Eng1p, an Endo-1,3-β-Glucanase Localized at the Daughter Side of the Septum, Is Involved in Cell Separation in Saccharomyces cerevisiae (original) (raw)
Abstract
ENG1 (YNR067c), a gene encoding a new endo-1,3-β-glucanase, was cloned by screening a genomic library with a DNA probe obtained by PCR with synthetic oligonucleotides designed according to conserved regions found between yeast exo-1,3-β-glucanases (Exg1p, Exg2p, and Ssg1p). Eng1p shows strong sequence similarity to the product of the Saccharomyces cerevisiae ACF2 gene, involved in actin assembly “in vitro,” and to proteins present in other yeast and fungal species. It is also related to plant glucan-binding elicitor proteins, which trigger the onset of a defense response upon fungal infection. Eng1p and Acf2p/Eng2p are glucan-hydrolyzing proteins that specifically act on 1,3-β linkages, with an endolytic mode of action. Eng1p is an extracellular, heavily glycosylated protein, while Acf2p/Eng2p is an intracellular protein with no carbohydrate linked by N-glycosidic bonds. ENG1 transcription fluctuates periodically during the cell cycle; maximal accumulation occurs during the M/G1 transition and is dependent on the transcription factor Ace2p. Interestingly, eng1 deletion mutants show defects in cell separation, and Eng1p localizes asymmetrically to the daughter side of the septum, suggesting that this protein is involved, together with chitinase, in the dissolution of the mother-daughter septum.
The yeast cell wall is a rigid structure that preserves the osmotic integrity of the cell and determines cellular morphology during the different stages of the life cycle. In Saccharomyces cerevisiae, the cell wall is essentially made up of highly mannosylated proteins and three different polysaccharide chains: (i) the predominant, linear 1,3-β-glucan, (ii) a minor, highly branched 1,6-β-glucan, and (iii) chitin. All of these components are covalently linked in vivo as part of a macromolecular structure composed of what has been called the “flexible building block,” in which mannoproteins are linked to the 1,3-β-glucan either directly (in the case of PIR proteins) or through a molecule of 1,6-β-glucan (glycosylphosphatidylinositol proteins) (reviewed in references 8, 43, and 57).
Glucans are the main components of the yeast cell wall and are responsible for the rigidity and mechanical strength of this structure. Although they do not undergo appreciable turnover during vegetative growth, it has been proposed that limited site-directed hydrolysis of the rigid skeletal wall β-glucans, mediated by endogenous β-glucanases, probably takes place during several morphogenetic processes, such as budding, wall growth, conjugation, and ascus formation (8, 42, 58).
Two broad classes of 1,3-β-glucanases occur in yeasts, the exoglucanases and the endoglucanases. As measured by their activity on the substrate laminarin (a linear 1,3-β-glucan), exo-1,3-β-glucanases account for the greater part of total glucanase activity in yeasts and hydrolyze the β-O-glycosidic linkages at the nonreducing end of the polymer chain, resulting in the release of glucose. These enzymes are not particularly specific because they usually also act on 1,6-β linkages (as measured with pustulan, a linear 1,6-β-glucan substrate), although with less efficiency, and even have β-glycosidase activity, since they hydrolyze synthetic glycosides such as _p_-nitrophenyl-β-d-glucoside and 4-methylumbelliferyl-β-d-glucoside. Endo-1,3-β-glucanases attack the linkages at intermediate points of the polymer chain, releasing a mixture of oligosaccharides, with glucose as a minor product.
In recent years, much work has focused on characterization of the genetic system governing 1,3-β-glucanase synthesis in yeasts. Three exo-1,3-β-glucanase-encoding genes (EXG1, EXG2, and SSG1) and one endo-1,3-β-glucanase-encoding gene (BGL2) in S. cerevisiae have been cloned and characterized. EXG1 (also known as BGL1 [28]) codes for a polypeptide whose differential glycosylation at the two potential N-glycosylation sites accounts for the two main extracellular exo-1,3-β-glucanases (ExoIa and ExoIb) detected in culture supernatants (38, 63). EXG2 codes for a highly glycosylated minor exo-1,3-β-glucanase (ExoII), with a C-terminal structure characteristic of polypeptides attached to the plasma membrane through a glycosylphosphatidylinositol anchor (8, 10). The SSG1 gene (also known as SPR1) specifies a sporulation-specific exo-1,3-β-glucanase, which is transcribed only in the latter stages of the process, beginning at the time of meiosis and reaching a maximum during spore formation (37, 54). The Exg1p, Exg2p, and Ssg1p proteins are extremely similar, with several highly conserved regions located in the same relative positions in all three polypeptides, which may be essential for their activity (8). BGL2 codes for a cell wall-associated glycoprotein that specifically binds to insoluble glucan and chitin (23, 36). Bgl2p does not show any similarity to these exo-1,3-β-glucanases; instead, it displays considerable similarity to plant 1,3-β-glucanases.
