Sulfolobus tengchongensis Spindle-Shaped Virus STSV1: Virus-Host Interactions and Genomic Features (original) (raw)
Abstract
A virus infecting the hyperthermophilic archaeon Sulfolobus tengchongensis has been isolated from a field sample from Tengchong, China, and characterized. The virus, denoted STSV1 (Sulfolobus tengchongensis spindle-shaped virus 1), has the morphology of a spindle (230 by 107 nm) with a tail of variable length (68 nm on average) at one end and is the largest of the known spindle-shaped viruses. After infecting its host, the virus multiplied rapidly to high titers (>1010 PFU/ml). Replication of the virus retarded host growth but did not cause lysis of the host cells. STSV1 did not integrate into the host chromosome and existed in a carrier state. The STSV1 DNA was modified in an unusual fashion, presumably by virally encoded modification systems. STSV1 harbors a double-stranded DNA genome of 75,294 bp, which shares no significant sequence similarity to those of fuselloviruses. The viral genome contains a total of 74 open reading frames (ORFs), among which 14 have a putative function. Five ORFs encode viral structural proteins, including a putative coat protein of high abundance. The products of the other nine ORFs are probably involved in polysaccharide biosynthesis, nucleotide metabolism, and DNA modification. The viral genome divides into two nearly equal halves of opposite gene orientation. This observation as well as a GC-skew analysis point to the presence of a putative viral origin of replication in the 1.4-kb intergenic region between ORF1 and ORF74. Both morphological and genomic features identify STSV1 as a novel virus infecting the genus Sulfolobus.
Viruses represent a huge source of biodiversity and possibly the largest part of the biomass on the planet (17, 60). This view has gained strong support from the work in the past 20 years or so on viruses in hot spring ecosystems, pioneered by Wolfram Zillig and colleagues (41, 42, 54). Nearly a dozen viruses have been identified from hyperthermophilic archaea of the genus Sulfolobus alone (54). Unique features, especially unusual morphologies, of these viruses have provided a basis for the introduction of four novel virus families, Fuselloviridae, Rudiviridae, Lipothrixviridae, and Guttaviridae (4, 63).
All members of the Fuselloviridae (SSV1, SSV2, SSV3, SSV RH, and SSV KI) are enveloped, spindle-shaped viruses (60 by 90 nm in size) with a circular double-stranded DNA genome of 14 to 17 kb (34, 58). The family Rudiviridae (SIRV1 and SIRV2) is characterized by a stiff 23- by 800- to 900-nm helical rod containing a 33- to 36-kb linear double-stranded DNA genome with covalently closed ends (12, 39). The lipothrixvirus SIFV is a 24- by 1,980-nm flexible rod with putative attachment fibers at both ends (5). The viral genome is a linear double-stranded DNA molecule of ≈42 kb. The fourth family is represented by SNDV, a droplet-shaped virus possessing a circular and modified double-stranded DNA genome estimated to be 20 kb (4). Recently, an icosahedral virus, named STIV for Sulfolobus turreted icosahedral virus, has been described, which probably represents the fifth category of Sulfolobus viruses (48). These viruses provide windows to genetic processes in Sulfolobus, a model organism for the study of Archaea, and are of great importance to the understanding of diversity and evolution of life on earth.
During a recent survey of Sulfolobus species and their extrachromosomal genetic elements in acidic hot springs in Tengchong, a geothermal area in southern China, we have isolated a crenarchaeotal virus infecting Sulfolobus tengchongensis (61). The virus, denoted STSV1 (Sulfolobus tengchongensis spindle-shaped virus 1), has the morphology of an unusually large spindle (230 by 107 nm) with a tail of variable length (≈68 nm on average) at one end. Virus-like particles resembling STSV1 in shape and size have also been found in hot springs in Yellowstone National Park (43, 47). STSV1 harbors a modified, double-stranded DNA genome of ≈75 kb. Notably, the genome sequence shares little similarity to those of the known virus families. STSV1 is the largest spindle-shaped virus that has been characterized.
MATERIALS AND METHODS
Sulfolobus strains and growth conditions.
Sulfolobus solfataricus strains P1 and P2 were generous gifts from K. Stedman (Portland, Oregon). Sulfolobus shibatae 51178 was purchased from the American Type Culture Collection (Rockville, MD). S. tengchongensis RT8-4 was described previously (61). Sulfolobus sp. strain H3-1 was isolated from acidic hot springs in Tengchong in this study. S. islandicus REY15A was isolated from a solfataric field in Iceland. All strains were grown aerobically with shaking in Zillig's medium (62) supplemented with 0.2% tryptone at 80°C and an initial pH of ≈3.3 (at 22°C).
Isolation of STSV1.
Samples collected from acidic hot springs and mud holes in Tengchong, Yunnan, China, were inoculated into the modified Brock's medium described by Zillig et al. (62). After shaking for 7 to 14 days at 80°C, the growing cultures were centrifuged (13,000 × g, 30 sec, 22°C). An aliquot of the supernatant of each culture was filtered through a 0.45-μm filter and spotted on the lawn of S. tengchongensis RT8-4, which was prepared as described previously (62). The plate was incubated for 4 to 5 days at 80°C to allow plaques to form. Gelrite pieces containing a plaque were recovered. Each gel piece was incubated for 1 h at 22°C in distilled water (50 μl) to yield a virus preparation. The virus was purified by isolating single plaques using plaque assays.
Plaque formation.
Serial dilutions (10 μl) of a virus preparation were mixed with a sample (200 μl) of the exponentially grown S. tengchongensis RT8-4 culture (109 cells). The mixture was incubated for 30 min at 22°C to allow the adsorption of the virus to the host cells. Immediately following the addition of 1.5 ml of Zillig's medium containing 0.25% Gelrite (80°C), the sample was layered over a premade 0.8% Gelrite plate (80°C). The plate was incubated for 3 days at 80°C.
Virus purification.
S. tengchongensis RT8-4 was grown overnight to an optical density at 600 nm (OD600) of 0.4 to 0.5. A sample (20 ml) of the culture (4.2 × 109 cells) was centrifuged at 6,000 × g for 15 min at 22°C, and the pellet was resuspended in Zillig's medium (0.4 ml). The cell suspension was mixed with 0.9 ml of an STSV1 preparation (2.1 × 109 PFU/ml). Following incubation for 30 min at 22°C, the mixture was transferred to Zillig's medium (400 ml). After shaking at 80°C for 3 to 4 days, the culture was centrifuged (6,000 × g, 20 min, 4°C). The supernatant was centrifuged again at 12,000 × g for 15 min at 4°C to remove cell debris. Virus particles were collected by precipitation with 10% (wt/vol) polyethylene glycol 6000 in the presence of 1 M NaCl and centrifugation at 12,000 × g for 15 min at 4°C. The pellet was resuspended in distilled water, and the suspension was clarified by centrifugation at 6,000 × g for 20 min at 4°C. Virus particles were purified by centrifugation in a CsCl density gradient (0.45 g/ml) at 125,000 × g for 24 h at 23°C.
For some experiments, STSV1 was prepared using a simplified method. After two rounds of centrifugation to remove host cells and cell debris, virus particles were collected from the supernatant by centrifugation at 120,000 × g for 2 h at 4°C. The pellet, which contained virus particles, was resuspended in Tris-EDTA (pH 8.0).
Electron microscopy.
Samples were stained with 2% (wt/vol) uranyl acetate and observed under an electron microscope (Hitachi H-600A).
Virus growth curve.
Exponentially growing S. tengchongensis RT8-4 cells were mixed with STSV1 at a multiplicity of infection of ≈3. After 30 min, the cells were washed with fresh Zillig's medium to remove free virus particles, diluted into preheated medium to an OD600 of ≈0.6, and incubated at 80°C. Samples were taken at intervals. The titer of virus particles released from the cells was estimated by serially diluting the culture supernatant and determining the highest dilution at which the sample formed plaques on an RT8-4 lawn. The amount of viral DNA associated with the host cells was measured by spotting cells on a nylon membrane and subsequent dot blotting.
