Legionella pneumophila Contains a Type II General Secretion Pathway Required for Growth in Amoebae as Well as for Secretion of the Msp Protease (original) (raw)
Abstract
We report the identification of a set of Legionella pneumophila genes that encode products with homology to proteins of the type II general secretion pathway of gram-negative bacteria. A strain containing a deletion-substitution mutation of two of these genes was unable to secrete the Msp protease. This strain was unable to multiply within the free-living amoeba Acanthamoeba castellanii yet was able to kill HL-60-derived macrophages. Because Msp is not required for growth in amoebae, other proteins which are important for growth in amoebae are likely secreted by this pathway.
Legionella pneumophila is the gram-negative facultative intracellular pathogen responsible for Legionnaires’ disease. L. pneumophila is able to infect and multiply within a variety of eukaryotic hosts, including human mononuclear phagocytes, and a wide variety of protozoa including the free-living amoeba Acanthamoeba castellanii. The bacteria are phagocytosed via a unique coiling mechanism and reside in a specialized phagosome that does not acidify or fuse with lysosomes. Following replication, the host cell lyses and the bacteria are released and are able to initiate a new infection cycle (for reviews, see references 1, 18, 46, and 47).
Identification of the lspFGHIJK genes.
As part of an effort to identify regulatory proteins of L. pneumophila, we attempted to complement a mutant gene product from Escherichia coli (20, 21). Maintenance and growth of E. coli and L. pneumophila and all DNA manipulations were carried out as described previously (39). A library of _Eco_RI-digested genomic DNA of L. pneumophila Philadelphia-1 cloned into the vector pMMB207 (31, 39) was used in a complementation screen. DNA sequencing of the vector-L. pneumophila genomic DNA junctions of a particular clone (plasmid pLM511) revealed homology to the DNA sequence encoding xcpS (gspF).
The xcp genes encode proteins whose products function in the main terminal branch (MTB) of the general secretion pathway (GSP) of Pseudomonas aeruginosa (3). The GSP is a type II protein secretion pathway that is highly conserved among gram-negative bacteria and was first described by Pugsley et al. 14, 15; for reviews, see references 34, 37, 38, and 42). Proteins secreted by the GSP have an initial _sec_-dependent step for export across the inner membrane. The proteins are then transported across the outer membrane via an apparatus consisting of the protein products of 12 to 15 genes. A well-studied paradigm for this pathway is the pullulanase secretion system of Klebsiella oxytoca (37).
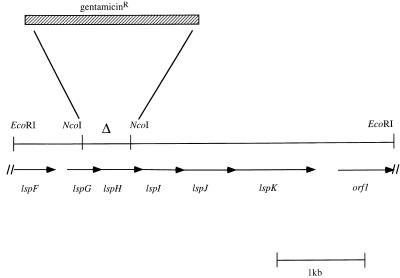
Plasmid pLM511 contains a single _Eco_RI fragment of L. pneumophila DNA, which is 4,279 bp in length. Sequence from both strands of DNA was generated (DNA Synthesis and Sequencing Facility of the Comprehensive Cancer Center, College of Physicians and Surgeons of Columbia University). There are two partial open reading frames and five complete open reading frames contained on the fragment (Fig. 1). Because of the homology to the GSP family of proteins, we named the open reading frames lsp for Legionella secretion pathway. The open reading frames containing homology to the GSP family of proteins were named lspFGHIJK, corresponding to the nomenclature of the pul operon homologs (37, 38, 42).
FIG. 1.
Schematic diagram of open reading frames (indicated by arrows) encoded on pLM511 and construction of a deletion-substitution that inactivates the lspGH genes. The orientation of the arrowheads denotes the direction of transcription. The _Eco_RI fragment contained in pLM511 is 4,279 bp in length and contains the complete DNA sequence encoding the lspGHIJK genes and the partial DNA sequence encoding the lspF and orf1 genes. For construction of a mutation in the lspGH genes, an internal 587-bp _Nco_I fragment was deleted (indicated by ▵) and a 2,118-bp gentamicin resistance cassette (hatched box) was inserted in its place.
