Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways (original) (raw)
Abstract
IFN-α/β plays an essential role in innate immunity against viral and bacterial infection. Among the proteins induced by IFN-α/β are the ubiquitin-like ISG15 protein and its E1- (Ube1L) and E2- (UbcH8) conjugating enzymes, leading to the conjugation of ISG15 to cellular proteins. It is likely that ISG15 conjugation plays an important role in antiviral response because a human virus, influenza B virus, inhibits ISG15 conjugation. However, the biological function of ISG15 modification remains unknown, largely because only a few human ISG15 target proteins have been identified. Here we purify ISG15-modified proteins from IFN-β-treated human (HeLa) cells by using double-affinity selection and use mass spectroscopy to identify a large number (158) of ISG15 target proteins. Eight of these proteins were subjected to further analysis and verified to be ISG15 modified in IFN-β-treated cells, increasing the likelihood that most, if not all, targets identified by mass spectroscopy are bona fide ISG15 targets. Several of the targets are IFN-α/β-induced antiviral proteins, including PKR, MxA, HuP56, and RIG-I, providing a rationale for the inhibition of ISG15 conjugation by influenza B virus. Most targets are constitutively expressed proteins that function in diverse cellular pathways, including RNA splicing, chromatin remodeling/polymerase II transcription, cytoskeleton organization and regulation, stress responses, and translation. These results indicate that ISG15 conjugation impacts nuclear as well as cytoplasmic functions. By targeting a wide array of constitutively expressed proteins, ISG15 conjugation greatly extends the repertoire of cellular functions that are affected by IFN-α/β.
Keywords: antiviral, innate immunity, mass spectroscopy
IFN-α/β plays an essential role in innate immunity against viral and bacterial infection (1). Among the proteins induced by IFN-α/β is the ISG15 protein, a ubiquitin-like protein that becomes conjugated to many cellular proteins (2, 3). ISG15 modification does not appear to target proteins for proteasomal degradation (4, 5), and the function of ISG15 modification remains unknown. Functional studies have been hampered by the fact that only a few human ISG15 target proteins have been identified.
Our interest in ISG15 originated in the course of experiments to elucidate the function of the NS1B protein of influenza B virus. We found that the NS1B protein binds ISG15 and inhibits its conjugation (6), indicating that ISG15 conjugation is likely to be an important part of the IFN-α/β-induced antiviral response. However, it was not evident how ISG15 conjugation might serve such a role. To address this issue and to elucidate the function of ISG15 conjugation, we first identified the E1 and E2 enzymes in the ISG15 conjugation pathway as Ube1L and UbcH8, respectively, both of which are induced by IFN-α/β (6, 7). These findings enabled us to develop a system for a proteomics-based identification of ISG15 target proteins, which is described in the present study.
We used this system to identify a large number (158) of ISG15 modified proteins in IFN-β-treated human (HeLa) cells. The identity of these ISG15 target proteins provides insights into the function of ISG15 modification. Several of the targets are IFN-α/β-induced antiviral proteins, providing a rationale for the inhibition of ISG15 conjugation by influenza B virus. Most targets are constitutively expressed human proteins that function in diverse cellular pathways, including RNA splicing, chromatin remodeling/polymerase II transcription, cytoskeleton organization and regulation, stress responses, and translation. These results indicate that ISG15 conjugation impacts nuclear as well as cytoplasmic functions and may have a role in regulating transcription and pre-mRNA splicing during the IFN-α/β response. Thus, by targeting this wide array of constitutively expressed proteins, ISG15 conjugation greatly extends the repertoire of cellular functions that are affected by IFN-α/β.
Materials and Methods
Plasmids. Plasmids containing the following PCR-generated reading frames were inserted into pcDNA3 vectors: Ube1L, UbcH8, His6-HA-ISG15, and His6–3xFLAG-ISG15. All of the cDNAs used for verifying ISG15 target proteins, except maspin, were generated by PCR by using a Human Leukocyte Matchmaker cDNA library (Clontech). The template for amplifying maspin was pEF-Maspin, provided by Zhang Min (Baylor School of Medicine, Houston). For the expression of V5-tagged target proteins, two modified pcDNA3 vectors containing the V5 epitope were constructed. The original BamHI site of pcDNA3 was eliminated and replaced by the V5 sequence followed by either a BamHI site (pcDNA3-V5-Bam) or a NotI site (pcDNA3-V5-Not). The PCR-generated reading frames for maspin, PTB-1, and thioredoxin reductase-1 (TrxR1) were cloned into pcDNA3-V5-Bam as BglII-BglII, BglII-R1, and BamH-R1 fragments, respectively. The PCR-generated reading frames for Hsp60 and moesin were cloned into pcDNA3-V5-Not as Not-XbaI and Not-EcoRI fragments, respectively. For the expression of 3xFLAG-RIG-I, its PCR-generated reading frame was inserted into the pCMV10 vector (Sigma).