Protein analysis.
CsCl-purified virus particles were dissolved in the loading buffer (20 μl) for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. After heating at 95°C for 3 min, the samples were subjected to electrophoresis in an SDS-polyacrylamide (12%) gel. The proteins were visualized by staining with Coomassie brilliant blue R-250. For protein identification, gel slices containing protein bands were excised from the stained gel. In-gel digestion of the proteins was performed as described previously (10). The digests were dissolved in 0.1% formic acid, and purified with ZipTipC18 pipette tips (Millipore, Bedford, MA) according to the manufacturer's procedure. The tryptic peptides were analyzed by two-dimensional chromatography coupled with ion trap mass spectrometry on a ProteomeX Workstation (ThermoFinnigan).
Lipid analysis.
Viral lipids were extracted and analyzed as described previously (5).
DNA isolation.
Total DNA from S. tengchongensis RT8-4 or that infected with STSV1 was isolated as described previously (61). To isolate STSV1 DNA from virus particles, SDS and proteinase K were added to a virus suspension to final concentrations of 1% and 1.25 mg/ml, respectively. After incubation for 30 min at 50°C, the mixture was extracted with phenol-chloroform, and the DNA was precipitated with ethanol.
Southern hybridization.
Purified STSV1 DNA and total DNA from S. tengchongensis RT8-4 infected with STSV1 were digested with a restriction endonuclease. The DNA fragments were resolved by electrophoresis in 0.8% agarose gel and transferred onto a Hybond N+ nylon membrane (Amersham Pharmacia Biotech). Southern hybridization was performed according to the procedure of Sambrook et al. (49). The probe was prepared by labeling the STSV1 DNA or a PCR fragment containing the viral int sequence (nucleotide positions 58818 to 60281) with [α-32P]dCTP using a Random Primer DNA labeling kit (TaKaRa). After hybridization, the membrane was exposed to X-ray film.
The amount of viral DNA associated with infected RT8-4 cells was measured by dot hybridization. A sample (1 ml) of a growing culture of RT8-4 infected with STSV1 was centrifuged, and the cells were resuspended in TE (80 μl). An aliquot (5 μl) of the cell suspension was spotted on a Hybond-N+ nylon membrane. The membrane was air dried, and placed successively for 5 min each on top of each of the following four filter papers that had been soaked in 10% SDS, 0.5 M NaOH; 1.5 M NaCl, 1 M Tris.HCl; 1.5 M NaCl (pH 7.4); and 2× SSC (0.3 M NaCl plus 30 mM sodium citrate), respectively. The membrane was then baked for 15 min at 80°C, and the DNA was cross-linked to the membrane by UV irradiation. Hybridization was performed as described above using the labeled int sequence as a probe. After hybridization, the membrane was dried and analyzed with a PhosphoImager (Molecular Dynamics, Inc.).
DNA sequencing and sequence analysis.
STSV1 genome was sequenced as described previously (50). Briefly, purified STSV1 DNA was mechanically sheared and treated with DNA modifying enzymes to generate blunt ends. Subsequently, 1.5- to 2.5-kb fragments were recovered from an agarose gel and ligated into pUC18 vector at the SmaI site. Plasmid DNAs were purified from shotgun clones using BioRobot 8000 (QIAGEN), and sequenced with the MegaBACE 1000 DNA Analysis System (Molecular Dynamics). A total of 1,365 sequence reads were generated, yielding an 8.3-fold coverage of the genome. The genome sequence of the STSV1 virus was assembled using the CONSED (22), and Sequencher 4.2 (Gene Codes Corp. Ann Arbor, MI) programs, and the sequence gaps were filled using custom primers. The sequence was determined for both strands, and any remaining sequence ambiguity was resolved by sequencing at least two clones.
A cumulative GC skew analysis of the genome was performed as described (23). Potential open reading frames (ORFs) were initially identified as coding regions using GenMark (13), and their start positions were determined by selecting for the largest possible ORFs with one of the three start codons (ATG, GTG, and TTG). Promoter, Shine-Dalgarno, and transcriptional terminator sequences were subsequently determined for individual ORFs as described previously (9, 45, 56). ORFs encoding proteins shorter than 50 amino acid residues were considered potential coding regions if an upstream promoter and/or Shine-Dalgarno sequence was found. The BlastP, SMART, and Pfam search tools (2, 31) were employed for sequence homology analysis.
Nucleotide sequence accession number.
The complete sequence of the STSV1 genome has been submitted to the DDBJ/EMBL/GenBank databases under accession number AJ783769.
RESULTS
Detection and isolation of STSV1.
A sample was collected from each of 38 hot springs and mud holes with temperatures ranging from 61 to 94°C and pHs from 2.0 to 6.0 in a major solfataric field in Tengchong, Yunnan, China. When inoculated into Zillig's medium, 27 of these samples resulted in apparent growth. To test for the presence of viruses in these samples, we prepared the supernatants from the enrichment cultures. Electron microscopic analysis showed that 8 out of the 10 tested samples contained a single type of virus-like particles. We also found that the filtrate of the supernatant obtained using a 0.45-μm membrane filter formed plaques on the lawn of S. tengchongensis RT8-4, which was isolated from the same area. Virus-like particles identical to those in the supernatant were recovered in the plaques. Based on this observation, we developed a plaque assay for the virus-like particles using S. tengchongensis RT8-4 as an indicator strain. The virus-like particles formed small (≤1 mm), clear plaques on the lawn. The virus purified from single plaques was denoted STSV1.
Morphology of STSV1.
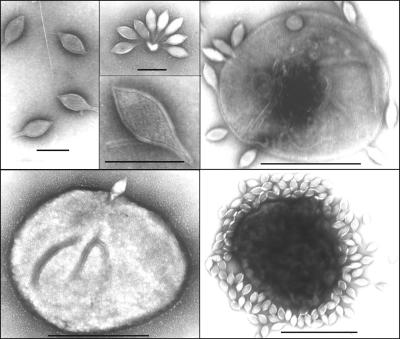
The STSV1 virion had the shape of a spindle with a tail at one end (Fig. 1). The spindle part of the virus was ≈230 by 107 nm in size, and the tail varied drastically in length from 0 to 133 nm, with an average of ≈68 nm. In a few cases, viral particles with tails at both ends were observed. STSV1 resembles SSV1 in shape, but the former is much larger. The virus particles of STSV1 often occurred singly but occasionally were clustered in a rosette-like pattern with tails attached to cell debris (Fig. 1). Close electron microscopic examination showed that the virus capsid was surrounded by a 10- to 15-nm envelope. Thin-layer chromatography revealed the presence of lipids in the chloroform/methanol extract of purified virus particles (Fig. 2). This is consistent with the finding that the virus lost its infectivity after exposure for 30 min to 1% (vol/vol) chloroform or diethyl ether.
FIG. 1.
Transmission electron micrographs of STSV1. Upper left panel, virus particles (bars, 200 nm); other panels, virus particles attached to S. tengchongensis RT8-4 cells (bars, 1 μm).
FIG. 2.