More recent work has allowed the identification of three sodium dodecyl sulfate (SDS)-extractable cell wall proteins (designated Scw3p, Scw4p, and Scw10p) that show significant similarity to glucanases (4). Although no experimental evidence was offered concerning the enzymatic activity of these proteins, it was suggested that they most likely represent potential endoglucanases or possibly transglycosidases. Furthermore, a search in the S. cerevisiae genome for other proteins related in sequence to the Scw proteins and to Exg1p and Bgl2p glucanases disclosed six additional genes (YBR056w, YGL028c, YIL123w, YKR042w, YJL116c, and YMR244c); it was therefore proposed that as many as 13 genes in the yeast genome may encode polypeptides with glucanase or related enzymatic activities. Finally, three new cell wall proteins (named Crh1p, Crh2p, and Crr1p) with similarity to bacterial β-glucanases and eukaryotic endo-transglycosidases have been identified (51).
This article reports the molecular cloning and characterization of two new S. cerevisiae genes (ENG1 and ACF2/ENG2) that code for proteins with detectable endo-1,3-β-glucanase activity and are not related in sequence to any of the aforementioned glucanases. Microscopic observation of eng1 deletion mutants revealed the presence of groups of cells that fail to separate, suggesting that the Eng1p endo-1,3-β-glucanase is involved in mother-daughter separation.
MATERIALS AND METHODS
Strains and culture conditions.
The S. cerevisiae strains used in this study are listed in Table 1. Yeast cells were grown routinely in YEPD or synthetic complete (SC) medium (55). Ura+ and Leu+ selections were carried out on SC medium without uracil or leucine, respectively. Transformants carrying the kanMX4 or the hphMX4 cassette were selected on YEPD plates containing geneticin (200 μg/ml) or hygromycin B (300 μg/ml), respectively. When 1,3-β-glucanase activity was determined on culture supernatants, the SC medium was buffered with 50 mM potassium phosphate (pH 6.5). To induce sporulation, cells were harvested from presporulation medium (0.5% yeast extract, 0.6% yeast nitrogen base, 0.5% peptone, 1% potassium acetate, 1% potassium biphthalate, pH 5.5) at 1 × 107 to 2 × 107 cells/ml, washed twice with sporulation medium (1% potassium acetate), and resuspended at 1.5 × 107 cells/ml in the same medium.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| α131-20 | _MAT_α ade2 ura3 leu1 can1 cyh2 | J. E. Haber |
| W303-1A | MATaura3 leu2 his3 trp1 ade2 can1 | S. Lindquist |
| YPA58 | W303-1A exg1_-Δ_1 exg2::LEU2 ssg1::URA3 | Our collection |
| YPA84 | W303-1A exg1_-Δ_1 exg2::ADE2 | Our collection |
| VB43 | W303-1A eng2::kanMX4 | This study |
| VB44 | W303-1A eng1::URA3 eng2::kanMX4 | This study |
| YPA24 | W303-1A × α131-20: MATa/_MAT_α ura3/ura3 leu2/+ his3/+ trp1/+ ade2/ade2 can1/can1 cyh2/+ | Our collection |
| VB28 | YPA24 eng1::URA3/eng1::URA3 | This study |
| VB53 | YPA24 eng2::kanMX4/eng2::kanMX4 | This study |
| VB54 | YPA24 eng1::URA3/eng1::URA3 eng2::kanMX4/eng2::kanMX4 | This study |
| LS61 | YPA24 ace2::hphMX4/ace2::hphMX4 | This study |
| LS64 | YPA24 swi5::kanMX4/swi5::kanMX4 | This study |
| LS67 | YPA24 ace2::hphMX4/ace2::hphMX4 swi5::kanMX4/swi5::kanMX4 | This study |
| LS79 | YPA24 cts1::hphMX4/cts1::hphMX4 | This study |
| LE30 | YPA24 eng1::URA3/eng1::URA3 cts1::hphMX4/cts1::hphMX4 | This study |
| LS93 | YPA24 egt2::kanMX4/egt2::kanMX4 | This study |
| LS94 | YPA24 eng1::URA3/eng1::URA3 egt2::kanMX4/egt2::kanMX4 | This study |
| AP1 a/α | MATa/_MAT_α ade1/+ ade2/ade2 ura1/+ his7/+ lys2/+ tyr1/+ +/ura3 +/leu1 +/cyh2 +/can1 gal1/+ | J. E. Haber |
| CEN.PK2 | MATa/_MAT_α ura3-52/ura3-52 leu2-3,112/_leu2-3,112 trp1_-289/_trp1_-_289 his3_-1/_his3_-1 | Euroscarf |
Cloning of ENG1 by degenerate PCR.
Specific probes from genomic DNA were obtained by PCR amplification with degenerate oligonucleotides designed according to the highly conserved regions in the S. cerevisiae exo-1,3-β-glucanases. The following oligonucleotides corresponding to the conserved amino acid sequences EPYITPS (oligonucleotide 35), IPIGYWA (oligonucleotide 36), HGAA/PGSQNG (oligonucleotide 37), and DHHHYQ/EVF (oligonucleotide 38) were synthesized: 5′-GAACC(A/T/G)TA(C/T)AT(C/T)AC(A/T/G)CC(A/T)TC-3′ (oligonucleotide 35); 5′-AT(C/T)CC(C/T)ATCGG(A/T)TA(C/T)TGGGC-3′ (oligonucleotide 36); 5′-CC(A/G)TTCTG(C/G)GA(A/T)CCAG(C/G)(A/G)GCACCATG-3′ (oligonucleotide 37); and 5′-AAA(C/G)ACTT(C/G)(A/G)TA(A/G)TGATGATGGTC-3′ (oligonucleotide 38). The sequences of oligonucleotides 37 and 38 are complementary to the coding strand.
