The chromatin remodelling factor Brg-1 interacts with β-catenin to promote target gene activation (original) (raw)
Abstract
Wnt-induced formation of nuclear Tcf–β-catenin complexes promotes transcriptional activation of target genes involved in cell fate decisions. Inappropriate expression of Tcf target genes resulting from mutational activation of this pathway is also implicated in tumorigenesis. The C-terminus of β-catenin is indispensable for the transactivation function, which probably reflects the presence of binding sites for essential transcriptional coactivators such as p300/CBP. However, the precise mechanism of transactivation remains unclear. Here we demonstrate an interaction between β-catenin and Brg-1, a component of mammalian SWI/SNF and Rsc chromatin-remodelling complexes. A functional consequence of reintroduction of Brg-1 into Brg-1-deficient cells is enhanced activity of a Tcf-responsive reporter gene. Consistent with this, stable expression of inactive forms of Brg-1 in colon carcinoma cell lines specifically inhibits expression of endogenous Tcf target genes. In addition, we observe genetic interactions between the Brg-1 and β-catenin homologues in flies. We conclude that β-catenin recruits Brg-1 to Tcf target gene promoters, facilitating chromatin remodelling as a prerequisite for transcriptional activation.
Keywords: Brg-1/β-catenin/chromatin remodelling/SWI/SNF/Tcf
Introduction
In organisms ranging from worms to mammals, Wnt signalling regulates a large array of developmental processes, including cell growth and differentiation, by altering gene expression patterns (Bienz and Clevers, 2000; Peifer and Polakis, 2000). Inappropriate expression of Wnt target genes resulting from deregulation of this pathway is also implicated in tumorigenesis.
In response to a Wnt signal, β-catenin levels in the cell increase, promoting a functional interaction with members of the Tcf/Lef family of transcription factors in the nucleus (Behrens et al., 1996; Molenaar et al., 1996; Hsu et al., 1998). This bipartite transcription factor complex is targeted to the promoters of specific genes via a sequence-specific DNA-binding domain in the Tcfs and mediates transcriptional activation by virtue of potent transactivation domains present in the C-terminus of β-catenin (van de Wetering et al., 1997; Hecht et al., 1999). In the absence of nuclear β-catenin, Tcf factors occupy target gene promoters in a complex with p300/CBP, CtBP and members of the Groucho family of corepressors to mediate transcriptional repression (Cavallo et al., 1998; Roose et al., 1998; Waltzer and Bienz, 1998; Brannon et al., 1999).
The mechanisms by which β-catenin promotes target gene activation are not well understood. β-catenin and its Drosophila counterpart Armadillo are composed of 12 imperfect protein interaction repeats (ARM repeats) flanked by unique N- and C-termini (Figure 1A) (Peifer et al., 1992, 1994). Both the N- and C-termini demonstrate transactivation potential in in vitro reporter assays, but the most potent transactivation domain is located at the C-terminus (van de Wetering et al., 1997; Hsu et al., 1998; Hecht et al., 1999). This region is also indispensable for Wingless signalling in vivo (van de Wetering et al., 1997; Cox et al., 1999), which probably reflects the presence of binding sites for essential transcriptional coactivators such as p300/CBP (Hecht et al., 2000; Takemaru and Moon, 2000). CBP is known to function as a transcriptional coactivator by connecting a variety of transcription factors to the basal transcription machinery and may alter local chromatin structure via its histone acetylase (HAT) activity to increase access of other transcription factors to target gene promoters (Ogryzko et al., 1996; Goldman, 1997). β-catenin could, therefore, be viewed as a docking molecule that recruits essential coactivators to Tcf target gene promoters.
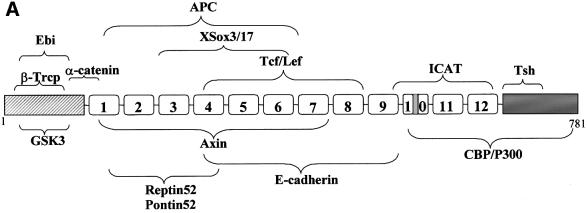
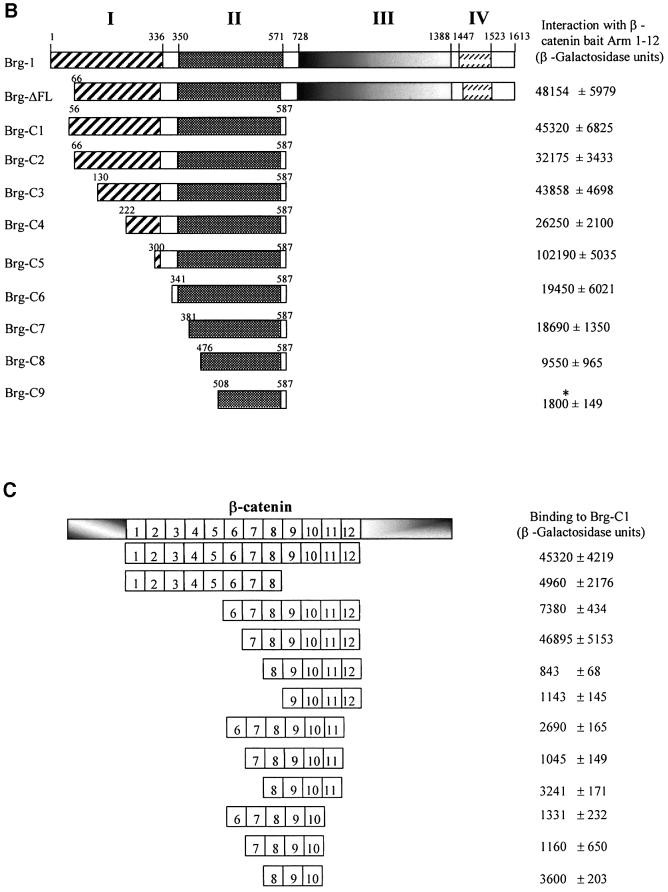
Fig. 1. β-catenin interacts specifically with Brg-1. (A) Schematic representation of the β-catenin domain structure. The N-terminal domain (grey stripes) contains four conserved serine/threonine phosphorylation sites for GSK-3β, which are essential for mediating destruction of free β-catenin. The central domain comprises 12 imperfect repeats of 42 amino acids (denoted Armadillo repeats 1–12; note the presence of an insertion within repeat 10), which are responsible for mediating many of the interactions between β-catenin and its binding partners. The C-terminal domain (shaded grey) contains potent transcriptional activation elements that are essential for the signalling activity of β-catenin. The regions of β-catenin responsible for mediating interaction with other proteins are indicated by curly brackets. (B) Mapping of the Brg-1 domain responsible for mediating interaction with β-catenin. I–IV denote regions of sequence conservation between Brg-1 and Drosophila brm (Khavari et al., 1993). Brg-1 deletion constructs were co-transformed with the β-catenin Arm1–12 bait into the Y190 reporter yeast strain and positive interactions quantified by measuring the activity of a β-galactosidase reporter gene. The asterisk denotes background β-galactosidase activity, as determined by co-transfection of empty prey vector with the β-catenin bait. (C) Mapping of Armadillo repeats mediating interaction with Brg-1. Baits comprising overlapping regions of the β-catenin Armadillo repeats were co-transformed with the Brg-C1 prey plasmid into the Y190 yeast strain and positive interactions quantified by measuring the activity of a β-galactosidase reporter gene.