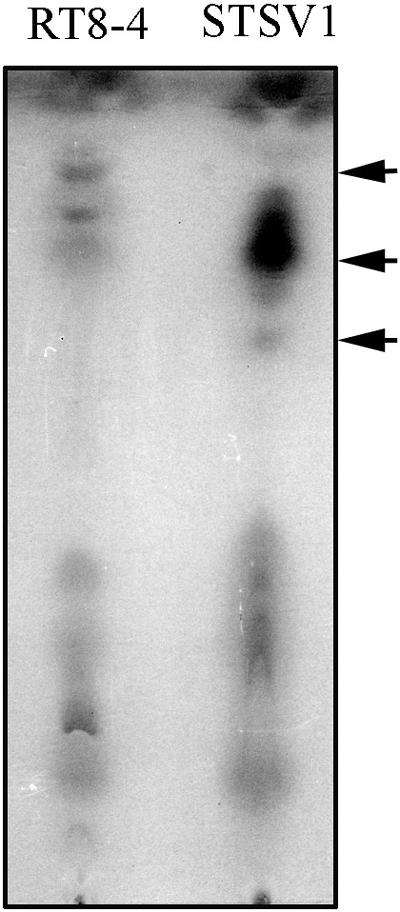
Thin-layer chromatography of lipids extracted from STSV1 particles and uninfected S. tengchongensis RT8-4 cells. The arrows indicate bands that occur differently in the two samples.
Viral proteins.
To analyze the protein composition of the virion, we subjected a purified virus sample to SDS-polyacrylamide gel electrophoresis. A very intense 18-kDa band as well as a number of minor bands were found on a Coomassie brilliant blue-stained gel (Fig. 3). To identify these proteins, gel slices containing the protein bands were excised. The proteins in the gel slices were subjected to in-gel digestion with trypsin, and the masses of the resulting peptides were determined by two-dimensional chromatography coupled with ion trap mass spectrometry.
FIG. 3.
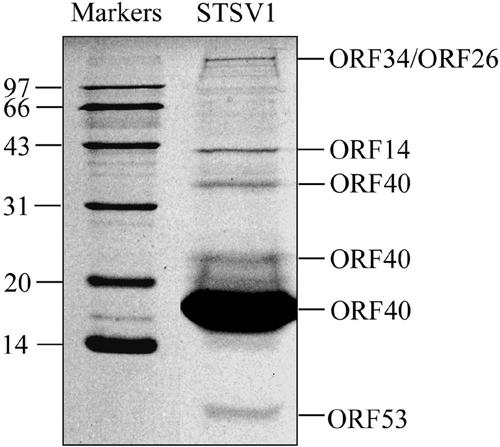
Analysis of proteins in the STSV1 virion. Purified STSV1 particles were subjected to SDS-polyacrylamide gel (12%) electrophoresis. After electrophoresis, the gel was stained with Coomassie brilliant blue R-250. The proteins in the bands were sliced and subjected to in-gel digestion with trypsin and identified by determining the masses of the tryptic peptides by two-dimensional chromatography coupled with ion trap mass spectrometry. ORFs encoding the proteins are indicated on the right, and molecular size standards (in kDa) are on the left.
A subsequent search of the sequence database of STSV1 (see below) using the tryptic mass results identified high-confidence matches to proteins encoded by STSV1 (Fig. 3). The protein in the major band is the product of ORF40 with a calculated molecular mass of 15.6 kDa and an isoelectric point of 9.9. The ORF40 product was also found in the band of approximately 34 kDa, suggesting that the protein formed dimers that were not completely dissociated by denaturation in SDS at 95°C. N-terminal amino acid sequencing analysis confirmed that no additional proteins comigrated with the ORF40 product in the major band. Therefore, the STSV1 virion appears to be dominated by a single protein. These results suggest that the ORF40 product is a coat protein that presumably serves a key architectural role in the assembly of the nucleocapsid of STSV1.
Four minor structural proteins are encoded by ORFs 14, 26, 34, and 53 (Fig. 3). The presence of multiple structural proteins in the virion is expected since STSV1 is a complex enveloped virus. The product of ORF34 is a large coiled-coil protein containing 2,308 amino acid residues. Since the protein has two transmembrane helices and shows sequence similarity to the large anchor protein of Staphylococcus (25) and other membrane proteins, including Lactococcus infection protein (19), it is presumably involved in the viral recognition of and/or attachment to the host cell. The ORF26 product was identified in the gel slice which also contained the ORF34 protein. Given its predicted molecular mass of 38 kDa, the protein presumably was covalently modified or formed a very stable complex, since no band was found where the monomeric form of an unmodified ORF26 product was supposed to migrate.
Viral multiplication.
STSV1 appeared to have a narrow host range. It did not infect Sulfolobus sp. strain H3-1, which was isolated from Tengchong (61), and shared > 99% similarity in 16S rDNA sequence to S. tengchongensis RT8-4. The virus was also unable to infect S. shibatae B12, S. islandicus REY15A, and S. solfataricus P1 and P2.
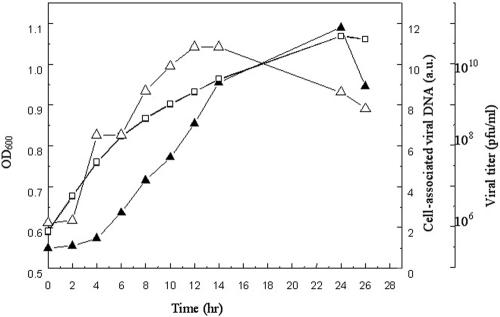
Infection by STSV1 significantly slowed the growth of the host cells. Under the growth conditions used in this study, the doubling time for cells in the infected culture was ≈30 h, compared to 11 h for those in the uninfected control in the exponential growth phase, although both cultures peaked at the same cell density. To determine the time required for the infecting STSV1 virus to initiate DNA replication and for the virus to release progeny particles, we infected exponentially growing host cells with the virus at a multiplicity of infection of ≈3 and followed the changes in cell-associated viral DNA as well as extracellular virus titer. We detected STSV1 DNA replication by measuring the cell-associated viral DNA instead of the viral DNA level in the cell since we were unable to remove virus particles attached to the host cells (see below).
As shown in Fig. 4, a jump in virus titer in the cell-free culture fluid occurred within 4 h of viral infection, which appeared to coincide with the onset of viral DNA replication, suggesting no significant delay in the release of STSV1 particles once they were produced. Like fuselloviruses, STSV1 was nonlytic. No cell lysis was observed, under the electron microscope, in cultures that actively produced virus particles, and an increase in virus release occurred concomitantly with an increase in cell density. After extruding from the host cell, virus particles often remained attached to the surface of the cell, and layers of virus particles were occasionally seen surrounding a single host cell (Fig. 1). At the peak, the number of virus particles (>1010 PFU) was at least 50 times greater than that of the host cells in an infected culture. The total DNA isolated from the culture at this point contained large amounts of viral DNA (Fig. 5).
FIG. 4.
Infection of S. tengchongensis RT8-4 by STSV1. The titer of STSV1 in the extracellular fluid, determined using a plaque assay, represents the highest dilution at which the sample was capable of forming plaques. The viral DNA associated with the host cells was quantitated by dot blotting. Symbols: □; cell density of an RT8-4 culture infected with STSV1; ▴, cell-associated viral DNA; ▵, virus titer in the extracellular fluid.
FIG. 5.
