The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain (original) (raw)
Abstract
Two proteins of the Hsp70 class (Ssb and Ssz1) and one of the J-type class (Zuo1) of molecular chaperones reside on the yeast ribosome, with Ssz1 forming a stable heterodimer with Zuo1. We designed experiments to address the roles of these two distantly related ribosome-associated Hsp70s and their functional relationship to Zuo1. Strains lacking all three proteins have the same phenotype as those lacking only one, suggesting that these chaperones all function in the same pathway. The Hsp70 Ssb, whose peptide-binding domain is essential for its in vivo function, can be crosslinked to nascent chains on ribosomes that are as short as 54 amino acids, suggesting that Ssb interacts with nascent chains that extend only a short distance beyond the tunnel of the ribosome. A ssz1 mutant protein lacking its putative peptide-binding domain allows normal growth. Thus, binding of unfolded protein substrates in a manner similar to that of typical Hsp70s is not critical for Ssz1's in vivo function. The three chaperones are present in cells in approximately equimolar amounts compared with ribosomes. The level of Ssb can be reduced only a few-fold before growth is affected. However, a 50- to 100-fold reduction of Ssz1 and Zuo1 levels does not have a substantial effect on cell growth. On the basis of these results, we propose that Ssbs function as the major Hsp70 chaperone for nascent chains on the ribosome, and that Ssz1 has evolved to perform a nonclassical function, perhaps modulating Zuo1's ability to function as a J-type chaperone partner of Ssb.
Within the crowded environment of the cell, proteins are particularly susceptible to aggregation before their folding or translocation across membranes (1). Molecular chaperones are able to protect unfolded proteins from aggregation by binding to exposed hydrophobic patches until the proteins are able to fold correctly (2, 3). During protein synthesis aggregation is a particular concern. Once a protein extends about 40 amino acids beyond the protective environment of the tunnel of the ribosome, it is exposed to the cytosol (4), but incapable of folding until an entire domain is exposed (about 50–300 amino acids) (5).
In both prokaryotes and eukaryotes, chaperones of the Hsp70- and J-types have been implicated in protein folding (6–8). Hsp70s work together with J-type chaperones (also referred to as Hsp40s or DnaJs) as part of a “chaperone machine”, functioning in a variety of cellular processes from protein folding to protein translocation across membranes (9, 10). Studies from several Hsp70s demonstrated that the C-terminal 28-kDa region of Hsp70s binds unfolded polypeptides (11). The highly conserved N-terminal 44-kDa ATPase domain regulates that binding through its interaction with adenine nucleotides. Hsp70 proteins have two conformational states: when an ADP molecule is bound to the nucleotide-binding site, the Hsp70 exhibits relatively stable polypeptide substrate binding; when ATP is bound, binding of substrate is relatively unstable. A polypeptide first interacts with an Hsp70 in the ATP-bound state, then hydrolysis of ATP to ADP stabilizes this interaction, which is subsequently destabilized by the exchange of ADP for ATP. This cycle of interaction of Hsp70s with unfolded proteins is facilitated by J-type chaperones, which contain a canonical J domain that interacts with the ATPase domain of Hsp70s (12). In addition, some, but not all, J-type proteins bind unfolded or partially folded polypeptides, preventing their aggregation, and targeting them to Hsp70s (13). Regardless of whether J-proteins themselves bind unfolded polypeptides, studies on organisms as diverse as human and bacteria and several different cellular compartments, indicate that Hsp70s and J-type chaperones function together.
In Saccharomyces cerevisiae, two types of Hsp70s (Ssb and Ssz) and one J-type chaperone (Zuo1) have been found stably associated with ribosomes and thus implicated in folding of newly synthesized polypeptide chains (14–17). Ssz and Ssb are both classified as Hsp70s on the basis of sequence comparisons, but they are quite divergent, having only 26% amino acid identity throughout their length. The Ssb class is composed of two proteins encoded by SSB1 and SSB2. Ssb1 and Ssb2 are greater than 99% identical, having only 4-amino acid differences between them. All data thus far indicate that the two proteins are identical in function and will be referred to as Ssb throughout this report. Ssb can be crosslinked to ribosome-associated nascent polypeptides, suggesting that it acts as a chaperone of these newly synthesized polypeptides (18). Consistent with this idea, Ssb lacking the peptide-binding domain is nonfunctional (19). Ssz, which is encoded by a single gene, SSZ1, is less well studied. Ssz1 forms a heterodimer with Zuo1 and is tethered to the ribosome through this interaction (17). This interaction, which is insensitive to nucleotide and high concentrations of salt, is unusually stable, unlike most Hsp70–J protein interactions, which are transient (20). However, a stable interaction between a J protein and an Hsp70 is not unprecedented, because a stable Hsp70–DnaJ complex has been found in an Archeabacteria (21).