However, several lines of evidence indicate that other cofactors are likely to be involved in β-catenin-mediated transactivation. First, β-catenin mutants unable to bind CBP are still capable of effecting transactivation (van de Wetering et al., 1997). Additionally, the cooperative effect of CBP on β-catenin transactivation is evident only for a subset of known Tcf target genes (Hecht et al., 2000). It is possible, therefore, that the transactivation potential of β-catenin may be tailored to suit the particular target gene or class of target gene through interaction with different cofactors.
The interaction between β-catenin and CBP, a protein with intrinsic HAT activity, may indicate a requirement for chromatin remodelling in Tcf target gene activation. It is generally believed that ‘closed’ chromatin functions to exclude transcription factors from their cognate binding sites in promoter regions (Adams and Workman, 1993; Blomquist et al., 1996) and inhibits access of the basal transcription machinery, including RNA polymerase II and TATA-box-binding protein (TBP), to the transcription initiation site (Laybourn and Kadonaga, 1992; Godde et al., 1995; Godde and Wolffe, 1996).
Genetic and biochemical studies, especially in yeast, have revealed two highly related yet distinct ATP-dependent chromatin-remodelling complexes, SWI/SNF and Rsc, implicated in transcriptional regulation. Both exist as 1.5–2 MDa multisubunit complexes, which are evolutionarily conserved, albeit with various subunit compositions in higher organisms (Wang et al., 1996a,b; Xue et al., 2000). A common feature of all SWI/SNF and Rsc complexes is the presence of an ATPase component, which is indispensable for its chromatin remodelling function. In humans, this ATPase activity is provided by either Brahma (Brm) or Brahma-related gene-1 (Brg-1) in the case of SWI/SNF (Wang et al., 1996b), or exclusively Brg-1 in the case of Rsc (termed PBAF in mammalian cells) (Xue et al., 2000).
Chromatin-remodelling complexes themselves do not appear to exhibit significant DNA-binding specificity, yet mutations in the yeast Brg-1/Brm homologue, Swi2, affect <6% of all known genes (Holstege et al., 1998). It is likely, therefore, that chromatin-remodelling activity is targeted to a subset of genes in vivo via interaction with sequence-specific transcription factors. For example, the glucocorticoid receptor recruits the SWI/SNF complex to the glucocorticoid receptor element (GRE), thereby facilitating chromatin remodelling within this region (Muchardt and Yaniv, 1993; Ostlund Farrants et al., 1997; Fryer and Archer, 1998). SWI/SNF is also recruited by the C/EBPβ transcription factor where it subsequently cooperates with c-Myb to activate transcription of myeloid genes (Kowenz-Leutz and Leutz, 1999).
Here we demonstrate an interaction between β-catenin and Brg-1. A functional consequence of reintroduction of Brg-1 into Brg-1-deficient cells is enhanced activity of a Tcf-responsive reporter gene. Consistent with this, stable expression of inactive forms of Brg-1 in colon carcinoma cell lines specifically inhibits expression of endogenous Tcf target genes. Reduction of brahma dosage in flies suppresses the rough eye phenotype caused by activated Armadillo, and enhances the wing margin defects due to Armadillo depletion, demonstrating a genetic interaction between Brg-1 and β-catenin in flies. We conclude that β-catenin recruits SWI/SNF or Rsc-like complexes via interaction with Brg-1 to Tcf target gene promoters, facilitating chromatin remodelling as a prerequisite for efficient transcriptional activation.
Results and discussion
β-catenin specifically interacts with the SWI/SNF and Rsc component Brg-1 in yeast
In a search for additional proteins which may interact with β-catenin to modulate Tcf target gene activity, we performed a two-hybrid screen of a human fetal brain cDNA library using a bait comprising Armadillo repeats 1–12 (Arm1-12). We screened ∼2 × 106 yeast clones and identified several interacting proteins, four of which corresponded to fragments of the SWI/SNF and Rsc component Brg-1. These partial clones encoded internal fragments of Brg-1 (Brg-C1–C4; Figure 1B). Additionally, there was a strong interaction between Arm1–12 and a Brg-1 construct lacking only the N-terminal 66 amino acids (ΔFL). No interactions were observed between the Brg-1 two-hybrid clones and a variety of other proteins, including hTcf-1, hTcf-4, mSox-4 and hAPC-2 (not shown). Of particular relevance is the lack of non-specific binding to Armadillo repeats present within the hAPC-2 bait protein, indicating that the interaction is not likely to be a general feature of Armadillo repeat proteins.
β-catenin Armadillo repeats 7–12 mediate binding to a conserved domain in Brg-1
To map the Brg-1 domain responsible for mediating the interaction with β-catenin, we generated a series of Brg-1 prey constructs and tested their ability to interact with the β-catenin bait in yeast (Figure 1B). The shortest fragment of Brg-1 found to retain full β-catenin-binding activity (Brg-C5) contains a region (designated domain II) that is conserved between human and Drosophila forms of Brg-1, but has no assigned function (Khavari et al., 1993). A similar domain exists in the yeast Swi2 homologue, but is considerably shorter (Khavari et al., 1993). The lack of conservation of this interaction domain may reflect the absence of β-catenin homologues in yeast. Deletion of the region adjacent to domain II in Brg-C5 diminished β-catenin binding, indicating that the functional domain may extend N-terminal to that defined by sequence homology.