Although some of the GSP family of proteins contain an additional two to four genes downstream of lspK (homologs of pulLMNO), the L. pneumophila lsp operon appears to end with lspK. No open reading frames encoding homologs of other proteins in this family were found on the contiguous 1 kb of DNA 3′ to lspK. A partial open reading frame (orf1) is encoded by the downstream region, but orf1 contains no homology to sequences in the databases. At this time, we cannot rule out the existence of an additional unlinked region encoding proteins with homology to pulLMNO located elsewhere on the L. pneumophila chromosome. However, in all other examples of genes encoding proteins in the GSP family, the genes are adjacent and in essentially the same order on the chromosome.
The lspFGHIJK genes encode proteins with significant homology to the gene products of the respective xcp family members (Table 1). Consistent with the cognate GSP homologs, each of the LspGHIJK proteins contains a putative signal sequence, and the predicted localization for all the protein products is in the inner membrane or periplasmic space. The signal sequence consensus site contained in the XcpTUVWX proteins for cleavage by XcpA/PilD (GFXXXE [11, 32]; also see reference [35]) is present in LspGHIJ (Table 1). Additionally, the LspGHIK proteins are devoid of cysteines, a trait common among GSP family members (2).
TABLE 1.
Characteristics of proteins encoded on pLM511
| Gene | No. of amino acids in open reading frame | Predicted molecular mass (kDa) | Presence of signal sequence (PSORT)a | Signal peptide consensus sequence (GFXXXE)b | Predicted location (PSORT)c | % Amino acid sequence identity to Xcp proteins (LALIGN)d |
|---|---|---|---|---|---|---|
| lspF | >126 | >13.7 | ? | ? | IM (1) | 40.8 |
| lspG | 140 | 15.4 | Yes (29) | GFSLIE | IM (1) | 59.4 |
| lspH | 161 | 17.8 | Yes (31) | GFSLIE | OM/PP | 28.8 |
| lspI | 125 | 14.1 | Yes (28) | GFSLIE | PP | 30 |
| lspJ | 205 | 23.7 | Yes (20) | GFSLIE | PP/OM | 28.9 |
| lspK | 322e | 36.1 | Yes (37) | NF | PP/OM | 25.6 |
| orf1 | >213 | >24.3 | No | IM (4) |
Construction of a strain containing a mutation in the lspGH genes.
To help identify protein products secreted by the L. pneumophila GSP, we constructed a mutation in the lspGH genes. The 4,279-bp _Eco_RI fragment from pLM511 was subcloned into pBR322 to generate pLM569. Plasmid pLM569 was digested with _Nco_I to remove an internal 587-bp fragment (Fig. 1). The larger 3,692-bp _Nco_I fragment containing the vector sequences was treated with Klenow enzyme, and a ligation was performed between the Klenow-treated fragment and a 2,118-bp _Hin_cII DNA fragment encoding gentamicin resistance (a gift from David Figurski). This resulted in plasmid pLM808 containing an gentamicin resistance cassette inserted within the _lspGH_-coding region (Fig. 1). Because the coding regions for the lspGHIJK genes overlap, such an insertion would likely be polar on the lspIJK genes and would therefore represent a null phenotype of the L. pneumophila GSP.
The 5,232-bp Eco_RV fragment containing lspFGH::Gentr_IJKorf1 from plasmid pLM808 was subcloned into the _Eco_RV site of the vector pLAW344 for allelic exchange (50). The resultant plasmid, pLM826, was electroporated into the wild-type strain L. pneumophila JR32. Allelic exchange of the lspGH::Gentr mutation onto the chromosome of JR32 was performed as described previously (50) and generated strain LM1520. Southern blot analysis confirmed the construction (data not shown).
A complementing plasmid, pLM828, was constructed by cloning the original 4,279-bp _Eco_RI fragment containing the lspFGHIJKorf1 genes from pLM511 into the vector pMMB207αc (a mobA Kans derivative of pMMB207αb-Km-14 [45]). Plasmid pLM828 was electroporated into strain LM1520, resulting in strain LM1559. Strain LM1558 is strain LM1520 containing the vector pMMB207αc.