Purification of ISG15 Conjugates. HeLa cells in each of five 150-mm culture dishes (total of 108 cells) were transfected by using Fugene 6 (Roche) with three plasmids: 12 μg of pcDNA3-His6-Flag-ISG15, 5 μg of pcDNA3-UbE1L, and 3 μg of pcDNA3-UbcH8. Transfection efficiency, as determined by using a GFP reporter plasmid, was ≈80%. Twenty-four hours posttransfection, 1,000 units/ml of human IFN-β (Berlex Biosciences, Richmond, CA) was added, and the cells were incubated for an additional 24 h. The untransfected control cells were incubated with 1,000 units/ml of human IFN-β for 24 h. Both sets of cells were lysed in a total of 5 ml of a buffer containing 8 M Urea, 50 mM Tris, pH 8.0, and 1.0% Triton X-100, followed by sonication for 40 sec with a Branson 450 Sonifier. Cell debris was removed by centrifugation at 18,000 × g for 30 min at 4°C. 2-Mercaptoethanol (β-ME) was then added to the resulting supernatant to a final concentration of 50 mM, and the mixture was incubated for 30 min at 4°C. Lysis buffer lacking Triton X-100 was then added to dilute the β-ME to 10 mM, and imidazole was added to 10 mM. The extracts were then subjected to the affinity selections described in Fig. 1_B_ and the text and analyzed on a 10% SDS-polyacrylamide gel.
Fig. 1.
Purification of ISG15 target proteins. (A) Establishing the conditions for optimal conjugation of plasmid-expressed tagged ISG15. HeLa cells were cotransfected with plasmids expressing His6-HA-ISG15, E1ISG15, and UbcH8, either without (lane 1) or with (lane 2) IFN-β treatment 24 h posttransfection. After another 24 h, cells were collected, and proteins were analyzed by immunoblotting with anti-HA antibody (Covance). (B) Scheme for purification of His6–3xFLAG-ISG15 conjugates from 108 HeLa cells. (C) Coomassie blue-stained gel of the proteins purified from IFN-β-treated cells transfected with plasmids expressing His6–3xFLAG-ISG15, E1ISG15, and UbcH8 (lane 2) and from untransfected, IFN-β-treated cells (lane 1).
Target Verification Experiments. Several strategies for verifying ISG15 targets were used. In one strategy for HuP56 and MxA, immunoblots with anti-HuP56 antibody (provided by Ganes Sen, Cleveland Clinic, Cleveland) or anti-MxA antibody (provided by Otto Haller, University of Freiburg, Freiburg, Germany) were carried out on either whole-cell extract from IFN-β-treated HeLa cells or affinity-purified ISG15 conjugates. Because the tagged ISG15 used in these experiments was His6-HA-ISG15, the purification procedure involved sequential affinity selection on anti-hemagglutinin (HA) antibody matrix (Covance, Richmond, CA) and Ni-NTA affinity matrix (Qiagen, Valencia, CA). In a second strategy for HuP56 and MxA, the whole-cell extract was immunoprecipitated with anti-HuP56, anti-MxA, or a control antibody, and the immunoprecipitate was immunoblotted with anti-ISG15 antibody or either anti-HuP56 or anti-MxA antibody. For the other ISG15 targets, cotransfection experiments were carried out by using ISG15 tagged with one epitope, and the target protein tagged with a different epitope, with or without cotransfection with plasmids expressing E1ISG15 and UbcH8, as described in the text and the legends to Figs. 2_C_ and 3.
Fig. 2.