Similarly, using a series of overlapping β-catenin Armadillo repeat region fragments, we were able to map the region mediating binding to Brg-1 as Armadillo repeats 7–12 (Figure 1C). Attempts to define this interaction domain further were unsuccessful, which may be due to the presence of multiple Brg-1 contact points within this Armadillo repeat region, or may simply reflect disruption of protein folding. This Brg-1 interaction domain overlaps with the C-terminal region of β-catenin recently shown to bind the transcriptional coactivator protein CBP/p300 (Hecht et al., 2000; Takemaru and Moon, 2000). CBP/p300 is considered to promote gene activation by virtue of its HAT activity, following its recruitment to target gene promoters via interaction with the activation domains of transcription factors (Cheung et al., 2000). Additionally, several regions of CBP have been shown to interact with general transcription factors such as TBP, TFIIB and the RNA polymerase II holoenzyme, suggesting that it functions as a coactivator in part by recruiting these proteins to the promoter (Kwon et al., 1994; Nakajima et al., 1997). However, β-catenin proteins containing extensively truncated C-terminal transactivation domains lacking the CBP-binding site still function as potent transactivators in vivo (van de Wetering et al., 1997; Hsu et al., 1998; Hecht et al., 1999). These β-catenin proteins also function in yeast cells that are deficient in CBP and p300 (Hecht et al., 1999). It is therefore likely that this C-terminal transactivation domain of β-catenin binds additional cofactors, such as Brg-1, which are necessary for mediating transactivation of target genes.
β-catenin and Brg-1 interact in vivo
To determine whether the interaction we observed between β-catenin and Brg-1 in the yeast two-hybrid system also occurs in vivo, we transiently co-transfected Flag-tagged Brg-C1 protein and Myc-tagged β-catenin into 293T cells. Immunoprecipitation of Brg-1 using anti-Flag antibody followed by immunoblotting with anti-Myc antibody revealed a 92 kDa band corresponding to β-catenin (Figure 2A). In addition, we could co-immunoprecipitate endogenous β-catenin using anti-haemagglutinin (HA) antibody following induced expression of full-length HA-tagged (K798R) Brg-1 in colon carcinoma cell line DLD1 stable transfectants (Figure 2B).
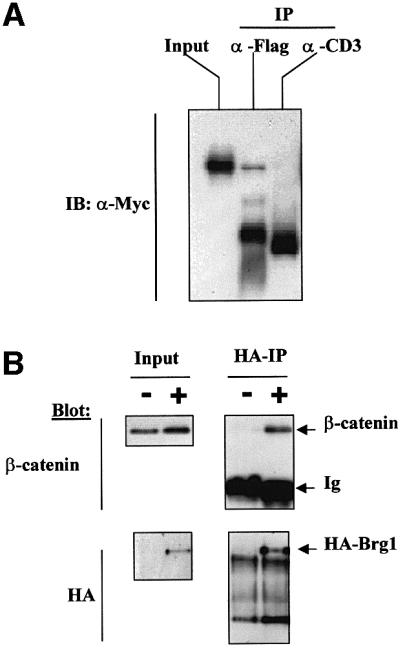
Fig. 2. β-catenin and Brg-1 interact in vivo. (A) 293T cells were transfected with plasmids expressing N-terminal Flag-tagged Brg-C1 clone (amino acids 56–587) and N-terminal Myc-tagged β-catenin. Whole-cell lysates were prepared 24 h later. Extracts were immuno precipitated with anti-Flag antibody or control anti-CD3 antibody as indicated. Precipitated protein was then immunoblotted with anti-Myc antibody to visualize the exogenous tagged β-catenin protein. (B) Expression of full-length K798R Brg-1 protein was induced in DK11 cells by treatment with doxycycline for 24 h. Non-induced (–) and induced (+) cells were then lysed and K798R protein immuno precipitated (IP) with an anti-HA-epitope monoclonal antibody. Precipitated protein was immunoblotted with either anti-HA or anti-β-catenin antibodies. Input lanes show that levels of β-catenin did not differ significantly between non-induced and induced samples, while induction of HA-tagged K798R Brg-1 is clearly visible in lysates before and after immunoprecipitation.
β-catenin and Brg-1 interact to promote transcriptional activation of Tcf-responsive reporter genes
An established function of Brg-1 as a component of the SWI/SNF and Rsc complexes is to mediate chromatin remodelling and, as a consequence, transcription initiation following recruitment to promoter regions of target genes (Biggar and Crabtree, 1999; Cheng et al., 1999; Cosma et al., 1999; Kowenz-Leutz and Leutz, 1999; Xue et al., 2000). The observed interaction between Brg-1 and a region of β-catenin implicated as containing auxiliary transactivation elements (Hecht et al., 1999) led us to consider the possibility that Brg-1-associated chromatin remodelling activity is targeted specifically to Tcf target genes in order to facilitate efficient transcriptional activation. We therefore assessed the effects of co-expressing β-catenin and wild-type Brg-1 on the activity of a Tcf-responsive reporter plasmid containing the Siamois promoter (Brannon et al., 1997), in the Brg-1/Brm-deficient cell line SW13. Expression of S33Yβ-catenin alone resulted in an ∼2- to 3-fold increase in reporter gene activity (Figure 3A). Significantly, this β-catenin-induced signalling activity was doubled by co-expression of wild-type Brg-1. Brg-1 expression in the absence of β-catenin did not affect Tcf reporter gene activity (Figure 3A), whereas Brg-1 alone efficiently enhanced glucocorticoid receptor-mediated transactivation of an MMTV reporter gene (data not shown). Importantly, the effects of β-catenin and Brg-1 on the Siamois reporter gene activity were strictly dependent upon the presence of optimal Tcf-binding sites within the Siamois promoter, indicating that binding of Tcf to target sites mediates recruitment of the β-catenin–Brg-1 complex (Figure 3A, compare wild-type with mutant _Siamois_-luc). The ATPase activity of Brg-1 is known to be essential for its chromatin-remodelling activity (Khavari et al., 1993; Wang et al., 1996b). Loss-of-function mutations in this domain have been shown to inhibit chromatin remodelling at promoters of glucocorticoid receptor target genes, inhibiting hormone-induced transcriptional activation. To assess the contribution of this Brg-1 ATPase activity towards Tcf target gene activation, we co-transfected β-catenin and a previously characterized ATPase mutant form of Brg-1, designated K798R (Khavari et al., 1993), or the partial Brg-C1 clone which lacks the entire ATPase domain together with the Siamois reporter gene, and determined reporter gene activity. As seen in Figure 3A, these mutant forms of Brg-1 are unable to cooperate with β-catenin in promoting transactivation of the Siamois reporter gene.
