Chromatin control of gene expression: Mixed-lineage leukemia methyltransferase SETs the stage for transcription (original) (raw)
Mixed-lineage leukemia recruitment absolutely depends on active transcription.
DNA is tightly packaged inside the nucleus into chromatin, an intricate assembly of nucleic acid, histones, and accessory proteins. Under normal circumstances, the higher-order structure of chromatin is a formidable obstacle for almost all biochemical processes that involve DNA. To enable transcription, replication, and repair, all eukaryotic cells possess a whole array of sophisticated molecular machines with the sole purpose of modifying chromatin structure to allow unhindered access to DNA. Next to ATP-powered units that literally push nucleosomes out of the way, there are many enzymes that catalyze the covalent modification of histones. The various methylations, acetylations, phosphorylations, and ubiquitinations either directly change the packaging properties of the histone octamer or mark these proteins for recognition by other proteins that help to read out this so-called “histone code” (1). A flurry of recent publications describes an ever-growing list of histone modifications, the associated enzymes, and the respective correlation with an “activated” or “repressed” chromatin state. Still, important questions remain. For example, it is well established that histones in actively transcribed regions are acetylated and have a certain methylation pattern. However, we have no clue how modification activities are specifically recruited to coding regions that need to be transcribed. We comprehend neither what happens if a gene must be switched off nor how an enzyme “knows” where a gene starts and where it ends. In this issue of PNAS, Milne et al. (2) begin to address these questions in an elegant system involving the histone H3 lysine 4-specific methyltransferase mixed-lineage leukemia (MLL).
The MLL gene has become notorious because of its frequent involvement in chromosomal translocations (11q23 translocations) that lead to the production of highly oncogenic fusion proteins in leukemias (3). The unaltered MLL protein contains a C-terminal SET domain, the signature sequence of an active methyltransferase (4). After posttranslational cleavage, the resulting MLLN and MLLC parts become incorporated into a high-molecular-weight protein complex (Fig. 1) that also includes the histone H4-specific acetyltransferase MOF (5).
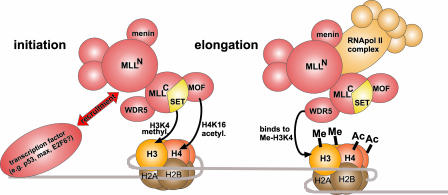
Fig. 1.
A model for the role of the MLL complex in transcriptional regulation. A sequence-specific transcription factor bound to a promoter recruits the MLL complex during transcriptional initiation. The C-terminal SET domain of MLL methylates lysine 4 of histone H3. Concomitantly, lysine 16 of histone H4 is acetylated by MOF. During transcriptional elongation, the MLL complex “rides” with RNApol II through an association of the RNApol II CTD with MLLN, MLLC, and menin. In addition, the WDR5 component specifically recognizes H3 methylated at lysine 4. In this way, the histone modification pattern spreads along an actively transcribed region. Note that not all known complex members are shown and that this diagram is not to scale.
The MLL complex is necessary to ensure proper transcription of the clustered homeobox (Hox) genes during embryogenesis and hematopoiesis (6). Milne et al. (2) exploited the differential expression of MLL-regulated genes (Hoxa9, Hoxc8, and Meis1) in diverse cellular environments to answer the question whether the MLL complex is permanently bound to target chromatin or occupies these genes only transiently when transcription is necessary. The results clearly indicate that MLL recruitment absolutely depends on active transcription. Despite the fact that Hoxc8 is under the direct control of MLL and MLL-precipitated Hoxc8 chromatin in embryonic fibroblasts where the gene is transcribed, the same gene was not bound by MLL in myeloblasts where the locus is silent. A similar situation was encountered for Hoxa9 that is represented by three different mRNAs. Again, MLL associated with Hoxa9 only in myeloblasts that actively transcribe this gene. Down-regulation of Hoxa9 during differentiation to neutrophils was paralleled by a loss of MLL binding, although MLL still was present in the cells. If MLL therefore is controlled by recruitment, then this immediately raises the question of what attracts this protein to target loci. The intrinsic DNA-binding domains of MLL cannot be responsible, because they are not sequence-specific and interact with sequences rich in AT bases and with unmethylated CpG dinucleotides (7, 8). More likely, sequence-specific transcription factors exist that enlist the MLL complex to aid in transcriptional activation. Alternatively, a promoter also could be “prepared” for MLL by particular histone modifications that can be “read” by the MLL complex. Support for both scenarios comes from recent publications describing the biochemical purification and characterization of the MLL complex. Proteomic analysis identified the transcription factors Max and E2F6 as potential complex members, and the tumor suppressor p53 seems to require MLL for efficient activation of the respective target genes. In addition, the WDR5 protein copurified with MLL. WDR5 specifically recognizes histone H3 methylated at lysine in position four (5, 9).
Mixed-lineage leukemia regulates gene expression at the stage of elongation.