PCR was performed with different combinations of oligonucleotides (35 and 37, 35 and 38, and 36 and 38) and genomic DNA from strain YPA58 (exg1 exg2 ssg1) as the template. The amplification products were cloned into the _Sma_I site of the Bluescript KS+ vector (Stratagene). ENG1 was identified by screening a yeast genomic library with a radioactively labeled 750-bp _Bam_HI-_Eco_RI fragment obtained from one of the PCR-derived clones (pD1).
Cloning of ACF2.
ACF2 (YLR144c) was cloned by PCR amplification of the corresponding chromosomal region from strain W303-1A with the XL PCR kit (Perkin Elmer) and primers that hybridized 545 bp upstream from the ATG start codon (5′-TAA_GGATCC_ACATCTTATAAACTCAGT-3′) and 383 bp downstream from the stop codon (5′-GAA_ATCGAT_ACTTTCCACCATCACATTGGA-3′) and introduced _Bam_HI and _Cla_I sites, respectively (italic). The 3.2-kb _Bam_HI-_Cla_I-restricted PCR product was cloned into the pRS426 vector (6) to generate plasmid pVB20.
Plasmid constructions.
Plasmid pSV16 contains an 8.9-kb _Sau_3A S. cerevisiae DNA fragment cloned into the _Bam_HI site of the YEp24 vector (5). Different DNA fragments from this insert were subcloned into the multicopy vector pRS425 (6) and used in 1,3-β-glucanase activity determinations. The resulting plasmids were pVB10 (6.2-kb _Hin_dIII insert), pVB11 (5.1-kb _Xho_I-_Hin_dIII insert), pVB12 (3.8-kb _Pst_I-_Hin_dIII insert), and pVB13 (2.6-kb _Xho_I-_Hin_dIII insert). Plasmid pVB34 is a pEG(KT)-derived plasmid (33) carrying an in-frame glutathione _S_-transferase (GST)-ACF2/ENG2 fusion under the control of the GAL1 promoter. Plasmids pVB35 and pVB36 (derived from pRS425) contain epitope-tagged versions of ENG1 and ACF2/ENG2, with a DNA fragment encoding three copies of the hemagglutinin (HA) epitope introduced in-frame at the C-terminal end of the ENG1 or the ACF2/ENG2 coding sequence.
The eng1::URA3 deletion allele (present in plasmid pVB32) was constructed by replacing the coding region (from +593 to +3318) with a 1.1-kb _Eco_RI-_Cla_I fragment containing the URA3 gene. The eng2::kanMX4 deletion cassette (plasmid pVB33) was constructed following the PCR recombinant method described by Wach (64), replacing the coding sequence (from +33 to +2282) with the kanMX4 module, which confers resistance to geneticin. Construction of the egt2::kanMX4 and swi5::kanMX4 deletion cassettes was performed with the same approach, in each case replacing the whole coding region with the selection marker. The ace2::hphMX4 and cts1::hphMX4 alleles, which confer resistance to hygromycin B, were constructed with the module created by Goldstein and McCusker (18). DNA fragments containing the deletion cassettes were used to transform haploid yeast strains, and their correct integration was checked by Southern blot or PCR amplification. Haploid strains were mated to construct the homozygous diploid deletion strains, isogenic to the wild-type YPA24.
RNA preparation and Northern analysis.
Total RNA was prepared by the method described by San Segundo et al. (54). When necessary, polyadenylated RNA was isolated with the mRNA purification kit (Amersham Pharmacia). The specific probes used to detect the different transcripts were the 1.8-kb _Pst_I-_Pst_I fragment from plasmid pVB10 for ENG1 and the 1.8-kb _Pst_I-_Cla_I fragment from plasmid pVB20 for ACF2/ENG2. Controls used in different experiments were CTS1 (a 0.73-kb fragment obtained by PCR amplification of the region from −41 to +774), SSG1 (a 1.2-kb _Sac_I-_Sal_I fragment of plasmid pPS23 [54]), ACT1 (the 1.7-kb _Bam_HI-_Hin_dIII fragment of plasmid pYact I [41]), HOP1 (the 1.3-kb _Bam_HI-_Hin_dIII fragment of plasmid pNH33-2 [22]), SPO12 (the 0.45-kb _Bam_HI-_Eco_RI fragment of plasmid pRE129 [31]), and SPS100 (a 0.75-kb fragment obtained by PCR amplification of the region spanning from −14 to +745).
Assay for β-glucanase activity.
1,3-β-Glucanase activity was assayed in cell extracts or in culture supernatants as previously described (54). For extract preparation, 109 cells were disrupted by shaking for 40 s with 1.5 g of glass beads (0.5-mm diameter) in a FastPrep FP120 (Bio101 Savant). Culture supernatants were concentrated about 20-fold by ultrafiltration (Amicon Diaflo PM-10 membranes) and dialyzed against acetate buffer (50 mM, pH 5.5) before performing the enzymatic assays. Determination of reducing sugars released in the reactions was performed by the methods of Somogyi (59) and Nelson (40). One unit of activity was defined as the amount of enzyme that catalyzed the release of reducing sugar groups equivalent to 1 μmol of glucose per h, and specific activity was expressed as U per milligram of protein or per milligram of dry cell weight. Protein determinations were carried out by the method of Peterson (45).
