Mutational Mechanisms of Williams-Beuren Syndrome Deletions (original) (raw)
Abstract
Williams-Beuren syndrome (WBS) is a segmental aneusomy syndrome that results from a heterozygous deletion of contiguous genes at 7q11.23. Three large region-specific low-copy repeat elements (LCRs), composed of different blocks (A, B, and C), flank the WBS deletion interval and are thought to predispose to misalignment and unequal crossing-over, causing the deletions. In this study, we have determined the exact deletion size and LCR copy number in 74 patients with WBS, as well as precisely defined deletion breakpoints in 30 of them, using LCR-specific nucleotide differences. Most patients (95%) exhibit a 1.55-Mb deletion caused by recombination between centromeric and medial block B copies, which share ∼99.6% sequence identity along 105–143 kb. In these cases, deletion breakpoints were mapped at several sites within the recombinant block B, with a cluster (>27%) occurring at a 12 kb region within the GTF2I/GTF2IP1 gene. Almost one-third (28%) of the transmitting progenitors were found to be heterozygous for an inversion between centromeric and telomeric LCRs. All deletion breakpoints in the patients with the inversion occurred in the distal 38-kb block B region only present in the telomeric and medial copies. Finally, only four patients (5%) displayed a larger deletion (∼1.84 Mb) caused by recombination between centromeric and medial block A copies. We propose models for the specific pairing and precise aberrant recombination leading to each of the different germline rearrangements that occur in this region, including inversions and deletions associated with WBS. Chromosomal instability at 7q11.23 is directly related to the genomic structure of the region.
Introduction
Williams or Williams-Beuren syndrome (WBS [MIM 194050]) is a segmental aneusomy syndrome that results from a heterozygous deletion of contiguous genes at 7q11.23 (Francke 1999). The great majority of patients display a deleted interval that has been estimated to encompass ∼1.5 Mb and to contain 25–30 genes (Peoples et al. 2000; Magano et al. 2001; DeSilva et al. 2002; Merla et al. 2002). However, a few exceptional patients with smaller deletions and either a full or partial phenotype have been reported (Korenberg et al. 2000; Pérez Jurado 2003). The WBS phenotype includes distinctive facial features, vascular stenoses (supravalvular aortic stenosis), and general cognitive deficits with a nonuniform profile (Burn 1986; Morris et al. 1988). Patients exhibit specific dissociations in higher cognitive functions with severe deficits in visuospatial construction but adequate performance in face processing and relatively unaffected language abilities (Bellugi et al. 2000; Mervis et al. 2000). The estimated prevalence of the disease ranges between 1/7,500 and 1/25,000 newborns, most cases being sporadic (Greenberg 1990; Stromme et al. 2002).
Large-scale sequencing of the WBS deleted and flanking regions is almost complete, and high-accuracy sequencing and annotation of the entire orthologous region in mouse has recently been reported (DeSilva et al. 2002). However, despite these advances in sequencing and gene identification, the specific contributions of most of these genes to the phenotype and the underlying pathogenic mechanisms of the disease are not fully understood.
Two mapping reports shed light on the complex genomic structure of the 7q11.23 region rearranged in patients with WBS (Peoples et al. 2000; Valero et al. 2000). Three large region-specific segmental duplications or low-copy repeat elements (centromeric, medial, and telomeric LCRs), each composed of three differentiated blocks called “A,” “B,” and “C,” flank the WBS common deletion region (Valero et al. 2000). Remarkably, the blocks of the centromeric and medial LCRs are in the same orientation (although in different order), whereas the third segmental duplication lies more telomeric, with the same order as the centromeric LCR but in the opposite orientation (fig. 1). Block B contains three genes in the medial location (Bm) (GTF2I [MIM 601674], NCF1 [MIM 233700], and GTF2IRD2), and the corresponding putative pseudogenes at the centromeric (Bc) (GTF2IP1, NCF1P1, and GTF2IRD2P1) and telomeric (Bt) blocks (GTF2IP2, NCF1P2, and GTF2IRD2P2). The common ∼1.5 Mb deletion is thought to occur by unequal crossing-over between directly oriented blocks within the centromeric and medial LCRs as revealed by deletion mapping and detection of common long-range restriction junction fragments (Pérez Jurado et al. 1996, 1998; Peoples et al. 2000; Valero et al. 2000). Haplotype analysis with polymorphic markers indicated that two-thirds of the deletions arise from crossover events between both chromosome 7 homologues during meiosis, whereas intrachromosomal rearrangements occur in one-third of cases (Urban et al. 1996; Baumer et al. 1998). A recent report has demonstrated the existence of a genomic rearrangement in 7q11.23 in some of the progenitors transmitting the WBS chromosome (Osborne et al. 2001). Interphase FISH analysis and detection of long-range restriction junction fragments revealed that ∼33% of such progenitors are heterozygous for a paracentric inversion estimated to be 1.5 Mb and encompassing the entire WBS region. As postulated by the authors, heterozygosity for such an inversion may lead to unequal chromosome pairing in meiotic prophase and thus may predispose to the WBS deletion (Osborne et al. 2001).
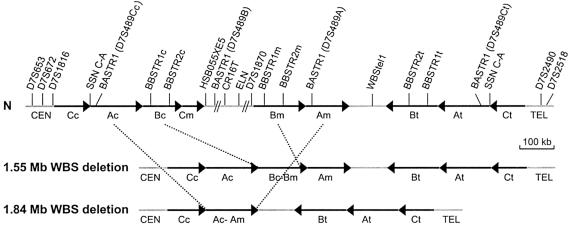
Figure 1.
Schematic representation of the 7q11.23 genomic region in normal chromosomes (N) and chromosomes with the WBS deletions. Blocks A, B, and C of centromeric (c), medial (m), and telomeric (t) LCRs are represented by black arrows that indicate their relative orientation. The single-copy regions between and outside the LCRs are depicted as gray lines. The limits of the common 1.55-Mb and the rarer 1.84-Mb deleted regions found in our patients with WBS are indicated by dotted lines. The few atypical nonrecurrent deletions that have been reported are not represented in this figure (reviewed by Korenberg et al. 2000; Pérez Jurado 2003). The locations of the relevant polymorphic markers used in this study are indicated.
Along with several other diseases recently reviewed (Mazzarela and Schlessinger 1998; Ji et al. 2000; Emanuel and Shaikh 2001; Stankiewicz and Lupski 2002), WBS is a prototype of genomic disorder caused by aberrant homologous recombination between region-specific LCRs. However, the precise mechanisms and the DNA sequences involved in the rearrangements leading to the deletions and inversions remain to be elucidated.
In this report, we present an extensive molecular study on the mutational mechanisms involved in WBS. We have characterized the deletion size in 74 patients with WBS and have precisely defined the sites of chromosomal breakage and strand exchange in 30 of them. In addition, we have confirmed the presence of inversions in some of the progenitors transmitting the rearranged chromosome. Specific models are proposed for aberrant chromosome pairing and recombination leading to inversions and deletions associated with WBS.
Subjects and Methods
Patients and DNA Isolation
Most patients with WBS were ascertained through the clinical genetics services at the Hospital del Mar in Barcelona and the Hospital La Paz in Madrid. Additional samples were received from other Spanish hospitals. Clinical criteria for inclusion within this study have been described elsewhere (Pérez Jurado et al. 1996). Genomic DNA was obtained from subjects with WBS and from available first-degree relatives under institutional review board–approved informed consent. DNA was isolated from peripheral blood lymphocytes by standard procedures.
Microsatellite and RFLP Analyses
The locations of all polymorphic markers used in this analysis are shown in figure 1. An RFLP within exon 20 of the elastin (ELN [MIM 130160]) gene was amplified as described and was digested with _Mva_I (Tromp et al. 1991). Primer information on loci HSB055XE5 (AFMb055xe5), D7S489 (also called “block A short tandem repeat 1” or “BASTR1”), D7S653, D7S672, D7S1816, D7S2518, and D7S1870 is available through the Genome Database. Sequence analyses of publicly available sequences allowed us to identify four new microsatellite markers: block B short tandem repeats 1 (BBSTR1) and 2 (BBSTR2), WBStel1, and CR16T. Heterozygosities for WBStel1 and CR16T are 88.9% and 82.5%, respectively. WBStel1 was genotyped following two rounds of PCR with external and nested primers, because the dinucleotide repeat is flanked by two Alu elements and it was not otherwise possible to obtain a specific amplicon of an appropriate size. Primers were designed with the program Primer3 and are as follows: BBSTR1-F, 5′-TAGAGACGGGGTTTCAGCAT-3′; BBSTR1-R, 5′-GTTGTACACCACAGCACCTG-3′; BBSTR2-F, 5′-CCCTCTGCCAATCCATAGAG-3′; BBSTR2-R, 5′-TTCCTTGTAACCATGAAAGGAT-3′; WBStel1-extF, 5′-GGGAATAGTACCCATCTCAAGG-3′; WBStel1-extR, 5′-AAGCCACACTTTCTCATCTGC-3′; WBStel1-nestedF, 5′-TGGGCAACAGAGCCAGAT-3′; WBStel1-nestedR, 5′-CAGCCTGGGTAACACAGTGA-3′; CR16T-F, 5′-CTCTGGGAGTTCCCAAATGC-3′ and CR16T-R, 5′-GAGGTTGCACTGAGCCAGA-3′. PCR was performed using standard protocols. All forward oligonucleotides were labeled with 5-FAM, HEX, or TET dyes, and samples were analyzed on an ABI PRISM310 Genetic Analyzer (PE Applied Biosystems). Estimation of the number of alleles at multilocus microsatellites was performed by comparing the relative ratios of the areas under the peaks from alleles of the same size in different samples, using the GeneScan 3.1 software (PE Applied Biosystems). Two possible distorting factors caused by the PCR technique were considered: (1) the effect of the allele size in the PCR yield (better PCR yield in general for smaller alleles) and (2) the presence of stutter peaks due to polymerase slippage that increase the area of real allele peaks. As an example, the number of alleles of patient 34.01 at BASTR1 (fig. 2_b_) was estimated by calculating a dosage quotient between the 173-bp and 144-bp alleles (10,261/23,413=0.44) and comparing it to that of the patient's father, 34.11 (12,347/23,547=0.52), and that of the patient's mother, 34.12 (1.03). These data indicate that, under the assumption that parents 34.11 and 34.12 have the normal number of alleles (two in the D7S489A range, four in the D7S489C range, and two in the D7S489B range), patient 34.01 displays only one allele at D7S489A and D7S489B.
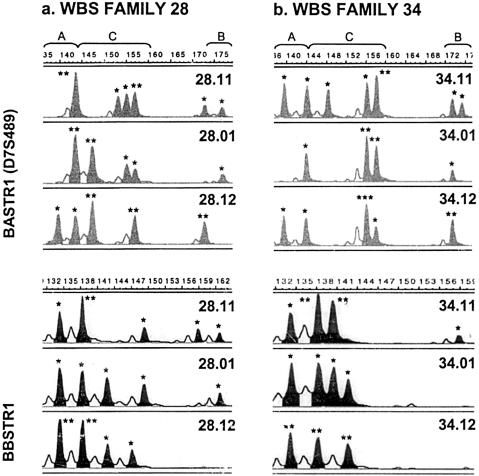
Figure 2.