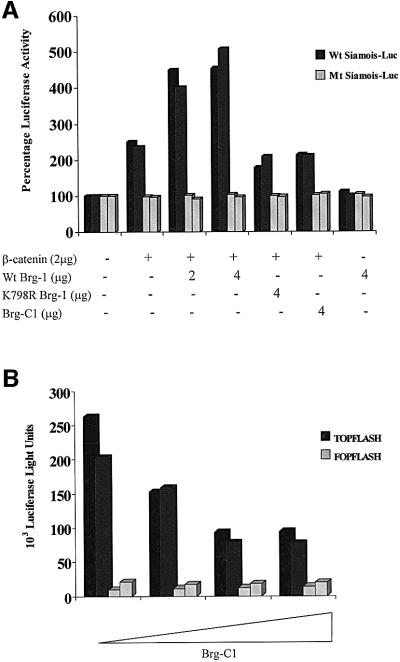
Fig. 3. Brg-1 enhances Tcf–β-catenin transcriptional activity. (A) SW13 cells were transfected with 1 µg of wild-type (black bars) or mutant Siamois reporter plasmid (grey bars) together with the expression constructs indicated. Cells were harvested after 48 h and luciferase activity determined. (B) A partial Brg-1 protein lacking the ATPase domain inhibits constitutive Tcf–β-catenin signalling in DLD1 colon carcinoma cells. A 1 µg aliquot of TOPFLASH (black bars) or FOPFLASH (grey bars) reporter plasmids was transfected into DLD1 cells in the presence or absence of Brg-C1 or full-length Brg-1 expression constructs. Luciferase activities were assayed 48 h later.
We conclude from these assays that a functional consequence of Brg-1–β-catenin complex formation is enhanced transcriptional activity of Tcf–β-catenin complexes on target gene promoters. The observed dependence on an intact ATPase domain of Brg-1 could indicate a role for active remodelling of the chromatin surrounding these promoter regions as a prerequisite for efficient transactivation.
Brg-1 mutants lacking ATPase activity inhibit Tcf–β-catenin signalling in a dominant-negative fashion
To explore further the potential requirement for the ATPase activity of Brg-1 in Tcf–β-catenin transcription, we co-transfected a Tcf reporter gene (TOPFLASH) containing five optimal Tcf-binding sites or the mutant control plasmid (FOPFLASH) into the colon carcinoma cell line DLD1. We previously have shown this cell line to have constitutive Tcf–β-catenin signalling activity resulting from the presence of a highly stable mutant form of β-catenin (Korinek et al., 1997; Morin et al., 1997). In this particular experiment, there was a 5-fold increase in TOPFLASH activity relative to the mutant reporter. Co-transfection of the partial two-hybrid clone Brg-C1, which efficiently binds β-catenin but lacks the ATPase domain, markedly reduces the activity of the TOPFLASH reporter in a dose-dependent manner (Figure 3B). We propose that this non-functional protein is acting to suppress the activity of endogenous Brg-1 by effectively competing for binding to β-catenin–Tcf complexes.
Inducible expression of Brg-1 mutants lacking ATPase activity specifically inhibits endogenous Tcf target gene activity
We recently have identified a set of ∼30 Tcf target genes in LS174T and DLD1 colon carcinoma cell lines (M.van de Wetering et al., in preparation). This was achieved using the T-REx™ system to generate colorectal cell lines with doxycycline-inducible expression of dominant-negative forms of Tcf-1 or Tcf-4 (dn Tcf) lacking the β-catenin-binding site. Induction of dn Tcf in these cell lines was found to down-regulate a relatively small number of gene transcripts, including the proto-oncogenes c-MYC (a previously described Tcf target gene) (He et al., 1998), c-ETS2, c-MYB and c-KIT, and an intestine-specific solute carrier, as assessed by DNA array analysis and confirmed by northern blotting. In an effort to obtain a more physiological readout of Brg-1 function on Tcf target gene activation, we used the same approach to generate colorectal cell lines with inducible expression of K798R Brg-1 (LK1 and LK3 derived from LS174T, and DK11 and DK19 derived from DLD1 cell lines; Figure 4A) and determined the effects of expression of this ATPase-inactive protein on endogenous target gene transcript levels. We observed down-regulation of c-MYC, c-ETS2, c-MYB, c-KIT and intestine-specific solute carrier transcript levels in LS174T and DLD1 cells following induction of K798R Brg-1 expression that mirrored the reduction observed following expression of dn Tcf (Figure 4B). Importantly, expression of genes whose activation is independent of Tcf–β-catenin activity, such as GAPDH, was not affected by expression of K798R Brg-1 (Figure 4B). We conclude from this that the ATPase activity of Brg-1 is required for expression of Tcf target genes, indicating a likely role for chromatin remodelling during transcriptional activation by Tcf–β-catenin.
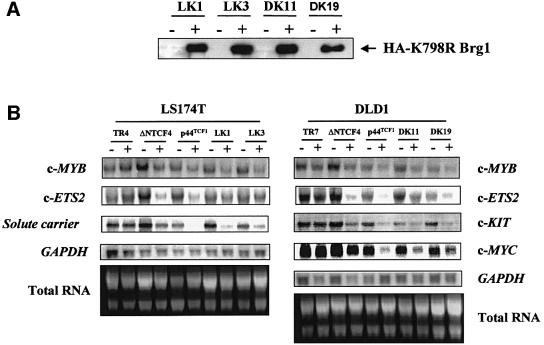
Fig. 4. (A) Induction of K798R Brg-1 expression in stable colon carcinoma cell line transfectants. Induction of K798R Brg-1 expression in LS174T (LK1 and LK3) and DLD1 (DK11 and DK19) transfectants by treatment with doxycycline for 24 h was assessed by western blotting using an anti-HA antibody directed against a C-terminal HA tag. (B) Stable expression of dominant-negative Tcf or K798R Brg-1 specifically down-regulates Tcf target genes. Cells with the indicated inducible expression constructs or parental LS174T TR4 and DLD1 TR7 cells were treated with doxycycline (+) or with vehicle alone (–) and RNA isolated 24 h later. RNA, resolved by electrophoresis and transferred to a nylon membrane, was probed with radiolabelled cDNA fragments of the indicated genes.