Identification of a protein secreted by the L. pneumophila GSP.
We were next interested in identifying a protein secreted by this system. A prime candidate is the major secretory protein (Msp) of L. pneumophila. Msp is a 38-kDa Zn2+ metalloprotease with caseinolytic and hemolytic activities and is the most abundant protein found in culture supernatants (17, 24). Msp contains homology to elastase, a Zn2+ metalloprotease which is secreted by the _xcp_-encoded GSP of P. aeruginosa (5, 25).
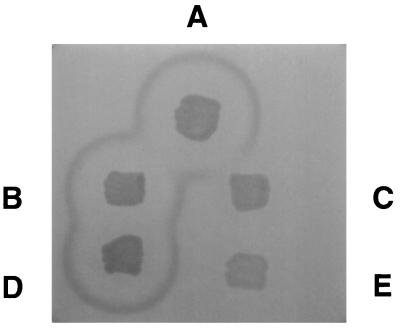
A simple test for the extracellular proteolytic activity of a msp+ strain is a ring of casein hydrolysis around a patch of wild-type L. pneumophila organisms grown on a agar plate containing casein (48). We tested the ability of the strain containing the mutation in lspGH to hydrolyze casein. Strains LM1558 and LM1559 were patched onto buffered yeast starch extract (BYSE) medium containing 10 g of casein per liter as described previously (48). As a control, we patched the wild-type strain JR32 onto the same plate. We also patched strains LS2102 (L. pneumophila mspA1::Tn_9_) and its cognate wild-type parent LS2029 (48) as Msp− and Msp+ controls, respectively.
A ring of hydrolysis was observed around the patch of JR32 growth but not around the patch of LM1558 growth (Fig. 2). This result indicates that the strain LM1558 cannot hydrolyze casein. Because the caseinolytic activity of Msp accounts for virtually all of the proteolytic activity of L. pneumophila (13, 48), we conclude that strain LM1558 is defective in the secretion of Msp. Plasmid pLM828 containing the wild-type lspFGHIJK genes (strain LM1559) is able to complement the inability to hydrolyze casein (Fig. 2). This result provides evidence that the Msp− phenotype of strain LM1558 is due to the loss of the L. pneumophila GSP and not to an extraneous mutation elsewhere in the genome.
FIG. 2.
Casein hydrolysis of the strain containing a mutation in the lspGH genes and the complemented strain. Various strains were patched onto a BYSE agar plate containing casein and incubated for 3 days at 37°C as described previously (48). Patches: A, Wild-type strain JR32; B, wild-type strain LS2029 (isogenic to LS2102); C, msp mutant strain LS2102 (LS2029 mspA1::Tn_9_); D, LM1558 (JR32 lspGH::Gentr pMMB207αc::lspFGHIJKorf1); E, LM1559 (JR32 lspGH::Gentr pMMB207αc).
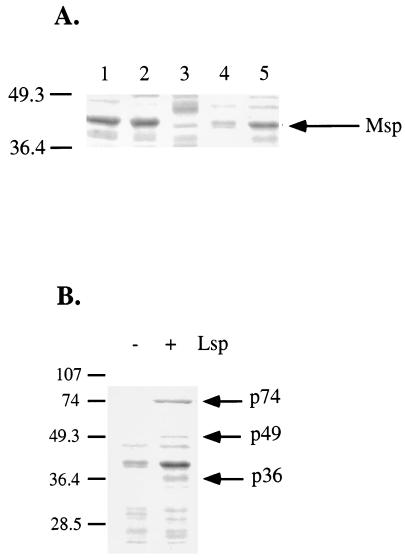
To confirm the results obtained in the casein hydrolysis experiment, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed to analyze the presence of Msp in L. pneumophila culture supernatants. Cell culture supernatants of the wild-type strain LS2029 and the Msp− strain LS2102 were analyzed for the presence of Msp, and the results compared to cell culture supernatants of strains JR32, LM1558, and LM1559. No Msp was observed in the culture supernatant of strain LS2102, as predicted (Fig. 3A). The culture supernatant from strain LM1558 contains low levels of Msp activity compared to that in the supernatant of the wild-type strain JR32 (Fig. 3A). Plasmid pLM828 containing the wild-type lspFGHIJK genes (strain LM1559) is able to complement the inability to secrete Msp into the culture supernatant (Fig. 3A). Taken together with the results from the casein hydrolysis experiment, these results confirm that Msp is a substrate for the L. pneumophila GSP. The fact that Msp requires secretion by the GSP confirms the functionality of the system in L. pneumophila. It has been shown that wild-type E. coli possesses a complete GSP operon, but this operon is not expressed during growth under laboratory conditions (19, 36).
