Human Immunodeficiency Virus Mutations during the First Month of Infection Are Preferentially Found in Known Cytotoxic T-Lymphocyte Epitopes (original) (raw)
Abstract
The full protein coding region of human immunodeficiency virus (HIV) genomes were sequenced using plasma collected from nine African-Americans prior to seroconversion and 7 to 28 days later. HIV mutations emerged in seven of these subjects at a genomewide rate of 2% per year. The location of nonsynonymous (NS) HIV mutations within these subjects was compared to their potential HLA-A and B types restricted CTL epitopes reported in the Los Alamos National Laboratory HIV immunology database. A statistically significant (P < 0.005) number of the early NS mutations (13.5%) were found within previously reported CTL epitopes. A virus sequencing and reported CTL epitopes database analysis therefore support a model where a significant proportion of very early nonsynonymous HIV mutations are selected by CTL.
Cytotoxic T-lymphocytes (CTL) play an important role in controlling viremia during primary human immunodeficiency virus (HIV) infection (6, 29). CTL recognize viral peptides presented in a complex with HLA class I molecules at the surface of infected cells, inducing T-cell activation and clonal expansions followed by lysis of infected target cells (37, 40). The resulting selection pressure on the replicating virus can lead to the emergence of CTL escape variants (7, 9, 18-20, 22, 25, 26, 36). CTL targeted epitopes have been reported in every HIV protein in the Los Alamos Database (28). The breadth of HIV-1-specific CTL response increases during chronic versus primary infection (4, 8, 13, 19). CTL responses appear to favor epitopes within regulatory and accessory proteins during primary infection, whereas later responses appear to preferentially recognize structural proteins (1, 8, 9).
CTL escape has been powerfully illustrated by the identification of an immunodominant Mamu-A*01-restricted Tat-specific CTL response rapidly selecting for viral escape variants during acute simian immunodeficiency virus (SIV) infection (3, 32). In the SIV model system, peptide reagents of the infecting SIV strain have allowed all CTL epitopes restricted by various HLA class I allele to be identified. As many as 64% of NS mutations selected over the entire course of SIV infection outside of the envelope gene have been located with known CTL epitopes (33). In HIV-1-infected subjects, viral mutations preventing CTL recognition were described in immunodominant epitopes and were correlated with a steady rise in viral load and disease progression (18, 25).
On a population basis, Moore et al. compared HIV protease and reverse transcriptase sequences of chronically infected patients to their presumably ancestral subtype consensus sequence and showed that consensus deviating sequences were overrepresented in known HLA-restricted CTL epitopes (31). When phylogenetically closely related but epidemiologically unlinked HIV genomes were analyzed positive selection in potential CTL target sites was also detected (35). Particular HLA types have also been associated with susceptibility or resistance to HIV infection (14, 30, 38) as well as with slow or rapid disease progression (10, 11, 16, 17, 21, 23, 24, 39).