Reduction of brahma dosage in Drosophila modifies mutant phenotypes caused by activation or depletion of Armadillo
We selected development in flies as a model system to gain further evidence for a functional interaction between Brg-1 and β-catenin in vivo, assuming that this interaction would be conserved between mammals and Drosophila. We thus asked whether reducing the gene dosage of brahma, the founder of the Brg-1 gene family (Tamkun et al., 1992), affected the mutant phenotypes caused by activation or depletion of Armadillo. First, we used a strain (GMR.Arm*) in which a constitutively activated form of Armadillo is overexpressed in the larval eye disc (Freeman and Bienz, 2001). The mutation in Arm* mimics the oncogenic point mutation S45F in the putative GSK3β phosphorylation site of β-catenin that renders the latter constitutively active (Polakis, 1999). Oncogenic forms of β-catenin such as this are potent transcriptional coactivators of Tcf (Morin et al., 1997). Flies bearing GMR.Arm* show rough and slightly glazed eyes whose size is reduced compared with the wild-type (Figure 5A and B), due to late onset of apoptosis in the pupal disc caused by Arm* and dTcf. This phenotype is independent of armadillo gene dosage, but is reversed considerably towards wild-type in dTcf heterozygotes whose gene dosage is reduced by half (Freeman and Bienz, 2001; Figure 5B and C). This rough eye phenotype was reversed even further towards wild-type in brahma heterozygotes (Figure 5D). Finally, a similar phenotypic suppression was observed in flies heterozygous for moira (Figure 5E), a gene encoding another component of the Brahma complex (Crosby et al., 1999). Importantly, heterozygosity of these genes did not affect the similar rough eye phenotype of GMR.Argos flies whose mitogen-activated protein (MAP) kinase signalling pathway is overactive, nor the rough eyes due to overexpression of GAL4 in the larval disc (Freeman and Bienz, 2001), indicating that the observed genetic interactions in the GMR.Arm* flies are specific. We conclude that the mutant eye phenotype caused by activated Armadillo is as sensitive to the levels of Brahma complex components as it is to dTcf levels, indicating that the Brahma complex is required for the activity of Arm*.
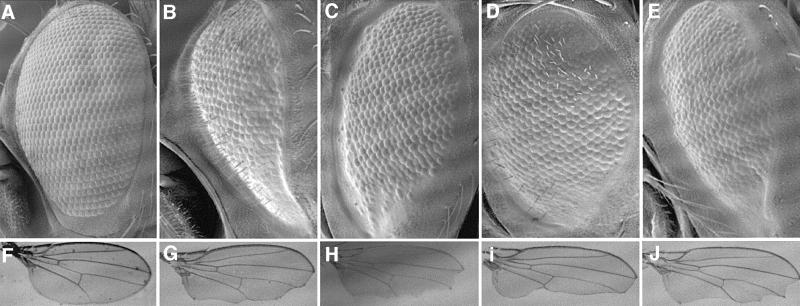
Fig. 5. Genetic interactions between Brahma complex components and Armadillo in Drosophila. Normal fly eye (A) and wing (F), compared with eyes from GMR.Arm* transformants (B–E) and wings from Engrailed.Gal4 UAS.Cad-I transformants (G–J) in different genetic backgrounds; (B and G) y w; (C and H) _dTC3_2/+; (D and I) _brm_2/+; (E and J) _mor_x/+. Note the strong suppression of the Arm* phenotype in the eye, and the strong enhancement of the Armunder phenotype in the wing due to reduced levels of endogenous Brahma. Similar though less pronounced modifications of the same phenotypes are caused by reducing dTcf and Moira levels.
We also asked whether heterozygosity of Brahma complex genes would affect the mutant wing phenotype caused by Armadillo depletion in the wing disc. In the wing, armadillo is required for the integrity of the margin, and sequestration of Armadillo at the membrane by overexpression of the intracellular domain of cadherin (Armunder) in the posterior wing disc causes extensive notches in the posterior wing (Sanson et al., 1996) (Figure 5F and G). This phenotype is worsened by heterozygosity for activating genes of the Wingless pathway, and suppressed by heterozygosity of antagonists of this pathway (Greaves et al., 1999). In particular, the posterior wing margin is completely absent, and the posterior wing area is much reduced, in Armunder flies heterozygous for armadillo (Sanson et al., 1996). Likewise, dTcf heterozygotes showed on average slightly narrower wings, and less residual posterior margin than Armunder controls (Figure 5G and H). This modifying effect of dTcf is much milder than that observed in the eye (see above), perhaps reflecting a dual function of dTcf in the wing margin (activating as well as repressing) similar to that observed in the embryonic cuticle (Cavallo et al., 1998). Significantly, brahma heterozygotes showed considerably narrower wings than the controls (Figure 5G and I). Indeed, brahma heterozygosity enhanced the wing margin phenotype as strongly as armadillo heterozygosity (not shown). Finally, we also observed a slight worsening of this phenotype in moira heterozygotes (Figure 5J). These genetic experiments in flies indicate functional interactions between Brahma complex genes and Armadillo/dTcf. Consistent with this, it has been reported that embryos derived from near-sterile brm transheterozygous mothers show reduced expression of dTcf target genes such as ultrabithorax and engrailed (Brizuela et al., 1994).
Taken together, these fly genetic data support the conclusions from our experiments in mammalian cells that the Brg-1 complex contributes to the activity of the β-catenin–Tcf transcription factor.