The presence of two types of Hsp70s on the ribosome raises the question of the functional relationship between them. Strains lacking each Hsp70 have the same phenotypes: sensitivity to cold temperatures, some protein synthesis inhibitors, and high osmotic strength, suggesting a functional relationship between them (14, 17). Ssz1 and Ssb might act very similarly to chaperones, or they might perform very different roles. Therefore, we set out to better understand how the two ribosome-associated Hsp70 proteins function on the ribosome. Strains lacking Ssb and Ssz1 have the same phenotypes as those lacking only one of the Hsp70s. In addition, we found that the C-terminal region of Ssz1, the putative peptide-binding domain, is dispensable for its function. Thus, Ssz1's critical role involves only its ATPase domain. We propose that Ssz1 has evolved to function in a unique fashion compared with other Hsp70 proteins that have been analyzed. A model in which Ssz1 functions to modulate the activity of the J-type chaperone Zuo1 that functions as the “J-partner” of the Hsp70 Ssb is discussed.
Materials and Methods
Yeast Strains.
All yeast strains used are isogenic with DS10 (his3–11, 15 leu2–3,112 lys1 lys2 Δ_trp1 ura3–52_) (19), with the exception mutations of the genes encoding SSB1, SSB2, SSZ1, and/or ZUO1. Deletions of ZUO1, SSB1, and SSB2 have been described (15, 19). A deletion of SSZ1 was made in which the sequence between −102 and +1,617 was replaced with the LYS2 gene. In all cases, absence of the protein encoded by the mutated gene was confirmed by immunoblot analysis. The strains used were: NL164_:_ Δ_ssb∷HIS3_ Δ_ssb2∷LEU2_; HE13, Δ_zuo1_∷HIS3; HE14, Δ_ssz1_∷LYS2; HE9, Δssb1∷HIS3, Δssb2∷LEU2, Δssz1∷LYS2; WW1, ssb1∷HIS3 ssb2∷LEU2 Δ_zuo1:URA3;_ HE12_,_ Δ_ssz1_∷LYS2 Δ_zuo1_∷HIS3; HE15, ssb1∷HIS3, ssb2∷LEU2 zuo1∷URA3 Δssz1∷LYS2. Strains were grown in 1% yeast extract/2% peptone/2% dextrose (YPD) or minimal (0.67% yeast nitrogen base without amino acids, 2% dextrose, supplemented with all amino acids and bases except those needed for selection) media.
Plasmid Construction.
To create the deletion constructs of SSZ1, the regions upstream and downstream of the deletion site were amplified by PCR from a pRS316 plasmid containing wild-type SSZ1 (22). The primers used created an _Xba_I site at the 3′ and 5′ ends of the amplified region. The PCR products were then cloned into the pRS316 plasmid allowing for removal of the coding region specified, but otherwise retaining wild-type SSZ1 sequence. The mutant SSZ1 genes include the following: Δ1–410 encodes a protein in which the N-terminal 410 amino acids are removed, Δ383–538 encodes a protein containing the first 382 amino acids of Ssz1 plus four amino acids (Ser-Arg-Ala-Leu) at the C terminus, Δ408–538 encodes a protein that expresses the first 407 amino acids of Ssz1 plus one amino acid (Phe) at the C terminus. Because of low expression from pRS316, both the Δ383–538 and Δ1–410 constructs were moved to pRS426 (23). Regions that were subjected to PCR were sequenced to verify that no additional mutations were created.
The CYC1-SSB1 plasmid was constructed by using the p416_CYC1_ plasmid (26). A _Bam_HI_-Sal_I fragment containing the entire coding region of SSB1 was obtained from a plasmid containing a SSB1 gene having a Bam_HI site immediately upstream of the initiating ATG (19) and cloned into the same sites of p416_CYC1. The CYC-SSZ1 plasmid was created by introducing Bam_HI and Sal_I sites by using PCR 100 nucleotides upstream and at the end of the SSZ1 gene, respectively, and cloning into the same sites of both the p416_CYC1 and p414_CYC1 plasmids. The GAL1-ZUO1 plasmid was constructed with the pYES2 plasmid (Invitrogen). A Bam_HI_-Xho_I fragment containing the entire coding region for ZUO1 was obtained from a p416_TEF-ZUO1 plasmid (pHE5; unpublished results) and placed into the same sites in pYES2. The pHE5 plasmid was created by PCR amplifying approximately the first 550 nucleotides of ZUO1 with a primer that introduced an _Bam_HI site at the 5′ end of the gene and by using the internal _Hin_dIII site. This region was sequenced to verify that no additional mutations were created and the remaining coding region of the ZUO1 gene was replaced by subcloning.
Generation of Ssz1-Specific Antibodies.
A fusion of the C-terminal 141 amino acids of Ssz1 to glutathione _S_-transferase was expressed in the Escherichia coli strain PK101 (24) lacking DnaK and DnaJ. The fusion protein was purified and injected into rabbits for antibody generation. To generate the fusion construct, SSZ1 was cleaved at an internal _Hin_dIII site, which was filled in to form a blunt end, and 3′ of the gene with _Sac_I, and cloned into the _Sma_I and _Sac_I sites of pGEX-KG (25). Antibody specific for the entirety of Ssz1 was provided by S. Moye-Rowley (Univ. of Iowa; 22).