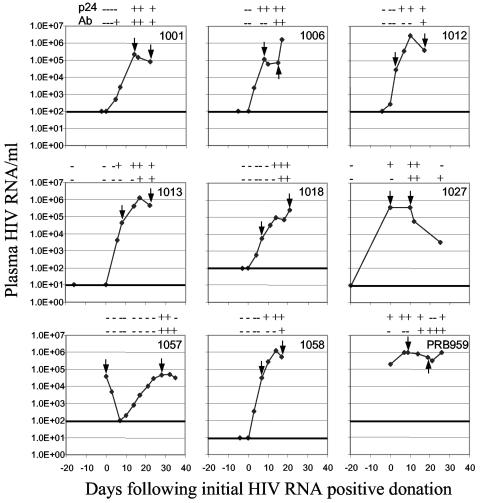
To directly identify HIV sequence changes and characterize HIV-1 regions undergoing possible selection very early following infection, we sequenced plasma HIV genomes of nine African-American plasma donors at two time points (Fig. 1). Subjects reported no symptoms of acute infection at the time of plasma collection and were not febrile. The initial donations were p24 antigen negative, anti-HIV-1 antibody negative, and HIV-1 RNA negative (using the National Genetics Institute UltraQual HIV-1 reverse transcription-PCR assay with a detection limit of five HIV-1 genome copies/ml) except for subjects 1057 and PRB959, who had an initial plasma viremia of 3.9 × 104 and 2 × 105 HIV-1 RNA copies/ml, respectively (Fig. 1). Longitudinally collected plasma samples were then quantified with the NGI SuperQuant HIV-1 reverse transcription-PCR assay. Anti-HIV antibodies were detected using the HIV-1 enzyme immunoassay (Abbott, Abbott Park, IL) and p24 antigenemia was detected with the Coulter HIV-1 p24 antigen assay (Beckman Coulter, Brea, CA) (Fig. 1). Antiretroviral therapies were not initiated during the period of study. Because donations were of plasma only, no peripheral blood mononuclear cells were available for functional analyses of the CTL response. These studies were approved by the University of California, San Francisco Committee on Human Research.
FIG. 1.
Viral load, p24 antigenemia, and anti-HIV-1 serology of acute infections. Nine African-Americans were characterized as undergoing primary infection. Day 0: first plasma sample with detectable viral RNA using the UltraQual assay or the SuperQuant assay (threshold of 5 and 100 RNA copies/ml, respectively). -/+: negative and positive reactivity, respectively, for p24 and for anti-HIV-1 antibodies.
Seroconversion was observed during the second or third week following initial RNA detection for seven subjects, during week 1 for 1001, and during the fourth week for 1057. The initial plasma donations of subject 1057 and PRB959 were already viremic but subsequent seroconversion confirmed primary infection status.
All open reading frames of plasma HIV-1 RNA from two time points separated by 7 to 28 days (arrows in Fig. 1) were sequenced. For genome amplification plasma HIV-1 RNA was purified, reverse transcribed, and subjected to nested PCR (5). The resulting half-genome amplicons were directly sequenced. The PCR and sequencing protocols have been described in detail (5). Sequences were manually edited using EditView, assembled into a single contig, and aligned using the Seqman and Megalign programs, respectively (DNAstar Inc., Madison, WI). The wide variation in the location of HIV mutations evolving within each subject is shown in Fig. 2.
FIG. 2.
Genomewide distribution of early HIV mutations. The vertical bar represent one nucleotide substitution.
Both complete and partial nucleotide sequence changes were observed by direct amplicon sequencing. Partial nucleotide changes were detected when either time point showed a mixed base consisting of two overlapping sequencing electrophoregram peaks (where the minority peak reached ≥25% of the dominant peak) while the other time point showed only a single base peak. These mixed bases are recorded using the IUB nomenclature in the deposited sequences (GenBank accession numbers AY331282 to AY331297, AY332236, and AY332237). Mixed bases, commonly seen when directly sequencing HIV amplicons, reflect polymorphisms in the viral quasispecies that are not yet fixed in the entire viral population. The short time interval between sampled plasma pairs is likely responsible for the frequent detection of such partial nucleotide changes. Partial mutations were analyzed in the same way as full nucleotide substitutions (i.e., positions changing from single into mixed residues, and vice versa, were counted as one mutation). No codon contained more than a single mixed base, and therefore all codons with a mixed base could be translated into only one or two amino acids.
