aging research through the American Foundation for Aging Research.
Author contributions: V.B.P., S.C., A.R.L.S., and R.G.R. designed research; V.B.P., G.-H.P., and Y.F. performed research; V.B.P., A.T., A.M.G., and B.T.C. contributed new reagents/analytic tools; V.B.P., S.C., G.-H.P., A.R.L.S., and R.G.R. analyzed data; and V.B.P., S.C., A.R.L.S., and R.G.R. wrote the paper.
Abbreviations: ChIP, chromatin immunoprecipitation; HAT, histone acetyltransferase; HDAC, histone deacetylase; NE, nuclear extract; NT, amino-terminal; poly(Q), polyglutamine; RNAi, RNA interference.
References
- 1.Zoghbi, H. Y. & Orr, H. T. (2000) Annu. Rev. Neurosci. 23**,** 217-247. [DOI] [PubMed] [Google Scholar]
- 2.Grote, S. K. & La Spada, A. R. (2003) Cytogenet. Genome Res. 100**,** 164-174. [DOI] [PubMed] [Google Scholar]
- 3.David, G., Abbas, N., Stevanin, G., Durr, A., Yvert, G., Cancel, G., Weber, C., Imbert, G., Saudou, F., Antoniou, E., et al. (1997) Nat. Genet. 17**,** 65-70. [DOI] [PubMed] [Google Scholar]
- 4.Feany, M. B. & La Spada, A. R. (2003) Neuron 40**,** 1-2. [DOI] [PubMed] [Google Scholar]
- 5.Panov, A. V., Gutekunst, C. A., Leavitt, B. R., Hayden, M. R., Burke, J. R., Strittmatter, W. J. & Greenamyre, J. T. (2002) Nat. Neurosci. 5**,** 731-736. [DOI] [PubMed] [Google Scholar]
- 6.Freiman, R. N. & Tjian, R. (2002) Science 296**,** 2149-2150. [DOI] [PubMed] [Google Scholar]
- 7.Martinez, E., Palhan, V. B., Tjernberg, A., Lymar, E. S., Gamper, A. M., Kundu, T. K., Chait, B. T. & Roeder, R. G. (2001) Mol. Cell. Biol. 21**,** 6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogryzko, V. V., Kotani, T., Zhang, X., Schiltz, R. L., Howard, T., Yang, X. J., Howard, B. H., Qin, J. & Nakatani, Y. (1998) Cell 94**,** 35-44. [DOI] [PubMed] [Google Scholar]
- 9.Grant, P. A., Schieltz, D., Pray-Grant, M. G., Steger, D. J., Reese, J. C., Yates, J. R., III, & Workman, J. L. (1998) Cell 94**,** 45-53. [DOI] [PubMed] [Google Scholar]
- 10.Pray-Grant, M. G., Schieltz, D., McMahon, S. J., Wood, J. M., Kennedy, E. L., Cook, R. G., Workman, J. L., Yates, J. R., III, & Grant, P. A. (2002) Mol. Cell. Biol. 22**,** 8774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sterner, D. E., Belotserkovskaya, R. & Berger, S. L. (2002) Proc. Natl. Acad. Sci. USA 99**,** 11622-11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brand, M., Yamamoto, K., Staub, A. & Tora, L. (1999) J. Biol. Chem. 274**,** 18285-18289. [DOI] [PubMed] [Google Scholar]
- 13.Vignali, M., Steger, D. J., Neely, K. E. & Workman, J. L. (2000) EMBO J. 19**,** 2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheel, H., Tomiuk, S. & Hofmann, K. (2003) Hum. Mol. Genet. 12**,** 2845-2852. [DOI] [PubMed] [Google Scholar]
- 15.Sanders, S. L., Jennings, J., Canutescu, A., Link, A. J. & Weil, P. A. (2002) Mol. Cell. Biol. 22**,** 4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmlinger, D., Hardy, S., Sasorith, S., Klein, F., Robert, F., Weber, C., Miguet, L., Potier, N., Van-Dorsselaer, A., Wurtz, J. M., et al. (2004) Hum. Mol. Genet. 13**,** 1257-1265. [DOI] [PubMed] [Google Scholar]
- 17.La Spada, A. R., Fu, Y. H., Sopher, B. L., Libby, R. T., Wang, X., Li, L. Y., Einum, D. D., Huang, J., Possin, D. E., Smith, A. C., et al. (2001) Neuron 31**,** 913-927. [DOI] [PubMed] [Google Scholar]
- 18.Kundu, T. K., Palhan, V. B., Wang, Z., An, W., Cole, P. A. & Roeder, R. G. (2000) Mol. Cell 6**,** 551-561. [DOI] [PubMed] [Google Scholar]
- 19.Chen, S., Peng, G. H., Wang, X., Smith, A. C., Grote, S. K., Sopher, B. L. & La Spada, A. R. (2004) Hum. Mol. Genet. 13**,** 53-67. [DOI] [PubMed] [Google Scholar]
- 20.Furukawa, T., Morrow, E. M., Li, T., Davis, F. C. & Cepko, C. L. (1999) Nat. Genet. 23**,** 466-470. [DOI] [PubMed] [Google Scholar]
- 21.Yoo, S. Y., Pennesi, M. E., Weeber, E. J., Xu, B., Atkinson, R., Chen, S., Armstrong, D. L., Wu, S. M., Sweatt, J. D. & Zoghbi, H. Y. (2003) Neuron 37**,** 383-401. [DOI] [PubMed] [Google Scholar]
- 22.Balasubramanian, R., Pray-Grant, M. G., Selleck, W., Grant, P. A. & Tan, S. (2002) J. Biol. Chem. 277**,** 7989-7995. [DOI] [PubMed] [Google Scholar]
- 23.Sterner, D. E., Grant, P. A., Roberts, S. M., Duggan, L. J., Belotserkovskaya, R., Pacella, L. A., Winston, F., Workman, J. L. & Berger, S. L. (1999) Mol. Cell. Biol. 19**,** 86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon, S. J., Pray-Grant, M. G., Schieltz, D., Yates, J. R., III, & Grant, P. A. (2005) Proc. Natl. Acad. Sci. USA 102**,** 8478-8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Spada, A. R. & Taylor, J. P. (2003) Neuron 38**,** 681-684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information