Preparation and Analysis of Cell Lysates and Ribosomal-Containing Fractions.
Lysates were prepared from cells grown overnight in minimal media lacking uracil to an OD600 of 4–5 at 30°C. The equivalent of 0.5 OD units of cells harvested by centrifugation were resuspended in 50 μl of water. Fifty microliters of 0.2 M NaOH was added, incubated for 5 min at room temperature, then pelleted and resuspended in 50 μl of SDS sample buffer, and boiled for 5 min (27). Equivalent amounts of extract were subjected to SDS/PAGE, transferred to nitrocellulose (Osmonics, Westborough, MA), and immunoblotted by using the enhanced chemiluminescence detection system (NEN).
Lysates for preparation of ribosome-containing fraction were prepared as described (14) from yeast cells grown in selective minimal media to an OD600 between 0.5 and 1.0. RNAguard (Amersham Pharmacia) was added at a dilution of 1:1,000. Approximately 20 OD260 units of lysate were loaded onto a 2-ml sucrose cushion containing CB buffer (20 mM Hepes, pH 7.5/10 mM KCl/1 mM EGTA/5 mM MgCl/10% glycerol/2 mM β-mercaptoethanol) plus 0.5 M sucrose and centrifuged at 50,000 rpm for 3 h in a TLA 100.3 rotor (Beckman Coulter) at 4°C. In some cases the resuspended pellets were directly subjected to SDS/PAGE and immunoblotting as described above.
To determine the salt sensitivity of the interaction of the ribosome with Ssz1 and Zuo1, the resulting pellets were resuspended in 50 μl CB/300 mM sorbitol. The resuspended pellets were then diluted into 500 μl CB/300 mM sorbitol and 50, 100, or 150 mM KCl and incubated at 4°C for 30 min. After incubation, the polysomes were loaded onto 2-ml sucrose cushions with the appropriate concentration of KCl and centrifuged as before. Pellets were resuspended in CB/300 mM sorbitol. Samples containing one tenth of the total loaded onto the cushion, supernatant, and pellet were subjected to SDS/PAGE and immunoblot analysis as described above.
Translation Experiments.
To produce the varying lengths of the precursor form of CoxIV (pCoxIV) nascent chain (96, 77, 54, and 34 amino acids), the COXIV template (28) was digested at the endogenous restriction sites (_Xho_II, _Cla_I, _Kpn_I, and _Dde_I, respectively). To obtain the 152-amino acid pCoxIV nascent chain, a _Sac_I restriction site was introduced with use of site-directed PCR mutagenesis, as were the mutations causing lysine to arginine alterations in pCoxIV. In vitro transcription, in vitro translation, and crosslinking procedures were performed essentially as described (18, 29). [35S]Methionine and 4-(3-tri-fluoromethyl-diazirino)benzoic acid (TBDA)-lysyl-tRNA were added to the translation along with an mRNA lacking a stop codon. Thus, the nascent chain, which was tethered to the ribosome, was radiolabeled and contained a photoactivatable crosslinker at lysine residues. After irradiation with light of wavelength >330 nm, the ribosome-nascent chain complexes were recovered by centrifugation. Resuspended ribosome-nascent chain complexes were subjected to immunoprecipitation. Radioactive bands were visualized by fluorography (Biomax-MS film with Biomax Transcreen Low Energy Intensifying Screen, Eastman Kodak).
Results
Strains Lacking Ssb, Ssz1, and Zuo1 Have the Same Phenotypes as Strains Lacking a Single Chaperone.
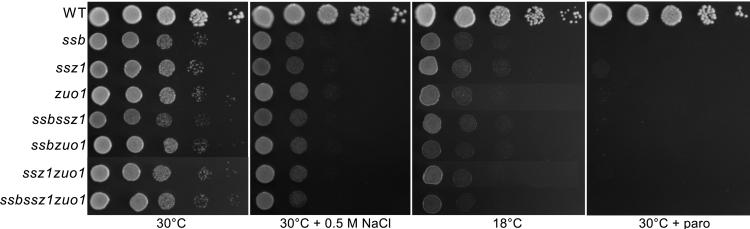
To begin to clarify the functional relationship between the two ribosome-associated Hsp70s, Ssb and Ssz1, we tested for genetic interactions between them. ssb and ssz1 strains grew very poorly at 18°C and were sensitive to the aminoglycoside paromomycin and high osmotic strength, as was a zuo1 strain (Fig. 1) (15, 17, 18). The ssb ssz1 strain showed the same growth defects. In addition, the ssb ssz1 zuo1 strain, lacking both ribosome-associated Hsp70s and the Hsp40 had essentially the same phenotype. The fact that the absence of a single chaperone has the same effect as the lack of all three suggested that they are involved in the same process and that disruption of either Hsp70 (or the Hsp40 Zuo1) disrupts that function.