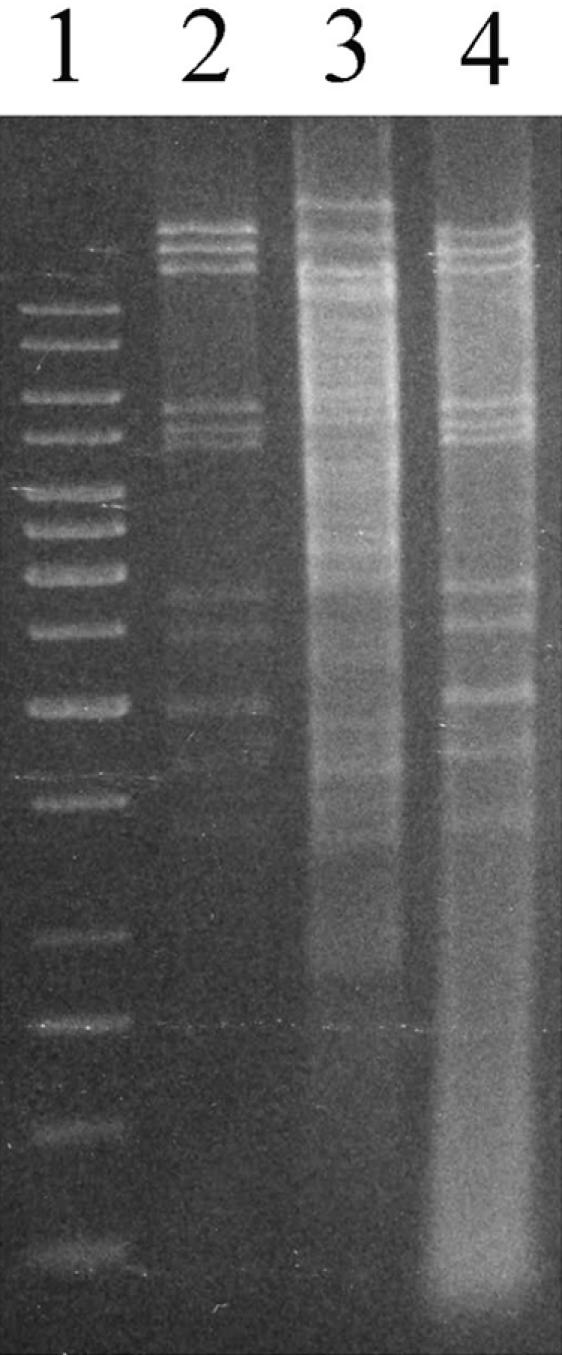
Restriction analysis of DNAs isolated from STSV1 particles, uninfected S. tengchongensis RT8-4 cells, and RT8-4 cells infected with STSV1. DNAs were isolated from virus particles and infected or uninfected cells and digested with EcoRI. The digests were electrophoresed in 0.8% agarose. Lane 1, size markers (from top to bottom: 10, 8, 6, 5, 4, 3.5, 3, 2.5, 2, 1.5, 1, 0.75, 0.5 and 0.25 kb); lane 2, purified STSV1 DNA; lane 3, DNA from uninfected RT8-4 cells; lane 4, DNA from RT8-4 cells infected with STSV1.
An infected RT8-4 culture was able to keep the virus after 10 transfers of pelleted cells in the stationary phase to new medium. To determine if the viral DNA was capable of integrating into the host genome, we first performed a Southern hybridization analysis on DNAs from both STSV1 and RT8-4 cells infected with the virus using either the entire viral genome sequence or the putative int gene as a probe. The int gene of STSV1 was used since an archaeal integrase gene may be partitioned into two fragments flanking the integrated viral genome during integration, as shown in SSV1 (46, 52, 53). The restriction patterns of the two DNA samples were identical with either probe (Fig. 6). No hybridization bands that would have suggested viral integration were found. It appears that STSV1 exists in a carrier state in the host cell.
FIG. 6.
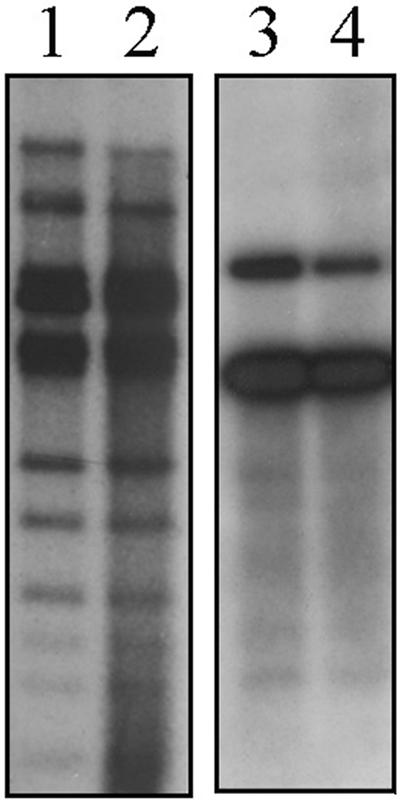
Southern blotting analysis of the restriction fragments of STSV1 DNA isolated from virus particles and S. tengchongensis RT8-4 cells infected with STSV1. Purified STSV1 DNA (lanes 1 and 3) and total DNA from RT8-4 cells infected with STSV1 (lanes 2 and 4) were digested with EcoRV, and the restriction digests were separated by electrophoresis in 0.8% agarose. Hybridization was performed using radiolabeled total viral DNA (left panel) or an _int_-containing fragment (right panel) as a probe.
Viral DNA modification.
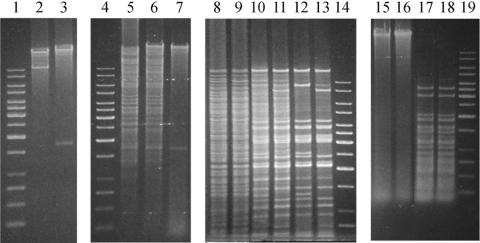
Digestion of the STSV1 DNA with EcoRI, EcoRV, XbaI, HindIII, BamHI, SacI, NcoI, or BglII produced a pattern expected from the viral genome sequence. However, cleavage with PstI, XhoI, or AvaI yielded fewer fragments than predicted (Fig. 7). For instance, although the genome has 14 PstI sites (CTGCAG), only two prominent fragments were found in the PstI digest of the viral DNA, suggesting that the viral DNA was modified at specific sites. To determine if the viral genome was modified in a Dam-like fashion, we cleaved the DNA with the isoschizomeric restriction enzymes Sau3AI, Dpn1, and MboI. Cleavage by Sau3AI is not affected by methylation of the adenine residue at the recognition site GATC, whereas restriction by DpnI depends on it and that by MboI is prevented by methylation of the adenine (35). STSV1 DNA was cleaved by MboI and Sau3AI but not DpnI (Fig. 7), excluding the possibility that the DNA was methylated in a Dam-like fashion.
FIG. 7.
Modification of STSV1 DNA. Purified STSV1 DNA and total DNAs from uninfected S. tengchongensis RT8-4 cells and from RT8-4 cells infected with STSV1 were digested with different restriction endonucleases and separated in 1% agarose gels. Lanes 1, 4, 14, and 19, size markers (from top to bottom: 10, 8, 6, 5, 4, 3.5, 3, 2.5, 2, 1.5, 1, 0.75, 0.5 and 0.25 kb); lane 2, STSV1 DNA digested with XhoI; lane 3, STSV1 DNA digested with PstI; lane 5, RT8-4 DNA digested with PstI; lane 6, DNA from infected RT8-4 cells digested with PstI; lane 7, STSV1 DNA digested with PstI; lane 8, RT8-4 DNA digested with MspI; lane 9, RT8-4 DNA digested with HpaII; lane 10, DNA from infected RT8-4 cells digested with MspI; lane 11, DNA from infected RT8-4 cells digested with HpaII; lane 12, STSV1 DNA digested with MspI; lane 13, STSV1 DNA digested with HpaII; lane 15, undigested STSV1 DNA; lane 16, STSV1 DNA digested with DpnI; lane 17, STSV1 DNA digested with Sau3AI; lane 18, STSV1 DNA digested with MboI.
We then tested the ability of a pair of cytosine methylation-sensitive isochizomeric endonucleases (HpaII and MspI) to cleave their recognition sequence, CCGG, in the STSV1 genome. Cleavage by HpaII and MspI is inhibited by methylation at the internal and external cytosine residues of the site, respectively (14). All of the CCGG sites in STSV1 DNA were cleaved by HpaII. However, some of the sites were resistant to cleavage by MspI, indicating methylation of specific cytosine residues. Given the observed inhibition of specific cleavage of the DNA by PstI, XhoI, and AvaI, which are sensitive to methylation of either cytosine or adenine residues in their restriction sites, additional forms of DNA modification (e.g., adenine methylation) may also occur in the viral genome.