Genotyping results of the multiple copy microsatellites BASTR1 (D7S489) and BBSTR1 in selected families (11: father, 01: patient and 12: mother). BASTR1 recognizes four loci, one within the commonly deleted region (D7S489B, allele range 168–180), one in block Am (D7S489A, allele range 136–144) and two different D7S489C loci (allele range 144–160) located in blocks Ac (D7S489Cc) and At (D7S489Ct). BBSTR1 recognizes three loci, one from each block B. Asterisks over each peak indicate the number of alleles, as predicted by dosage analysis. a, In family 28, the patient shows six alleles corresponding to loci D7S489A and D7S489C, but only one allele within the range of D7S489B (upper panel) and five alleles at BBSTR1 (lower panel). Therefore, this patient lacks one block B but none of the block A copies, indicating that he bears the typical 1.55-Mb deletion that arose as a result of crossing-over between blocks Bc and Bm. Both parents have a normal number of alleles at each locus. b, In family 34, the son affected with WBS displays only one allele at D7S489A and D7S489B (upper panel) and only four alleles at BBSTR1 (lower panel), whereas both parents have a normal number of alleles at each locus. Therefore, the patient has a larger deletion (1.84 Mb) that includes two blocks B (Bc and Bm) and one block A, most probably because of recombination between blocks Ac and Am.
Sequence Analysis
Searches of genomic sequences were carried out in a number of databases, including High Throughput Genomic Sequences (HTGS), by use of the BLAST software (Altschul et al. 1990). Consensus sequences for LCRs were established by use of the SeqMan program (DNASTAR) and were aligned and compared to each other by use of MegAlign (DNASTAR) and ClustalW (Thompson et al. 1994). Identity and homology distribution among the sequences were calculated by use of DNAsp 3.60 (Rozas and Rozas 1999). RepeatMasker was run to identify interspersed repeat sequences.
Site-Specific Nucleotide (SSN) Assays
Locations of the site-specific nucleotide differences or SSNs among blocks Bc, Bm, and/or Bt that were used in this study are shown in figure 3. PCR primers, amplimer sizes, and detection procedures are listed in table 1. Typically, for each SSN assay, PCR reactions (25 μl) were set up with 50 ng of genomic DNA, 10 pmols of each primer, and 0.2 U of Taq polymerase (Ecogen) in the manufacturer’s buffer. PCR was performed on a thermal cycler (GeneAmp System 9600, PE Applied Biosystems) as follows: 94°C for 5 min; 25 cycles of 94°C for 40 s, 50°C–60°C (depending on the melting temperature of primers in the reaction) for 30 s, and 72°C for 40 s; and a final extension of 72°C for 10 min. Most differences correspond to single-nucleotide changes that were detected by use of restriction enzymes, according to the manufacturer’s instructions (New England Biolabs, Roche), followed by size fractionation on 1%–3% agarose gels. In two cases (SSNs 4 and 13), the difference consists of a small insertion/deletion that could be detected in 10% polyacrylamide gels.
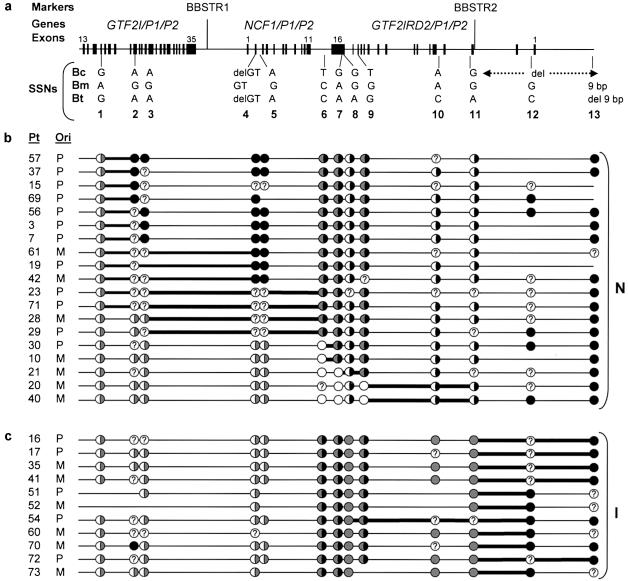
Figure 3.
Mapping the region of exchange in block B in 30 WBS unrelated patients with the common 1.55-Mb deletion. a, Detailed scheme representing the entire ∼143-kb length of block B. Note that the last 38 kb (dotted arrow) are absent in block Bc. Polymorphic microsatellite markers and genes are shown on the top. Exons are depicted as black boxes and numbers at the top indicate the first and last exons within the block. Locations and sequence differences of the 13 SSNs used in this study are indicated below the line. b and c, Each row represents the inferred recombinant block B in the deleted chromosome of each individual with WBS (Pt: patient; Ori: parental origin of the deletion, P: paternal, M: maternal). The predicted genotypes at each position are depicted as circles: white circles, Bc-type; black circles, Bm-type; gray circles, Bt-type; half-white/half-gray circles, either Bc-type or Bt-type; half-white/half-black circles, either Bc-type or Bm-type; half-gray/half-black circles, either Bm-type or Bt-type. A question mark is used when the number of Bc, Bm, or Bt copies cannot be determined because a polymorphism in this position is detected in at least one of the parents. Patients without the inversion (N) are grouped in b, whereas patients with predicted inversion (I) are displayed in c. For each patient, mapping of the region of exchange was narrowed between two SSNs as depicted by a thicker line. Note that in 7–12 of 19 patients without the inversion, the transition from Bc (half-white/half-gray circle, because Bt can not be ruled out) to Bm (black circle) occurs somewhere within the 12 kb between SSNs 1 and 2 (b). On the other hand, all patients with the inversion undergo the transition from Bt (gray circle) to Bm (black circle) somewhere between SSNs 11 and 13, at the end of the block (c).
Table 1.
SSNs Used for Mapping the Deletion and Inversion Breakpoints
| Primer | |||||
|---|---|---|---|---|---|
| SSN | Positiona | Forward | Reverse | Detection Procedure | Polymorb |
| 1 | 5,603 bp, GTF2I (exon 16) | 5′-GAGCACTACAGATTCAATGATTT-3′ | 5′-TTATTTCATGTGGAAGGTAACAT-3′ | _Eco_RI; Bc and Bt: 176, 30, 1486; Bm: 176, 30, 1341, 145 | 1/114 (0.9%) |
| 2 | 16,572 bp, GTF2I (exon 21) | 5′-CTCAAGCTCTTGGACTCAC-3′ | 5′-ATCCCAGGAGGCAAGTAGGAAAT-3′ | _Bst_UI; Bm: 151, 246, 759; Bc and Bt: 397, 759 | 25/110 (22.7%) |
| 3 | 17,512 bp, GTF2I (intron 21) | 5′-CTCAAGCTCTTGGACTCAC-3′ | 5′-ATCCCAGGAGGCAAGTAGGAAAT-3′ | _Rsa_I; Bm: 34, 293, 115, 438, 281; Bc and Bt: 34, 293, 115, 438, 209, 72 | 15/118 (12.7%) |
| 4 | 48,967 bp, NCF1 (exon 2) | 5′-TCCCCCGACTCTGGCTTTC-3′ | 5′-GGAACTCGTAGATCTCGGTGA-3′ | delGT; Bc and Bt: 103; Bm: 105 | 11/122 (9%) |
| 5 | 49,344 bp, NCF1 (intron 2) | 5′-TCCCCCGACTCTGGCTTTC-3′ | 5′-GGGGAGCTTGAGGTCATCAG-3′ | _Taq_I; Bc and Bt: 980, 1168, 40; Bm: 419, 561, 1168, 40 | 10/102 (9.8%) |
| 6 | 63,859 bp, _NCF1_-GTF2IRD2 | 5′-CTCACCCCTTCTGCTTGTTC-3′ | 5′-CAGATGATAGGGAGGGGACA-3′ | _Sma_I; Bc: 552, 1754; Bm and Bt: 552, 632, 1122 | 1/110 (1%) |
| 7 | 68,364 bp, GTF2IRD2 (exon 16) | 5′-ATGAATAGTGAGGCATACAATG-3′ | 5′-CCATAGATTGGATCCGAGACCT-3′ | _Bst_UI; Bc: 499, 222, 147, 243; Bm and Bt: 868, 243 | 0/70 (0%) |
| 8 | 69,422 bp, GTF2IRD2 (exon 16) | 5′-CCCGGATCAGGTCATACATC-3′ | 5′-GGGAAAACGCAAGATAGAC-3′ | _Bst_UI; Bt: 209, 516; Bc and Bm: 209, 356, 160 | 2/106 (1.9%) |
| 9 | 75,076 bp, GTF2IRD2 (exon 14) | 5′-TTGGACAAATTTTTGCAGAGT-3′ | 5′-CCGTCACAGGTGAAGCAAT-3′ | _Hae_III; Bc: 149, 314; Bm and Bt: 149, 197, 117 | 2/116 (1.7%) |
| 10 | 94,627 bp, GTF2IRD2 (intron 4) | 5′-TGCAAGGTCGTAATTCTCAGG-3′ | 5′-TGTCTTTCCATAGGCATGAAGA-3′ | _Msp_I; Bt: 42, 201, 753; Bc and Bm: 42, 954 | 7/110 (6.4%) |
| 11 | 104,958 bp, GTF2IRD2 (intron 3) | 5′-TTGTAAAATGGTGTTTATTTTAGG-3′ | 5′-GCCCCACAAACTTGGATCTG-3′ | _Tru_9I; Bt: 20, 242, 51, 156; Bc and Bm: 20, 100, 142, 51, 156 | 7/122 (5.7%) |
| 12 | 124,648 bp, GTF2IRD2 (intron 1) | 5′-TCCCCCTAGAACTCGAAACC-3′ | 5′-CTTTCGGAACGTCCTCCTCC -3′ | _Rsa_I; Bm: 272, 76, 182; Bt: 272, 258 | 19/88 (21%) |
| 13 | 142,983 bp | 5′-CTGGGTCGAGACAGGGTAAG-3′ | 5′-AAATGTGTCTATGGACCCGC-3′ | 9-bp del; Bm: 103; Bt: 94 | 11/116 (9.5%) |
| C-A | Junction between blocks C and A | 5′-CCGAACTACGCCTATGGAAC-3′ | 5′-GCGGCTCACTTCTGTAATCC-3′ | _Hin_fI; C-Ac: 100, 161, 305; C-At: 100, 161, 123, 182 | 6/70 (8.5%) |
Estimation of the Relative Numbers of Block Bc, Bm, and Bt Copies
For each SSN assay, a digital image of the gel was captured at varying exposure times, to ensure that the bands were not saturated. Then, intensities of bands corresponding to presumed blocks Bc, Bm, and/or Bt were quantified by use of the Volume Tool from the Quantity One software package (Bio-Rad). Relative intensities were calculated by means of a dosage quotient for the block that can be distinguished in a particular SSN assay relative to the other amplified blocks. This block B dosage quotient was calculated for all patients, as well as for their parents as reference values in the same experiment. A final ratio, called the “patient/progenitor block B ratio,” was thereby calculated by relating the dosage obtained for each WBS patient to the mean dosage value of his or her progenitors. Reproducibility of results was evaluated by repeating each experiment at least twice with a different number of PCR cycles.