No mutations were observed in subjects 1006 and 1027, who were sampled only 7 and 12 days apart (Fig. 1). For the seven other genome pairs, a total of 107 nucleotide substitutions were detected, resulting in intrasubject mutation frequencies of 0.08% to 0.37%. The accumulation of nucleotide substitutions occurred at an average yearly rate of 2.05%. The genomewide dN/dS ratios, calculated using the SNAP program from the HIV sequence database [www.hiv.lanl.gov (27)], ranged from 0.249 to 0.87, reflecting overall purifying selection against amino acid changes. The gene distribution of NS mutations varied widely between donors. Overall the highest number of NS mutations was located in env (n = 19), pol (n = 15), gag (n = 14), nef (n = 6), vif (n = 3), and vpr (n = 3). In these subjects no NS mutations were detected in the early expressed accessory genes tat, rev, and vpu. When adjusted for gene length, the highest density of NS mutations was found with the vpr, nef, and gag genes (The high mutation rate for the short open reading frame of vpr was the result of a single NS mutation in three subjects [1013, 1018, and 1057] located within Vpr amino acids 34 to 40).
The total number of nucleotide substitutions was correlated with both the time interval between the two sequenced time points and the time interval between the initial detection of HIV RNA and the second sequenced time point (P = 0.05, _r_2 = 0.32 and P = 0.02, _r_2 = 0.48, respectively). The number of nucleotide substitutions was also positively correlated with changes in plasma viremia between the two sequenced time points (P = 0.04, _r_2 = 0.37) indicating that increasing viral loads were associated with a higher number of mutations. Similar trends were observed using NS mutations only (P = 0.09, _r_2 = 0.24; P = 0.006, _r_2 = 0.62; and P = 0.06, _r_2 = 0.3, respectively).
HIV-1 evolution is influenced by numerous virus-host interactions including changes in antibody and CTL selection pressures upon infection of a new host. In order to evaluate the influence of the HLA class I types on the location of early NS mutations, high-resolution HLA-A and -B types were determined by PCR and sequencing of human DNA purified from plasma samples. DNA was isolated in a clean room designed for pre-PCR manipulation using the QIAamp DNA mini kit (QIAGEN, Valencia, CA). Locus-specific PCR amplifications were performed for exons 2 and 3; exon 4 was amplified as required to resolve ambiguities (Perkin Elmer Life Sciences, Shelton, CT) (L. A. Baxter-Lowe, unpublished). Ambiguities involving polymorphism in the leader peptide (HLA-A*7401/02 and B*2705/13) were not resolved, because these mutations are not predicted to alter the peptide binding characteristics of the protein product. These are listed in Table 1 as HLA-A*7401 and HLA-B*2705. For subject 1013, there was insufficient DNA to resolve the exon 4 ambiguity of HLA-A*2301/07N. This has been listed in Table 1 as HLA-A*2301 as HLA-A*2307N is extremely rare. Eighteen different HLA-A and -B types were detected among the seven plasma donors whose HIV evolved between the sampled time points (Table 1). Many of these types are frequently observed in African-Americans (10).
TABLE 1.
High-resolution HLA class I typing
| Subject | HLA-A | HLA-B |
|---|---|---|
| 1001 | 0202 0301 | 0702 1516 |
| 1006 | 2601 6802 | 1510 1510 |
| 1012 | 6601 6802 | 4201 5301 |
| 1013 | 2301 3001 | 4201 5301 |
| 1018 | 6801 7401 | 1503 5802 |
| 1027 | 3002 3201 | 2703 2705 |
| 1057 | 0301 2301 | 0702 1503 |
| 1058 | 2301 6801 | 5801 5802 |
| PRB959 | 0308 3001 | 0705 4501 |
When published CTL epitopes for these 18 HLA class I molecules were sought in the existing literature only 13 of 18 HLA types had any CTL epitopes described (Los Alamos HIV immunology database, January 2005). A total of 62 HIV CTL epitopes with a range of 1 to 16 underlined epitopes per HLA type were identified. Eight of the 59 HIV NS mutations (13.5%) were located within reported HLA class I restricted epitopes (Table 2). These eight NS mutations, found in three subjects, were within epitopes restricted by one or in two cases by two of their host's class I molecules (Table 2).
TABLE 2.