To characterize the mode of action of the enzymes, two substrates were used. The first was periodate-oxidized laminarin, which is not degraded by enzymes with an exo-hydrolytic mechanism and is therefore the substrate employed for the identification of endo-1,3-β-glucanases (19). The second was _p_-nitrophenyl-β-d-glucoside, which is degraded only by exo-1,3-β-glucanases. For activity against _p_-nitrophenyl-β-d-glucoside, the amount of _p_-nitrophenol released was determined spectrophotometrically by measuring optical density at 410 nm (39). One unit of enzyme catalyzed the release of 1 μmol of _p_-nitrophenol per h under the reaction conditions used. Substrate specificity was tested with other glucose polymers, such as pustulan (1,6-β-glucan), carboxymethyl cellulose (1,4-β-glucan), dextran (1,6-α-glucan), starch (1,4-α:1,6-α-glucan), lichenan (1,3-β:1,4-β-glucan), and nigeran (1,3-α:1,4-α-glucan). Reaction products were detected as reducing sugar groups released.
Purification of endo-1,3-β-glucanases.
For partial Eng1p purification, culture supernatants from strain YPA84 (exg1 exg2) carrying plasmid pSV16 were concentrated in an Amicon unit equipped with a PM10 membrane, dialyzed against water, and buffered with 50 mM acetate buffer (pH 5.5). Samples (250 mg of protein) were then applied to a DEAE-Bio-Gel A ion-exchange column (20 by 1.5 cm) equilibrated with acetate buffer, which retained the β-glucanase activity, and eluted with a linear NaCl gradient (0 to 0.5 M in acetate buffer; total volume, 200 ml). Fractions with β-glucanase activity were pooled, concentrated, dialyzed, applied to a BioGel A column, and eluted with buffer containing 0.1 M NaCl. Fractions with enzymatic activity were pooled and used for β-glucanase assays.
For Acf2p/Eng2p, a _GST_-ACF2/ENG2 fusion was constructed, expressed in S. cerevisiae under the control of the _GAL1_-inducible promoter, and the fusion protein was purified following the protocol described by Mitchell et al. (33) with minor modifications. In brief, strain YPA84 transformed with plasmid pVB34 or vector pEG(KT) was grown to a density of 2 × 107 cells/ml in SC−Ura selective medium with raffinose as the carbon source. Cultures were induced by adding galactose to a final concentration of 4% and harvested when the cells had doubled in density twice, resuspended in sorbitol buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, and 10 mM Tris-HCl, pH 7.4), and disrupted by shaking with glass beads. Cellular debris was removed by centrifugation at 7,000 × g for 5 min. The supernatant was loaded into a glutathione-Sepharose column and washed with sorbitol buffer, and bound proteins were eluted with 15 mM glutathione.
Immunoblot analysis.
Cells were grown in SC−Leu to mid-log phase (1.5 × 107 cells/ml), washed, disrupted with glass beads in lysis buffer (100 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 7.5), and centrifuged for 10 min at 14,000 × g to remove cell debris. Culture supernatants were concentrated 50-fold by ultrafiltration (Amicon Diaflo PM-10 membranes). Samples were mixed with an equal volume of twofold-concentrated Laemmli sample buffer (53) and boiled for 5 min before loading onto 3 to 15% polyacrylamide gradient gels containing SDS. After electrophoresis, proteins were blotted onto nitrocellulose membranes and probed with anti-HA antibodies (monoclonal antibody 12CA5; Roche), and the immune complexes were detected by chemiluminescence (ECL kit; Amersham). Endoglycosidase H treatment was performed following the instructions of the manufacturer (Roche).
Microscopic observation and indirect immunofluorescence.
Fresh cells were mounted directly in growing medium for microscopic observation. When required, cells were fixed in 4% formaldehyde, incubated at room temperature with gentle shaking for 2 h, and then washed twice with phosphate-buffered saline. Calcofluor White staining was performed as described by Pringle (49). Indirect immunofluorescence was essentially performed as outlined by Pringle et al. (48), except that cells were subjected to a mild treatment (3 min) with zymolyase 20T (12.5 μg/μl) and glusulase (0.02%) or were not treated, depending on the experiment. For detection of Eng1p-HA, cells were incubated overnight at 4°C with primary antibody (mouse monoclonal HA.11; Babco), followed by a 3-h incubation at room temperature with Alexa Fluor 594-conjugated goat anti-mouse immunoglobulin antibody (Molecular Probes). Preparations were photographed with a Leica DMRXA microscope equipped with a Photometrics Sensys charge-coupled device camera and analyzed with the Leica QFISH software.
Nucleotide sequence accession numbers.
The accession number of the protein sequences described in this paper are as follows: Saccharomyces cerevisiae Eng1p (P53753) and Acf2/Eng2p (CAA97716); Schizosaccharomyces pombe SpEng1p (Q9UT45) and SpEng2p (Q09850); Candida albicans Eng1p (Q9UR03) and CaEng2p (Q9UUY9); Aspergillus fumigatus AfEngl1p (AAF13033); and the Bacillus halodurans protein (Q9KG76). The plant glucan-binding elicitor proteins were from Glycine max (BAA11407), Phaseolus vulgaris (AAF19265), and Arabidopsis thaliana (Q9LPQ0 and Q9LFT3).