Detection of De Novo Junction Fragments
We designed a PCR experiment to amplify a putative de novo junction fragment of 3.4 kb between SSNs 1 and 2. The forward primer (BK1F1: 5′-CTCCCTCCTCATCCGCACCTT-3′) was designed to anneal specifically to the block Bc and contains two mismatches (shown in boldface) with respect to consensus sequences of blocks Bm and Bt. The reverse primer (BK1R: 5′-TGCTGGCCTTTGTGTTATCATC-3′) is complementary to blocks Bm and Bt, with two mismatches at the 3′ end with respect to block Bc sequence. A second forward primer (BK1F2: 5′-TCATGTGGACAAATTTCACTT-3′) that amplifies a 2.0-kb fragment from blocks Bm and Bt, in combination with BK1R, was used as an internal positive control. PCR reactions with 50 ng of DNA, 5 pmols of BK1F1, 2.5 pmols of BK1F2, and 7.5 pmols of BK1R were subjected to 30 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 2.5 min. All DNA samples were also amplified with two primers complementary to the Bm and Bt blocks that give rise to the same 3.4-kb fragment (BK1F1mt: 5′- CTCCATCCTCATCCGCACCTC-3′ and BK1R), to check the DNA quality.
FISH Analysis
Chromosome spreads for interphase FISH analysis were prepared from peripheral blood lymphocytes using standard methodology. Three-color FISH was performed with one BAC located centromeric to the WBS deleted region (RP11-421b22), one BAC from the GTF2IRD1 locus (RP4-665p05), and one PAC containing the STX1 locus (RP11-622p13), labeled with digoxigenin, biotin, and a 1:1 mix of biotin and digoxigenin, respectively. Purified BAC DNAs were labeled by nick-translation (Roche), and unincorporated nucleotides were removed by passage through a sephadex-G50 column (Roche). Hybridization was performed by use of standard protocols and was detected with anti-digoxigenin-fluorescein (green), avidin-rhodamine (red), or both (yellow) (Roche). Nuclei were counterstained with DAPI/antifade (Q-biogene).
Slides were visualized under a fluorescent microscope (AH3, Olympus) and images were analyzed with the Cytovision 3.1 software (Applied Imaging Ltd.). At least 20 interphase nuclei where all three probes could be identified in close alignment with each other were scored for each individual.
Results
Deletion Mapping and Parental Origin in Patients with WBS
A total of 74 patients with WBS and 7q11.23 submicroscopic deletions documented by FISH analysis, Southern blotting, and/or microsatellite typing were included in the study. Deletion mapping was performed by genotyping multiple polymorphic markers from the region. In agreement with previous reports (Pérez Jurado et al. 1996; Robinson et al. 1996), all patients were hemizygously deleted at HSB055XE5 and D7S1870 (lack of parental inheritance or quantitative dosage reduction), which represent the internal limits of the deleted interval (fig. 1). Biparental inheritance at D7S1816 and/or D7S672 in the centromeric side and at D7S2490 and/or D7S2518 in the telomeric side roughly defined the external limits of the deletions in all informative cases (fig. 1). A novel additional marker (WBStel1) situated in the single-copy region between the medial and telomeric blocks of LCRs was not deleted in any patient. Deletions were de novo in all informative cases, of maternal origin in 30 patients (45%) and paternal origin in 37 (55%). The parental origin of the deletion could not be established in seven patients, because DNA from the parents was not available.
The analysis of multiple-copy STR markers located in blocks A and B allowed us to establish the specific LCRs involved in the unequal crossover event. Dinucleotide repeats located in the intergenic region between GTF2I/ GTF2IP1/GTF2IP2 and NCF1/NCF1P1/NCF1P2 (BBSTR1) and intron 3 of the GTF2IRD2/GTF2IRD2P1/GTF2IRD2P2 gene (BBSTR2) identify six alleles in control individuals, two from each of the block B copies (fig. 1). The microsatellite marker AFM136xe3 (BASTR1) identifies a locus within the commonly deleted region (D7S489B) and three additional loci, each located in one of the block A copies (D7S489A, D7S489Cc, and D7S489Ct) (fig. 1) (Pérez Jurado et al. 1996; Robinson et al. 1996). The vast majority of patients (70 of 74) displayed only five block B alleles with BBSTR1 and BBSTR2 but the normal six alleles from block A, indicating that the deletion was caused by recombination between blocks Bc and Bm (figs. 1 and 2_a_). The approximate size of this deletion by detailed analysis of the publicly available sequence is ∼1.55 Mb. Four patients (5.5%) exhibited four alleles with BBSTR1 and BBSTR2, two fewer than their parents, suggesting the absence of two block B copies in the rearranged chromosome (fig. 2_b_). As expected, these four patients showed only five block A alleles with hemizygosity at the D7S489A locus (fig. 2_b_), indicating that the deletion extended further (∼1.84 Mb), and was most likely caused by misalignment and aberrant recombination between blocks Ac and Am (fig. 1).
Refinement of Deletion Breakpoint Regions in Block B Using SSN Assays
We assembled and integrated all GenBank sequence data available from genomic BAC and PAC clones spanning blocks Bc (CTA-269p13, RP11-396k3, RP11-450o3, and RP11-483g21), Bm (239c10, RP4-771p04, RP11-813J7, and CTA-350l10) and Bt (RP11-729p19 and RP11-219m8). Site specificity of the clones was defined by differential anchoring points at least at one of their ends. Sequence comparisons revealed that blocks Bm and Bt share a ∼143-kb region of continuous homology (99.7% identity), whereas block Bc aligns only with the first ∼105 kb (99.6% identity to both Bm and Bt). A consensus sequence was established for each block; then, a total of 442 individual nucleotide differences including small gaps were found between the consensus of blocks Bc and Bm. Theoretically, these differences could be used to classify the recombinant block B sequence in patients with WBS bearing the typical 1.55-Mb deletion as either Bc, Bm, or Bt type at that site. In practice, some sites that differ between the available sequences of the genomic clones appeared to be identical in control individuals. In addition, some sites appeared to have a relatively high degree of polymorphism or lack of copy specificity among control individuals and, therefore, were not used to determine the site of chromosomal exchange.
We tested a total of 34 putative block B (Bc/Bm/Bt) SSNs in 10 control samples, and selected 13 on the basis of their high degree of site specificity and ease of typing (table 1, fig. 3_a_). We used them to further localize the breakpoints within this block in 30 patients with the common 1.55-Mb deletion. The locations and the copy-specific nucleotides at the 13 SSNs are shown in figure 3_a_. In most PCR assays (SSNs 1–11), six copies of block B sequence (two Bc, two Bm, and two Bt) were amplified in normal individuals, five copies (one Bc, one Bm, two Bt and one recombinant Bc-Bm) in patients with the typical 1.55-Mb deletion, and four copies in those patients with larger deletions (one Bc, one Bm, and two Bt). After PCR amplification, the SSN analysis was performed as described in the “Subjects and Methods” section (table 1). Then, a dosage analysis was performed to elucidate the relative number of Bc, Bm, and/or Bt copies and to predict the copy type in the recombinant block B in patients. At SSNs 1, 2, 3, 4, and 5, block Bm displays a specific nucleotide that can be distinguished from Bc and Bt. Progenitors were expected to display a Bm:(Bc+Bt) ratio of 2:4, whereas patients in whom the breakpoint occurred distally should have a 1:4 ratio and those in whom the site of exchange is proximal should show a 2:3 ratio. Therefore, the patient/progenitor block B ratios expected in these cases were 0.5 (1:4/2:4) and 1.33 (2:3/2:4), respectively. Consequently, when, in our experiments, this final ratio was <0.915, a 1 Bm:4 (Bc+Bt) dosage for the patient was deduced, whereas, if it was >0.915, it was considered as a 2 Bm:3 (Bc+Bt) dosage. The same kind of calculations and inferences were performed for SSNs 6, 7, and 9, in which block Bc can be distinguished from blocks Bm and Bt. In these assays, a patient/progenitor block B ratio of 0.5 suggests that the breakpoint is proximal, whereas the ratio is 1.33 if the breakpoint is distal. Finally, in SSN assays 12 and 13 only Bt and Bm copies are amplified, and, thus, a patient/progenitor block B ratio of 1 (2:2/2:2, Bt:Bm) indicates a breakpoint proximal to this site, whereas a ratio of 2 (2:1/2:2, Bt:Bm) indicates that it is distal.
For SSNs 1–7, 9, 12, and 13 consistent values for patient/progenitor block B ratios were obtained in most cases, with means very close to the expected 0.5, 1, or 1.33 values, and standard deviations of <30% of the mean value (table 2). These results allowed us to define the relative number of block B-type copies at each position analyzed and then to infer whether the recombinant block B was Bm type (breakpoint proximal or centromeric to the position) or Bc type (breakpoint distal or telomeric) (fig. 3_b_). Abnormal relative dosage results were obtained in some progenitors at several SSNs, suggesting the occasional presence of non–site-specific sequence in LCR blocks (see table 1 for polymorphism degree in each SSN), which prevented definition of the copy type in the recombinant block B of the patient (represented by circles filled with a question mark in fig. 3). Most of the SSNs only distinguish one copy with respect to the other two, but this information may be enough to define the entire recombinant block with consistency. As an example, patient 20.01 shows one Bm copy at SSN4 (fig. 4_a_) and this is indicated in figure 3_b_ with a half-white, half-gray circle, because SSN4 distinguishes block Bm from blocks Bc and Bt and thus, strictly speaking, the results are compatible with the recombinant Bc-Bm block being Bc-type or Bt-type. This same patient 20.01 displays two Bc copies at SSN9 (fig. 4_b_), and this is denoted by a white circle in figure 3_b_. Both results indicate that all the partially informative positions in the recombinant block B between SSN4 and 9 must be Bc type so that the deletion breakpoint is located distal to SSN9 in this patient.
Table 2.