Mutations located in reported CTL epitopes
| Subject no. | Position (HxB2)a | Defined CTL epitope sequence | Subject's HIV sequenceb | Mutation | Restricting HLA |
|---|---|---|---|---|---|
| 1001 | Env 211 | VSFEPIPPHYCA | VSFKPIPIHYCA | K to E | A2 serotype |
| Env 379 (381) | SFNCGGEFF | SFNCGGEFF | E to G | B1516 | |
| KNCGGEFFYCNS | FNCGGEFFYCNT | E to G | A2 serotype | ||
| Env 501 (504) | VKIEPLGVAPTKAKRRVVQR | VKIEPLGVAPTRAKRKVVQR | K to R | A2 serotype | |
| 1057 | p6 39/Gag 492 | YPLTSLRSLF | YPLTSLRSLF | T to A | B7 serotype |
| p6 42/Gag 494 | YPLTSLRSLF | YPLTSLRSLF | R to K | B7 serotype | |
| Vpr 40 | FPRIWLHGL | FPRIWLHGL | H to Y | B0702 | |
| Nef 184 | WRFDSRLAF | WKFDSRLAL | K to E | B1503 | |
| DPEKEVLQWK | DSEGEVLQWK | K to E | B7 serotype | ||
| 1058 | Env 588 | RYLKDQQLL | RYLKDQQLL | K to E | A2301 |
We calculated the probability of 8 out of 59 NS mutations occurring within previously reported CTL epitopes for the seven subjects with HIV mutations. All 62 reported epitopes restricted by their hosts' class I HLA-A and HLA-B molecules were compiled from the database. The total numbers of amino acids included within reported CTL epitopes were calculated individually for each of the seven primary infection subjects and then added up (the same amino acids found in overlapping epitopes within the same subject were counted once). A total of 943 amino acids were found within reported CTL epitopes. The total number of amino acids encoded by HIV in these seven subjects was 22,048 (Table 3). The fraction of virally encoded amino acids within reported CTL epitopes was therefore 943/22,048 (4.3%). The proportion of NS mutations found within these reported CTL epitopes was 8/59 (13%), which was significantly greater than expected by chance, considering the low density (4.3%) of reported CTL epitopes (P < 0.005, test of proportion and Fisher’s exact test). This result indicated that NS mutations evolving within the first month of infection were significantly more likely to be found within than outside a reported CTL epitope.
TABLE 3.
Odds ratios of NS mutations within reported CTL epitopea
| Subject no. | No. of NS mutations | No. of NS mutations in reported CTL epitopes | % of NS mutations in reported CTL epitopes | Total no. of aa in genome | Total no. of aa within reported CTL epitopes | % of total aa within reported CTL epitopes | Odds ratio for NS mutations being within reported CTL epitopes | 95% confidence interval |
|---|---|---|---|---|---|---|---|---|
| 1001 | 7 | 3 | 42.9 | 3,163 | 237 | 7.5 | 5.7 | 2.4, 13.6 |
| 1012 | 5 | 0 | 0 | 3,155 | 134 | 4.25 | 0 | |
| 1013 | 13 | 0 | 0 | 3,131 | 92 | 2.94 | 0 | |
| 1018 | 10 | 0 | 0 | 3,150 | 88 | 2.8 | 0 | |
| 1057 | 11 | 4 | 36.36 | 3,147 | 293 | 9.3 | 3.9 | 1.8, 8.6 |
| 1058 | 8 | 1 | 12.5 | 3,155 | 80 | 2.5 | 4.9 | 0.8, 31.2 |
| 959 | 5 | 0 | 0 | 3,147 | 19 | 0.6 | 0 | |
| Total | 59 | 8 | 13.5 | 22,048 | 943 | 4.3 | 3.1 | 3.0, 11.1 |
We also calculated separately for each subject and for all subjects together the odds ratio of finding an NS mutation in a predicted CTL epitope (Table 3). Odds ratio values in three subjects were greater than one, ranging from 3.9 to 5.7, reflecting the preferential location of NS mutations within reported CTL epitopes. An odds ratio of 3.1 was calculated using all seven subjects with HIV mutations.