Figure 1.
ssb, zuo1, and ssz1 mutant phenotypes are not additive. Approximately equal numbers of cells of the indicated genotypes were subjected to a 10-fold serial dilution and spotted onto rich media containing the indicated additions. Plates were incubated for the following number of days: YPD 30°C, 2 days; YPD + 0.5 M NaCl, 3 days; YPD + 50 μg/ml paromomycin, 2 days; YPD 18°C, 5 days.
Ssb Can Be Crosslinked to Nascent Chains as Short as 54 Amino Acids.
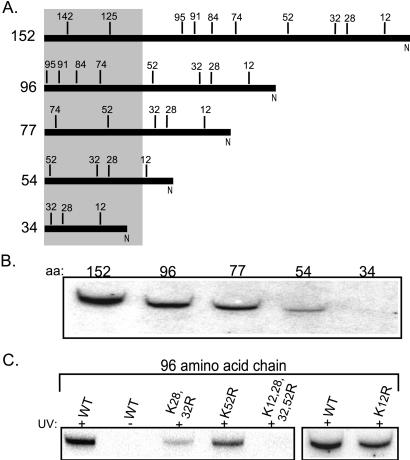
To better understand the relationship between these two Hsp70s, we set out to analyze interactions with the ribosome-associated nascent chains. Ssb has been crosslinked to ribosome-associated nascent chains produced in a yeast translation extract (18). However, the chains tested had a limited size distribution. To try to delineate more specifically the sites of interaction of Ssb, mRNAs were produced that encoded pCoxIV polypeptides of 152, 96, 77, 54, or 34 amino acids (Fig. 2A). These translated chains remained tethered to the ribosome because of the absence of a stop codon. In vitro translation was performed in the presence of [35S]methionine and lysyl-tRNAs charged with lysines bearing a photoactivatable crosslinker, TBDA. Labeled nascent chains bearing this photoactivatable probe can form covalent attachments with interacting proteins, which can be identified by immunoprecipitation. The chains of 152, 96, 77, and 54 amino acids could be crosslinked to Ssb, but we were unable to detect crosslinking to the shortest chain that was 34 amino acids (Fig. 2B). These results are consistent with Ssb interacting with chains extending beyond the ribosome tunnel, but not those sequestered within it (30, 31). The signal was significantly weaker in the 54-amino acid chain compared with the longer chains. Assuming the ribosome tunnel sequesters 40 amino acids, the only lysine expected to be exposed in this nascent chain would be the lysine at position 12, which would be very close to the exit site.
Figure 2.
Ssb can be crosslinked to short nascent chains. (A) Diagram of the nascent polypeptide pCoxIV chains used in B and C. The number to the left indicates the number of amino acids in each chain. The numbers above the line indicate the position of lysine residues. The shaded box represents the region of each nascent chain that would be within the ribosome tunnel, based on the assumption that 40 amino acids are sequestered. (B) Truncated mRNAs transcribed from COXIV DNA and encoding 152, 96, 77, 54, or 34 amino acids were translated in a yeast translation extract in the presence of [35S]methionine and TBDA-lysyl tRNA. The translation products were irradiated (+) or, as a control, not irradiated (−). The ribosome-nascent chain complexes were then pelleted, resuspended, and subjected to immunoprecipitation with antibodies specific for Ssb. Equal amounts of radioactive material were immunoprecipitated for each sample. The immunoprecipitations were subjected to SDS/PAGE and exposed to film. (C) The mRNA encoding the wild-type (WT) 96-aa pCoxIV and chains of the same length having lysine-to-arginine alterations at the indicated positions were translated and subjected to the procedure described in B.
To better understand the interaction of Ssb with the nascent chain, we further characterized the 96-amino acid chain. This polypeptide contains 8 lysine residues, 4 of which would be expected to extend beyond the ribosome tunnel. To assess the contribution of these 4 lysines, at positions 12, 28, 32, and 52, to the crosslinking signal, we made constructs that changed the codons encoding these lysines to those encoding arginines. As expected, if Ssb was being crosslinked to the nascent chain extending beyond the ribosome, no signal was detected if the lysines at positions 12, 28, 32, and 52 were removed. Alterations at positions 12, 28/32, or 52 gave intermediate amounts of crosslinking, suggesting that Ssb may interact at a variety of sites along the chain.
Because our goal was to understand the functional relationship between the two Hsp70s and the Hsp40 on the ribosome, we performed crosslinking experiments to attempt to detect interaction of Zuo1 and Ssz1 with nascent chains. No interaction was detected (data not shown). Although this result is not conclusive, it is consistent with Ssb being the predominant chaperone interacting with the nascent chain on the ribosome.
The C-terminal Domain of Ssz1, Which Encodes the Putative Peptide-Binding Domain, Is Not Required in Vivo.