FIG. 3.
SDS-PAGE analysis of L. pneumophila culture supernatants. Supernatants of L. pneumophila overnight cultures were filtered through a 0.45-μm filter (Millipore), and the proteins from the supernatant were precipitated in 10% trichloroacetic acid. SDS loading buffer was added to the protein pellet, and an aliquot was electrophoresed on a 12% polyacrylamide gel. The proteins were visualized by Commassie blue staining. Molecular mass markers (in kilodaltons) are indicated on the left. (A) Lanes: 1, wild-type strain JR32; 2, wild-type strain LS2029 (isogenic to LS2102); 3, msp mutant strain LS2102 (LS2029 mspA1::Tn_9_); 4, LM1558 (JR32 lspGH::Gentr pMMB207αc); 5, LM1559 (JR32 lspGH::Gentr pMMB207αc::lspFGHIJKorf1). (B) Culture supernatants of Lsp− (LM1558) and Lsp+ (LM1559) strains. The three proteins which are present in Lsp+ but not in Lsp− culture supernatants, with approximate molecular masses of 74, 49, and 36 kDa, are indicated on the right.
During this analysis, we noticed that the protein profiles of the culture supernatants of the Lsp− and Lsp+ strains differed significantly (Fig. 3B). The proteins from an SDS-PAGE gel were electroblotted onto a polyvinyl difluoride membrane (Millipore), and three bands (p74, p49, and p36) were excised from the membrane. The samples were subjected to N-terminal sequencing for 8 cycles each (Protein Chemistry Core Facility, Columbia University). In this manner, the N-terminal amino acid sequences were obtained for p74 (AQPTACVN), p49 (YYTSQGSI), and p36 (KDVYEIKH). The sequence databases do not contain any proteins with homology to these three amino acid sequences. The proteins p74, p49, and p36 represent examples of additional proteins that are likely secreted by the L. pneumophila GSP (Fig. 3B).
Analysis of the intracellular growth phenotype of the strain containing a mutation in the lsp genes.
We were then interested in examining the ability of the strain containing a mutation in the lspGH genes to replicate within eukaryotic hosts. We first tested the ability of strain LM1520 for cytotoxicity of HL-60-derived macrophages. The assay was performed as described previously (27, 28). The cytotoxicity of strain LM1520 was compared with the results obtained from the L. pneumophila wild-type strain, JR32, and the mutant strain 25D (22). Strain LM1520 was able to kill macrophages in a manner identical to that of wild-type strain JR32 (data not shown). This finding indicates that the putative secretion system encoded by the lsp operon, or a protein secreted by it, is not required for killing of a macrophage-like cell line.