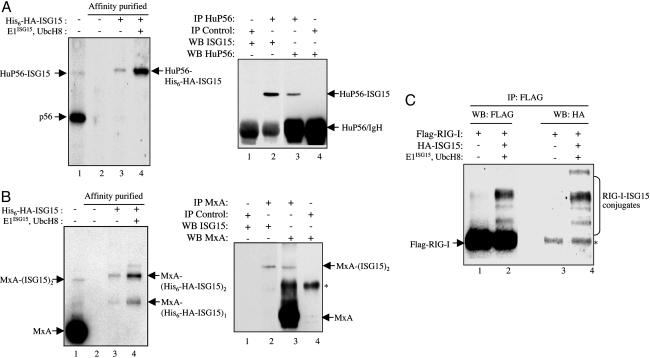
Validation of HuP56, MxA, and RIG-I as ISG15 targets. (A) HuP56 validations. (Left) Whole-cell extract from untransfected IFN-β-treated HeLa cells was immunoblotted with anti-HuP56 antibody (lane 1). Cell extracts from the other HeLa cell samples shown (lanes 2–4) were subjected to sequential affinity selections on anti-HA antibody matrix and Ni-NTA resin. These cells were either untransfected (lane 2) or transfected with a plasmid expressing His6-HA-ISG15 (lane 3) or with this plasmid plus the plasmids expressing E1ISG15 and UbcH8 (lane 4). Twenty-four hours posttransfection, the cells were treated with IFN-β and incubated another 24 h. The affinity-purified proteins were immunoblotted with anti-HuP56 antibody. The band corresponding in size to ISG15-modified HuP56 is indicated, based on molecular weight standards not shown. (Right) Whole-cell extract from untransfected IFN-β-treated HeLa cells were immunoprecipitated with either anti-HuP56 antibody (lanes 2 and 3) or a control antibody (lanes 1 and 4). The immunoprecipitates were immunoblotted with either anti-ISG15 antibody (lanes 1 and 2) or anti-HuP56 antibody (lanes 3 and 4). The bands corresponding to HuP56-ISG15 and HuP56 are denoted. (B) MxA validations. The experiments for the Left and Right were carried out as described in A, except that anti-MxA antibody was used. As indicated, the predominant MxA conjugate contains two molecules of ISG15. *, Nonspecific band. (C) RIG-I verification. HeLa cells were transfected with a plasmid expressing 3xFLAG-RIG-I. One set of cells was also cotransfected with plasmids expressing HA-ISG15, E1ISG15, and UbcH8 (lanes 2 and 4). Control cells were not cotransfected with these plasmids (lanes 1 and 3). Twenty-four hours posttransfection, the cells were treated with IFN-β and incubated another 24 h. Cell extracts were immunoprecipitated with anti-FLAG antibody and were immunoblotted with either anti-FLAG (lanes 1 and 2) or anti-HA (lanes 3 and 4) antibody. The predominant ISG15 conjugate corresponds in size to RIG-I-(ISG15)3, based on molecular weight standards not shown.
Fig. 3.
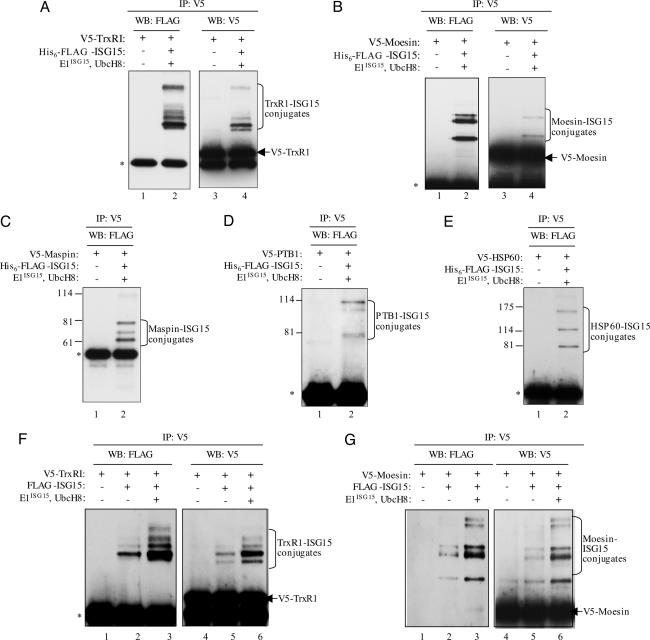
Verification of five constitutively expressed proteins as ISG15 targets. (A–E) HeLa cells were transfected either with only the plasmid expressing the indicated V5-tagged protein (lanes 1 and 3 in A and B; lane 1 in C–E) or with this plasmid plus the plasmids expressing His6–3xFLAG-ISG15, E1ISG15, and UbcH8 (lanes 2 and 4 in A and B; lane 2 in C–E). Twenty-four hours posttransfection, the cells were treated with IFN-β and incubated another 24 h. Cell extracts were immunoprecipitated with anti-V5 antibody (Serotec) and immunoblotted with anti-FLAG antibody (Sigma). For TrxR1 and moesin (A and B), the immunoprecipitates were also immunoblotted with anti-V5 antibody (lanes 3 and 4). Based on molecular weight standards (not shown for TxR1 and moesin), the ISG15 conjugates correspond in size to species containing one to five His-6–3xFLAG-ISG15 molecules. Some of the bands may correspond to target proteins modified by both His6–3xFLAG-ISG15 and untagged endogenous ISG15. (F and G) HeLa cells were transfected with only a plasmid expressing V5-TrxR1 or V5-moesin (lanes 1 and 4), with this plasmid plus a plasmid expressing 3xFLAG-ISG15 (lanes 2 and 5), or with these three plasmids plus the plasmids expressing E1ISG15 and UbcH8 (lanes 3 and 6). The cells were then treated with IFN-β. Cell extracts were immunoprecipitated with anti-V5 antibody and immunoblotted with either anti-FLAG or anti-V5 antibody, as indicated.