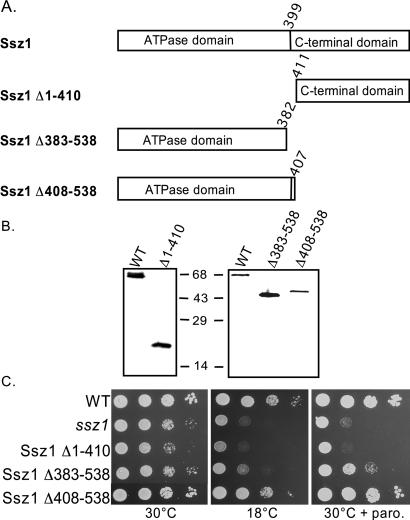
Because we could find no evidence for the interaction of Ssz1 with the nascent chain, and because of its unusually stable interaction with Zuo1, we wanted to determine the regions of Ssz1 important for its function. Three deletion mutants were constructed: a deletion of the 44-kDa ATPase domain (Ssz1Δ1–410), a C-terminal truncation of the putative peptide-binding domain (Ssz1Δ408–538), and a larger C-terminal truncation removing sequences encoding the putative peptide-binding domain and 17 amino acids of the ATPase domain (Ssz1Δ383–538) (Fig. 3A). In each case the sequences were completely removed from the constructs to alleviate concerns about translational readthrough. These constructs were transformed into an ssz1 strain. Deletion of the ATPase domain resulted in a null phenotype, indicating the importance of this region. However, cells expressing only the ATPase domain of Ssz1 grew as well as wild-type cells, both at 18°C and in the presence of paromomycin (Fig. 3C). The deletion that removed 17 amino acids from the ATPase domain and the putative peptide-binding domain was unable to rescue the ssz1 phenotypes. However, both constructs expressed protein at nearly normal levels (Fig. 3B). Thus, the peptide-binding domain is not required for the function of Ssz1, at least under the laboratory conditions tested, whereas the ATPase domain is critical for function.
Figure 3.
The C terminus of Ssz1 is dispensable. (A) Diagram of SSZ1 mutants used in this study. Unexpressed regions were completely deleted from the expression plasmid. (B) Immunodetection of mutant SSZ1 proteins. Cell extracts were prepared from the indicated strains and subjected to electrophoresis and immunoblotting. Antibodies used were specific for the C-terminal domain (Left) or full-length (Right) Ssz1. (C) Cell cultures used for preparation of extracts used in B were subjected to a 10-fold serial dilution and spotted onto minimal media lacking uracil. Plates were incubated for 2 days at 30°C or 4 days at 18°C.
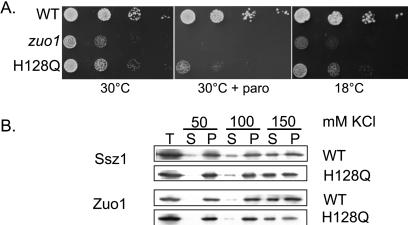
In addition, we wanted to address whether the J domain of Zuo1 was critical for function. This conserved domain has been shown to be essential for the in vivo function of several J-type proteins and to be critical for interactions with their Hsp70 partners (32). We had shown that a mutant protein containing a complete deletion of the J domain was not functional (15). However, because such a deletion might have profound effects on the structure of the remaining domains of the protein, we analyzed the effect of a single amino acid alteration in the J domain, H128Q. This alteration of a highly conserved histidine has been shown to affect function of other J-type proteins (33, 34). Strains expressing this mutant Zuo1 grew slowly and showed sensitivity to paromomycin, but were not as defective as mutants completely lacking the protein (Fig. 4A). The mutant protein was still associated with ribosomes, as was Ssz1 (Fig. 4B). In addition, the salt sensitivity of the interaction of Ssz1 and the mutant Zuo1 suggested that the interaction of Zuo1 with both the ribosome and Ssz1 was not significantly affected.
Figure 4.
The J domain of Zuo1 is required for function, but does not destabilize the Ssz1:Zuo1 interaction. (A) Growth phenotype of strains expressing Zuo1 with an alteration (H128Q) in the J domain. A 10-fold dilution series of wild-type (WT) cells or cells carrying the H128Q mutation in ZUO1 was plated onto minimal medium with the indicated additions and incubated at 30°C for 2 days or 18°C for 7 days. (B) Salt sensitivity of Ssz1-ribosome interaction. A ribosome fraction obtained from WT and zuo1 H128Q cells were separated on sucrose cushions containing the indicated salt concentrations. Equivalent samples of the supernatant and pellet fractions were subjected to electrophoresis and immunoblot analysis with Zuo1- or Ssz1-specific antibodies.
Very Low Levels of Zuo1 and Ssz1, Unlike Ssb, Are Sufficient for Normal Growth.