We also studied if the genomic DNA of RT8-4 cells infected with STSV1 was modified in the same fashion as the viral DNA. As shown in Fig. 7, the viral DNA in a sample of total DNA prepared from the infected cells was modified, judging by the pattern of PstI cleavage. By contrast, the PstI cleavage pattern of the host genomic DNA from the infected cells was identical to that from uninfected cells. There are two possible interpretations for this observation. In the first interpretation, the modification system is host encoded, so both the host and the infecting viral DNAs are modified. In the second interpretation, the modification system is encoded by the virus. In this scenario, the uninfected host genome is not modified, and the viral modification system only methylates hemimethylated sites which are generated during the replication of fully methylated DNA.
The latter possibility appears more plausible for the following reasons. First, the host strain was found to harbor a 20-kb plasmid which contains five PstI sites and one XhoI site (unpublished results). These sites were all cleaved by the cognate enzymes. Second, the size distribution of PstI fragments of the total DNA from the uninfected cells does not suggest extensive modification at PstI sites in the host genome, which would occur if the modification system was encoded by the host. Third, the viral genome encodes several putative modification enzymes (see below).
Genomic features.
The complete STSV1 genome contains 75,294 base pairs, which is much larger than the largest crenarchaeal viral genome (56 kb) that has been reported (54). The G+C content of the STSV1 genome is 35%, falling within the range of those of the known Sulfolobus genomes (28, 51). A total of 74 ORFs were identified in the STSV1 genome (see the supplemental material). Most genes appear to exist as singlets: 76% of the ORFs are preceded by a putative promoter and 40% are followed by a T stretch of more than six bases, which presumably functions as a transcriptional terminator (45). Thus, the genomic structure of STSV1 differs from that of known crenarchaeal viruses and plasmids, in which genes are generally organized in operons.
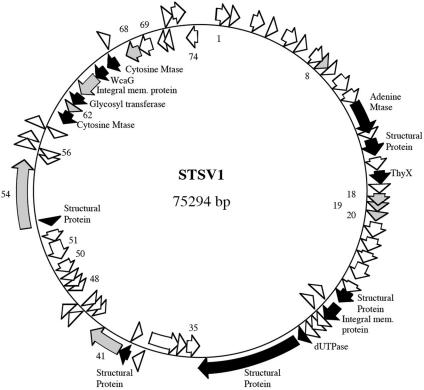
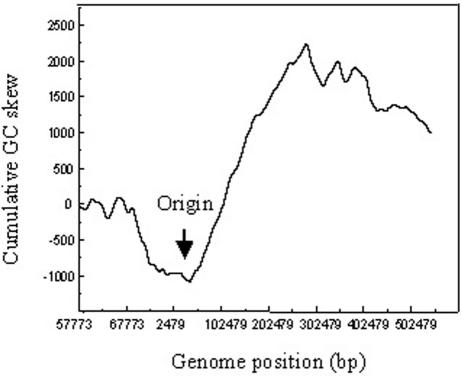
Remarkably, the STSV1 genome is highly asymmetric: the genes in one half of the genome are transcribed in one direction, and two thirds of the genes in the other half are arranged in the opposite direction (Fig. 8). The biased gene distribution points to the possibility that the origin and terminus of STSV1 DNA replication are located between ORF1 and ORF74 and between ORF34 and ORF35, respectively. This possibility is supported by a cumulative GC skew analysis of the genome (Fig. 9). Intriguingly, a 1.4-kb intergenic region exists between ORF74 and ORF1. This region has a GC content of 22%, which is substantially lower than that for the entire viral genome (35%). Runs of consecutive A and T are scattered throughout the region.
FIG. 8.
Organization of the STSV1 genome. Putative ORFs are shown as arrows along the line representing the genome. ORFs encoding putative enzymes or proteins are indicated in black, and those resembling hypothetical ORFs from Sulfolobus species or their viruses are in grey.
FIG. 9.
Cumulative GC skew of the STSV1 genome. The putative origin of viral DNA replication is indicated by an arrow.
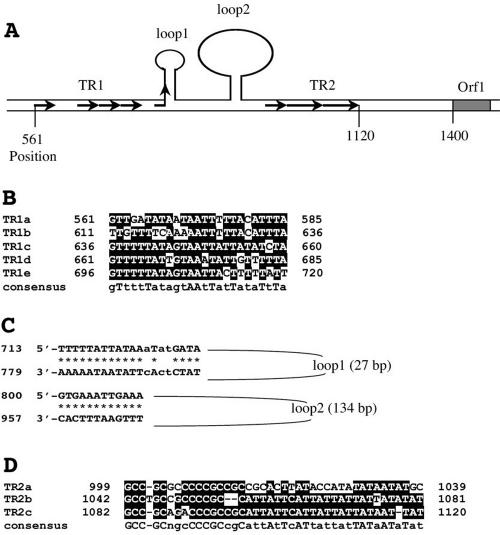
Since known origins of DNA replication typically contain sequence repeats and AT-rich sequences, we performed a detailed analysis of the sequence of the intergenic region. As shown in Fig. 10, two sets of tandem repeats were found in the region: a 25-bp tandem repeat of five and half copies (TR1) and a 40-bp tandem repeat of two and half copies (TR2). The latter is located proximal to ORF1. Interestingly, TR2 is divided into an upstream GC-rich half and a downstream AT-rich half. There are also two sets of inverted repeats with the potential to form stem-loop structures, which are located between TR1 and TR2. These sequence features argue plausibly for the notion that the origin of STSV1 DNA replication is located in the intergenic region.
FIG. 10.
Sequence features of the region containing the putative origin of STSV1 DNA replication. The positions of the cis elements in the region and the start codon of ORF1 are numbered with reference to the complete sequence of STSV1. (A) An illustration of the putative replication origin. TR1, tandem repeat 1; TR2, tandem repeat 2; loop1, stem-loop 1; loop2, stem-loop 2. The figure was not drawn to scale. (B) Sequence alignment of the 25-bp tandem repeats (TR1). Bases conserved in at least three repeats are highlighted in black. The consensus sequence is shown at the bottom. (C) Inverted repeats forming stem-loop structures. Mismatches are in lowercase letters. (D) Multiple sequence alignment of the 40-bp tandem repeats.
Analysis of putative ORFs.
A search of a local Sulfolobus virus database with the STSV1 ORFs revealed only two high-similarity matches to known viral ORFs, both of which were from the genomes of the Sulfolobus rudiviruses SIRV1 and SIRV2. STSV1 ORF18 was a homologue of SIRV2 ORF310 (or SIRV1 ORF306), which encodes a hypothetical protein, whereas STSV1 ORF33 was similar to SIRV1 ORF158b (or SIRV2 ORF158b), which has been shown to encode dUTPase (39, 40).
Although STSV1 is similar in shape to fuselloviruses, none of the STSV1 ORFs showed high similarity to those of known fuselloviruses. Weak similarity was detected for ORF41 and ORF69: the former displayed 20% and 37% sequence identity and similarity, respectively, to SSV2 ORF809, which is conserved in the SSV viruses (55, 58); and the latter was 28% identical and 52% similar to SSV2 ORF233, which is highly conserved among fuselloviruses as well as the satellite virus pSSVx and has been suspected to play a role in viral packaging (3). These results show that STSV1 is not closely related to any of the known archaeal viruses in general and fuselloviruses in particular.
Searches of the public databases using BlastP, SMART, and Pfam search tools (2, 7, 31) revealed that several of the STSV1 ORFs have significant sequence similarity to known or putative enzymes involved in polysaccharide biosynthesis, nucleotide metabolism and DNA modification (Fig. 8; see the supplemental material). Four ORFs (ORFs 29, 63, 64, and 65) implicated in polysaccharide biosynthesis presumably function in the biosynthesis of virus-specific lipids in the viral envelope. ORF29 encodes a putative integral membrane protein with 12 transmembrane helices and appears to belong to a family of polysaccharide biosynthesis proteins (7). Multiple genes encoding homologues of this protein have been identified in Sulfolobus genomes (28, 51).