Statistical Analysis of the Values Obtained when Estimating the Relative Number of Block Bc, Bm, and Bt Copies in Patients with WBS by Means of SSN Assays
| Value | ||||||
|---|---|---|---|---|---|---|
| SSN (Block Copies)and Patient/ProgenitorBlock Ratios | Threshold | Mean | Maximum | Minimum | SD | SD/Mean |
| SSN 1 (Bm:[Bc+Bt]): | ||||||
| 1:4/2:4 = 0.5 | <0.915 | .52 | .65 | .37 | .07 | .13 |
| 2:3/2:4 = 1.33 | >0.915 | … | … | … | … | … |
| SSN 2 (Bm:[Bc+Bt]): | ||||||
| 1:4/2:4 = 0.5 | <0.915 | .64 | .77 | .38 | .13 | .20 |
| 2:3/2:4 = 1.33 | >0.915 | 1.49 | 1.82 | 1.19 | .24 | .16 |
| SSN 3 (Bm:[Bc+Bt]): | ||||||
| 1:4/2:4 = 0.5 | <0.915 | .57 | .68 | .32 | .12 | .21 |
| 2:3/2:4 = 1.33 | >0.915 | 1.31 | 2.27 | 1.02 | .37 | .28 |
| SSN 4 (Bm:[Bc+Bt]): | ||||||
| 1:4/2:4 = 0.5 | <0.915 | .51 | .66 | .41 | .07 | .14 |
| 2:3/2:4 = 1.33 | >0.915 | 1.48 | 1.73 | 1.11 | .17 | .11 |
| SSN 5 (Bm:[Bc+Bt]): | ||||||
| 1:4/2:4 = 0.5 | <0.915 | .49 | .54 | .43 | .04 | .08 |
| 2:3/2:4 = 1.33 | >0.915 | 1.35 | 1.68 | 1.17 | .16 | .12 |
| SSN 6 (Bc:[Bm+Bt]): | ||||||
| 1:4/2:4 = 0.5 | <0.915 | .53 | .70 | .37 | .07 | .13 |
| 2:3/2:4 = 1.33 | >0.915 | 1.39 | 1.63 | 1.26 | .17 | .12 |
| SSN 7 (Bc:[Bm+Bt]): | ||||||
| 1:4/2:4 = 0.5 | <0.915 | .52 | .70 | .44 | .10 | .20 |
| 2:3/2:4 = 1.33 | >0.915 | 1.55 | 1.67 | 1.44 | .12 | .08 |
| SSN 8 (Bt:[Bc+Bm]): | ||||||
| 2:3/2:4 = 1.33 | <2.165 | 1.37 | 1.74 | 1.17 | .13 | .10 |
| 3:2/2:4 = 3 | >2.165 | 3.05 | 4.01 | 2.41 | .45 | .15 |
| SSN 9 (Bc:[Bm+Bt]): | ||||||
| 1:4/2:4 = 0.5 | <0.915 | .49 | .59 | .40 | .04 | .09 |
| 2:3/2:4 = 1.33 | >0.915 | 1.36 | 1.51 | 1.22 | .15 | .11 |
| SSN 10 (Bt:[Bc+Bm]): | ||||||
| 2:3/2:4 = 1.33 | <2.165 | 1.43 | 1.75 | 1.09 | .23 | .16 |
| 3:2/2:4 = 3 | >2.165 | 3.83 | 4.55 | 2.95 | .53 | .14 |
| SSN 11 (Bt:[Bc+Bm]): | ||||||
| 2:3/2:4 = 1.33 | <2.165 | 1.22 | 1.68 | 1.00 | .21 | .17 |
| 3:2/2:4 = 3 | >2.165 | 3.44 | 4.62 | 2.62 | .61 | .18 |
| SSN 12 (Bt:Bm): | ||||||
| 2:2/2:2 = 1 | <1.5 | 1.00 | 1.19 | .70 | .11 | .15 |
| 2:1/2:2 = 2 | >1.5 | … | … | … | … | … |
| SSN 13 (Bt:Bm): | ||||||
| 2:2/2:2 = 1 | <1.5 | 1.04 | 1.21 | .70 | .14 | .13 |
| 2:1/2:2 = 2 | >1.5 | … | … | … | … | … |
| SSN C-A (Ct-At:Cc-Ac): | ||||||
| 2:2/2:2 = 1 | <2 | 1.00 | 1.32 | .62 | .17 | .17 |
| 3:1/2:2 = 3 | >2 | … | … | … | … | … |
Figure 4.
Analysis of samples from patients with WBS and their progenitors with block B SSNs to map deletion breakpoints. Relative block dosages calculated from band intensities, as described in the “Methods” section, are depicted bellow each lane. a, Representative results of the SSN 4 assay, which amplifies exon 2 of the NCF1 gene and NCF1P1/NCF1P2 pseudogenes. Both pseudogenes in blocks Bc and Bt have a 2-bp deletion at the beginning of exon 2 (delGT) that is not present in the NCF1 gene in Bm. Patients 20.01 and 21.01 show one gene copy versus four pseudogenes, suggesting that the deletion breakpoint is telomeric to this point. Patients 3.01 and 61.01 display two gene copies versus three pseudogenes, indicating that the crossing-over occurred proximal to this position. Patients 17.01 and 35.01 have a 1 Bm:4 (Bc+Bt) ratio, most likely because of the presence of three block Bt copies (see fig. 3_c_). Progenitor 21.12 has three gene-type copies and three pseudogenes, most probably as a result of gene conversion. b, Results of the SSN 9 assay in a few patients. A nucleotide change in exon 14 of GTF2RD2/GTF2RD2P1/GTF2RD2P2 creates a restriction site for _Hae_III in blocks Bm and Bt that is not present in Bc, as represented in the scheme. Patients 20.01 and 40.01 show three digested copies versus two nondigested copies, indicating that unequal crossing-over occurred distal to this position. On the contrary, patients 71.01 and 37.01 have three digested copies versus two nondigested copies, showing that the breakpoint is proximal to this nucleotide. Patients 17.01 and 35.01 display a 1 Bc:4 (Bc+Bt) ratio, most likely because of the presence of three block Bt copies (see panel c). c, SSN 11 assay allows detection of the inversion in patients with WBS. As represented in the scheme, a nucleotide change in block Bt destroys a _Tru9_I site that is present in blocks Bc and Bm. Patients 7.01, 71.01, and 42.01 show three digested copies (blocks Bc and Bm) versus two nondigested copies (block Bt), suggesting the existence of two block Bt copies, one in each chromosome. In patients 17.01 and 35.01, we observe a reduction of the intensity of bands 3 and 4 (Bc and Bm) and an increase of the intensity of band 1 (Bt). The patient/progenitor block B ratio is close to 3 in both cases, suggesting the existence of three block Bt copies (one in the normal chromosome and two in the WBS chromosome) but only two blocks Bc and Bm (both in the normal chromosome). The 3 Bt:2 (Bc+Bm) ratio suggests that the WBS chromosome arose in a progenitor heterozygous for the inversion. Dosage calculations in sample 23.11 indicate that he has only one Bt-like copy versus five Bc and Bm blocks, most probably as a result of gene conversion. His son, patient 23.01, inherited this polymorphism.
In 19 patients, the transition from block Bc to Bm was localized between two SSNs, allowing the precise localization of the deletion breakpoints (fig. 3_b_). These deletion breakpoints were mapped within the GTF2I/GTF2IP1 gene or the GTF2I/GTF2IP1-NCF1/NCF1P1 intergenic region in 10–14 cases (53%–74%), in the NCF1/NCF1P1 gene in 2–6 cases (11%–31%), and within GTF2IRD2/GTF2IRD2P1 in 3 cases (16%). Therefore, breakpoints appear to occur at any position within block B, although they tend to concentrate at the beginning, in a 12-kb region between SSNs 1 and 3, located at exons 16 and intron 21 of the GTF2I/GTF2IP1 gene, respectively (7–12 of 19 patients).
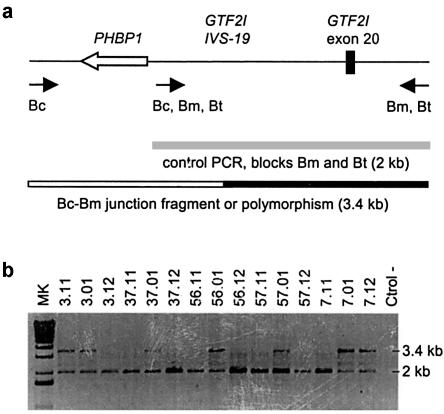
Detection of a Recurrent Deletion Junction Fragment
On the basis of the SSN data, we designed a PCR assay to specifically amplify a putative 3.4-kb de novo deletion-junction fragment between SSNs 1 and 2 using a centromeric-specific forward primer and a medial and telomeric-specific reverse primer (fig. 5_a_). This assay was performed in the 12 WBS samples (57.01, 37.01, 15.01, 69.01, 56.01, 3.01, 7.01, 61.01, 19.01, 42.01, 23.01, and 71.01), in which the SSN results suggest that the deletion breakpoint is located between SSNs 1 and 2 (fig. 3_b_), as well as in their parents and in several control DNA samples. The 3.4-kb amplification product between introns 19 and 20 of the GTF2I/P1 gene was obtained in patients 56.01, 57.01, 37.01, 3.01, 7.01, 19.01, 23.01, and 71.01 (fig. 5_b_). The same amplification product was also obtained in 20% of parents and controls, probably because of polymorphic variants present in some normal chromosomes. Both parents of patients 56.01, 57.01, and 37.01 were negative for the 3.4-kb amplification product, indicating that the 3.4-kb band present in this patients corresponds to a de novo deletion-junction fragment. No final information is available for samples 69.01 and 19.12, because the inability to amplify the 2-kb control PCR product suggests technical problems due to DNA degradation. Parents 3.11, 7.12, 23.11, and 71.11 were positive for the 3.4-kb amplification product, raising the possibility that patients 3.01, 7.01, 23.01, and 71.01 are PCR positive because they carry an inherited polymorphism in the region and not a de novo deletion-junction fragment (fig. 5_b_). Haplotype analysis of the unaffected sisters demonstrated that patient 3.01 has not inherited the polymorphic variant from his father (3.11) and thus that the 3.4-kb amplification product represents indeed a de novo junction fragment. On the contrary, haplotype data could not discard the 3.4-kb amplification fragment being a polymorphism in family 7 (data not shown). No siblings were available for patients 23.01 and 71.01, and thus, in these cases, we cannot establish whether the 3.4-kb amplification product corresponds to a de novo Bc-Bm junction fragment or to an inherited polymorphism. Sequencing of the 3.4-kb PCR product did not shed any light on the problem either. On the basis of publicly available sequence, there are only four nucleotide differences between blocks Bm and Bc/Bt in this 3.4-kb fragment. Unfortunately, all four positions display a Bc-type nucleotide in all the PCR fragments analyzed from patients (56.01, 57.01, 7.01, 23.01, and 71.01) and parents (7.12, 23.11, and 71.11), regardless of whether they correspond to proved or putative de novo junction fragments (data not shown). Therefore, a specific junction fragment in this region has been obtained in at least 4 (and maybe up to 8) of 19 individuals with a recombinant Bc/Bm block.
Figure 5.