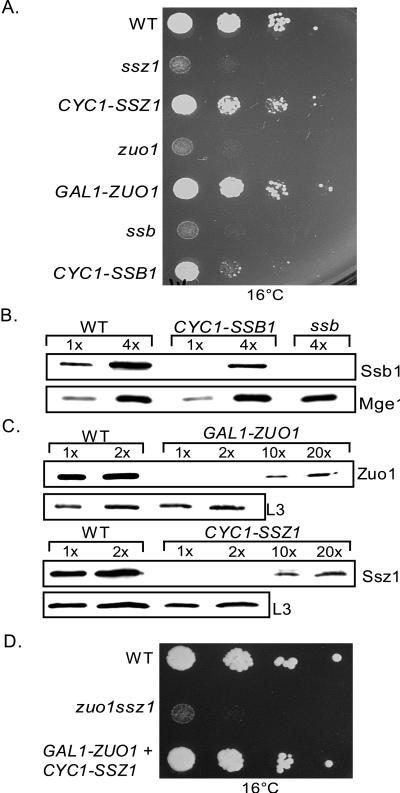
The Ssz1:Zuo1:ribosome ratio in vivo is ≈1:1:1 (15, 17). The number of Ssb molecules in cells is somewhat higher, with the ratio of Ssb to ribosomes being between 2 and 4 (18). This stoichiometry between chaperones and ribosomes suggests that each ribosome requires a molecule of each chaperone protein for normal function. To test this idea we asked what levels of these three proteins are required for wild-type function. Ssb1, Ssz1, and Zuo1 were expressed from heterologous promoters. Reducing expression of Ssb to 25% of normal (Fig. 5 A and B), a ratio of 0.5–1 Ssb molecules per ribosome resulted in severely compromised growth at 16°C.
Figure 5.
Ssz1 and Zuo1 levels, but not Ssb, levels can be substantially reduced without affecting growth. (A) Wild-type (WT), ssz1, ssz1 expressing SSZ1 under control of the CYC1 promoter (CYC1-SSZ1), zuo1, zuo1 expressing ZUO1 from the glucose-repressed GAL1 promoter (GAL1-ZUO1). ssb or ssb expressing SSB1 under control of the CYC1 promoter (CYC1-SSB1) cells were grown in minimal media lacking uracil. Cells were diluted and spotted on the same media and incubated at 16°C for 8 days. (B) Cell extracts were prepared from an equivalent number of cells of the strains described in A. One or four times the amount of the indicated extracts were subjected to electrophoresis and immunoblotting with antibodies specific to Ssb or as a control Mge1. (C) Ribosome-containing pellets were obtained by centrifugation through sucrose cushions. Equal amounts of the ribosomal pellets (1×) or twice that amount (2×), 10 times (10×), and 20 times (20×) were subjected to electrophoresis and immunoblotted with antibody specific for Zuo1, Ssz1, or ribosomal protein L3 as a control. (D) Wild-type (WT), zuo1 ssz1, and zuo1 ssz1 expressing ZUO1 and SSZ1 from the repressed GAL1 and CYC1 promoters, respectively (GAL1-ZUO1 + CYC1-SSZ1) cells were grown in minimal selective media, subjected to a 10-fold dilution series, and spotted onto the same media. Plates were incubated for 8 days at 16°C.
When Ssz1 and Zuo1 were similarly reduced in abundance, no effect on growth was observed (data not shown). Therefore, we made expression constructs that would result in production of even lower levels of these two proteins. Because the levels of expression from these constructs were so low and because of the association of Zuo1 and Ssz1 with ribosomes, we used purified ribosomes to determine the relative levels of Zuo1 and Ssz1 in these strains. Two strains, one expressing Zuo1 from the GAL1 promoter under repression by glucose and one expressing Ssz1 from the CYC1 promoter were found to express between 1 and 2% of the normal level of Zuo1 and Ssz1, respectively (Fig. 5C). Both of these strains grew very similarly to a wild-type strain at low temperatures, and in the presence of aminoglycoside antibiotics (Fig. 5A and data not shown). Therefore, expression of these two proteins at very low levels is sufficient to perform their function. In summary, Ssb is needed at a stoichiometry of at least 0.5–1 molecules per ribosome, whereas only 1–2 molecules of Ssz1 or Zuo1 per 100 ribosomes is required.
To determine whether Ssz1 and Zuo1 could be reduced to low levels simultaneously and allow wild-type growth, a ssz1 zuo1 strain was transformed with plasmids expressing Zuo1 and Ssz1 under control of the repressed GAL1 and the CYC1 promoters, respectively. This strain was able to grow, as well as a wild-type strain at low temperatures (Fig. 5D), indicating that both proteins are only required in low amounts for normal growth under the conditions tested.
Discussion
The yeast ribosome has two associated Hsp70s, Ssb and Ssz1. Evidence presented here and elsewhere supports the idea that Ssb acts as a chaperone for nascent polypeptides chains associated with the ribosome. Ssb can be crosslinked to short segments of nascent chains extending beyond the exit site of the ribosome. In addition, its peptide-binding domain is essential for its function (19), and its association with ribosomes is more resistant to salt in the presence of nascent chain (18). This enhanced stability is reduced under two conditions expected to destabilize the interaction of Ssb with the nascent chain, in the presence of ATP and in the presence of an amino acid alteration in the peptide-binding cleft (19).