ORF63 contains the COG0438 domain of group I glycosyltransferases, which are conserved among bacteria, eucarya, and archaea, including the crenarchaeal virus AFV1 (11). ORF65 shows high similarity to nucleoside diphosphate-sugar epimerases (WcaG) and has a closely related homologue in Methanosarcina acetivorans (18). The M. acetivorans protein has been suggested to play a role in the biosynthesis of the cell envelope. ORF64 has no significant matches to proteins of known function. However, since it encodes a putative integral membrane protein and is located between ORF63 and ORF65 in the genome, ORF64 may also be involved in polysaccharide biosynthesis.
STSV1 encodes two key enzymes (ORF16 and ORF33) in thymine metabolism that may alter the pool sizes of dUTP and dTMP in host cells. Both genes have homologues in the two sequenced Sulfolobus genomes (28, 51). The ORF16 product is a putative thymidylate synthase, ThyX (Thy1 in Dictyostelium discoideum) (16, 37). ThyX has a wide but sporadic phylogenetic distribution, almost exclusively limited to microbes lacking thyA (30). The protein converts dUMP into thymidylate in a flavin-dependent fashion. ORF33 is a homologue of the dUTPase of SIRV (40).
Three STSV1 ORFs (ORFs 12, 61, and 66) encode putative DNA methyltransferases (MTases). ORF12 shows significant similarity to adenine-specific MTases containing a zinc ribbon (33). Its homologues are widespread among sequenced bacterial and archaeal genomes. Since ORF12 contains motifs conserved among N-MTases (motifs I and IV), and carries the signature sequence DPPY in motif IV, it appears to encode an N6-adenine MTase (32). ORF61 is 23/40% and 22/40% identical/similar, respectively, to the PspGI methylase (M.PspGI) of Pyrococcus sp. GI-H and the MvaI methylase (M.AvaI) encoded by a plasmid from Micrococcus variabilis, respectively (36). Like M.PspGI and M.AvaI, the STSV1 protein contains a set of nine motifs conserved both in sequence and location in the α-subgroup of N-MTases. The presence of the signature sequence SPPY in its motif IV identifies the ORF61 product as a putative N4-cytosine MTase (32, 36).
ORF66 also possesses the nine conserved sequence motifs but, based on their relative location, belongs to the β-subgroup of N-MTases. The ORF66 protein shares sequence homology with M.Cfr91, an N4-cytosine MTase from Citrobacter freundii RFL9 (20/34% identity/similarity) as well as M.MboII and M.HpaI, N6-adenine MTases from Moraxella bovis (22/39% identity/similarity) and Haemophilus parainfluenzae (22/40% identity/similarity), respectively. However, since motif IV in the ORF66 protein has the SPPY sequence, the protein is more likely a N4-cytosine MTase. Consistent with this prediction, ORF66 is 55% identical or 77% similar to an ORF in the S. acidocaldarius genome (Chen et al., unpublished data), which is presumably responsible for the GGCC-specific N4-cytosine methylating activity identified previously (24). The presence of viral encoded restriction-modification systems is consistent with the observed modification at specific sites in the STSV1 genome.
Domain analysis has provided clues into the functions of additional ORFs. The putative product of ORF56 shows significant similarity to tyrosine integrases (38). However, the STSV1 protein contains only 187 amino acid residues and is much smaller than all other known archaeal integrases (the SSV- and pNOB8-type integrases are approximately 350 and 440 amino acids in size, respectively) (52, 53). ORF50 encodes a protein containing a ParB-like domain that is most similar to ParB of Brucella melitensis (15). Biochemical studies have shown that the ParB protein of the broad-host-range IncPα plasmid RK2 is a nuclease which preferentially cleaves single-stranded DNA and nicks supercoiled DNA at sites of potential single-strandedness (26). The protein appears to be involved in plasmid RK2 stabilization, possibly by playing a role in the resolution of a Holliday intermediate in the process of plasmid decatenation.
DISCUSSION
Spindle-shaped viruses are widespread among and unique to Archaea. They have been isolated from both crenarchaeotes (the genera Sulfolobus and Acidianus) and euryarchaeotes (the genera Pyrococcus, Haloarcula, and Thermococcus) (8, 20, 21, 42, 43, 47, 48). Electron microscopic data suggest that spindle-shaped viruses or virus-like particles occur much more frequently in hyperthermophiles than in mesophiles or moderate thermophiles (1). Spindle-shaped viruses vary in size and possess either a single tail at one end or two tails, one at each end of the virus particle. Members of the family Fuselloviridae (SSV1, SSV2, SSV3, SSV RH, and SSV KI) are the best-characterized spindle-shaped viruses (34, 58). In the present study, we have isolated and characterized STSV1, a novel spindle-shaped virus of Sulfolobus. STSV1 is similar in shape to but much larger than known fuselloviruses. Interestingly, none of the ORFs of STSV1 show high similarity to those of fuselloviruses. In fact, STSV1 is closely related to none of the known viruses at the genomic level. Based on its morphological and genomic features, we propose that STSV1 represents a novel virus family.
In this study, we examined under the electron microscope the supernatants of 10 enrichment cultures established with samples collected from various spots in the Tengchong area under growth conditions that had been previously used for the isolation of Sulfolobus viruses. Virus particles were observed in eight of the samples. Interestingly, all of these viruses had the morphology of STSV1. This is unexpected since a number of different viral morphotypes were encountered in hot spring samples from Yellowstone National Park and Iceland (47, 62). The absence of an SSV-like virus in Tengchong is especially intriguing since the small spindle-shaped fuselloviruses appear to exist in abundance in acidic hot springs around the globe. Although the sample size in this study does not permit a reliable estimate of viral diversity in Tengchong, our results suggest that STSV1 is a predominant Sulfolobus virus in the region. In their surveys of viruses in hot springs in Yellowstone National Park, Rice et al. (47) and Rachel et al. (43) found virus-like particles that bore resemblance to STSV1 in shape and size. Therefore, it is likely that STSV1-like viruses are also present in acidic hot springs in other locations.
Like all crenarchaeotal viruses known thus far, STSV1 is a nonlytic virus. When host cells were infected in the exponential growth phase with the virus, they started to release mature progeny virus particles in less than 4 h. In an infected culture, the virus titer normally peaked at >1010 particles/ml, a level comparable to that of SSV1 in a UV-induced culture (34). Unlike fuselloviruses, STSV1 apparently did not integrate into the host genome. However, the infected host cells kept the virus after repeated transfers, suggesting that STSV1 existed in a carrier state in S. tengchongensis RT8-4. This is somewhat surprising in view of the presence in the viral genome of a gene encoding putative tyrosine integrase. A possibility exists that the STSV1 integration occurs in strains other than RT8-4 in the Tengchong springs. It is equally possible that the virally encoded putative integrase is inactive since the protein is significantly smaller than its archaeal homologues (52, 53). We are now in the process of testing this possibility.
Analysis of the protein composition of the STSV1 virion identified a major protein (ORF40) and four minor structural proteins (ORFs 14, 26, 34, and 53). Given its abundance, the major protein (ORF40) is probably the coat protein in the spindle part of the virion. No similar dominance in viral structure by a single protein was observed in known fuselloviruses (44, 58). In addition, the ORF40 protein shows no sequence similarity to VP1 or VP2, the coat proteins of fuselloviruses (44). It would be of interest to investigate if these similarly shaped viral morphotypes reflect deep divergence from a single ancestor or represent independently originated viruses. The minor structural proteins are conceivably components of the envelope or tail of the STSV1 virion, presumably serving architectural, connecting roles and/or interacting with host.