Mass Spectroscopy. The 11 slices from both lanes of the gel shown in Fig. 1_C_ were subjected to in-gel trypsin digestion (8). The resulting peptide fractions were loaded onto 100-μm internal diameter fused silica columns packed in-house with C18-resin (Michrom Bioresources, Auburn, CA) and separated over a 25-min gradient of increasing acetonitrile concentration (2.5–97.4%). Upon elution from the column, peptides were introduced into a LTQ mass spectrometer (Thermo Electron, San Jose, CA). The instrument was set to cycle between acquiring a full MS scan (m/z range 375–1,275) and then 10 subsequent tandem MS spectra of the 10 most abundant precursor ions observed in the preceding MS scan. Data obtained in this manner were analyzed by using the human National Center for Biotechnology Information database in conjunction with the sequest search algorithm (9). These searches were performed allowing for the following possible modifications: 16.0 Da on methionines (oxidation), 114.1 Da on lysines (ISG15 remnant created upon trypsinolysis at ISG15-modified sites), and 174.1 Da on cysteines (acrylamide adduct). Accepted peptide matches required both a dCN of 0.075 and the following XCorr cutoff values for 1+,2+, and 3+ peptides, respectively: 1.8, 2.1, and 2.8. If a given protein was found by two or more peptides to be unique to the preparation from the transfected cells, it was concluded to be a putative target of ISG15. The categorization of identified proteins shown in Fig. 4 and Tables 1 and 2, which are published as supporting information on the PNAS web site, was based on information from bioknowledge retriever (proteome.incyte.com).
Fig. 4.
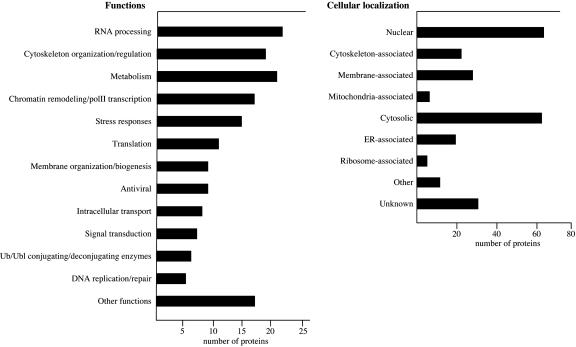
Categorization of ISG15 targets by function (Left) and intracellular localization (Right).
Results
Our system for the identification of ISG15 target proteins was based on the transfection of plasmids expressing a double-affinity-tagged ISG15, Ube1L/E1ISG15, and UbcH8 into human (HeLa) cells. In initial experiments, the tagged ISG15 was His6-HA-ISG15. As shown in Fig. 1_A_, His6-HA-tagged ISG15 conjugates were produced in the transfected cells (lane 1), and the level of conjugates was increased ≈5-fold after IFN-β treatment (lane 2). These results suggested that at least some of the ISG15 ligases and target proteins are constitutively expressed, but that (i) at least some factors required for ISG15 conjugation, perhaps ISG15 E3 ligases, are IFN-β-induced proteins; and/or (ii) some ISG15 target proteins are themselves IFN-β-induced proteins.
We carried out pilot purifications of ISG15 conjugates by using either His6-HA-ISG15 or His6–3xFLAG-ISG15 and found that a higher yield of purified conjugates was obtained by using the latter substrate. The scheme for the purification of His6–3xFLAG-ISG15 conjugates from 108 HeLa cells is outlined in Fig. 1_B_. The conjugates were purified by sequential affinity selections on Ni-NTA resin and anti-FLAG antibody matrix. Because efficient elution from the anti-FLAG antibody matrix required boiling in 1% SDS, which might also elute proteins that were nonspecifically bound to the matrix, we carried out a second Ni-NTA resin selection after the anti-FLAG column. The strongly denaturing conditions used in the two rounds of Ni-NTA selection would be expected to remove any proteins that were not themselves ISG15 modified but might have been in complexes with ISG15 protein targets. As a control, extracts from untransfected IFN-β-treated cells were subjected to the same purification procedure. Fig. 1_C_ shows the Coomassie blue-stained SDS gel of the proteins purified from IFN-β-treated cells transfected with plasmids expressing His6–3xFLAG-ISG15, E1ISG15, and UbcH8 (lane 2) and from untransfected IFN-β-treated cells (lane 1). Both lanes contain heavy and light IgG chains, originating from SDS elution of the anti-FLAG affinity matrix. Lane 2 contains a large number of Coomassie-stained bands in the molecular weight region expected for ISG15 conjugates, whereas few or no bands were seen in this region of the gel in the control lane (lane 1).