RESULTS
Cloning of β-glucanase-encoding genes.
Taking advantage of the high degree of similarity exhibited by the S. cerevisiae Exg1p, Exg2p, and Ssg1p exo-1,3-β-glucanases, the cloning of additional putative 1,3-β-glucanase-encoding genes from this yeast was carried out by PCR amplification. Degenerate oligonucleotides designed according to the conserved regions (oligonucleotides 35, 36, 37, and 38) were used as primers to synthesize specific probes from genomic DNA obtained from a triple exg1 exg2 ssg1 deletion mutant (YPA58) to avoid amplification of these three exoglucanases. From all the combinations used, only primers 35 and 38 were able to amplify two DNA fragments of the expected size (around 750 bp), which were cloned in the Bluescript KS+ vector and sequenced. Only one of them (clone pD1) contained a continuous open reading frame, and this PCR product was used to clone the full-length gene by screening a genomic S. cerevisiae DNA library constructed in the multicopy vector YEp24.
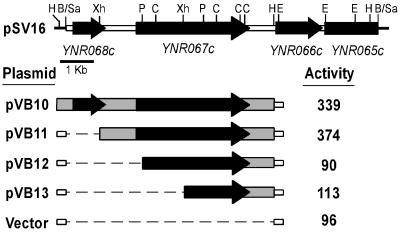
One positive isolate was obtained after screening approximately 30,000 colonies, which contained a plasmid (pSV16, Fig. 1) with a DNA insert of about 8.9 kb carrying three open reading frames (YNR066c, YNR067c, and YNR068c) and part of the YNR065c gene. To check that the cloned fragment did indeed contain a β-glucanase-encoding gene, an exg1 exg2 double mutant (strain YPA84) was transformed with plasmid pSV16, and enzymatic activity was determined in cell extracts with laminarin as the substrate. The 1,3-β-glucan-degradative capacity of cells transformed with this plasmid was about threefold higher than that of strains harboring the vector alone (around 300 versus 90 U), demonstrating that the DNA insert coded for a polypeptide with 1,3-β-glucanase activity.
FIG. 1.
Physical and functional maps of ENG1 (YNR067c). At the top is a map of plasmid pSV16, with arrows indicating the 5′ to 3′ orientation of the different coding sequences. Deletion constructs used to test β-glucanase activity are indicated below, with solid boxes designating the sequences present in each plasmid and dashed lines representing the fragments absent from each construct. To the right are the levels of 1,3-β-glucanase activity (in milliunits per milligram of protein) against laminarin detected in whole-cell lysates in the exg1 exg2 strain YPA84. Restriction sites: B, _Bam_HI; C, _Cla_I; E, _Eco_RI; H, _Hin_dIII; P, _Pst_I; Sa, Sau3A; Xh, _Xho_I.
The sequence of the PCR product used as the probe matched an internal region of YNR067c, suggesting that this gene could be responsible for the increase in β-glucanase activity detected. To confirm this observation, the ability of different DNA fragments cloned in multicopy plasmids to complement the 1,3-β-glucanase-deficient strain was assayed (Fig. 1). As shown, plasmid pVB11 contained all the necessary information to elicit β-glucanase activity levels similar to those produced by cells carrying plasmid pVB10, whereas in the other transformants (containing pVB12 or pVB13), the enzymatic activity against laminarin was similar to that of strains harboring the vector alone. Therefore, YNR067c was named ENG1 (for endo-1,3-β-glucanase; see below). The complete sequence of the 5.1-kb _Xho_I-_Hin_dIII region present in plasmid pVB11 was determined and was found to be identical to the genomic sequence present in the databases.
Eng1p is a member of a family of proteins present in other fungal species and plants.
Eng1p codes for a 1,117-amino-acid protein with a predicted mass of 121,077 Da. Comparison of its amino acid sequence with those of proteins in the databases identified four polypeptides with high similarity to the last two-thirds of the Eng1p sequence, from residue 400 to the C terminus. One of them is the product of the S. cerevisiae YLR144c gene, known as ACF2, which has been shown to code for a 779-amino-acid protein required for cortical actin assembly in an in vitro assay with permeabilized cells (30). The other three proteins were present in the fission yeast Schizosaccharomyces pombe, encoded by eng1+ and eng2+, and in the pathogenic fungus Aspergillus fumigatus, Engl1p. Additionally, by using degenerate oligonucleotides designed according to conserved regions in those proteins, we were able to clone two homologous genes from the dimorphic yeast Candida albicans (Esteban et al., unpublished data); these were designated ENG1 (accession number AJ251464) and ENG2 (AJ251465).
Moreover, a group of plant proteins showed a lower but significant degree of similarity to Eng1p. These proteins, present in Glycine max (GmGBP), Phaseolus vulgaris (PvGBP), and Arabidopsis thaliana (GBP1 and GBP2), have been described as β-glucan elicitor binding proteins, signal transducers located in the plasma membrane of root cells that bind short β-glucan molecules (the elicitors) and trigger the signaling events necessary for the onset of the defense against fungal attacks (34, 62). The last sequence related to Eng1p is a protein of bacterial origin and unknown function that has been identified in the Bacillus halodurans genome sequence project.