By contrast, no evidence indicates that Ssz1 acts as a chaperone. The C-terminal segment of Ssz1 extending beyond the ATPase domain, which encodes the peptide-binding domain of Hsp70s, is not required for rescue of the cold temperature and drug sensitivity found in the null mutant. Therefore, the simplest explanation is that Ssz1 does not require interaction with unfolded polypeptides to perform its function, which, to our knowledge, is the only example in which an Hsp70's in vivo function does not depend on its peptide-binding domain. Eukaryotic Hsp70s of mitochondria, the endoplasmic reticulum and cytosol, and those of the cytosol of prokaryotes, have been shown to require the peptide-binding domain (19, 35). In several cases single-amino acid changes within the peptide-binding cleft itself results in loss of function (19, 36, 37; P. D'Silva and E. Craig, unpublished results). A precedent exists for a “truncated Hsp70”. A 45-kDa protein related to the ATPase domain of Hsp70s, called Stch, has been found in human, rat, and Caenorhabditis elegans (38, 39). In addition, an apparently alternatively spliced transcript encoding a version of the major mammalian cytosolic Hsp70, called Hsc54, which lacks a substantial portion of the peptide-binding domain, as well as the C-terminal 10-kDa domain, has been found in human cells (40). No data exists concerning the in vivo function of these “truncated” Hsp70s. It is conceivable that the ATPase domain has a peptide-binding site that has hereto been unrecognized on any Hsp70. However, we consider it much more likely that these proteins, like Ssz1, have evolved functions that are independent of direct binding to unfolded polypeptides.
But, our data do not address whether Ssz1 is capable of binding unfolded polypeptides and functioning as a chaperone. Even though the absence of the peptide-binding domain has no obvious effect under the conditions that we tested, it is quite possible that Ssz1 is able to bind peptide and does so in vivo, and that this function is critical under some growth conditions, or in the absence of another chaperone. The amino acid similarity to other Hsp70s is so low in the putative peptide-binding domain (22% amino acid identity between Ssz1 and Ssa1 or 23% between Ssz1 and Ssb1, respectively) that it is difficult to predict whether the C-terminal region of Ssz1 would form a domain similar in structure to other Hsp70s. Regardless, the ability of Ssz1 to function in the absence of its putative peptide-binding domain should raise a note of caution. Because the ATPase domain is the most conserved domain among all Hsp70s, the presence of sequences C-terminal to such a domain is often sufficient to consider that the protein is able to bind unfolded proteins and function as a chaperone. Although true in many cases, other exceptions to the rule may well exist.
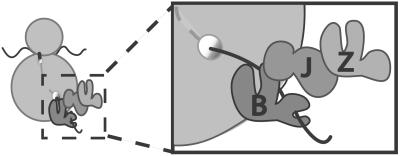
How then might these three proteins function together on the ribosome? The genetic evidence strongly suggests that Ssb, Ssz1, and Zuo1 function together, because the phenotype of each mutant lacking a single protein is very similar to that of a mutant lacking all three. In addition, Zuo1 likely functions as a J-type protein, because a single-amino acid alteration in the J domain has a substantial affect on function in vivo. On the basis of these data we suggest the following model (Fig. 6): Ssb functions as a molecular chaperone on the ribosome, binding to nascent polypeptide chains soon after their emergence from the ribosome. Zuo1 is the partner J-type protein for Ssb, facilitating its interaction with polypeptide substrates, with the J domain playing a critical role in this process, as expected. Ssz1 forms a heterodimer with Zuo1 (17). Rather than acting as a molecular chaperone by binding unfolded polypeptides, its critical role involves modulating the activity of Zuo1. The ATPase domain is required for this regulation, but the mechanism by which it occurs is not known. This model is consistent with the data of Rospert and colleagues (42). In their system, the Ssz1/Zuo1 heterodimer is required for Ssb to be crosslinked to nascent chains, suggesting that Zuo1 is the “J partner” for Ssb, even though it is stably associated with another Hsp70.
Figure 6.
Model of Ssb, Ssz1, and Zuo1 on ribosome. The three chaperones interact near the exit site of the ribosome. Ssb (B) and Zuo1 (J) interact as a “typical” Hsp70:J-protein pair, with Ssb binding the nascent chain. Ssz1 (Z) associates with the ribosome by its interaction with Zuo1, modulating the interaction of this J-protein with Ssb. This atypical function of Ssz1 does not require its putative binding domain. Whether Ssz1 binds unfolded proteins under any circumstance is unknown.
The ability to reduce Zuo1 or Ssz1 to a very low level relative to its normal expression is surprising. But J-type chaperones are often found in significantly lower amounts than their partner Hsp70 and are known to be able to act catalytically (41). However, this difference in levels required for Ssb compared with Zuo1 raises interesting mechanistic questions, because all of Zuo1 is usually found associated with ribosomes (15, 17). Perhaps only a small subset of ribosomes (1–2%) usually have Zuo1 bound, and these are sufficient for performing the required function. Alternatively, in some manner, Zuo1/Ssz1 cycles among ribosomes, servicing all the ribosomes containing Ssb. Regardless of the mechanism, understanding how this unique complex functions with the Hsp70 Ssb should provide additional insights into facilitation of folding of newly synthesized proteins.
Acknowledgments
We thank Scott Moye-Rowley for Ssz1 antibody and plasmids, Sabine Rospert for communication of results before publication, and Jarek Marszalek and Chris Pfund for helpful suggestions on the manuscript. This work was supported by National Institutes of Health Grants GM31107 (to E.A.C.) and HL60889 (to M.W.) and the DeWitt Wallace Fund (to M.W.).