Amplification of a deletion-junction fragment in some patients. a, PCR strategy for amplifying a deletion-junction fragment containing the prohibitin (PHBP1) pseudogene and part of introns 19 and 20 of the GTF2I gene. Specific primers for blocks Bc and Bm/Bt allow the amplification of a recombinant Bc-Bm fragment of 3.4 kb. A nested primer that anneals to all three blocks and amplifies a 2.0-kb product from blocks Bm and Bt was used as an internal positive PCR control. b, The PCR assay detects a de novo 3.4-kb fragment in patients 37.01, 56.01, and 57.01 not present in their healthy parents. The same fragment is also found in patients 3.01 and 7.01 but also in one of their parents, probably because of the presence of a polymorphism.
Detection of Inversions
SSN assays 8, 10, and 11 allow the identification of Bt copies versus Bc and Bm copies. Patients were expected to have ratios of 2:3 (Bt:[Bc+Bm]), whereas parents should have the normal 2:4 ratios, and thus a patient/progenitor block B ratio of 1.33 (2:3/2:4) was predicted. However, 11 of 30 patients (37%) consistently displayed a patient/progenitor block B ratio close to 3 (3:2/2:4) at these three SSNs, consistent with the presence of three block Bt–type copies of the five total block B copies (figs. 3_c_ and 4_c_). Since the copy ratio was normal in their parents (2:4), the only logical explanation for this finding was the presence of two Bt-type blocks in the rearranged chromosome 7, keeping the intact structure (Bc-Bm-Bt) on the other chromosome.
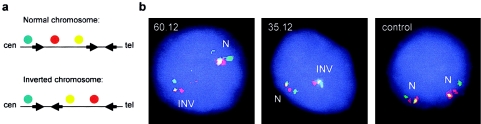
In agreement with recent data reported by Osborne et al. (2001), we postulated that the WBS transmitting progenitors of those patients showing a triple dosage of the Bt allele were carriers of an inversion. The WBS deletion would be the result of meiotic unequal exchange between the inverted chromosome (with telomeric-type LCRs at the centromeric site) and its non-inverted homologue. Confirmation of the existence of an inversion was obtained in all four transmitting progenitors of the patients showing a triple dosage of the Bt allele that were analyzed by three-color interphase FISH analysis (samples 35.12, 51.11, 54.11, and 60.12). When using two probes from the WBS commonly deleted region and one probe centromeric to the deletion, we found the normal disposition of the signals in one chromosome but a different order in the other chromosome, indicating a change in the orientation of the WBS region relative to the centromeric DNA (fig. 6).
Figure 6.
Detection of the 7q11.23 inversion polymorphism in WBS transmitting progenitors by three-color interphase FISH. a, Order of the probes along a normal chromosome and a chromosome with the 7q11.23 inversion polymorphism. The probes, from centromere to telomere, correspond to RP11-421b22, RP11-622p13, and RP4-665p05. Centromeric, medial, and telomeric LCRs are depicted as black arrows. b, On interphase nuclei from two transmitting mothers showing a triple dosage of the Bt allele (60.12 and 35.12), two different chromosomes can be distinguished: one with the signals in the expected order (N) and another in which the yellow signal appears between the red and the green, indicating an inversion of the region (INV). A nucleus from a control individual with two normal chromosomes is also shown.
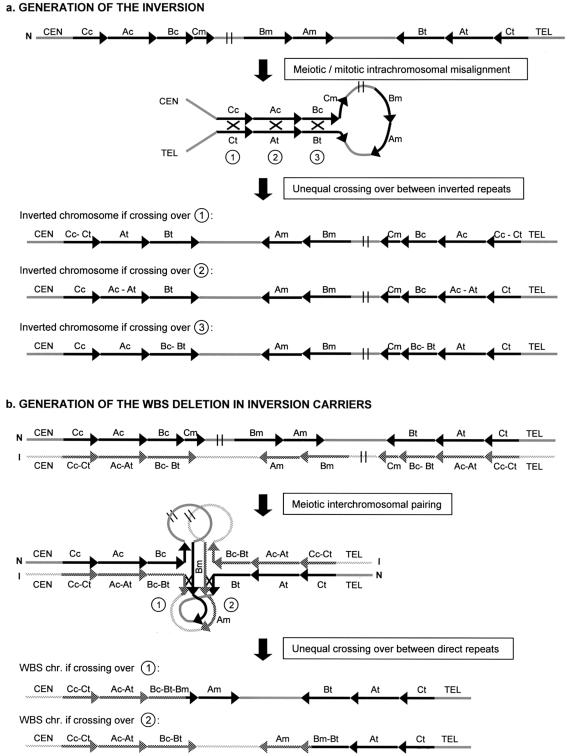
These paracentric inversions in parents most likely originated through intrachromosomal recombination between the centromeric and telomeric LCR blocks, which are in an opposite orientation (fig. 7_a_). To investigate whether the crossing-over leading to the inversion occurred at block C, A, or B, we analyzed SSN C-A, located between blocks C and A (fig. 1 and table 1), in the 11 patients with rearranged chromosomes (inversion plus deletion). Six patients showed the normal 2 Cc-Ac:2 Ct-At ratio, indicating that the exchange events leading to the original inversions had happened at either block A or block B (crossovers 2 or 3 in fig. 7_a_) (data not shown). Thus, in these patients the inverted interval encompasses a genomic fragment ranging from 1.79 Mb (recombination at the end of block B) to 2.34 Mb (recombination at the beginning of block A). Five patients showed a Cc-Ac versus Ct-At ratio of 1:3, although the presence of the same abnormal dosage Cc-Ac:Ct-At ratio in one of their progenitors prevents from drawing any final conclusion.
Figure 7.
a, Mechanism for the origin of the inversion polymorphism. During chromosome pairing in cell division, the large inverted segmental duplications containing centromeric and telomeric blocks A, B, and C may favor the partial refolding of one chromosome, allowing intrachromatid or unequal sister chromatid synapsis. Nonallelic homologous recombination (black X) can occur in block C, A, or B, resulting in a paracentric inversion ranging in size from 2.34 to 1.79 Mb. b, Mechanism for the generation of the WBS deletion in parents heterozygous for the inversion polymorphism. Asynapsis at the inverted region promotes the folding of both chromosomes, to attempt pairing of homologous sequences. Note that the centromeric and telomeric segmental duplications are identical in length at blocks C and A, but block Bc is 38 kb shorter than block Bt, which is identical to Bm. In this model, all sequences of the region could undergo perfect matching with sequences from the homologous chromosome. The WBS chromosome is the result of unequal crossing-over (black X) either between block Bt from the inverted chromosome and Bm from the normal one (type 1) or between block Bm from the inverted allele and block Bt of the normal one (type 2). Recombination events at any other site within the loops would result in either acentric or dicentric chromosomes 7 (most likely nonviable). N: normal chromosome, I: inverted chromosome.
Generation of the WBS Deletion in Carriers of an Inversion Polymorphism
Heterozygosity for the inversion polymorphism should invariably lead to abnormal chromosome pairing in meiotic prophase. During pairing, the first regions of sequence divergence occur at the internal edges of both inverted LCRs, because Bc is 38 kb shorter than Bt. In figure 7_b,_ we illustrate how both chromosomes could fold onto themselves to achieve an optimal or even complete sequence pairing. On the basis of this model, only unequal crossing-over events occurring between either block Bt of the inverted chromosome and block Bm of the noninverted homologue (type 1), or block Bm of the inverted allele and Bt of the normal one (type 2), would result in either a WBS-deleted chromosome or the reciprocal duplication. This WBS chromosome would contain telomeric block B sequences from the inverted chromosome at the centromeric side. Therefore, a patient bearing such a WBS chromosome would display three copies of telomeric-type alleles (one from the normal chromosome and two from the rearranged chromosome) in the fragment between the ancestral inversion breakpoint in the parental chromosome and the de novo deletion breakpoint. Recombination at any other site within the loops would lead to acentric and dicentric chromosomes 7, both most likely nonviable.
Our model for the generation of the deletion in inversion carriers predicts that the interchromosomal exchange event must take place within the last 38 kb of blocks Bm and Bt (fig. 7_b_). In agreement with this, we found a gain of a Bt-type block in the SSNs proximal to this region (SSNs 8, 10, and 11), suggesting that the deletion breakpoint is located distally (figs. 3_c_ and 4_c_). In addition, we detected a normal 2Bm:2Bt ratio in all patients with the inverted-deleted chromosome when performing the SSN assays 12 and/or 13 (fig. 3_c_). These data indicate that the unequal crossing-over causing the deletion in inversion carriers is always localized between SSNs 11 and 13, in the 38-kb region only present in blocks Bm and Bt, as predicted by our model.
The dosage assay at SSN 11 was performed in the remaining samples of patients with WBS who had 7q11.23 deletions to assess the presence of inversions. At this position at the very end of the ∼105-kb alignment between Bc and Bm/Bt, there should always be a gain of a Bt-type sequence and loss of a Bc-type sequence if the rearranged WBS chromosome originated in an inversion carrier (fig. 7_b_). Comparative dosage analysis was suggestive of the presence of an inverted chromosome in 10 of 44 additional cases, for a total of 21 of 74 (28%). The degree of polymorphic variation (non–site specificity) at this site, among normal individuals (WBS parents and controls), was 5.7%. This variation could lead to misinterpretation of the results in some cases, although comparison of the block B dosages obtained from patients and their parents should prevent this error.
Occurrence of Inter- versus Intrachromosomal Rearrangements
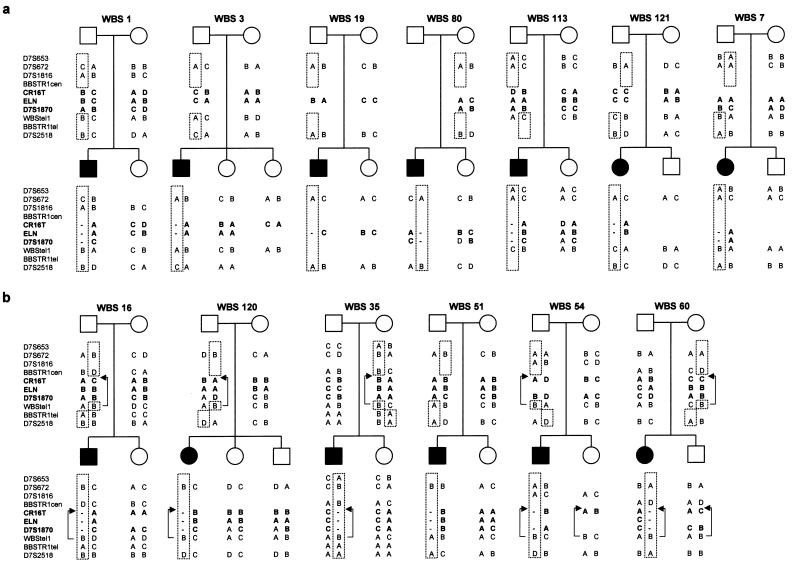
We investigated in 13 families whether a meiotic interchromosomal recombination event had occurred in the critical or flanking regions during the generation of the WBS chromosome. Patients and available first-degree relatives (parents and siblings) were genotyped at several chromosome 7 loci, located either centromeric (D7S653, D7S672, D7S1816, and BBSTR1cen), within (CR16T, ELN, and D7S1870) or telomeric (WBStel1, BBSTR1tel, and D7S2518) to the critical WBS deletion (fig. 1). Haplotype analysis of unaffected siblings enabled us to establish the phase of the alleles on the parental and WBS chromosomes (fig. 8) with an estimated error rate of 1%–2%, which corresponds to the probability of a random recombination event within the region in the meiosis leading to the sibling’s chromosomes.