The number of CTL epitopes restricted by the HLA class I molecules of these African American plasma donors in the Los Alamos databases is likely to be a large underestimate of potential epitopes, because many of these HLA types have not been studied in detail (i.e., no CTL epitopes were reported for 5 of the 18 HLA types). Furthermore most studies mapping epitopes have done so using only a subset of HIV proteins, and epitopes have typically been identified using reference or subtype consensus strains rather than autologous strains. Mutational escape from CTL can also occur at different steps during peptide processing and HLA binding. Viral protein degradation by the proteasome, transport of peptides into the endoplasmic reticulum, peptide trimming, and peptide loading may all be affected by sequence changes flanking rather than within the final antigenic peptide (2, 12, 15). Finally, some NS mutations linearly distant from the CTL epitope may also be selected as compensatory mutations to reduce the fitness cost imposed by CTL escape mutations (34).
The immature state of the immune response so early following primary infection may also have limited the amount of CTL selection pressure on the replicating quasispecies although in the SIV model system immunodominant epitope escape mutations have been seen within 4 weeks of infection (3, 32). The percentage of early NS mutations reported here that may affect CTL epitopes may therefore be a minimum value. Indeed we noted that the two subjects with the highest number of HLA-A and -B types restricted CTL epitopes in the database (1001 and 1057) were among the three subjects with an overabundance of NS mutations within reported epitopes (Tables 2 and 3). As more CTL epitopes to less frequent HLA-A and -B types are reported, a greater proportion of very early NS mutations may therefore be found within CTL epitopes.
In summary, the location of NS mutations occurring within the first month of acute HIV infection varied greatly among subjects. The influence of the host's class I HLA type was reflected within weeks of infection by the preferential distribution of 13.5% of NS mutations within previously reported CTL epitopes.
Acknowledgments
Support for this study was provided to E.L.D. by the UC Universitywide AIDS Research Program (ID02-SF-075) and NIAID (AI47320 and AI43261).
REFERENCES
- 1.Addo, M. M., M. Altfeld, E. S. Rosenberg, R. L. Eldridge, M. N. Philips, K. Habeeb, A. Khatri, C. Brander, G. K. Robbins, G. P. Mazzara, P. J. Goulder, and B. D. Walker. 2001. The HIV-1 regulatory proteins Tat and Rev. are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc. Natl. Acad. Sci. USA 98**:**1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., M. Altfeld, X. G. Yu, K. M. O'Sullivan, M. Lichterfeld, S. Le Gall, M. John, B. R. Mothe, P. K. Lee, E. T. Kalife, D. E. Cohen, K. A. Freedberg, D. A. Strick, M. N. Johnston, A. Sette, E. S. Rosenberg, S. A. Mallal, P. J. Goulder, C. Brander, and B. D. Walker. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 78**:**7069-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407**:**386-390. [DOI] [PubMed] [Google Scholar]
- 4.Altfeld, M., E. S. Rosenberg, R. Shankarappa, J. S. Mukherjee, F. M. Hecht, R. L. Eldridge, M. M. Addo, S. H. Poon, M. N. Phillips, G. K. Robbins, P. E. Sax, S. Boswell, J. O. Kahn, C. Brander, P. J. Goulder, J. A. Levy, J. I. Mullins, and B. D. Walker. 2001. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J. Exp. Med. 193**:**169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardin, F., B. L. Herring, L. Peddada, and E. L. Delwart. 2003. Primary infection of a male plasma donor with divergent HIV variants from the same source followed by rapid fluctuations in their relative frequency and viral recombination. AIDS Res. Hum. Retroviruses 19**:**1009-1015. [DOI] [PubMed] [Google Scholar]
- 6.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68**:**6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3**:**205-211. [DOI] [PubMed] [Google Scholar]
- 8.Cao, J., J. McNevin, S. Holte, L. Fink, L. Corey, and M. J. McElrath. 2003. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J. Virol. 77**:**6867-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, J., J. McNevin, U. Malhotra, and M. J. McElrath. 2003. Evolution of CD8+ T cell immunity and viral escape following acute HIV-1 infection. J. Immunol. 171**:**3837-3846. [DOI] [PubMed] [Google Scholar]