This region of both gel lanes was cut into 11 slices, each of which was subjected to in-gel trypsin digestion (8). The resulting peptides were extracted from the gel and loaded onto a reverse-phase column. By using a gradient of increasing acetonitrile concentration, bound peptides were separated and eluted directly into a LTQ mass spectrometer. Once in the instrument, both MS (precursor mass) and tandem MS (sequencing) information was acquired for the eluting peptides. Data obtained in this manner were then used by the sequest search algorithm in conjunction with the human protein database to characterize the various peptides present (9). Identified peptides were subjected to stringent search standards, including high XCorr values. If a given protein was identified by two or more different peptides and was found only in the His6–3xFLAG-ISG15 transfected cell sample (Fig. 1_C_, lane 2) and not in the control sample (lane 1), the protein was concluded to be a putative substrate of ISG15 and is tabulated in Table 1. This list of putative ISG15 targets contains 158 proteins. Proteins identified in both samples, or only in the control sample, were considered to be false positives and are tabulated in Table 2. Only 16 proteins are listed in this category, only seven of which are found solely in the control sample. Most of the proteins that were found in both samples were detected by a much larger number of peptides in the transfected sample compared with the control sample, indicating they are also likely to be ISG15 targets (e.g., the DHX15 splicing factor, p54nrb, and DNA topoisomerase1). However, we have not included these proteins in the list of putative ISG15 targets. Because there was very little overlap between the transfected and control samples, we can conclude that the purification procedure has removed most contaminating proteins lacking His6–3xFLAG-ISG15 modification.
Twelve of the putative ISG15 target proteins are IFN-α/β-induced proteins, accounting at least in part for the increase in ISG15 conjugates resulting from IFN-β treatment. Nine of these IFN-α/β-induced proteins can be classified as antiviral proteins: PKR, MxA, GBP-1, HuP56, HuP54, HuP60, HuP58, RIG-I, and STAT1 (10). STAT1, the transcription factor required for the synthesis of IFN-α/β-induced mRNAs, was previously identified as an ISG15 target (4). We chose three of the IFN-α/β-induced antiviral proteins (HuP56, MxA, and RIG-I) and verified that they are ISG15-modified in IFN-β-treated cells. HuP56 binds to the eIF3 translation initiation factor and inhibits translation and virus replication (10), MxA is a large GTPase that inhibits the replication of a wide spectrum of viruses (10), and RIG-I is an RNA helicase that responds to RNA virus infection by triggering the activation of the transcription factors (IRF-3 and NF-κB) that are required for the production of IFN-β (11, 12).
HuP56 and MxA are abundant proteins, because they are highly induced in response to IFN-α/β (10). For this reason, we were able to establish that endogenous HuP56 and MxA are conjugated to ISG15 by the endogenous conjugation system. Suggestive evidence for conjugation was provided by direct immunoblotting of whole-cell extract from IFN-β-treated HeLa cells with anti-HuP56 or -MxA antibody. These immunoblots detected a minor HuP56-containing species that migrated at a position corresponding to a mono-ISG15 conjugate, and two minor MxA-containing species that migrated at positions corresponding to mono- and di-ISG15 conjugates, with the latter predominating (Fig. 2 A and B Left, lane 1). We took two approaches to definitively show that these bands corresponded to ISG15 conjugates. First, cells were transfected with a plasmid expressing His6-HA-ISG15 (in the absence of plasmids expressing E1ISG15 and UbcH8), and conjugates were purified by sequential affinity selection on anti-HA antibody and Ni-NTA affinity matrices. His6-HA-ISG15-containing proteins were then immunoblotted with either anti-HuP56 (Fig. 2_A_ Left, lane 3) or anti-MxA antibody (Fig. 2_B_ Left, lane 3). These immunoblots detected a HuP56 protein species migrating at the expected position for HuP56-His6-HA-ISG15 and two MxA species migrating at the positions expected for MxA- (His6-HA-ISG15)1 and MxA-(His6-HA-ISG15)2 (with the latter predominating), demonstrating that endogenous HuP56 and MxA are ISG15 modified. It should be noted that the His6-HA-ISG15 conjugates migrated slightly slower than the conjugates containing endogenous untagged ISG15 (Fig. 2 A and B Left, compare lanes 1 and 3). In the presence of plasmid-expressed E1ISG15 and UbcH8, increased amounts of the His6-HA-ISG15 conjugates were produced, but the pattern of ISG15 conjugation was not changed by the enhanced expression of these two enzymes (Fig. 2 A and B Left, compare lanes 3 and 4).