Groucho corepressor proteins, which repress Tcf target gene activity in the absence of Wnt signalling, are known to recruit histone deactylases and are likely to effect repression by altering chromatin structure (Peifer et al., 1992; Cavallo et al., 1998; Roose et al., 1998; Chen et al., 1999). Additionally, a recent study demonstrated a role for SWI/SNF-mediated chromatin remodelling of Tcf target gene promoters in ensuring effective repression of gene activity in the absence of β-catenin during fly development (Collins and Treisman, 2000). Potentially, Groucho proteins in complex with Tcf could recruit Brahma complexes to target gene promoters through an interaction mediated by the histone deacetylase rpd3 (Zhang et al., 2000). The data presented here support a mechanism in which β-catenin accumulation following Wnt signalling promotes the formation of β-catenin–SWI/SNF (or –Rsc) complexes in the nucleus, in competition with Groucho repressor complexes. In cooperation with the histone-acetylating activity of β-catenin-bound p300/CBP, Brg-1-associated complexes would then remodel the chromatin structure of target gene promoters into a conformation more accessible to the basal transcription machinery, enhancing transactivation of target genes and leading to cellular responses. The initially paradoxical observation that chromatin-remodelling complexes are required for both the activation and repression of perhaps the same set of target genes can be resolved by the finding that in vitro, SWI/SNF and Rsc can catalyse both forward and reverse nucleosome remodelling reactions (Logie and Peterson, 1997; Lorch et al., 1998).
Materials and methods
Plasmids
All β-catenin bait constructs were generated by cloning the indicated regions in-frame with the DNA-binding domain of yeast Gal4 in the pMD4 vector (a gift of L.van’t Veer). hAPC2, hSOX-4, hTcf-1 and hTcf-4 two-hybrid constructs were gifts of Johan van Es and Annette Baas.
Brg-C5 (amino acids 300–587) and Brg-C6 (amino acids 381–587) were generated by restriction digestion. Brg-C7 (amino acids 341–587) was generated by high fidelity PCR. Brg-C8 (amino acids 476–587) and Brg-C9 (amino acids 508–587) were isolated from an independent two-hybrid screen (A.Hurlstone). All constructs were checked by sequencing.
Flag-tagged Brg-C1 was generated by subcloning the library-derived Brg-C1 (amino acids 56–587) cDNA in-frame with an N-terminal Flag-epitope tag in pcDNA3 (Invitrogen).
Expression plasmids encoding human Brg-1 (pCMV-BRG1) and the ATPase-defective variant of Brg-1 K798R (pCMV-K798R) have been described previously (Murphy et al., 1999), and were a kind gift of Daniel A.Engel (Department of Microbiology and Cancer Center, University of Virginia School of Medicine, Charlottesville, VA). The cDNA insert encoding K798R was shuttled from pCMV-K798R to pcDNA4/TO (Invitrogen) to generate pcDNA4/TO-K798R. pCIneo-S33Yβ-cat has been described previously (Morin et al., 1997). An N-terminal Myc epitope-tagged version of wild-type β-catenin was generated in pcDNA3 by Mascha van Noort.
Yeast two-hybrid screen
The pMD4 β-catenin (Arm1–12) bait was transformed into the HF7C reporter yeast strain using a standard small-scale transformation protocol (Clontech). This bait strain was transformed subsequently with 75 µg of a Matchmaker human fetal brain cDNA library according to the manufacturer’s protocol (Clontech). Positive interacting clones were isolated and identified as previously described (Molenaar et al., 1996).
For two-hybrid analysis of interactions between Brg-1 and β-catenin or control bait proteins, the yeast strain Y190 was co-transformed with bait and prey recombinant vectors in the presence of 20 µg of herring testis carrier DNA. Positive interactions were determined as previously described (Molenaar et al., 1996). Protein interactions were quantified by measuring the activity of a β-galactosidase reporter gene.
Generation of stable cell-lines
LS174T and DLD1 cell lines expressing the Tet repressor from an integrated pcDNA6/TR expression plasmid (part of the T-REx™ system, Invitrogen) (LS174T TR4 and DLD1 TR7, respectively) were gifts of Marc van de Wetering. These cell lines were re-electroporated with pcDNA4/TO-K798R and subclones selected with resistance to blasticidin and Zeocin™ (Invitrogen). Positive clones were identified first by immunohistochemical staining for the in-frame C-terminal HA epitope tag and subsequently by western blotting for the HA epitope. Two subclones each of LS174 TR4 and DLD1 TR7 showing inducible expression of K798R (LK1 and LK3, and DK11 and DK19) were used for further analysis. The construction of cell lines with inducible expression of N-terminal deleted Tcf4 (ΔNTcf4) or the p44 isoform of Tcf1 (p44Tcf1), both of which lack a β-catenin interaction domain, is described elsewhere (M.van de Wetering et al., in preparation) and were also kind gifts of Marc van de Wetering.
Immunoprecipitation and immunoblotting
293T cells were transfected with expression vectors using FuGENE 6 (Boehringer) and harvested 24 h later. Whole-cell extracts were prepared in Triton X-100 lysis buffer (20 mM Tris–HCl pH 8.0, 1% Triton X-100, 140 mM NaCl, 10% glycerol) containing protease inhibitors (Roche). Immunoprecipitations were performed with anti-FlagM2 mouse monoclonal antibody (Sigma catalogue no. A1205) or, as a negative control, an isotype-matched anti-CD3 monoclonal antibody and protein A–Sepharose (Sigma). Beads were washed three times in lysis buffer and immunoprecipitated proteins detected by western blotting with anti-Myc epitope mouse monoclonal antibody (9E12) and subsequently with horseradish peroxidase-conjugated rabbit anti-mouse IgG (Pierce). Immunoreactive proteins were visualized by enhanced chemiluminescence (ECL plus; Amersham Pharmacia Biotech)
DK11 cells were induced to express K798R by incubation with doxycycline (1 µg/ml final concentration) for 24 h or treated with vehicle (ethanol) alone. Cells were then harvested and sonicated in lysis buffer [phosphate-buffered saline (PBS) containing 0.1% IGEPAL, 2 mg/ml bovine serum albumin (BSA), 2 mM dithiothreitol (DTT) and protease inhibitors]. Immunoprecipitation was performed using anti-HA epitope monoclonal antibody (12CA5) and protein A–Sepharose. Precipitated protein was washed three times in lysis buffer and detected by western blotting for the HA epitope tag or endogenous β-catenin using 12CA5 or anti-β-catenin monoclonal antibody (Transduction Laboratories no. C19220), respectively.
Detection of endogenous gene expression
Cell lines harbouring doxycycline-inducible expression constructs or parental clones LS174T TR4 and DLD1 TR7 were incubated with doxycycline (1 µg/ml final concentration) or vehicle (ethanol) alone for 24 h. Total cellular RNA was prepared in TRIZol reagent (Life Technologies) and 10 µg resolved by electrophoresis in a formaldehyde-containing agarose gel before transfer to a Hybond-N nylon membrane (Amersham Pharmacia Biotech). Membranes were probed with radiolabelled human cDNA fragments from the indicated genes. Membranes were washed three times in 0.2× SSC/0.1% SDS at 65°C and signal detected by exposing membranes to a Phosphor Screen read on a Molecular Dynamics PhosphorImager.