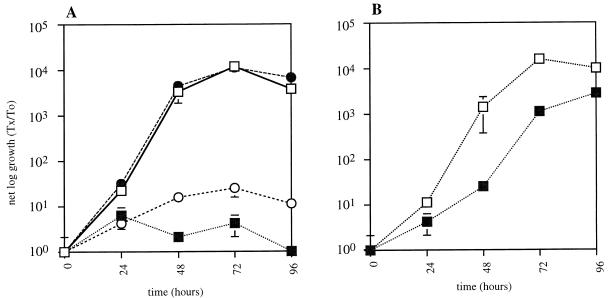
L. pneumophila also has the ability to multiply intracellularly within the free-living amoeba A. castellanii (12, 30, 40, 41). Therefore, strain LM1558 was tested for its ability to replicate within amoebae. Growth and maintenance of A. castellanii was carried out as described previously (12, 30). The assay for replication within amoebae was based on previously described methods (12, 30). L. pneumophila at a multiplicity of infection (MOI) of 10 was added to an adherent monolayer of 1.2 × 105 amoebae. After incubation for 30 min at 37°C to allow for infection, the wells were washed three times with 0.5 ml of Acanthamoeba medium buffer to remove extracellular bacteria. A sample of the infection supernatant was removed once every 24 h for 4 days. Colony forming units (CFUs) of extracellular bacteria were quantitated on ACES-buffered charcoal yeast extract (ABCYE) plates. Wild-type strain JR32 replicated 104-fold within 72 h while the mutant strain 25D did not (Fig. 4A). Strain LM1558 containing a mutation in the lspGH genes is clearly defective for replication within amoebae. The 4,279-bp _Eco_RI fragment was able to complement the growth defect in A. castellanii (Fig. 4A). Therefore, either the secretion apparatus itself or, more likely, another protein that is secreted by this system is required for replication within protozoa. In order to rule out Msp as this protein, we tested the ability of strains LS2029 (Msp+) and LS2102 (Msp−) to replicate within A. castellanii. The results show that both strains replicate approximately 104-fold in amoebae (Fig. 4B). This result indicates that the growth defect of strain LM1558 in amoebae is not due to the inability of this strain to secrete Msp and provides evidence for the existence of other secreted proteins that are important for growth in amoebae. An alternative explanation for our results is that the deletion-substitution mutation is polar on expression of orf1, and it is the orf1 gene product that is required for growth within amoebae. However, we do not believe that orf1 contributes to this phenotype because the open reading frames of the lspGHIJK genes overlap and there is considerable distance between the end of lspK and the beginning of orf1 (314 bp). Additionally, orf1 does not have homology to genes in the GSP family, further suggesting that it is not a member of this operon. Therefore, we believe that it is unlikely that a mutation in lspGH would affect the function, if any, of orf1.
FIG. 4.
Intracellular growth of strains in A. castellanii. The net log growth of the various strains is plotted as a function of time. Error bars represent standard deviation and may not be visible. (A) Wild-type L. pneumophila strain JR32 (open squares) and the mutant 25D (closed squares) are the controls. Strain LM1558 (open circles) is strain LM1520 (JR32 lspGH::Gentr) containing the vector pMMB207αc. Strain LM1559 (closed circles) is strain LM1520 containing the plasmid pMMB207αc::lspFGHIJKorf1. (B) Wild-type L. pneumophila strain LS2029 (open squares) and the isogenic msp mutant strain LS2102 (filled squares).
In summary, our analysis shows that the L. pneumophila GSP or, a protein secreted by it, is required for growth within amoebae. Many of the proteins secreted by the GSP of gram-negative bacteria play a role in pathogenesis, as many of the organisms from which these components originate are plant or human pathogens. Our evidence indicates that Msp is secreted by the L. pneumophila GSP. Indeed, it was postulated that Msp might be a virulence factor (4, 6–10, 13, 29, 48). However, Msp is not required for L. pneumophila to either kill macrophages (29, 48) or multiply within A. castellanii (reference 29 and this work). Therefore, it is likely that one or more proteins other than Msp secreted by the L. pneumophila GSP are required for replication within A. castellanii. Several organisms that have a GSP secrete more than one protein, and L. pneumophila secretes many other exoenzymes, including acid and alkaline phosphatases and lipases (16). Further work is needed to identify other proteins that are secreted by the GSP and to determine the precise requirements for replication within A. castellanii. The observation that a set of genes (encoding the L. pneumophila GSP) is absolutely required for growth within A. castellanii but not in a macrophage-like cell line supports the protozoan host as a more restrictive model of L. pneumophila intracellular growth.