As found in other sequenced genomes of crenarchaeotal viruses, a majority of the ORFs in the STSV1 genome do not encode a known function. Only 14 of the 74 predicted viral ORFs can be assigned a function with confidence. The virus encodes two enzymes that are presumably involved in the biosynthesis of dTTP: dUTPase (ORF33) and thymidylate synthase (ORF16). Since Sulfolobus is known to lack the ability to utilize exogenous thymine for DNA biosynthesis (40), the two enzymes may function to increase the pool size of dTTP in host cells to support the rapid multiplication of the virus which has an AT-rich genome (35% in GC content).
DNA modification has been shown to occur in both crenarchaeotal and euryarchaeotal viruses (4, 59). A fraction of the genomes of ΦCh1, a head/tail phage from the haloalkaliphilic archaeon Natrialba magadii, is methylated by virally encoded methylase in a Dam-like fashion (6, 29, 59). Phage ΦN of Halobacterium salinarum was also shown to contain a fully cytosine-methylated genome (57). The Sulfolobus SNDV virus is the only crenarchaeotal virus with a modified genome that has been described (4). Like the genomes of ΦCh1, the SNDV DNA is modified by Dam-like methylation. The methylase is most likely viral encoded and recognizes only hemimethylated GATC sites of the SNDV genome (4).
In the present study, we show that the genome of STSV1 is modified, presumably by virally encoded proteins. Three genes encoding putative MTases have been identified in the viral genome. However, the modification pattern of STSV1 DNA was different from those observed in other archaeal viruses. The STSV1 DNA was clearly not modified in a Dam-like fashion. Modification of cytosine residues was detected, and additional forms of modification are likely. The STSV1 modification system appeared to be capable of distinguishing between viral and host DNAs, as only viral DNA was modified. The viral DNA-specific modification may be achieved through recognition by the viral enzymes of hemimethylated sites produced during the replication of fully methylated viral DNA. The roles of DNA modification in the STSV1-host interaction are unclear, but they may include protection of the viral genome from enzymatic attack by the host and selective regulation of viral replication and gene expression.
Interestingly, the STSV1 genome is highly asymmetric and divides into equal halves with respect to gene orientation. A similar bias in gene orientation has been observed in many bacterial and eukaryotic genomes (27). In comparison, the genomes of fuselloviruses are also highly organized but separated into two unequal parts (at a size ratio of ≈1:3) of opposite gene orientation. The biased gene orientation as well as the strand compositional asymmetry in the STSV1 genome, as revealed by the cumulative GC skew, suggest that the origin and terminus of viral replication are located between ORF74 and ORF1 and between ORF34 and ORF35, respectively. Identification of the intergenic region between ORF74 and ORF1 as a candidate replication origin is reinforced by an unusually high AT content and the presence of two sets of tandem repeats as well as two sets of inverted repeats in the region. Based on the above results, we propose that replication of the STSV1 genome commences from the proposed origin and proceeds bidirectionally in the θ mode.
Supplementary Material
[Supplemental material]
Acknowledgments
We are very grateful to Kenneth M. Stedman for the generous gift of Sulfolobus solfataricus strains P1 and P2. We thank Qinghe Bian for assistance in electron microscopy and Kim Bügger for maintaining a local Sulfolobus virus database.
This work was supported by grants 39925001 and 30030010 from the National Natural Science Foundation of China (NSFC), 2004CB719603 from National Basic Research Program of China to L.H., and grants 30328002 from NSFC and 26-03-0042 from the Danish Technical Research Council and Archaea Centre from the Danish Natural Sciences Research Council to Q.S.
Footnotes
REFERENCES
- 1.Ackermann, H. W. 2001. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 146**:**843-857. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, H. P., Q. She, H. Phan, K. Stedman, D. Prangishvili, I. Holz, J. K. Kristjansson, R. Garrett, and W. Zillig. 1999. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol. Microbiol. 44**:**217-226. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, H. P., U. Ziese, and W. Zillig. 2000. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology 272**:**409-416. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, H. P., W. Zillig, U. Ziese, I. Holz, M. Crosby, T. Utterback, J. F. Weidmann, J. K. Kristjanson, H. P. Klenk, K. E. Nelson, and C. M. Fraser. 2000. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267**:**252-266. [DOI] [PubMed] [Google Scholar]
- 6.Baranyi, U., R. Klein, W. Lubitz, D. H. Kruger, and A. Witte. 2000. The archaeal halophilic virus-encoded Dam-like methyltransferase M. phiCh1-I methylates adenine residues and complements dam mutants in the low salt environment of Escherichia coli. Mol. Microbiol. 35**:**1168-1179. [DOI] [PubMed] [Google Scholar]
- 7.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30**:**276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bath, C., and M. L. Dyall-Smith. 1998. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J. Virol. 72**:**9392-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell, S. D., and S. P. Jackson. 2001. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4**:**208-213. [DOI] [PubMed] [Google Scholar]
- 10.Beranova-Giorgianni, S., and D. M. Desiderio. 2000. Mass spectrometry of the human pituitary proteome: identification of selected proteins. Rapid Commun. Mass. Spectrom. 14**:**161-167. [DOI] [PubMed] [Google Scholar]
- 11.Bettstetter, M., X. Peng, R. A. Garrett, and D. Prangishvili. 2003. AFV1, a novel virus infecting hyperthermophilic archaea of the genus Acidianus. Virology 315**:**68-79. [DOI] [PubMed] [Google Scholar]
- 12.Blum, H., W. Zillig, S. Mallok, H. Domdey, and D. Prangishvili. 2001. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology 281**:**6-9. [DOI] [PubMed] [Google Scholar]
- 13.Borodovsky, M., and J. McIninch. 1993. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 17**:**123-133. [Google Scholar]
- 14.Butkus, V., L. Petrauskiene, Z. Maneliene, S. Klimasauskas, V. Laucys, and A. Janulaitis. 1987. Cleavage of methylated CCCGGG sequences containing either N4-methylcytosine or 5-methylcytosine with MspI, HpaII, SmaI, XmaI and Cfr9I restriction endonucleases. Nucleic Acids Res. 15**:**7091-7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99**:**443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dynes, J. L., and R. A. Firtel. 1989. Molecular complementation of a genetic marker in Dictyostelium using a genomic DNA library. Proc. Natl. Acad. Sci. USA 86**:**7966-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forterre, P. 2001. New viruses for the new millennium. Trends Microbiol. 9**:**114. [DOI] [PubMed] [Google Scholar]
- 18.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12**:**532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geller, B. L., R. G. Ivey, J. E. Trempy, and B. Hettinger-Smith. 1993. Cloning of a chromosomal gene required for phage infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 175**:**5510-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geslin, C., M. Le Romancer, G. Erauso, M. Gaillard, G. Perrot, and D. Prieur. 2003. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, “_Pyrococcus abyssi_”.J. Bacteriol. 185**:**3888-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geslin, C., M. Le Romancer, M. Gaillard, G. Erauso, and D. Prieur. 2003. Observation of virus-like particles in high temperature enrichment cultures from deep-sea hydrothermal vents. Res. Microbiol. 154**:**303-307. [DOI] [PubMed] [Google Scholar]
- 22.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8**:**195-202. [DOI] [PubMed] [Google Scholar]