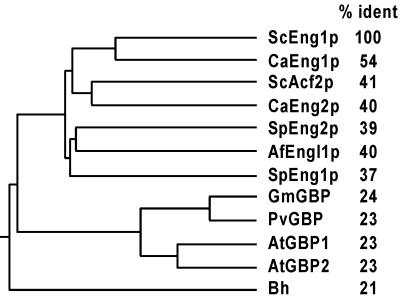
The Clustal program was used to create a multiple alignment of the most conserved domain of the 12 sequences, from amino acids 400 to 1117 in Eng1p, and the derived dendrogram is shown in Fig. 2. The sequences clustered according to their phylogenetic origin, with one branch including the fungal proteins, another containing the plant proteins, and the bacterial protein being the most distantly related member of the family. In the yeast and fungal group, S. cerevisiae Eng1p and C. albicans Eng1p were the most similar pair, with 54% identity in the conserved region. S. cerevisiae Eng1p and G. max GBP showed 24% identity, and S. cerevisiae Eng1p and the B. halodurans protein constituted the two most divergent proteins of the group (21% identity in the conserved domain).
FIG. 2.
Eng1p belongs to a family of conserved proteins. The dendrogram was generated by the Clustal program from the alignment of the protein sequences of yeast and fungal glucanases, including the S. cerevisiae Eng1p (ScEng1p, amino acids 400 to 1117) and Acf2p (ScAcf2p, amino acids 75 to 779), C. albicans Eng1p (CaEng1p, from 428 to 1145) and Eng2p (CaEng2p, from 51 to 734), S. pombe Eng1p (SpEng1p, from 1 to 738) and SpEng2p proteins, and the A. fumigatus protein Engl1p (AfEngl1p). The plant β-glucan elicitor binding proteins were from G. max (GmGBP), P. vulgaris (PvGBP), and A. thaliana (AtGBP1 and AtGBP2). The sequence from the B. halodurans protein (Bh) is also included. The percent identity between Eng1p and each protein (determined by pairwise alignments) is shown in the right column.
Eng1p is an endoglucanase highly specific for 1,3-β-glucans.
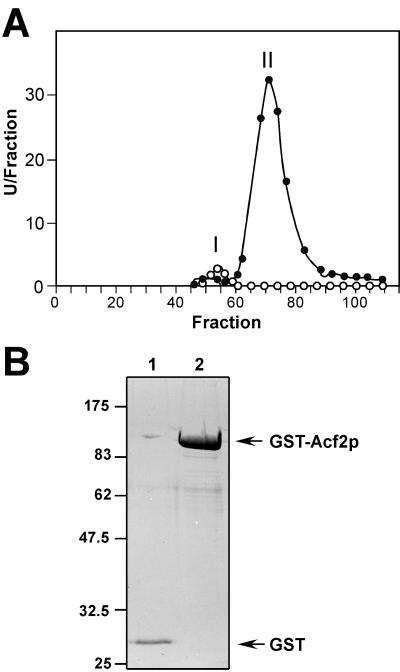
The fact that Eng1p showed little sequence similarity to other known 1,3-β-glucanases prompted us to characterize its substrate specificity and mechanism of action. For this purpose, Eng1p was partially purified from culture supernatants of an exg1 exg2 double mutant (strain YPA84) transformed with a high-copy-number plasmid containing ENG1 (plasmid pVB10). The purification procedure consisted of the use of an anion exchange column (DEAE-Biogel A) followed by filtration chromatography (Biogel A 1.5). As a control, culture supernatants from strain YPA84 carrying the vector alone were subjected to the same treatments. The elution pattern of the filtration chromatography is shown in Fig. 3A. As can be seen, 1,3-β-glucanase activity from the control strain carrying the vector alone (open circles) was very low and eluted as a single peak (labeled I in the figure), while in the fractions from the transformant overexpressing ENG1 (solid circles), an additional peak (peak II) was detected, accounting for about 90% of the total 1,3-β-glucanase activity.
FIG. 3.
Purification of 1,3-β-glucanases encoded by ENG1 and ACF2/ENG2. (A) β-Glucanase activity against laminarin assayed in the fractions eluted from gel filtration chromatography on Biogel A 1.5. The plot represents the elution profile of the β-glucanase activity produced in strain YPA84 (exg1 exg2) carrying ENG1 on high-copy-number plasmid pVB10 (solid circles) or vector alone (open circles), expressed as units per fraction. (B) Purification of the GST-Acf2p fusion protein. A GST-Acf2p fusion protein (lane 2) or GST (lane 1) was expressed in S. cerevisiae under the control of a galactose-inducible promoter and purified on glutathione-Sepharose columns. Proteins used for enzymatic activity determination were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie blue. Sizes are shown in kilodaltons.