- 10.Carrington, M., and R. E. Bontrop. 2002. Effects of MHC class I on HIV/SIV disease in primates. AIDS 16(Suppl. 4)**:**S105-114. [DOI] [PubMed] [Google Scholar]
- 11.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283**:**1748-1752. [DOI] [PubMed] [Google Scholar]
- 12.Craiu, A., T. Akopian, A. Goldberg, and K. L. Rock. 1997. Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc. Natl. Acad. Sci. USA 94**:**10850-10855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalod, M., M. Dupuis, J. C. Deschemin, C. Goujard, C. Deveau, L. Meyer, N. Ngo, C. Rouzioux, J. G. Guillet, J. F. Delfraissy, M. Sinet, and A. Venet. 1999. Weak anti-HIV CD8(+) T-cell effector activity in HIV primary infection. J. Clin. Investig. 104**:**1431-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Sorrentino, A. H., K. Marinic, P. Motta, A. Sorrentino, R. Lopez, and E. Illiovich. 2000. HLA class I alleles associated with susceptibility or resistance to human immunodeficiency virus type 1 infection among a population in Chaco Province, Argentina. J. Infect. Dis. 182**:**1523-1526. [DOI] [PubMed] [Google Scholar]
- 15.Draenert, R., S. Le Gall, K. J. Pfafferott, A. J. Leslie, P. Chetty, C. Brander, E. C. Holmes, S. C. Chang, M. E. Feeney, M. M. Addo, L. Ruiz, D. Ramduth, P. Jeena, M. Altfeld, S. Thomas, Y. Tang, C. L. Verrill, C. Dixon, J. G. Prado, P. Kiepiela, J. Martinez-Picado, B. D. Walker, and P. J. Goulder. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199**:**905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344**:**1668-1675. [DOI] [PubMed] [Google Scholar]
- 17.Geczy, A. F., H. Kuipers, M. Coolen, L. J. Ashton, C. Kennedy, G. Ng, R. Dodd, R. Wallace, T. Le, C. H. Raynes-Greenow, W. B. Dyer, J. C. Learmont, and J. S. Sullivan. 2000. HLA and other host factors in transfusion-acquired HIV-1 infection. Hum. Immunol. 61**:**172-176. [DOI] [PubMed] [Google Scholar]
- 18.Geels, M. J., M. Cornelissen, H. Schuitemaker, K. Anderson, D. Kwa, J. Maas, J. T. Dekker, E. Baan, F. Zorgdrager, R. van den Burg, M. van Beelen, V. V. Lukashov, T. M. Fu, W. A. Paxton, L. van der Hoek, S. A. Dubey, J. W. Shiver, and J. Goudsmit. 2003. Identification of sequential viral escape mutants associated with altered T-cell responses in a human immunodeficiency virus type 1-infected individual. J. Virol. 77**:**12430-12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412**:**334-338. [DOI] [PubMed] [Google Scholar]
- 20.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3**:**212-217. [DOI] [PubMed] [Google Scholar]
- 21.Hendel, H., S. Caillat-Zucman, H. Lebuanec, M. Carrington, S. O'Brien, J. M. Andrieu, F. Schachter, D. Zagury, J. Rappaport, C. Winkler, G. W. Nelson, and J. F. Zagury. 1999. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J. Immunol. 162**:**6942-6946. [PubMed] [Google Scholar]
- 22.Jones, N. A., X. Wei, D. R. Flower, M. Wong, F. Michor, M. S. Saag, B. H. Hahn, M. A. Nowak, G. M. Shaw, and P. Borrow. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 200**:**1243-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2**:**405-411. [DOI] [PubMed] [Google Scholar]
- 24.Keet, I. P., J. Tang, M. R. Klein, S. LeBlanc, C. Enger, C. Rivers, R. J. Apple, D. Mann, J. J. Goedert, F. Miedema, and R. A. Kaslow. 1999. Consistent associations of HLA class I and II and transporter gene products with progression of human immunodeficiency virus type 1 infection in homosexual men. J. Infect. Dis. 180**:**299-309. [DOI] [PubMed] [Google Scholar]
- 25.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193**:**375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koenig, S., A. J. Conley, Y. A. Brewah, G. M. Jones, S. Leath, L. J. Boots, V. Davey, G. Pantaleo, J. F. Demarest, C. Carter, et al. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1**:**330-336. [DOI] [PubMed] [Google Scholar]
- 27.Korber, B. HIV signature and sequence variation analysis, p. 55-72. In A. G. Rodrigo and G. H. Learn (ed.), Computational analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 28.Korber, B. T. M., C. Brander, B. F. Haynes, R. A. Koup, C. Kuiken, J. P. Moore, B. D. Walker, and D. I. Watkins. 2002. HIV molecular immunology, LA-UR 03-5816 ed. Los Alamos National Library Theoretical Biology and Physics, Los Alamos, N.Mex.