It has been shown previously that L. pneumophila possesses another type II secretion system involved in type IV pilus biogenesis. Liles et al. (26) reported the identification of the L. pneumophila homologs of the P. aeruginosa pilBCD genes. We note that the lsp operon reported herein does not encode the homolog of the prepilin peptidase, PulO, that is required for the processing of the signal sequences present on several of the Pul proteins in K. oxytoca (33). The xcp operon of P. aeruginosa also lacks the O homolog (3; also see references 23 and 43). In the xcp system, the prepilin peptidase PilD (of the pilus biogenesis system) substitutes for the function of the missing O protein in the xcp system, in effect performing the signal peptide cleavage and modification processes for both the pil and xcp gene products (3, 32). Unless L. pneumophila encodes an as-yet-undiscovered unlinked O homolog, the L. pneumophila pilD gene product (26) may function for both pilus biogenesis (pil) and the GSP (lsp) in the same manner as that observed for P. aeruginosa. It remains to be determined if the pilBCD system is required for the pathogenesis of L. pneumophila. The dot-icm gene products are proposed to function as a novel secretion system that is required for inhibition of phagosome-lysosome fusion and for intracellular multiplication (44, 49). L. pneumophila is not unique in this aspect as multiple secretion systems have also been found in other prokaryotes, most notably bacterial pathogens. Therefore, it seems likely that L. pneumophila is typical in its acquisition of some common types of secretion systems for use in pathogenesis in eukaryotic hosts.
Nucleotide sequence accession number.
The L. pneumophila lspFGHIJK sequence has been deposited in the GenBank database under accession no. AF111940.
Acknowledgments
We thank Tony Pugsley for insightful discussions; Pat Higgins for E. coli NH757, the strain that was used in the complementation experiment; David Figurski for the gentamicin resistance cassette; and Carmen Rodriguez for laboratory maintenance.
L.M.H. was supported in part by NIH training grant AI-07161 and by NRSA grant AI-09718. This work was supported by NIH grant AI-23549 to H.A.S.
ADDENDUM IN PROOF
Liles et al. (M. R. Liles, P. H. Edelstein, and N. P. Cianciott, Mol. Microbiol. **31:**959–970, 1999) recently showed that a pilD mutant strain is defective in the secretion of Msp, which further supports our hypothesis that PilD functions as the prepilin peptidase in the Legionella pneumophila GSP.
REFERENCES
- 1.Abu Kwaik Y, Gao L Y, Stone B J, Venkataraman C, Harb O S. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol. 1998;64:3127–3133. doi: 10.1128/aem.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm R A, Mattick J S. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial bionesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178:3809–3817. doi: 10.1128/jb.178.13.3809-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol. 1992;6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 4.Baskerville A, Conlan J W, Ashworth L A, Dowsett A B. Pulmonary damage caused by a protease from Legionella pneumophila. Br J Exp Pathol. 1986;67:527–536. [PMC free article] [PubMed] [Google Scholar]
- 5.Black W J, Quinn F D, Tompkins L S. Legionella pneumophila zinc metalloprotease is structurally and functionally homologous to Pseudomonas aeruginosa elastase. J Bacteriol. 1990;172:2608–2613. doi: 10.1128/jb.172.5.2608-2613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blander S J, Breiman R F, Horwitz M A. A live avirulent mutant Legionella pneumophila vaccine induces protective immunity against lethal aerosol challenge. J Clin Investig. 1989;83:810–815. doi: 10.1172/JCI113962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blander S J, Horwitz M A. Vaccination with the major secretory protein of Legionella pneumophila induces cell-mediated and protective immunity in a guinea pig model of Legionnaires’ disease. J Exp Med. 1989;169:691–705. doi: 10.1084/jem.169.3.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blander S J, Horwitz M A. Vaccination with Legionella pneumophila membranes induces cell-mediated and protective immunity in a guinea pig model of Legionnaires’ disease. J Clin Investig. 1991;87:1054–1059. doi: 10.1172/JCI115065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blander S J, Horwitz M A. Vaccination with the major secretory protein of Legionella induces humoral and cell-mediated immune responses and protective immunity across different serogroups of Legionella pneumophila and different species of Legionella. J Immunol. 1991;147:285–291. [PubMed] [Google Scholar]