In the second approach, untransfected IFN-β-treated cells were used to identify endogenous HuP56 and MxA conjugates containing only untagged endogenous ISG15. Whole-cell extract from IFN-β-treated HeLa cells was immunoprecipitated with either anti-HuP56 or -MxA antibody, followed by immunoblotting with anti-ISG15 antibody (Fig. 2 A and B Right). The ISG15 immunoblot detected a protein with the mobility expected for HuP56-ISG15 (Fig. 2_A_ Right, lane 2), and an immunoblot with anti-HuP56 antibody confirmed that this species contains HuP56 (lane 3). This species was not present when the extract was immunoprecipitated with a control antibody (lanes 1 and 4). The MxA immunoblot detected a protein with the mobility expected for MxA-(ISG15)2 (Fig. 2_B_ Right, lane 2), and an immunoblot with anti-MxA antibody confirmed that this species contains MxA (lane 3). We conclude that endogenous HuP56 and MxA in IFN-β-treated HeLa cells are conjugated to endogenous ISG15 by the endogenous conjugation system.
For the validation of RIG-I, HeLa cells were transfected with plasmids expressing FLAG-RIG-I, E1ISG15, UbcH8, and HA-ISG15, followed by IFN-β treatment (Fig. 2_C_). The control transfection left out the plasmid expressing HA-ISG15. Cell extracts were immunoprecipitated with anti-FLAG antibody and immunoblotted with either anti-FLAG antibody (lanes 1 and 2) or anti-HA antibody (lanes 3 and 4). RIG-I modified with one or more ISG15 molecules was detected in the cells expressing HA-ISG15 (lanes 2 and 4). The predominant conjugate migrated at the expected position of RIG-I-(ISG15)3.
Most of the targets we identified by mass spectroscopy are constitutively expressed proteins. We chose five of these proteins and verified that they are ISG15 targets: TrxR1, an enzyme that protects against oxidative stress (13); maspin, a serpin that plays a role in normal development and malignant transformation of mammary glands (14); moesin, part of a three-protein complex that links plasma membrane proteins with the actin cytoskeleton (15); heat-shock protein 60 (Hsp60), a chaperonin that assists protein folding and protects proteins from denaturation after stress (16); and polypyrimidine tract-binding protein 1, an RNA-binding protein that regulates alternative splicing (17). The strategy for verification was similar to that used above for RIG-I, except that the target proteins were tagged with V5, and ISG15 was tagged with FLAG. The target protein was immunoprecipitated by using anti-V5 antibody, followed by immunoblots with anti-FLAG antibody to detect ISG15 conjugates (Fig. 3 A–E). Each of these five proteins was conjugated to one or more ISG15 molecules. The anti-V5 immunoprecipitate was also immunoblotted with anti-V5 antibody to verify that the target protein was ISG15 modified. As shown for TrxR1 and moesin (Fig. 3 A and B), a small fraction of these two proteins was ISG15 modified. Similar results were obtained for the other three proteins (data not shown). ISG15 conjugation of TrxR1 and moesin was also observed in the absence of plasmid-expressed E1ISG15 and UbcH8 (Fig. 3 F and G), demonstrating that the endogenous ISG15 conjugation system modifies these two proteins. In the presence of plasmid-expressed E1ISG15 and UbcH8, increased amounts of the ISG15 conjugates of TrxR1 and moesin were produced, but the pattern of ISG15 conjugation was not changed.
All eight proteins that we tested have been verified to be ISG15 modified in IFN-β-treated cells, increasing the likelihood that most, if not all, the targets identified by mass spectroscopy are bona fide ISG15 targets. These validation experiments indicate that ISG15, like SUMO (18), is typically conjugated to only a small fraction of the total pool of any given target protein (4, 19).
Discussion
Our identification of IFN-α/β-induced antiviral proteins as ISG15 targets provides a rationale for the inhibition of the ISG15 conjugation pathway mediated by the NS1B protein of influenza B virus. It is reasonable to postulate that ISG15 modification of these IFN-α/β-induced antiviral proteins increases their stability, activity, and/or interaction with other proteins, because one might expect two IFN-α/β-induced proteins (the antiviral protein and ISG15) to be working in concert to mediate the biologic effect of IFN-α/β. In this scenario, the small fraction of the target protein that is ISG15 modified would represent an activated species or a species with a specific modified activity. It is more difficult to imagine how modification of a small fraction of these antiviral proteins might inactivate the entire pool of these proteins. Future experiments may determine how ISG15 conjugation affects the functions of these IFN-α/β-induced antiviral proteins, and how such effects may impact the antiviral response.