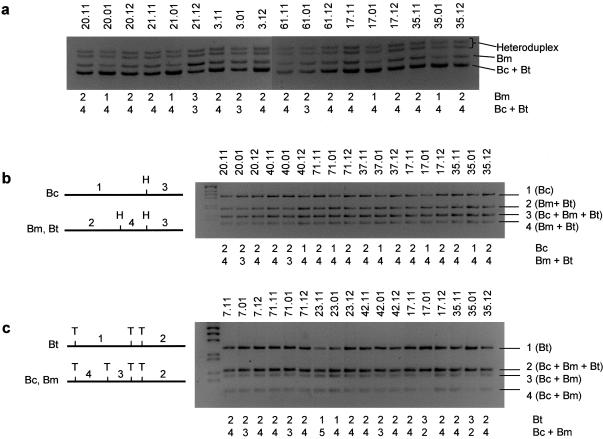
Figure 8.
Haplotype analyses of 13 families with WBS, performed using polymorphic markers centromeric to (D7S653, D7S672, D7S1816, and BBSTR1cen), within (CR16T, ELN, and D7S1870), and telomeric to (WBStel1, BBSTR1tel, and D7S2518) the WBS deleted region. Absence or presence of recombination between centromeric and telomeric markers indicates that the deletion arose as a result of an intra- or interchromosomal recombination event, respectively. In the inverted chromosomes, absence or presence of recombination between centromeric markers and WBStel1 indicates that the deletion arose through a type 1 (patient 51.01) or type 2 (patients 16.01, 120.01, 35.01, 54.01, and 60.01) interchromosomal crossing-over event, respectively (fig. 7_b_). The brackets encompass the inverted interval with the arrows indicating the position of the WBStel1 marker in chromosomes with an inversion (.11: father, .01: patient, .02/.03: unaffected sibs, and .12: mother).
In four (1.01, 3.01, 19.01, and 80.01) of seven patients without the inversion, the deletion arose through an intrachromosomal event, as determined by the absence of recombination between centromeric and telomeric markers, whereas, in three cases (7.01, 113.01, and 121.01), it was due to an interchromosomal exchange (fig. 8_a_).
On the other hand, haplotype data demonstrated crossing-over between centromeric and telomeric markers in all six patients with an inferred inversion in the deleted chromosome (16.01, 35.01, 51.01, 54.01, 60.01, and 120.01), suggesting that the deletion derived from an interchromosomal recombination event (fig. 8_b_). These results indicate that, in the progenitors heterozygous for the inversion, deletions arise from crossover events between both chromosome 7 homologues during meiosis I. On the basis of our model, there are two specific sites of nonallelic pairing where unequal crossing-over may lead to the WBS chromosome in carriers of paracentric inversions: between block Bt of the chromosome with the inversion and block Bm of the normal chromosome (type 1) and between block Bm of the chromosome with the inversion and block Bt of the normal chromosome (type 2) (fig. 7_b_). Genotypes at the WBStel1 locus allowed us to infer the type of unequal crossing-over that led to the WBS chromosome. No recombination between centromeric markers and WBStel1 was observed in five cases (16.01, 35.01, 54.01, 60.01, and 120.01), indicating a type 2 interchromosomal exchange that resulted in a WBS chromosome harboring a small inversion of the region between medial and telomeric LCRs. One case (51.01) showed recombination between centromeric markers and WBStel1, indicating a type 1 rearrangement resulting in a WBS chromosome without additional inversion of single-copy sequences.
Search for Sequence Features that Promote Homologous Recombination
We searched for sequence elements that could be responsible for the preferential use of block B as the substrate for homologous recombination and the clustering of deletion breakpoints in specific block B regions in patients without the inversion (fig. 3_b_). Blocks Bc and Bm show an overall sequence identity of 99.6% with no major gaps, whereas blocks Ac and Am display a lower degree of identity (98.2%) including two large deletions of 15 and 26 kb in block Am. The total percentage of repetitive elements in block B, calculated by RepeatMasker, was estimated to encompass 49.7% of the sequence, which is significantly higher than the average 34% predicted for DNA with similar GC content (Smit 1996). Short interspersed nuclear elements (SINEs) account for 41.1% of the sequence and show a quite uniform distribution along the block (data not shown). On the basis of these facts, although a high density of Alu elements is thought to contribute to a high level of unequal homologous recombination (Batzer and Deininger 2002), they do not appear to be related to the clustering of the deletion breakpoints at the beginning of the block B. In contrast, the distribution of the sequence differences between blocks Bc and Bm is asymmetric, with 99.8% identity in the first half (50 kb) and 99.4% in the second half. Therefore, there is a correlation between higher degree of sequence identity and higher frequency of aberrant recombination leading to the deletions. In addition, 7–12 of 19 deletion breakpoints cluster in a 12-kb region between exon 16 and intron 21 of the GTF2I gene and GTF2IP1 pseudogene (fig. 3_b_) and a 3.4-kb de novo junction fragment was amplified by PCR in 4–8 cases (fig. 5). Interestingly, a 1.1-kb region presenting 83% identity to the prohibitin cDNA (PHB [MIM 176705]) is located in this interval, within intron 19 of the GTF2I gene and GTF2IP1/GTF2IP2 pseudogenes but in the opposite transcriptional orientation (fig. 5_a_). The lack of an ORF in this sequence suggests that it corresponds to a processed pseudogene generated by retrotransposition of a prohibitin mRNA. At least two ESTs with identical sequence obtained from fetal brain and stomach cancer have been identified, indicating that the prohibitin pseudogenes may be transcribed in some tissues. No other obvious recombination-promoting sequences found in association with recurrent genomic mutations were identified in the region.
Discussion
Recurrent Deletion Events between Specific Blocks of 7q11.23 LCRs
We have dissected the molecular mechanisms leading to the genomic mutations that occur in the 7q11.23 region by studying 74 patients with typical WBS. As previously suggested, WBS deletions arise as a consequence of misalignment mediated by highly homologous LCRs that flank the critical region followed by unequal crossing-over. On the basis of the organization and orientation of the LCR blocks along with some experimental data, it was previously predicted that WBS deletions must arise by recombination between blocks located in the same orientation in the centromeric and medial location, either block B or block A (Peoples et al. 2000; Valero et al. 2000). We have shown here that most patients with WBS (95%) exhibit a 1.55-Mb deletion caused by unequal crossing-over between the centromeric (usually Bc, but Bt in the case of an inversion) and medial block B copies. A larger deletion (∼1.84 Mb) mediated by aberrant recombination between blocks Ac and Am accounts for only 5% of deletion cases (fig. 1). Preferential implication of specific LCRs has also been reported in other genomic disorders, such as DiGeorge/velocardiofacial syndromes (DGS [MIM 188400]/VCFS [MIM 192430]) affecting 22q11.2 (Shaikh et al. 2001; McDermid and Morrow 2002), Prader-Willi and Angelman syndromes (PWS [MIM 176270]/AS [MIM 105830]) on 15q11-q13 (Amos-Landgraf et al. 1999; Christian et al. 1999), and Smith-Magenis syndrome (SMS [MIM 182290]) on 17p11.2 (Chen et al. 1997). In the 7q11.23 rearrangements causing WBS, two main factors might favor the involvement of block B copies: (1) the degree of sequence homology is significantly higher between blocks Bc and Bm (99.6% overall identity with no large gaps) than between blocks Ac and Am (98.2% identity and two large gaps corresponding to deletions of 15 and 26 kb in Am); and (2) the length of the genomic interval between blocks Bc and Bm (1.55 Mb) is shorter than that between blocks Ac and Am (∼1.84 Mb).
Uneven Distribution of the Sites of Chromosome Breakage in the WBS Deletions
We have also addressed the question whether deletion breakpoints in the recombinant block B localize to specific sites or whether they are widely dispersed within the extent of the block. A single 557-bp region located close to a Mariner transposase sequence within a 27-kb LCR is the recombination hotspot associated with the Charcot-Marie-Tooth disease type 1A (CMT1A [MIM 118220]) and hereditary neuropathy with liability to pressure palsies (HNPP [MIM 162500]) rearrangements on chromosome 17p11.2 (Kiyosawa et al. 1995; Lopes et al. 1996; Reiter et al. 1998). Another recombination hotspot exists in a 2-kb fragment containing a chi-like sequence located within each of the flanking repeats that mediate microdeletions in some patients with neurofibromatosis type 1 (NF1 [MIM 162200]) (López-Correa et al. 2001). However, the precise sites of chromosome breakage and strand exchange in other genomic disorders with larger and more complex LCRs are not known. The structural complexity, copy number, and high homology of the LCRs in 7q11.23 has also hampered the precise definition of WBS deletion breakpoints. Crossovers that lead to the WBS deletion cannot be detected by standard Southern analysis, since the recombinant restriction fragments would be identical in size to those in the original blocks because of the high degree of sequence identity. Long-range restriction (pulsed field gel electrophoresis) approaches have only succeeded when targeting restriction sites located outside the LCRs (Pérez Jurado et al. 1998; Peoples et al. 2000). We attempted an alternative approach to define deletion breakpoints using several SSNs throughout block B that are detected by PCR, restriction analysis, and comparison of band intensities. The results obtained by this method allowed an accurate estimation of block Bc/Bm/Bt ratios in patients, as demonstrated by the mean values and standard deviations (table 2), and were consistent in each patient because a single region of either Bc-Bm or Bt-Bm transition was detected (with the only exception being patient 70.01). Moreover, the method was validated by the finding of a de novo Bc-Bm junction fragment in some patients in the region pinpointed by the SSN analyses. A key point in the PCR-based SSN assays was the analysis of patients’ progenitors in the same experiment to use them as reference values and also to identify putative polymorphisms that could lead to misinterpretations.
SSN analyses have enabled us to refine, within intervals of ∼5–30 kb, the region of Bc/Bm exchange in 19 patients with the common 1.55-Mb deletion (fig. 3_b_). Although the exact site of chromosome breakage and strand exchange could not be further narrowed down in many cases, because of the high sequence homology, 84% of them (16 of 19 cases) are located at the proximal half of the block that harbors the GTF2I/GTF2IP1 and NCF1/NCF1P1 genes. Moreover, in 7–12 of these 19 patients (37%–63%), the deletion breakpoints cluster in a 12-kb region between exon 16 and intron 21 of GTF2I/GTF2IP1 gene, which represents only an 11.4% of the total sequence of block B.