- 10.Blander S J, Szeto L, Shuman H A, Horwitz M A. An immunoprotective molecule, the major secretory protein of Legionella pneumophila, is not a virulence factor in a guinea pig model of Legionnaires’ disease. J Clin Investig. 1990;86:817–824. doi: 10.1172/JCI114779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleves S, Voulhoux R, Michel G, Lazdunski A, Tommassen J, Filloux A. The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family) Mol Microbiol. 1998;27:31–40. doi: 10.1046/j.1365-2958.1998.00653.x. [DOI] [PubMed] [Google Scholar]
- 12.Bozue J A, Johnson W. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect Immun. 1996;64:668–673. doi: 10.1128/iai.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conlan J W, Baskerville A, Ashworth L A. Separation of Legionella pneumophila proteases and purification of a protease which produces lesions like those of Legionnaires’ disease in guinea pig lung. J Gen Microbiol. 1986;132:1565–1574. doi: 10.1099/00221287-132-6-1565. [DOI] [PubMed] [Google Scholar]
- 14.d’Enfert C, Chapon C, Pugsley A P. Export and secretion of the lipoprotein pullulanase by Klebsiella pneumoniae. Mol Microbiol. 1987;1:107–116. doi: 10.1111/j.1365-2958.1987.tb00534.x. [DOI] [PubMed] [Google Scholar]
- 15.d’Enfert C, Ryter A, Pugsley A P. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 1987;6:3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowling J N, Saha A K, Glew R H. Virulence factors of the family Legionellaceae. Microbiol Rev. 1992;56:32–60. doi: 10.1128/mr.56.1.32-60.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyfus L A, Iglewski B H. Purification and characterization of an extracellular protease of Legionella pneumophila. Infect Immun. 1986;51:736–743. doi: 10.1128/iai.51.3.736-743.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 19.Francetic O, Pugsley A P. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J Bacteriol. 1996;178:3544–3549. doi: 10.1128/jb.178.12.3544-3549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuner K, Hacker J, Brand B C. The alternative sigma factor ς28 of Legionella pneumophila restores flagellation and motility to an Escherichia coli fliA mutant. J Bacteriol. 1997;179:17–23. doi: 10.1128/jb.179.1.17-23.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickey E K, Cianciotto N P. Cloning and sequencing of the Legionella pneumophila fur gene. Gene. 1994;143:117–121. doi: 10.1016/0378-1119(94)90615-7. [DOI] [PubMed] [Google Scholar]
- 22.Horwitz M A. Characterization of avirulent mutant Legionella pneumophila that survive but do not multiply within human monocytes. J Exp Med. 1987;166:1310–1328. doi: 10.1084/jem.166.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard S P, Critch J, Bedi A. Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J Bacteriol. 1993;175:6695–6703. doi: 10.1128/jb.175.20.6695-6703.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keen M G, Hoffman P S. Characterization of a Legionella pneumophila extracellular protease exhibiting hemolytic and cytotoxic activities. Infect Immun. 1989;57:732–738. doi: 10.1128/iai.57.3.732-738.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazdunski A, Guzzo J, Filloux A, Bally M, Murgier M. Secretion of extracellular proteins by Pseudomonas aeruginosa. Biochimie. 1990;72:147–156. doi: 10.1016/0300-9084(90)90140-c. [DOI] [PubMed] [Google Scholar]
- 26.Liles M R, Viswanathan V K, Cianciotto N P. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marra A, Blander S J, Horwitz M A, Shuman H A. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marra A, Horwitz M A, Shuman H A. The HL-60 model for the interaction of human macrophages with the Legionnaires’ disease bacterium. J Immunol. 1990;144:2738–2744. [PubMed] [Google Scholar]