Three of the IFN-α/β-induced ISG15 targets identified by mass spectroscopy are Ub/Ubl conjugation enzymes. It is not surprising that two of these enzymes are UbE1L and UbcH8, the E1 and E2 enzymes in the ISG15 conjugation pathway. Proteomic analyses of SUMO- and Ub-conjugated proteins also identified several components of the conjugation systems for these proteins (20, 21). The third IFN-α/β-induced Ub/Ubl conjugation enzyme in our proteomic analysis was CEB1/Herc5, a HECT domain E3 ligase (22). CEB1/Herc5 may be a Ub E3 ligase that is ISG15 modified, analogous to the Nedd8 modification of cullins that activates cullin-based Ub ligases (23). Alternatively, CEB1/Herc5 may function as an E3 enzyme in ISG15 conjugation, particularly because at least some HECT domain E3 ligases have the ability to conjugate ISG15 to proteins in vitro (7). Our proteomic analysis also identified a constitutively expressed protein that functions in Ub conjugation, Ubc13, an E2 enzyme that catalyzes the synthesis of lysine-63 polyubiquitin chains in cooperation with Mms2 (24). Two SUMO-specific constitutively expressed enzymes were also identified, Pc2, a SUMO E3 ligase, and SENP1, a SUMO protease (18). These results suggest a possible crosstalk between the ISG15 pathway and the Ub and SUMO pathways at the level of conjugating/deconjugating enzymes. Interestingly, five of the identified ISG15 targets are also known SUMO targets (designated by ** in Table 1).
The ISG15 targets identified by mass spectroscopy were categorized with respect to function and intracellular localization, as depicted in Fig. 4. Some ISG15 targets fall into more than one functional category. A relatively large number of the ISG15 targets are functionally or structurally associated with the cytoskeleton, including a number of major intermediate filament proteins such as vimentin. This likely explains a previous immunofluorescence study that localized ISG15 to intermediate filaments (25). In addition, two ISG15 targets, moesin and ezrin, are part of a tripartite complex that links the plasma membrane to the underlying actin cytoskeleton (15). Because this protein complex is essential for efficient cell-to-cell spread of Listeria and probably other bacteria (26), it is reasonable to hypothesize that ISG15 modification of one or more of the proteins in this complex may play a role in the host defense against these bacterial infections. Proteins that function in stress responses were also identified as targets, including key enzymes involved in oxidative stress, TrxR1, glutathione _S_-transferase (13), and several heat-shock proteins. A significant number of metabolic enzymes were identified as ISG15 targets by mass spectroscopy, although the significance of this finding in unclear.
Perhaps the most surprising finding was that a large number of ISG15 target proteins function in RNA processing or in chromatin remodeling/RNA polymerase II transcription. These proteins account for most of the nuclear targets of ISG15, which represent ≈30% of the total ISG15 targets. The chromatin remodeling/polymerase II transcription factors range from a component of the basal polII transcription machinery (TAFII150) to sequence-specific transcription factors (Smad4 and FUBP3), to chromatin remodeling factors (CHD-1, RBBP4, and Sin3a). The proteins that function in RNA processing include many splicing factors, which range from protein components of the essential U2 (DDX42, SAP49, and SAP62) and U5 snRNPs (U5–200 kD) to an alternative splicing factor (PTB-1), as well as several of the heterogeneous nuclear ribonucleoprotein proteins that are bound to nuclear premRNA before splicing (27). These results show that ISG15 conjugation impacts nuclear functions and may have a role in regulating transcription and premRNA splicing during the IFN-α/β response.
IFN-α/β up-regulates the transcription of ≈100 genes in epithelial cells, and the encoded proteins affect several cellular pathways (28, 29). Among the genes induced by IFN-α/β are ISG15 and several, if not all, of the enzymes required for ISG15 conjugation. As a result of the conjugation of ISG15 to a wide array of cellular proteins, the ISG15 conjugation pathway greatly extends the repertoire of functions that are affected by IFN-α/β.
Supplementary Material
Supporting Tables
Acknowledgments
This work was supported by National Institutes of Health Grants AI11772 (to R.M.K.), CA72943 (to J.M.H.), and HG00041 and GM67945 (to S.G.).
Author contributions: C.Z., C.D., J.M.H., S.G., and R.M.K. designed research; C.Z. and C.D. performed research; C.D. and S.G. contributed new reagents/analytic tools; C.Z., C.D., J.M.H., S.G., and R.M.K. analyzed data; and C.Z., C.D., J.M.H., and R.M.K. wrote the paper.
Abbreviations: HA, hemagglutinin; TrxR1, thioredoxin reductase-1.