Therefore, although breakpoints can occur at any position, they show an uneven distribution with a tendency to cluster in the proximal half of the block Bc/Bm, where there is an apparent hotspot narrowed to a 3.4–12-kb fragment. Interestingly, blocks Bc and Bm show a higher percentage of identity (99.8%) at the first half compared with the second half (99.4%). In addition, five of the seven existing stretches of >2 kb of continuous identity are located in the first half of the block, and two of them in the 12-kb hotspot region. The length of these regions of uninterrupted identity exceeds by far the 200–300 bp of perfect sequence identity that constitutes the minimum efficient processing segment (MEPS) required by the cellular machinery for strand exchange during somatic recombination in mammalian cells (Waldman and Liskay 1988). Direct sequencing of recombination products has suggested that a similar or slightly greater length of complete identity (300–500 bp) may be required during unequal exchanges in meiosis (Vnencak-Johns and Phillips 1990; Reiter et al. 1998). Therefore, it is clear that unequal crossing-over leading to the 1.55-Mb WBS deletion preferentially occurs in regions of extremely high sequence identity. In addition, active transcription of the GTF2I and GTF2IP1 genes in all tissues examined, including testes and blastocysts (Pérez Jurado et al. 1998; Wang et al. 1998), suggests that both copies may be transcribed in germ cells and thus could also facilitate recombination by means of opening the chromatin structure. The processed prohibitin pseudogene located within the 3.4–12-kb hotspot region appears also to be transcribed, which might further facilitate recombination. No other sequence elements thought to promote homologous recombination, such as transposase-like elements or AT-rich palindromic sequences were found in the entire region. A significant sex bias (_P_=.03) was observed in favor of paternal origin of the deletions among those cases with breakpoints located at the hotspot region. It will be interesting to determine whether differential transcription of the GTF2I/GTF2IP1 genes and/or the prohibitin pseudogenes during spermatogenesis and oogenesis could be responsible for the observed bias.
Inversions of the WBS Region
Genomic polymorphisms in the organization of the WBS region and flanking LCRs could predispose some individuals to the genomic rearrangements associated with WBS. Recent data demonstrated the existence of an inversion of the whole WBS interval in approximately one-third of the progenitors transmitting the WBS chromosome (Osborne et al. 2001). By SSN assays that distinguish block Bt versus Bc and Bm, we predict that 28% of our patients (21 of 74) bear the inversion in the rearranged chromosome. A definitive proof that the progenitor transmitting the rearranged chromosome carries an inversion was obtained, by FISH in all four cases analyzed and by haplotype data in five families. Interestingly, the genomic organization of centromeric and telomeric LCRs consists of inverted repeat blocks (C, A, and B) flanking a 1.79-Mb region that includes the WBS deleted interval. This genomic structure may allow flipping of the DNA fragment in between, thus resulting in an inverted orientation of the whole region with preservation of all genes involved. As postulated in figure 7_a,_ the crossing-over leading to such inversion might occur between centromeric and telomeric blocks A, B, or C. Therefore, the size of the inverted interval is larger than previously estimated (Osborne et al. 2001). It encompasses a genomic fragment ranging from 1.79 (recombination at the end of block B) to 2.56 Mb (recombination at the beginning of block C). The inversion breakpoints always lie externally to the WBS deletion and do not disrupt any of the genes commonly deleted in patients with WBS, which is in agreement with the absence of abnormal phenotype in all inversion carriers studied by us.
On the basis of the current knowledge about the genomic structure of the 7q11.23 region, additional genomic rearrangements leading to either polymorphic variation or disease can be predicted to occur, such as deletions or duplications of some LCR blocks, reciprocal duplications of the entire WBS region, or smaller inversions between medial and telomeric segmental duplications.
Inter- and Intrachromosomal Events: Precise Interchromosomal Rearrangements in Inversion Carriers
The crossover events that led to the deletion in progenitors carrying the inversion polymorphism are all located at the end of block B, within the last 38 kb that are absent in block Bc. This finding is due to the fact that the final 38 kb of blocks Bt and Bm are most likely the only regions of paralogous misalignment. During meiosis I in inversion carriers, an “active search” for homology would force both the inverted and normal chromosomes 7 into forming a structural microloop in the middle of the inverted segment (fig. 7_b_). Based on this model, complete sequence pairing could be achieved even in the presence of a paracentric inversion in one of the homologues. Even without complete pairing, homologous sequences located in the same direction within the loops, such as the 38 kb of blocks Bt and Bm, would tend to establish nonallelic pairing. In any case, only meiotic crossovers scored as type 1 or 2 in figure 7_b_ would result in a WBS chromosome. This region harbors the promotor and first exons of GTF2IRD2, another ubiquitously expressed gene (O. de Luis and L. A. Pérez Jurado, unpublished results) that might also favor local recombination events. The products of an uneven number of recombination events at any other position within the loops, in case there is complete pairing, would be expected to result in acentric fragments and dicentric anaphase bridges.
Our haplotype data (fig. 8) demonstrate that WBS deletions in patients bearing the inversion are indeed the result of unequal crossing-over between chromosome 7 homologues during meiosis I. Moreover, the genotypes at WBStel1 indicate that the strand exchange occurred between block Bt of the chromosome with the inversion and block Bm of the normal chromosome in one case (type 1), whereas it happened the other way around, between block Bm of the inverted chromosome and block Bt of the normal chromosome, in five cases (type 2). Regarding the parental origin of the deletion, no sex bias was observed in inversion carriers.
On the other hand, deletions arose through either interchromosomal (three of seven) or intrachromosomal (four of seven) events in patients without the inversion, as determined by the presence or absence of recombination between centromeric and telomeric markers. When patients with and without the inversion are taken into account, our data are in agreement with previous family studies of de novo WBS deletions, which report that approximately two-thirds of the cases are mediated by interchromosomal rearrangements, whereas one-third occur intrachromosomally, regardless of the parental origin of the deletion (Urban et al. 1996; Baumer et al. 1998).
Conversion Events between LCRs
Many of the block B theoretically defined site-specific changes (SSNs or _cis_-morphisms) appeared to have a certain degree of non–site specificity or _trans_-morphism (confusing variation at the two alleles of one or several loci) (Lupski 2003) among both unaffected and affected individuals, leading to some ambiguity in the results. Despite that, consistent results allowing the identification of a single deletion and inversion breakpoints were obtained in all patients except one (patient 70.01, fig. 3_c_). The single contradictory result at SSN2 in this patient might represent either the presence of _trans_-morphisms at more than one locus in both parents or a de novo conversion event in the patient. It is worth mentioning that coincidental _trans_-morphisms were frequently found in clusters encompassing several kilobases in some progenitors, most likely in the same allele. This loss of site specificity suggests the existence of sequence exchange among LCR copies, possibly through gene-conversion events, and points to a mechanism for preserving gene function and maintaining the high sequence identity among the LCRs. Interestingly, SSN 4 detects a GT insertion (NCF1) or deletion (NCF1P1 and NCF1P2) at the beginning of exon 2, which is the only relevant change that predicts a lack of function in the NCF1P1 and NCF1P2 pseudogenes (Görlach et al. 1997). Its analysis revealed that several individuals (9%) bear more than two copies of the putatively functional NCF1 gene variant (fig. 4_a_). This phenomena may have implications for the diagnosis of both patients and carriers of an autosomal recessive form of chronic granulomatous disease (CGD), which is caused, in 90% of cases, by the deletion of this dinucleotide GT in both copies of the NCF1 gene (Casimir et al. 1991; Görlach et al. 1997; Heyworth et al. 2002). The autosomal recessive inheritance of this disease implies that all copies (centromeric, medial, and telomeric) would have to be mutated.
Diagnostic Considerations
We have shown that the genotyping of SSNs and multiple-copy STRs is well suited as a supplement to conventional diagnostic techniques. Three PCR-based assays that are informative in most cases (BASTR1, BBSTR1, and SSN 11) provide quite precise diagnostic information about the size of the deletion and the putative presence of genomic variants on 7q11.23 in the progenitors. Genotyping the multiple-copy STRs within blocks A and B permits to easily define the blocks involved in the rearrangements in patients with WBS and then to infer the size (either 1.55 or 1.84 Mb) of the deletion. In addition, the dosage analysis of SSN 11 at the edge of the alignment between blocks Bc and Bm in individuals with WBS allows to predict if the deletion has occurred in a progenitor heterozygous for the 7q11.23 paracentric inversion.
Implications for Genetic Counseling
WBS is almost always sporadic, with a few cases of documented vertical transmission and concordant MZ twins (Morris et al. 1993; Castorina et al. 1997). Although two sets of second cousins with WBS have been reported, the finding of deletions in unrelated chromosomes suggested independent mutational events in both sets (Pérez Jurado et al. 1996). A single occasion of recurrent WBS in two siblings with deletions on the maternally inherited haploidentical chromosome suggested a premeiotic intrachromosomal event responsible for gonadal mosaicism in the mother (Kara-Mostefa et al. 1999). Therefore, the recurrence risk in the sibship of a proband with WBS with unaffected parents is expected to be negligible, close to the population risk (Baumer et al. 1998). However, some considerations derived from our and previous data are worth mentioning:
- 1.
If the deletion is found to occur intrachromosomally, the rare possibility of a premeiotic event with gonadal mosaicism should be considered (Kara-Mostefa et al. 1999). - 2.
Parents heterozygous for an inversion at 7q11.23 may have an increased risk of meiotic misalignment and aberrant recombination leading to gametes with unbalanced rearrangements. This recurrence risk will depend on the likelihood that the specific misalignment followed by unequal recombination at the misaligned region will occur. Since in this region a recombination rate of 1% approximately corresponds to the genome average of 1 Mb, the frequency of recombination in the nonallelic misaligned fragments (two fragments of 38 kb each between blocks Bm and Bt, for a total of 76 kb) would be relatively low, about 10−3. This low recombination rate may explain why the recurrence risk for WBS appears not to be significantly increased in some families despite the finding of these “predisposing” alleles. - 3.
In carriers of the paracentric inversion, the formation of either one or two chromosomal loops encompassing a 1.79-Mb region may occur, leading to complete pairing. If recombination take place within the loops (∼2%), it would lead to either an acentric or dicentric chromosome 7, both most likely nonviable. However, it is still unknown whether there is an slightly increased rate of abortion in families carrying the inversion polymorphism.
In summary, a variety of mutational mechanisms, all mediated by the local segmental duplications, have been found in the 7q11.23 region. Results presented in this paper should contribute to a better understanding of the nature and mechanism of formation of these recurrent genomic mutations. Further studies will focus on an exhaustive search for genotype-phenotype correlations in patients with different deletion breakpoints, with and without the inversion, as well as to determine any potential variation of recurrence risk for WBS depending on the parental alleles. It will also be interesting to define the evolutionary steps that led to the genomic organization of LCRs in 7q11.23 and to establish the actual prevalence of genomic polymorphisms in different populations. Rearrangements mediated by similar mechanisms during mitosis may also be involved in human somatic diseases.