Luciferase assay
SW13 cells (2 × 105) were transfected using lipofectamine 2000 (Life Technologies) with a total of 6 µg of the various plasmid combinations: 1 µg of the Siamois promoter reporter plasmid (wild-type pS01234-luc or a derivative mutant version, pS24-luc) (Brannon et al., 1997); 0.05 µg of internal control pRL-TK (Promega); the indicated amounts of S33Yβ-catenin and Brg-1 expression vectors; and empty pcDNA3 vector as stuffer. DLD1 cells (2 × 105) were transfected using lipofectin (Life Technologies) with a total of 6 µg of the various plasmid combinations: 1 µg of reporter plasmid (pTOPFLASH or pFOPFLASH) (van de Wetering et al., 1997); 0.05 µg of internal control pRL-TK; the indicated amounts of the Flag-Brg-C1 expression vector; and empty pcDNA3 vector as stuffer. Luciferase activities were measured 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega).
Cell culture
293T, DLD1 and LS174T cell lines were cultured in Dulbecco’s modified Eagle’s medium (Life Technologies), supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, penicillin and streptomycin. Blasticidin (10 µg/ml) and Zeocin™ (250 µg/ml) (Invitrogen) were included for selection of K798R- or dn-Tcf-expressing subclones. SW13 cells were purchased from ATCC and cultured in Leibowitz medium (Life Technologies) supplemented with 10% FCS, 2 mM glutamine, penicillin and streptomycin.
Drosophila strains and phenotypic analysis
The following fly transformants were used: GMR.Arm* (Freeman and Bienz, 2001); GMR.Argos, GMR.Gal4 (Casci et al., 1999); and Engrailed.GAL4 UAS.cad-I (Sanson et al., 1996). All mutant strains used are described in Flybase (http://flybase.bio.indiana.edu/). All crosses were performed at 25°C.
Flies were prepared for scanning electron microscopy, and wings were mounted in Euparal for viewing under bright-field illumination, as described (Riese et al., 1997).
Acknowledgments
Acknowledgements
We would like to thank Dr Marc van de Wetering for advice on the manuscript. A.H was supported by Wellcome Trust fellowship 054 945.
References
- Adams C.C. and Workman,J.L. (1993) Nucleosome displacement in transcription. Cell, 72, 305–308. [DOI] [PubMed] [Google Scholar]
- Behrens J., von Kries,J.P., Kuhl,M., Bruhn,L., Wedlich,D., Grosschedl,R. and Birchmeier,W. (1996) Functional interaction of β-catenin with the transcription factor LEF-1. Nature, 382, 638–642. [DOI] [PubMed] [Google Scholar]
- Bienz M. and Clevers,H. (2000) Linking colorectal cancer to Wnt signaling. Cell, 103, 311–320. [DOI] [PubMed] [Google Scholar]
- Biggar S.R. and Crabtree,G.R. (1999) Continuous and widespread roles for the Swi–Snf complex in transcription. EMBO J., 18, 2254–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomquist P., Li,Q. and Wrange,O. (1996) The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J. Biol. Chem., 271, 153–159. [DOI] [PubMed] [Google Scholar]
- Brannon M., Gomperts,M., Sumoy,L., Moon,R.T. and Kimelman,D. (1997) A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev., 11, 2359–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M., Brown,J.D., Bates,R., Kimelman,D. and Moon,R.T. (1999) XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development, 126, 3159–3170. [DOI] [PubMed] [Google Scholar]
- Brizuela B.J., Elfring,L., Ballard,J., Tamkun,J.W. and Kennison,J.A. (1994) Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics, 137, 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casci T., Vinos,J. and Freeman,M. (1999) Sprouty, an intracellular inhibitor of Ras signaling. Cell, 96, 655–665. [DOI] [PubMed] [Google Scholar]
- Cavallo R.A., Cox,R.T., Moline,M.M., Roose,J., Polevoy,G.A., Clevers,H., Peifer,M. and Bejsovec,A. (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature, 395, 604–608. [DOI] [PubMed] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.W., Davies,K.P., Yung,E., Beltran,R.J., Yu,J. and Kalpana,G.V. (1999) c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nature Genet., 22, 102–105. [DOI] [PubMed] [Google Scholar]
- Cheung P., Tanner,K.G., Cheung,W.L., Sassone-Corsi,P., Denu,J.M. and Allis,C.D. (2000) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell, 5, 905–915. [DOI] [PubMed] [Google Scholar]
- Collins R.T. and Treisman,J.E. (2000) Osa-containing Brahma chromatin remodelling complexes are required for the repression of wingless target genes. Genes Dev., 14, 3140–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma M.P., Tanaka,T. and Nasmyth,K. (1999) Ordered recruitment of transcription and chromatin remodelling factors to a cell cycle- and developmentally regulated promoter. Cell, 97, 299–311. [DOI] [PubMed] [Google Scholar]
- Cox R.T. et al. (1999) Membrane-tethered Drosophila Armadillo cannot transduce Wingless signal on its own. Development, 126, 1327–1335. [DOI] [PubMed] [Google Scholar]
- Crosby M.A., Miller,C., Alon,T., Watson,K.L., Verrijzer,C.P., Goldman-Levi,R. and Zak,N.B. (1999) The trithorax group gene moira encodes a brahma-associated putative chromatin-remodelling factor in Drosophila melanogaster. Mol. Cell. Biol., 19, 1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. and Bienz,M. (2001) EGF receptor/Rolled MAP kinase signalling protects cells against activated Armadillo in the Drosophila eye. EMBO Rep., 2, 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C.J. and Archer,T.K. (1998) Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature, 393, 88–91. [DOI] [PubMed] [Google Scholar]
- Godde J.S. and Wolffe,A.P. (1996) Nucleosome assembly on CTG triplet repeats. J. Biol. Chem., 271, 15222–15229. [DOI] [PubMed] [Google Scholar]
- Godde J.S., Nakatani,Y. and Wolffe,A.P. (1995) The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res., 23, 4557–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M.A. (1997) Executive decision: chromatin structure and gene regulation. Trends Genet., 13, 387–388. [DOI] [PubMed] [Google Scholar]