References
- 1.Biron, C. A. & Sen, G. C. (2001) in Field Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Wiliams, & Wilkins, Philadelphia), Vol. 1, pp. 321-51. [Google Scholar]
- 2.Haas, A. L., Ahrens, P., Bright, P. M. & Ankel, H. (1987) J. Biol. Chem. 262**,** 11315-11323. [PubMed] [Google Scholar]
- 3.Loeb, K. R. & Haas, A. L. (1992) J. Biol. Chem. 267**,** 7806-7813. [PubMed] [Google Scholar]
- 4.Malakhov, M. P., Kim, K. I., Malakhova, O. A., Jacobs, B. S., Borden, E. C. & Zhang, D. E. (2003) J. Biol. Chem. 278**,** 16608-16613. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz, D. C. & Hochstrasser, M. (2003) Trends Biochem. Sci. 28**,** 321-328. [DOI] [PubMed] [Google Scholar]
- 6.Yuan, W. & Krug, R. M. (2001) EMBO J. 20**,** 362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao, C., Beaudenon, S. L., Kelley, M. L., Waddell, M. B., Yuan, W., Schulman, B. A., Huibregtse, J. M. & Krug, R. M. (2004) Proc. Natl. Acad. Sci. USA 101**,** 7578-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gygi, M. P., Licklider, L. J., Peng, J. & Gygi, S. P. (2003) in Protein Analysis: A Laboratory Manual., ed. Simpson, R. (Cold Spring Harbor Lab. Press, Woodbury, NY), pp. 514-519.
- 9.Eng, J., McCormack, A. L. & Yates, J. R. (1994) J. Am. Soc. Mass Spectrom. 5**,** 976-989. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar, S. N. & Sen, G. C. (2004) Pharmacol. Ther. 103**,** 245-259. [DOI] [PubMed] [Google Scholar]
- 11.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S. & Fujita, T. (2004) Nat. Immunol. 5**,** 730-737. [DOI] [PubMed] [Google Scholar]
- 12.Sumpter, R., Jr., Loo, Y. M., Foy, E., Li, K., Yoneyama, M., Fujita, T., Lemon, S. M. & Gale, M., Jr. (2005) J. Virol. 79**,** 2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mustacich, D. & Powis, G. (2000) Biochem. J. 346 Pt 1, 1-8. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, M. (2004) Front. Biosci. 9**,** 2218-2226. [DOI] [PubMed] [Google Scholar]
- 15.Morales, F. C., Takahashi, Y., Kreimann, E. L. & Georgescu, M. M. (2004) Proc. Natl. Acad. Sci. USA 101**,** 17705-17710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bukau, B. & Horwich, A. L. (1998) Cell 92**,** 351-366. [DOI] [PubMed] [Google Scholar]
- 17.Wagner, E. J. & Garcia-Blanco, M. A. (2001) Mol. Cell. Biol. 21**,** 3281-3288.11313454 [Google Scholar]
- 18.Johnson, E. S. (2004) Annu. Rev. Biochem. 73**,** 355-382. [DOI] [PubMed] [Google Scholar]
- 19.Hamerman, J. A., Hayashi, F., Schroeder, L. A., Gygi, S. P., Haas, A. L., Hampson, L., Coughlin, P., Aebersold, R. & Aderem, A. (2002) J. Immunol. 168**,** 2415-2423. [DOI] [PubMed] [Google Scholar]
- 20.Peng, J., Schwartz, D., Elias, J. E., Thoreen, C. C., Cheng, D., Marsischky, G., Roelofs, J., Finley, D. & Gygi, S. P. (2003) Nat. Biotechnol. 21**,** 921-926. [DOI] [PubMed] [Google Scholar]
- 21.Wohlschlegel, J. A., Johnson, E. S., Reed, S. I. & Yates, J. R., 3rd (2004) J. Biol. Chem. 279**,** 45662-45668. [DOI] [PubMed] [Google Scholar]
- 22.Hochrainer, K., Mayer, H., Baranyi, U., Binder, B., Lipp, J. & Kroismayr, R. (2005) Genomics 85**,** 153-164. [DOI] [PubMed] [Google Scholar]
- 23.Cope, G. A. & Deshaies, R. J. (2003) Cell 114**,** 663-671. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann, R. M. & Pickart, C. M. (1999) Cell 96**,** 645-653. [DOI] [PubMed] [Google Scholar]
- 25.Loeb, K. R. & Haas, A. L. (1994) Mol. Cell. Biol. 14**,** 8408-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pust, S., Morrison, H., Wehland, J., Sechi, A. S. & Herrlich, P. (2005) EMBO J. 24**,** 1287-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurica, M. S. & Moore, M. J. (2003) Mol. Cell 12**,** 5-14. [DOI] [PubMed] [Google Scholar]
- 28.Der, S. D., Zhou, A., Williams, B. R. & Silverman, R. H. (1998) Proc. Natl. Acad. Sci. USA 95**,** 15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiss, G. K., Carter, V. S., He, Y., Kwieciszewski, B. K., Holzman, T., Korth, M. J., Lazaro, C. A., Fausto, N., Bumgarner, R. E. & Katze, M. G. (2003) J. Virol. 77**,** 6367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Tables