References
- 1.Minton A. Curr Opin Struct Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B, Deuerling E, Pfund C, Craig E A. Cell. 2000;101:119–122. doi: 10.1016/S0092-8674(00)80806-5. [DOI] [PubMed] [Google Scholar]
- 3.Frydman J. Annu Rev Biochem. 2001;70:603–649. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 4.Malkin L I, Rich A J. J Mol Biol. 1967;26:329–346. doi: 10.1016/0022-2836(67)90301-4. [DOI] [PubMed] [Google Scholar]
- 5.Jaenicke R. Biochemistry. 1991;30:147–161. [Google Scholar]
- 6.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Nature (London) 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 7.Teter S A, Houry W A, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulus C, Hartl F U. Cell. 1999;97:755–765. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, Schilke B, Craig E, Horwich A. Proc Natl Acad Sci USA. 1998;95:12860–12865. doi: 10.1073/pnas.95.22.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig E A. Science. 1993;260:1902–1903. doi: 10.1126/science.8100364. [DOI] [PubMed] [Google Scholar]
- 10.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 11.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 12.Greene M, Maskos K, Landry S. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langer T, Lu C, Echols H, Flanagan J, Hayer M K, Hartl F U. Nature (London) 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 14.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 15.Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig E A. EMBO J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallstrom T C, Moye-Rowley W S. Mol Microbiol. 2000;36:402–413. doi: 10.1046/j.1365-2958.2000.01858.x. [DOI] [PubMed] [Google Scholar]
- 17.Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S. Proc Natl Acad Sci USA. 2001;98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke B A, Lopez-Buesa P, Walter W A, Wiedmann M, Craig E A. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfund C, Huang P, Lopez-Hoyo N, Craig E. Mol Biol Cell. 2001;12:3773–3782. doi: 10.1091/mbc.12.12.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer M P, Laufen T, Paal K, McCarty J S, Bukau B. J Mol Biol. 1999;289:1131–1144. doi: 10.1006/jmbi.1999.2844. [DOI] [PubMed] [Google Scholar]
- 21.Motohashi K, Taguchi H, Ishii N, Yoshida M. J Biol Chem. 1994;269:27074–27079. [PubMed] [Google Scholar]
- 22.Hallstrom T C, Katzmann D J, Torres R J, Sharp W J, Moye-Rowley W S. Mol Cell Biol. 1998;18:1147–1155. doi: 10.1128/mcb.18.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang P J, Craig E A. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan K L, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 26.Mumberg D, Muller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 27.Kushnirov V V. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 28.Wiedmann B, Sakai H, Davis T, Wiedmann M. Nature (London) 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- 29.Gorlich D, Kurzchalia T V, Wiedmann M, Rapoport T A. Methods Cell Biol. 1991;34:241–262. doi: 10.1016/s0091-679x(08)61684-2. [DOI] [PubMed] [Google Scholar]
- 30.Gabashvili I, Gregory S, Valle M, Grassucci R, Worbs M, Wahl M, Dahlberg A, Frank J. Mol Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- 31.Hardesty B, Kramer G. Prog Nucleic Acid Res Mol Biol. 2000;66:41–66. doi: 10.1016/s0079-6603(00)66026-9. [DOI] [PubMed] [Google Scholar]
- 32.Cheetham M E, Caplan A J. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan W, Craig E A. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wall D, Zylicz M, Georgopoulos C. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 35.Tokunaga M, Kato S, Kawamura-Watabe A, Tanaka R, Tokunaga H. Yeast. 1998;14:1285–1295. doi: 10.1002/(SICI)1097-0061(1998100)14:14<1285::AID-YEA329>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 36.Montgomery D, Morimoto R, Gierasch L. J Mol Biol. 1999;286:915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- 37.Mayer M P, Schroder H, Rudiger S, Paal K, Laufen T, Bukau B. Nat Struct Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- 38.Otterson G A, Flynn G C, Kratzke R A, Coxon A, Johnston P G, Kaye F J. EMBO J. 1994;13:1216–1225. doi: 10.1002/j.1460-2075.1994.tb06371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otterson G, Kaye F. Gene. 1997;199:287–292. doi: 10.1016/s0378-1119(97)00383-1. [DOI] [PubMed] [Google Scholar]
- 40.Tsukahara F, Yoshioka T, Muraki T. Mol Pharm. 2001;58:1257–1263. doi: 10.1124/mol.58.6.1257. [DOI] [PubMed] [Google Scholar]
- 41.Liberek K, Wall D, Georgopoulos C. Proc Natl Acad Sci USA. 1995;92:6224–6228. doi: 10.1073/pnas.92.14.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gautschi M, Mun A, Ross S, Sabine R. Proc Natl Acad Sci USA. 2002;99:4209–4214. doi: 10.1073/pnas.062048599. [DOI] [PMC free article] [PubMed] [Google Scholar]