- 23.Grigoriev, A. 1988. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26**:**2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grogan, D. W. 2003. Cytosine methylation by the SuaI restriction-modification system: implications for genetic fidelity in a hyperthermophilic archaeon. J. Bacteriol. 185**:**4657-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101**:**9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, E. P., T. Mincer, H. Schwab, A. B. Burgin, and D. R. Helinski. 1999. Plasmid RK2 ParB protein: purification and nuclease properties. J. Bacteriol. 181**:**6010-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlin, S. 1999. Bacterial DNA strand compositional asymmetry. Trends Microbiol. 7**:**305-308. [DOI] [PubMed] [Google Scholar]
- 28.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 8**:**123-140. [DOI] [PubMed] [Google Scholar]
- 29.Klein, R., U. Baranyi, N. Rossler, B. Greineder, H. Scholz, and A. Witte. 2002. Natrialba magadii virus phiCh1: first complete nucleotide sequence and functional organization of a virus infecting a haloalkaliphilic archaeon. Mol. Microbiol. 45**:**851-863. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn, P., S. A. Lesley, I. I. Mathews, J. M. Canaves, L. S. Brinen, X. Dai, A. M. Deacon, M. A. Elsliger, S. Eshaghi, R. Floyd, A. Godzik, C. Grittini, S. K. Grzechnik, C. Guda, K. O. Hodgson, L. Jaroszewski, C. Karlak, H. E. Klock, E. Koesema, J. M. Kovarik, A. T. Kreusch, D. McMullan, T. M. McPhillips, M. A. Miller, M. Miller, A. Morse, K. Moy, J. Ouyang, A. Robb, K. Rodrigues, T. L. Selby, G. Spraggon, R. C. Stevens, S. S. Taylor, H. van den Bedem, J. Velasquez, J. Vincent, X. Wang, B. West, G. Wolf, J. Wooley, and I. A. Wilson. 2002. Crystal structure of thy1, a thymidylate synthase complementing protein from Thermotoga maritima at 2.25 A resolution. Proteins 49**:**142-145. [DOI] [PubMed] [Google Scholar]
- 31.Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz, R. Mott, F. Ciccarelli, R. R. Copley, C. P. Ponting, and P. Bork. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30**:**242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253**:**618-632. [DOI] [PubMed] [Google Scholar]
- 33.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. “CDD: a curated Entrez database of conserved domain alignments”.Nucleic Acids Res. 31**:**383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, A., S. Yeats, D. Janekovic, W.-D. Reiter, W. Aicher, and W. Zillig. 1984. SAV1, a temperate U.V. inducible DNA virus like particle from the archaebacterium Sulfolobus acidocaldarius B12. EMBO J. 3**:**2165-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McClelland, M., M. Nelson, and E. Raschke. 1994. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 22**:**3640-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan, R., J. Xiao, and S. Xu. 1998. Characterization of an extremely thermostable restriction enzyme, PspGI, from a Pyrococcus strain and cloning of the PspGI restriction-modification system in Escherichia coli. Appl. Environ. Microbiol. 64**:**3669-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myllykallio, H., G. Lipowski, D. Leduc, J. Filee, P. Forterre, and U. Liebl. 2002. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297**:**105-107. [DOI] [PubMed] [Google Scholar]
- 38.Nunes-Duby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26**:**391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng, X., H. Blum, Q. She, S. Mallok, K. Brugger, R. A. Garrett, W. Zillig, and D. Prangishvili. 2001. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291**:**226-234. [DOI] [PubMed] [Google Scholar]
- 40.Prangishvili, D., H. P. Klenk, G. Jakobs, A. Schmiechen, C. Hanselmann, I. Holz, and W. Zillig. 1998. Biochemical and phylogenetic characterization of the dUTPase from the archaeal virus SIRV. J. Biol. Chem. 273**:**6024-6029. [DOI] [PubMed] [Google Scholar]
- 41.Prangishvili, D., K. Stedman, and W. Zillig. 2001. Viruses of the extremely thermophilic archaeon Sulfolobus. Trends Microbiol. 9: 39-43. [DOI] [PubMed]
- 42.Prangishvili, D. 2003. Evolutionary insights from studies on viruses of hyperthermophilic archaea. Res. Microbiol. 154**:**289-294. [DOI] [PubMed] [Google Scholar]
- 43.Rachel, R., M. Bettstetter, B. P. Hedlund, M. Haring, A. Kessler, K. O. Stetter, and D. Prangishvili. 2002. Remarkable morphological diversity of viruses and virus-like particles in hot terrestrial environments. Arch. Virol. 147**:**2419-2429. [DOI] [PubMed] [Google Scholar]
- 44.Reiter, W. D., P. Palm, A. Henschen, F. Lottspeich, W. Zillig, and B. Grampp. 1987. Identification and characterization of the genes encoding three structural proteins of the Sulfolobus virus-like particle SSV1. Mol. Gen. Genet. 206**:**144-153. [Google Scholar]
- 45.Reiter, W. D., P. Palm, and W. Zillig. 1988. Analysis of transcription in the archaebacterium Sulfolobus indicates that archaebacterial promoters are homologous to eukaryotic pol II promoters. Nucleic Acids Res. 16**:**1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiter, W. D., P. Palm, and S. Yeats. 1989. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 17**:**1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice, G., K. Stedman, J. Snyder, B. Wiedenheft, D. Willits, S. Brumfield, T. McDermott, and M. J. Young. 2001. Viruses from extreme thermal environments. Proc. Natl. Acad. Sci. USA 98**:**13341-13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice, G., L. Tang, K. Stedman, F. Roberto, J. Spuhler, E. Gillitzer, J. E. Johnson, T. Douglas, and M. Young. 2004. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc. Natl. Acad. Sci. USA 101**:**7716-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.She, Q., H. Phan, R. A. Garrett, S.-V. Albers, K. M. Stedman, and W. Zillig. 1998. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles 2**:**417-425. [DOI] [PubMed] [Google Scholar]
- 51.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeote Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98**:**7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.She, Q., K. Brugger, and L. Chen. 2002. Archaeal integrative elements and their impact on evolution. Res. Microbiol. 153**:**325-332. [DOI] [PubMed] [Google Scholar]
- 53.She, Q., B. Shen, and L. Chen. 2004. Archaeal integrases and mechanisms of gene capture. Biochem. Soc. Trans. 32**:**222-226. [DOI] [PubMed] [Google Scholar]
- 54.Snyder, J. C., K. Stedman, G. Rice, B. Wiedenheft, J. Spuhler, and M. J. Young. 2003. Viruses of hyperthermophilic Archaea. Res. Microbiol. 154**:**474-482. [DOI] [PubMed] [Google Scholar]
- 55.Stedman, K. M., Q. She, H. Phan, H. P. Arnold, I. Holz, R. A. Garrett, and W. Zillig. 2003. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res. Microbiol. 154**:**295-302. [DOI] [PubMed] [Google Scholar]
- 56.Tolstrup, N., C. W. Sensen, R. A. Garrett, and I. G. Clausen. 2000. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles 4**:**175-179. [DOI] [PubMed] [Google Scholar]
- 57.Vogelsang-Wenke, H., and D. Oesterheld. 1988. Isolation of a halobacterial phage with a fully cytosine-methylated genome. Mol. Gen. Genet. 211**:**407-414. [Google Scholar]
- 58.Wiedenheft, B., K. Stedman, F. Roberto, D. Willits, A. K. Gleske, L. Zoeller, J. Snyder, T. Douglas, and M. Young. 2004. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J. Virol. 78**:**1954-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Witte, A., U. Baranyi, R. Klein, M. Sulzner, C. Luo, G. Wanner, D. H. Kruger, and W. Lubitz. 1997. Characterization of Natronobacterium magadii phage phi Ch1, a unique archaeal phage containing DNA and RNA. Mol. Microbiol. 23**:**603-616. [DOI] [PubMed] [Google Scholar]
- 60.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64**:**69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiang, X., X. Dong, and L. Huang. 2003. Sulfolobus tengchongensis sp. nov., a novel thermoacidophilic archaeon isolated from a hot spring in Tengchong, China. Extremophiles 7**:**493-498. [DOI] [PubMed] [Google Scholar]
- 62.Zillig, W., A. Kletzin, C. Schleper, I. Holz, D. Janekovic, J. Hain, M. Lanzendorfer, and J. K. Kristjansson. 1994. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst. Appl. Microbiol. 16**:**609-628. [Google Scholar]
- 63.Zillig, W., H. P. Arnold, I. Holz, D. Prangishvili, A. Schweier, K. Stedman, Q. She, H. Phan, R. Garrett, and J. K. Kristjansson. 1998. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles 2**:**131-140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]