Unexpectedly, Milne et al. (2) also could show that MLL binding is not restricted to the TATA box area, where transcriptional initiation takes place. Instead, they found MLL spread across the whole transcriptional unit in a pattern essentially mimicking the distribution of RNA polymerase II (RNApol II) itself. As could be shown in pull-down assays, indeed, the C-terminal domain (CTD) of RNApol II interacted directly with MLLN, MLLC, and menin (10, 11), another MLL complex member. This is an intriguing observation, because studies in yeast have shown that the evolutionarily related complex containing the Saccharomyces histone H3 lysine 4 methyltransferase Set1p also binds to and likely travels alongside RNApol II. This association depends on the Paf1 elongation complex, and Paf1 deletion abrogates H3 lysine 4 methylation (12, 13). Milne et al. (2) found more evidence for a close link between MLL-dependent histone methylation and elongation. In _Mll_-/- cells that can be grown in culture despite the fact that _Mll_-/- animals die in utero, RNApol II seemed to be stalled on MLL-controlled genes. In these cells, RNApol II phosphorylated on serine 2 within the CTD accumulated around the TATA box. A switch from serine 5 to serine 2 phosphorylation characterizes the transition from initiation to the elongation phase, and, concomitantly, serine 2 phosphorylated RNApol II should be found more evenly distributed along the coding region. Reintroduction of MLL into _Mll_-/- cells restored the normal spreading pattern of RNApol II and also reestablished histone H3 lysine 79 methylation, a mark that normally is introduced during elongation. Because elongation is a general process, the question arises whether MLL is necessary for all productive transcription inside a cell and therefore should be regarded as a general transcription factor, as suggested recently (14). According to Milne et al. (2), this seems highly unlikely, because several loci could be identified in Mll knockout cells that were clearly occupied and transcribed by RNApol II despite the absence of Mll. In contrast, genes known to be under the control of MLL also lost RNApol II binding if Mll was not available.
In summary, the results of Milne et al. (2) suggest that MLL regulates gene expression at the stage of elongation. In this regard, it joins a growing group of transcriptional regulators with importance in hematopoiesis like c-Myc and NF-κB that also exert their influence on transcription, at least in part by modulating elongation (15, 16). These molecules influence the activity of pTEF-b, the kinase responsible for phosphorylation of the RNApol II CTD. It will be interesting to see whether crosstalk between MLL and pTEF-b-dependent regulation of elongation exists. Finally, as always in science, several tough questions remain to be answered. First, sequence-specific transcription factors that recruit the MLL complex await discovery. This will probably be facilitated by a better knowledge of direct MLL target genes. Second, it needs to be resolved how histone modification, in particular the methylation mark, is erased once MLL has left a gene that needs to be switched off. The newly discovered demethylating enzymes might be a potential lead here. Third, it is completely unknown how histone methylation expedites transcriptional elongation. And, finally, it would be of utmost interest to work out how the aggressively oncogenic MLL fusion derivatives differ in their transactivation mechanism from MLL and whether one can interfere with this process to develop therapeutic tools.
See companion article on page 14765.
References
- 1.Khorasanizadeh, S. (2004) Cell 116**,** 259-272. [DOI] [PubMed] [Google Scholar]
- 2.Milne, T. A., Dou, Y., Martin, M. E., Brock, H. W., Roeder, R. G. & Hess, J. L. (2005) Proc. Natl. Acad. Sci. USA 102**,** 14765-14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daser, A. & Rabbitts, T. H. (2004) Genes Dev. 18**,** 965-974. [DOI] [PubMed] [Google Scholar]
- 4.Milne, T. A., Briggs, S. D., Brock, H. W., Martin, M. E., Gibbs, D., Allis, C. D. & Hess, J. L. (2002) Mol. Cell 10**,** 1107-1117. [DOI] [PubMed] [Google Scholar]
- 5.Dou, Y., Milne, T. A., Tackett, A. J., Smith, E. R., Fukuda, A., Wysocka, J., Allis, C. D., Chait, B. T., Hess, J. L. & Roeder, R. G. (2005) Cell 121**,** 873-885. [DOI] [PubMed] [Google Scholar]
- 6.Yu, B. D., Hess, J. L., Horning, S. E., Brown, G. A. & Korsmeyer, S. J. (1995) Nature 378**,** 505-508. [DOI] [PubMed] [Google Scholar]
- 7.Birke, M., Schreiner, S., Garcia-Cuellar, M. P., Mahr, K., Titgemeyer, F. & Slany, R. K. (2002) Nucleic Acids Res. 30**,** 958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeleznik-Le, N. J., Harden, A. M. & Rowley, J. D. (1994) Proc. Natl. Acad. Sci. USA 91**,** 10610-10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wysocka, J., Swigut, T., Milne, T. A., Dou, Y., Zhang, X., Burlingame, A. L., Roeder, R. G., Brivanlou, A. H. & Allis, C. D. (2005) Cell 121**,** 859-872. [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama, A., Wang, Z., Wysocka, J., Sanyal, M., Aufiero, D. J., Kitabayashi, I., Herr, W. & Cleary, M. L. (2004) Mol. Cell. Biol. 24**,** 5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milne, T. A., Hughes, C. M., Lloyd, R., Yang, Z., Rozenblatt-Rosen, O., Dou, Y., Schnepp, R. W., Krankel, C., Livolsi, V. A., Gibbs, D., et al. (2005) Proc. Natl. Acad. Sci. USA 102**,** 749-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krogan, N. J., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M. A., Dean, K., Ryan, O. W., Golshani, A., Johnston, M., et al. (2003) Mol. Cell 11**,** 721-729. [DOI] [PubMed] [Google Scholar]
- 13.Ng, H. H., Robert, F., Young, R. A. & Struhl, K. (2003) Mol. Cell 11**,** 709-719. [DOI] [PubMed] [Google Scholar]
- 14.Guenther, M. G., Jenner, R. G., Chevalier, B., Nakamura, T., Croce, C. M., Canaani, E. & Young, R. A. (2005) Proc. Natl. Acad. Sci. USA 102**,** 8603-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barboric, M., Nissen, R. M., Kanazawa, S., Jabrane-Ferrat, N. & Peterlin, B. M. (2001) Mol. Cell 8**,** 327-337. [DOI] [PubMed] [Google Scholar]
- 16.Kanazawa, S., Soucek, L., Evan, G., Okamoto, T. & Peterlin, B. M. (2003) Oncogene 22**,** 5707-5711. [DOI] [PubMed] [Google Scholar]