Substrate specificity was assayed with laminarin, pustulan, carboxymethyl cellulose, starch, nigeran, lichenan, and dextran. The hydrolytic mode of action was determined with the synthetic compound _p_-nitrophenyl-β-d-glucoside, which is only hydrolyzed by exoglucanases, and periodate-oxidized laminarin, which can only be degraded by endoglucanases. As can be seen in Table 2, Eng1p specifically hydrolyzed molecules containing 1,3-β glycosidic bonds and had an endohydrolytic mode of action, because it was able to degrade periodate-oxidized laminarin but not _p_-nitrophenyl-β-d-glucoside. The slight degradation of lichenan observed can be taken as an indication that Eng1p is much less effective at hydrolyzing 1,3-β bonds adjacent to 1,4-β bonds in this mixed-linkage glucan. Together, these results show that the protein encoded by ENG1 is a glucanase, specifically active on 1,3-β-linkages and with an endo-type mechanism of hydrolysis.
TABLE 2.
Substrate specificity of the Eng1p and Eng2p glucanases
| Substratea | Main linkage type | Enzymatic activityb (% of control) | |
|---|---|---|---|
| Eng1p | Eng2p | ||
| Laminarin | 1,3-β | 100 | 100 |
| IO4-oxidized laminarin | 1,3-β | 95 | 80 |
| Pustulan | 1,6-β | <0.5 | <0.5 |
| Carboxymethyl cellulose | 1,4-β | <0.5 | <0.5 |
| Dextran | 1,6-α | <0.5 | <0.5 |
| Lichenan | 1,3-β:1,4-β | 2.5 | 2 |
| Starch | 1,4-α:1,6-α | <0.5 | <0.5 |
| Nigeran | 1,3-α:1,4-α | <0.5 | <0.5 |
| PNPG | <0.1 | <0.1 |
The polypeptide encoded by ACF2 is also an endo-1,3-β-glucanase.
As mentioned above, ACF2 has previously been identified as a factor required for the assembly of cortical actin in vitro, suggesting that the encoded polypeptide has an intracellular localization (30). Despite this, the strong sequence similarity to Eng1p was an indication that Eng2p might also display β-glucanase activity. To further investigate this possibility, ACF2 was PCR amplified and cloned in the high-copy-number plasmid pRS426 to create plasmid pVB20. This plasmid was used to transform exg1 exg2 cells, and β-glucanase activity in cell extracts was determined with laminarin as the substrate. Activity was about five times higher in transformants harboring plasmid pVB20 than in control cells carrying only the vector (524 versus 93 U). This result suggests that the protein encoded by ACF2 is a β-glucanase.
To exclude the possibility that the increase in β-glucanase activity was due to an indirect effect, a GST-Acf2p fusion protein expressed under the control of the GAL1 promoter was constructed (plasmid pVB34) and transformed into exg1 exg2 cells (strain YPA84). As a control, the same strain was transformed with the original vector, expressing GST only. After culture induction by the addition of galactose to the medium, enzymatic activity determination in cell extracts revealed that the 1,3-β-glucan-degradative ability of transformants carrying the GST-Acf2p fusion was about 22-fold higher than that found in cells transformed with the vector (3,500 versus 154 U). Treatment of the extracts with glutathione-Sepharose beads showed that a large fraction of the 1,3-β-glucanase activity produced by cells expressing the GST-Acf2p fusion was retained in the bead fraction, whereas no activity was detected when extracts from the control strain were used (1,500 versus 0 U). When the bound proteins were eluted from the beads and subjected to gel electrophoresis, only one band of the expected size for the _GST_-_ACF2_-encoded polypeptide was detected (Fig. 3B).
To test for substrate specificity and the hydrolytic mode of action of Acf2p, affinity-purified GST-Acf2p was incubated with different substrates. The results obtained (Table 2) indicated that Acf2p was able to cleave internal 1,3-β linkages, since enzymatic activity was only detected against laminarin and periodate-oxidized laminarin. These data indicate that the protein encoded by ACF2 is an endoglucanase specific for β-1,3 linkages (and will also be referred to as ENG2 in this report).
Eng1p and Acf2p/Eng2p have different cellular localizations and carbohydrate contents.
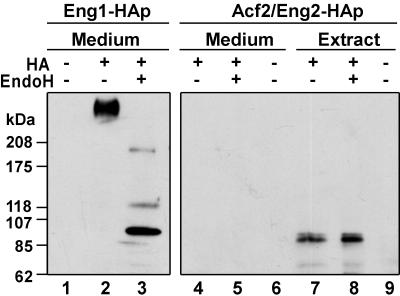
The two endo-1,3-β-glucanases described here show different structural characteristics at the N terminus, suggesting different localizations in the cell. Thus, while Eng1p contains a hydrophobic region with the characteristics of a secretion signal, symptomatic of an extracellular localization (membrane or cell wall), no evident signal sequence could be detected in Acf2p/Eng2p. As a first approach to analyzing the cellular localization of these two proteins, β-glucanase activity was measured with laminarin as the substrate in cell extracts and in culture supernatants of transformants containing ENG1 or ACF2/ENG2 on high-copy-number plasmids (Table 3). A large fraction of the total β-glucanase activity produced by ENG1 was present in the culture medium (around 57%), an indication that the enzyme was being secreted. By contrast, the β-glucanase activity produced by cells overexpressing ACF2/ENG2 was present only in cell extracts, and no enzymatic activity was detected in the culture supernatants. The previous observations were confirmed by Western analysis with two epitope-tagged versions of the endo-1,3-β-glucanases (Eng1p-HA and Eng2p-HA). With anti-HA antibodies, it was confirmed that Eng1p was present in culture supernatants, where it appeared as a diffuse, high-molecular-weight band suggesting that it is a heavily glycosylated protein (Fig. 4, lane 2). However, Eng2p-HA was present only in cell extracts, not in the culture supernatants, and appeared as a 90-kDa protein (lane 7).