- Greaves S., Sanson,B., White,P. and Vincent,J.P. (1999) A screen for identifying genes interacting with armadillo, the Drosophila homolog of β-catenin. Genetics, 153, 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T.C., Sparks,A.B., Rago,C., Hermeking,H., Zawel,L., da Costa,L.T., Morin,P.J., Vogelstein,B. and Kinzler,K.W. (1998) Identification of c-MYC as a target of the APC pathway. Science, 281, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Hecht A., Litterst,C.M., Huber,O. and Kemler,R. (1999) Functional characterization of multiple transactivating elements in β-catenin, some of which interact with the TATA-binding protein in vitro. J. Biol. Chem., 274, 18017–18025. [DOI] [PubMed] [Google Scholar]
- Hecht A., Vleminckx,K., Stemmler,M.P., van Roy,F. and Kemler,R. (2000) The p300/CBP acetyltransferases function as transcriptional coactivators of β-catenin in vertebrates. EMBO J., 19, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Hsu S.C., Galceran,J. and Grosschedl,R. (1998) Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with β-catenin. Mol. Cell. Biol., 18, 4807–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari P.A., Peterson,C.L., Tamkun,J.W., Mendel,D.B. and Crabtree,G.R. (1993) BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature, 366, 170–174. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker,N., Morin,P.J., van Wichen,D., de Weger,R., Kinzler,K.W., Vogelstein,B. and Clevers,H. (1997) Constitutive transcriptional activation by a β-catenin–Tcf complex in APC–/– colon carcinoma. Science, 275, 1784–1787. [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E. and Leutz,A. (1999) A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell, 4, 735–743. [DOI] [PubMed] [Google Scholar]
- Kwon H., Imbalzano,A.N., Khavari,P.A., Kingston,R.E. and Green,M.R. (1994) Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature, 370, 477–481. [DOI] [PubMed] [Google Scholar]
- Laybourn P.J. and Kadonaga,J.T. (1992) Threshold phenomena and long-distance activation of transcription by RNA polymerase II. Science, 257, 1682–1685. [DOI] [PubMed] [Google Scholar]
- Logie C. and Peterson,C.L. (1997) Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J., 16, 6772–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., Cairns,B.R., Zhang,M. and Kornberg,R.D. (1998). Activated RSC–nucleosome complex and persistently altered form of the nucleosome. Cell, 94, 29–34. [DOI] [PubMed] [Google Scholar]
- Molenaar M., van de Wetering,M., Oosterwegel,M., Peterson-Maduro,J., Godsave,S., Korinek,V., Roose,J., Destree,O. and Clevers,H. (1996) XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell, 86, 391–399. [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks,A.B., Korinek,V., Barker,N., Clevers,H., Vogelstein,B. and Kinzler,K.W. (1997) Activation of β-catenin–Tcf signaling in colon cancer by mutations in β-catenin or APC. Science, 275, 1787–1790. [DOI] [PubMed] [Google Scholar]
- Muchardt C. and Yaniv,M. (1993) A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J., 12, 4279–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.J., Hardy,S. and Engel,D.A. (1999) Human SWI–SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol., 19, 2724–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T., Uchida,C., Anderson,S.F., Lee,C.G., Hurwitz,J., Parvin,J.D. and Montminy,M. (1997) RNA helicase A mediates association of CBP with RNA polymerase II. Cell, 90, 1107–1112. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Ostlund Farrants A.K., Blomquist,P., Kwon,H. and Wrange,O. (1997) Glucocorticoid receptor–glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell. Biol., 17, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M. and Polakis,P. (2000) Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science, 287, 1606–1609. [DOI] [PubMed] [Google Scholar]
- Peifer M., McCrea,P.D., Green,K.J., Wieschaus,E. and Gumbiner,B.M. (1992) The vertebrate adhesive junction proteins β-catenin and plakoglobin and the Drosophila segment polarity gene armadillo form a multigene family with similar properties. J. Cell Biol., 118, 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Berg,S. and Reynolds,A.B. (1994) A repeating amino acid motif shared by proteins with diverse cellular roles. Cell, 76, 789–791. [DOI] [PubMed] [Google Scholar]
- Polakis P. (1999) The oncogenic activation of β-catenin. Curr. Opin. Genet. Dev., 9, 15–21. [DOI] [PubMed] [Google Scholar]
- Riese J., Yu,X., Munnerlyn,A., Eresh,S., Hsu,S.C., Grosschedl,R. and Bienz,M. (1997) LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell, 88, 777–787. [DOI] [PubMed] [Google Scholar]
- Roose J., Molenaar,M., Peterson,J., Hurenkamp,J., Brantjes,H., Moerer,P., van de Wetering,M., Destree,O. and Clevers,H. (1998) The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature, 395, 608–612. [DOI] [PubMed] [Google Scholar]
- Sanson B., White,P. and Vincent,J.P. (1996) Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature, 383, 627–630. [DOI] [PubMed] [Google Scholar]
- Takemaru K.I. and Moon,R.T. (2000) The transcriptional coactivator CBP interacts with β-catenin to activate gene expression. J. Cell Biol., 149, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J.W., Deuring,R., Scott,M.P., Kissinger,M., Pattatucci,A.M., Kaufman,T.C. and Kennison,J.A. (1992) brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell, 68, 561–572. [DOI] [PubMed] [Google Scholar]
- van de Wetering M. et al. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell, 88, 789–799. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz,M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- Wang W. et al. (1996a) Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. EMBO J., 15, 5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xue,Y., Zhou,S., Kuo,A., Cairns,B.R. and Crabtree,G.R. (1996b) Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev., 10, 2117–2130. [DOI] [PubMed] [Google Scholar]
- Xue Y., Canman,J.C., Lee,C.S., Nie,Z., Yang,D., Moreno,G.T., Young,M.K., Salmon,E.D. and Wang,W. (2000) The human SWI/SNF-B chromatin-remodelling complex is related to yeast Rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl Acad. Sci. USA, 97, 13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.S., Gavin,M., Dahiya,A., Postigo,A.A., Ma,D., Luo,R.X., Harbour,J.W. and Dean,D.C. (2000) Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell, 101, 79–89. [DOI] [PubMed] [Google Scholar]