- 29.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68**:**4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald, K. S., K. R. Fowke, J. Kimani, V. A. Dunand, N. J. Nagelkerke, T. B. Ball, J. Oyugi, E. Njagi, L. K. Gaur, R. C. Brunham, J. Wade, M. A. Luscher, P. Krausa, S. Rowland-Jones, E. Ngugi, J. J. Bwayo, and F. A. Plummer. 2000. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 181**:**1581-1589. [DOI] [PubMed] [Google Scholar]
- 31.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296**:**1439-1443. [DOI] [PubMed] [Google Scholar]
- 32.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8**:**493-499. [DOI] [PubMed] [Google Scholar]
- 33.O'Connor, D. H., A. B. McDermott, K. C. Krebs, E. J. Dodds, J. E. Miller, E. J. Gonzalez, T. J. Jacoby, L. Yant, H. Piontkivska, R. Pantophlet, D. R. Burton, W. M. Rehrauer, N. Wilson, A. L. Hughes, and D. I. Watkins. 2004. A dominant role for CD8+-T-lymphocyte selection in simian immunodeficiency virus sequence variation. J. Virol. 78**:**14012-14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peyerl, F. W., D. H. Barouch, W. W. Yeh, H. S. Bazick, J. Kunstman, K. J. Kunstman, S. M. Wolinsky, and N. L. Letvin. 2003. Simian-human immunodeficiency virus escape from cytotoxic T-lymphocyte recognition at a structurally constrained epitope. J. Virol. 77**:**12572-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piontkivska, H., and A. L. Hughes. 2004. Between-host evolution of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1: an approach based on phylogenetically independent comparisons. J. Virol. 78**:**11758-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94**:**1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rammensee, H. G., J. Bachman, and S. Stevanovich. 1997. MHC ligands and peptide motifs. Landes Biosciences., Georgetown, Texas.
- 38.Rowland-Jones, S. L., T. Dong, K. R. Fowke, J. Kimani, P. Krausa, H. Newell, T. Blanchard, K. Ariyoshi, J. Oyugi, E. Ngugi, J. Bwayo, K. S. MacDonald, A. J. McMichael, and F. A. Plummer. 1998. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Investig. 102**:**1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, J., C. Costello, I. P. Keet, C. Rivers, S. Leblanc, E. Karita, S. Allen, and R. A. Kaslow. 1999. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retroviruses 15**:**317-324. [DOI] [PubMed] [Google Scholar]
- 40.Zinkernagel, R. M., and P. C. Doherty. 1979. MHC-restricted cytotoxic T cells: studies on the biological role of polymorphic major transplantation antigens determining T-cell restriction-specificity, function, and responsiveness. Adv. Immunol. 27**:**51-177. [DOI] [PubMed] [Google Scholar]