- 29.Moffat J F, Edelstein P H, Regula D P, Jr, Cirillo J D, Tompkins L S. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol Microbiol. 1994;12:693–705. doi: 10.1111/j.1365-2958.1994.tb01057.x. [DOI] [PubMed] [Google Scholar]
- 30.Moffat J F, Tompkins L S. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect Immun. 1992;60:296–301. doi: 10.1128/iai.60.1.296-301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morales V M, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 32.Nunn D N, Lory S. Components of the protein-excretion apparatus of Pseudomonas aeruginosa are processed by the type IV prepilin peptidase. Proc Natl Acad Sci USA. 1992;89:47–51. doi: 10.1073/pnas.89.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pugsley A P. Processing and methylation of PuIG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol. 1993;9:295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 34.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pugsley A P, Dupuy B. An enzyme with type IV prepilin peptidase activity is required to process components of the general extracellular protein secretion pathway of Klebsiella oxytoca. Mol Microbiol. 1992;6:751–760. doi: 10.1111/j.1365-2958.1992.tb01525.x. [DOI] [PubMed] [Google Scholar]
- 36.Pugsley A P, Francetic O. Protein secretion in Escherichia coli K-12: dead or alive? Cell Mol Life Sci. 1998;54:347–352. doi: 10.1007/s000180050162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pugsley A P, Francetic O, Hardie K, Possot O M, Sauvonnet N, Seydel A. Pullulanase: model protein substrate for the general secretory pathway of Gram-negative bacteria. Folia Microbiol. 1997;42:184–192. doi: 10.1007/BF02818976. [DOI] [PubMed] [Google Scholar]
- 38.Pugsley A P, Francetic O, Possot O M, Sauvonnet N, Hardie K R. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in Gram-negative bacteria—a review. Gene. 1997;192:13–19. doi: 10.1016/s0378-1119(96)00803-7. [DOI] [PubMed] [Google Scholar]
- 39.Purcell M, Shuman H A. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect Immun. 1998;66:2245–2255. doi: 10.1128/iai.66.5.2245-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowbotham T J. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J Clin Pathol. 1983;36:978–986. doi: 10.1136/jcp.36.9.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowbotham T J. Current views on the relationships between amoebae, legionellae and man. Isr J Med Sci. 1986;22:678–689. [PubMed] [Google Scholar]
- 42.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 43.Sandkvist M, Michel L O, Hough L P, Morales V M, Bagdasarian M, Koomey M, DiRita V J. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal G, Purcell M, Shuman H A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segal G, Shuman H A. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol Microbiol. 1998;30:197–208. doi: 10.1046/j.1365-2958.1998.01054.x. [DOI] [PubMed] [Google Scholar]
- 46.Shuman H A, Horwitz M A. Legionella pneumophila invasion of mononuclear phagocytes. Curr Top Microbiol Immunol. 1996;209:99–112. doi: 10.1007/978-3-642-85216-9_6. [DOI] [PubMed] [Google Scholar]
- 47.Shuman H A, Purcell M, Segal G, Hales L, Wiater L A. Intracellular multiplication of Legionella pneumophila: human pathogen or accidental tourist? Curr Top Microbiol Immunol. 1998;225:99–112. doi: 10.1007/978-3-642-80451-9_6. [DOI] [PubMed] [Google Scholar]
- 48.Szeto L, Shuman H A. The Legionella pneumophila major secretory protein, a protease, is not required for intracellular growth or cell killing. Infect Immun. 1990;58:2585–2592. doi: 10.1128/iai.58.8.2585-2592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel J P, Andrews H L, Wong S K, Isberg R R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 50.Wiater L A, Sadosky A B, Shuman H A. Mutagenesis of Legionella pneumophila using Tn903dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol Microbiol. 1994;11:641–653. doi: 10.1111/j.1365-2958.1994.tb00343.x. [DOI] [PubMed] [Google Scholar]