Acknowledgments
We thank the Spanish Williams Syndrome Association, patients and families for their invaluable collaboration and support, Drs. Rodríguez-Criado, Ruiz-Cabello, del Castillo, Toledo and Duque for referring patients, Drs M.A. Pujana, L. Armengol and M. Del Campo for helpful discussions, Roser Corominas for technical support, Drs F. Calafell and J. Rozas for assistance in sequence analysis and Dr. L. Osborne for assistance with FISH. This work was supported by grants from the Spanish Ministries of Health (FIS01/1129, ISCIII Networks C03/07 and X03/07) and Science and Technology (SAF3941/2001), and la Foundation Jerôme Lejèune. L.F.M is a recipient of a FIS studentship.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST search engine, http://www.ncbi.nlm.nih.gov/BLAST/
- ClustalW, http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html
- DNAsp 3.51, http://www.ub.es/dnasp/
- GenBank, http://www.ncbi.nih.gov/GenBank/ (for BACs CTA-269p13 [accession number AC005080], RP11-396k3 [accession number AC006995], RP11-450o3 [accession number AC105418, RP11-483g21] [accession number AC00416], 239c10 [accession number AC004883], RP4-771p04 [accession number AC083884], RP11-813J7 [accession number AC005098], CTA-350l10 [accession number AC124781], RP11-729p19 [accession number AC027219], and RP11-219m8 [accession number AC124781]; prohibitin-related ESTs from fetal brain [accession number AA076811]; and stomach cancer [accession number AW814764])
- Genome Database, http://gdbwww.gdb.org/ for primer information on HSB055XE5 [ID number 609780], D7S489 [ID number 188049], D7S653 [ID number 199574], D7S672 [ID number 199800], D7S1816 [ID number 684408], D7S2518 [ID number 612411], and D7S1870 [ID number 377150])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for WBS, GTF2I, NCF1, ELN, prohibitin cDNA, DGS, VCFS, PWS, AS, SMS, CMT1A, HNPP, and NF1)
- Primer3, http://www-genome.wi.mit.edu/genome_software/other/primer3.html
- RepeatMasker, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65:370–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3:370–379 [DOI] [PubMed] [Google Scholar]
- Baumer A, Dutly F, Balmer D, Riegel M, Tükel T, Krajewska-Walasek M, Schinzel AA (1998) High level of unequal meiotic crossovers at the origin of the 22q11.2 and 7q11.23 deletions. Hum Mol Genet 7:887–984 [DOI] [PubMed] [Google Scholar]
- Bellugi U, Lichtenberger L, Jones W, Lai Z, St George M (2000) The neurocognitive profile of Williams Syndrome: a complex pattern of strengths and weaknesses. J Cogn Neurosci Suppl 12:7–29 [DOI] [PubMed] [Google Scholar]
- Burn J (1986) Williams syndrome. J Med Genet 23:389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimir CM, Bu-Ghanim HN, Rodaway AR, Bentley DL, Rowe P, Segal AW (1991) Autosomal recessive chronic granulomatous disease caused by deletion at a dinucleotide repeat. Proc Natl Acad Sci USA 88:2753–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castorina P, Selicorni A, Bedeschi F, Dalpra L, Larizza L (1997) Genotype-phenotype correlation in two sets of monozygotic twins with Williams syndrome. Am J Med Genet 69:107–111 [DOI] [PubMed] [Google Scholar]
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH (1999) Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet 8:1025–1037 [DOI] [PubMed] [Google Scholar]
- DeSilva U, Elnitski L, Idol JR, Doyle JL, Gan W, Thomas JW, Schwartz S, Dietrich NL, Beckstrom-Sternberg SM, McDowell JC, Blakesley RW, Bouffard GG, Thomas PJ, Touchman JW, Miller W, Green ED (2002) Generation and comparative analysis of approximately 3.3 Mb of mouse genomic sequence orthologous to the region of human chromosome 7q11.23 implicated in Williams syndrome. Genome Res 12:3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel BS, Shaikh TH (2001) Segmental duplications: an expanding role in genomic instability and disease. Nat Rev Genet 2:791–800 [DOI] [PubMed] [Google Scholar]
- Francke U (1999) Williams syndrome: genes and mechanisms. Hum Mol Genet 8:1947–1954 [DOI] [PubMed] [Google Scholar]
- Görlach A, Lee PL, Roesler J, Hopkins PJ, Christensen B, Green ED, Chanock SJ, Curnutte JT (1997) A p47-phox pseudogene carries the most common mutation causing p47-phox- deficient chronic granulomatous disease. J Clin Invest 100:1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg F (1990) Williams syndrome professional symposium. Am J Med Genet 6:85–88 [Google Scholar]
- Heyworth PG, Noack D, Cross AR (2002) Identification of a novel NCF-1 (p47-phox) pseudogene not containing the signature GT deletion: significance for A47 degrees chronic granulomatous disease carrier detection. Blood 100:1845–1851 [DOI] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD (2000) Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res 10:597–610 [DOI] [PubMed] [Google Scholar]
- Kara-Mostefa A, Raoul O, Lyonnet S, Amiel J, Munnich A, Vekemans M, Magnier S, Ossareh B, Bonnefont JP (1999) Recurrent Williams-Beuren syndrome in a sibship suggestive of maternal germ-line mosaicism. Am J Hum Genet 64:1475–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa H, Lensch MW, Chance PF (1995) Analysis of the CMT1A-REP repeat: mapping crossover breakpoints in CMT1A and HNPP. Hum Mol Genet 4:2327–2334 [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Chen XN, Hirota H, Lai Z, Bellugi U, Burian D, Roe B, Matsuoka R (2000) Genome structure and cognitive map of Williams syndrome. J Cogn Neurosci Suppl 12:89–107 [DOI] [PubMed] [Google Scholar]
- Lopes J, LeGuern E, Gouider R, Tardieu S, Abbas N, Birouk N, Gugenheim M, Bouche P, Agid Y, Brice A (1996) Recombination hot spot in a 3.2-kb region of the Charcot-Marie-Tooth type 1A repeat sequences: new tools for molecular diagnosis of hereditary neuropathy with liability to pressure palsies and of Charcot-Marie-Tooth type 1A. Am J Hum Genet 58:1223–1230 [PMC free article] [PubMed] [Google Scholar]
- López-Correa C, Dorschner M, Brems H, Lázaro C, Clementi M, Upadhyaya M, Dooijes D, Moog U, Kehrer-Sawatzki H, Rutkowski JL, Fryns JP, Marynen P, Stephens K, Legius E (2001) Recombination hotspot in NF1 microdeletion patients. Hum Mol Genet 10:1387–1392 [DOI] [PubMed] [Google Scholar]
- Lupski JR (2003) 2002 Curt Stern Award Address. Genomic disorders: recombination-based disease resulting from genomic architecture. Am J Hum Genet 72:246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magano LF, Bayés M, Flores R, Pérez Jurado LA (2001) Towards a complete transcription map of the Williams-Beuren deletion region. Eur J Hum Genet Suppl 9:244 [Google Scholar]
- Mazzarella R, Schlessinger D (1998) Pathological consequences of sequence duplications in the human genome. Genome Res 8:1007–1021 [DOI] [PubMed] [Google Scholar]
- McDermid HE, Morrow BE (2002) Genomic disorders on 22q11. Am J Hum Genet 70:1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merla G, Ucla C, Guipponi M, Reymond A (2002) Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum Genet 110:429–438 [DOI] [PubMed] [Google Scholar]
- Mervis CB, Klein-Tasman BP (2000) Williams syndrome: cognition, personality, and adaptive behavior. Ment Retard Dev Disabil Res Rev 6:148–158 [DOI] [PubMed] [Google Scholar]
- Morris CA, Demsey SA, Leonard CO, Dilts C, Blackburn BL (1988) Natural history of Williams syndrome: physical characteristics. J Pediatr 113:318–326 [DOI] [PubMed] [Google Scholar]
- Morris CA, Thomas IT, Greenberg F (1993) Williams syndrome: autosomal dominant inheritance. Am J Med Genet 47:478–481 [DOI] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW (2001) A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 29:321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples R, Franke Y, Wang YK, Pérez-Jurado L, Paperna T, Cisco M, Francke U (2000) A physical map, including a BAC/PAC clone contig, of the Williams-Beuren syndrome deletion region at 7q11.23. Am J Hum Genet 66:47–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Jurado LA, Peoples R, Kaplan P, Hamel BC, Francke U (1996) Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am J Hum Genet 59:781–792 [PMC free article] [PubMed] [Google Scholar]
- Pérez Jurado LA, Wang YK, Peoples R, Coloma A, Cruces J, Francke U (1998) A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Hum Mol Genet 7:325–334 [DOI] [PubMed] [Google Scholar]
- Pérez Jurado LA (2003) Williams-Beuren syndrome: a model of recurrent genomic mutation. Horm Res 59 Suppl 1:106–113 [DOI] [PubMed] [Google Scholar]
- Reiter LT, Hastings PJ, Nelis E, De Jonghe P, Van Broeckhoven C, Lupski JR (1998) Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am J Hum Genet 62:1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WP, Waslynka J, Bernasconi F, Wang M, Clark S, Kotzot D, Schinzel A (1996) Delineation of 7q11.2 deletions associated with Williams-Beuren syndrome and mapping of a repetitive sequence to within and to either side of the common deletion. Genomics 34:17–23 [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 2:174–175 [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS (2000) Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet 9:489–501 [DOI] [PubMed] [Google Scholar]
- Smit AF (1996) The origin of interspersed repeats in the human genome. Curr Opin Genet Dev 6:743–748 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 [DOI] [PubMed] [Google Scholar]
- Stromme P, Bjornstad PG, Ramstad K. (2002) Prevalence estimation of Williams syndrome. J Child Neurol 17:269–271 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromp G, Christiano A, Goldstein N, Indik Z, Boyd C, Rosenbloom J, Deak S, Prockop D, Kuivaniemi H (1991) A to G polymorphism in ELN gene. Nucleic Acids Res 19:4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban Z, Helms C, Fekete G, Csiszar K, Bonnet D, Munnich A, Donis-Keller H, Boyd CD (1996) 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover. Am J Hum Genet 59:958–962 [PMC free article] [PubMed] [Google Scholar]
- Valero MC, de Luis O, Cruces J, Pérez Jurado LA (2000) Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: the low-copy repeats that flank the Williams-Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s). Genomics 69:1–13 [DOI] [PubMed] [Google Scholar]
- Vnencak-Jones CL, Phillips JA 3rd (1990) Hot spots for growth hormone gene deletions in homologous regions outside of Alu repeats. Science 250:1745–1748 [DOI] [PubMed] [Google Scholar]
- Waldman AS, Liskay RM (1988) Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol Cell Biol 8:5350–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YK, Pérez Jurado LA, Francke U (1998) A mouse single-copy gene, Gtf2i, the homolog of human GTF2I, that is duplicated in the Williams-Beuren syndrome deletion region. Genomics 48:163–170 [DOI] [PubMed] [Google Scholar]