TABLE 3.
1,3-β-Glucanase activity levels in S. cerevisiae strains overexpressing ENG1 or ACF2/ENG2
| Plasmid | Gene | 1,3-β-Glucanase activitya (mU/mg of dry cell weight) | |
|---|---|---|---|
| Cell extracts | Culture supernatants | ||
| pRS425 | Vector | 58 | 72 |
| pVB10 | ENG1 | 178 (43) | 235 (57) |
| pVB20 | ACF2/ENG2 | 252 (100) | 69 (0) |
FIG. 4.
Cellular localization and carbohydrate content of the Eng1p and Acf2p/Eng2p proteins. Strain YPA84 (exg1 exg2) was transformed with plasmids containing epitope-tagged versions of ENG1 and ACF2/ENG2 in which three copies of the HA epitope were inserted at the C terminus immediately before the stop codon. Culture supernatants (Medium, lanes 1 to 6) or whole-cell extracts (Extract, lanes 7 to 9) prepared from cells bearing ENG1-HA (lanes 2 and 3) or ACF2/ENG2-HA (lanes 4 to 5 and 7 to 8) were treated with endoglycosidase H (EndoH, lanes 3, 5, and 8) and separated by 3 to 15% gradient SDS-PAGE. Proteins were transferred to nitrocellulose membranes and probed with monoclonal anti-HA antibodies. Lane 1 contains supernatants from mutant cells carrying the wild-type ENG1 gene, and lanes 6 and 9 contain supernatants or whole-cell extracts, respectively, from transformants bearing wild-type ACF2/ENG2.
Cell wall and extracellular proteins usually undergo extensive carbohydrate modifications during their transit through the secretory pathway (24). To investigate whether the endoglucanases were N-glycosylated, culture supernatants or cell extracts from cells expressing the tagged proteins were treated with endoglycosidase H to remove the N-linked carbohydrate (Fig. 4, lanes 3 and 8). For Eng1p-HA, a major band of approximately 90 kDa and two minor forms of 120 and 190 kDa were detected (lane 3), indicating that Eng1p is a glycoprotein with a high content of carbohydrate bound by N-glycosidic bonds. By contrast, the electrophoretic mobility of Acf2p/Eng2p was not altered by endoglycosidase H treatment (lane 8), as expected for an intracellular protein. These results confirm that the two endo-1,3-β-glucanases have different localizations in the cell and that they also have different carbohydrate contents.
ENG1 transcription requires Ace2p.
To assess the size of the transcript and the expression level of ENG1 and ACF2/ENG2 in vegetatively growing cells, Northern analyses were performed. A single 2.5-kb transcript was found when total yeast RNA was hybridized with a radioactively labeled probe specific for ACF2/ENG2. No transcript for ENG1 was observed in these conditions, but a 3.6-kb mRNA molecule was detected when polyadenylated RNA was used, suggesting that the ENG1 expression level is much lower than that of ENG2 (data not shown). The size of both transcripts was in good agreement with the expected size of the gene and indicates that the genes are expressed at different levels during vegetative growth.
DNA microarray analysis of the transcription pattern during the cell cycle has allowed the identification of several groups of coregulated genes (61). ACF2/ENG2 shows no periodic fluctuation during the cell cycle, but ENG1 is one of the genes included in the “SIC1 cluster,” whose expression peaks at the M/G1 transition. Transcription of this group of genes is dependent on the transcriptional activators Ace2p and/or Swi5p. In a recent study, it has been shown that expression of the SIC1 cluster genes has different requirements for these activators: Ace2p is the main regulator of CTS1, SCW11, YHR143w, and YER124c, whereas expression of PIR1, YPL158c, and YNL046w is dependent on Swi5p alone. EGT2, SIC1, and ASH1 require both regulators for maximal transcript accumulation (12). However, no reliable data for ENG1 could be gathered in that study, in concordance with the low level of expression described here.
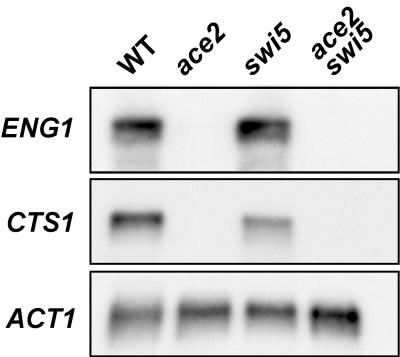
To analyze which of these transcriptional regulators (Ace2p, Swi5p, or both) controls ENG1 expression during the cell cycle, transcription was measured by Northern analysis in isogenic wild-type, ace2, swi5, and ace2 swi5 deletion mutants. As can be seen in Fig. 5, accumulation of ENG1 mRNA was completely dependent on the presence of Ace2p and not Swi5p, since no transcripts were detected in the ace2 or ace2 swi5 mutants. This observation clearly indicates that ENG1 is a cell cycle-regulated gene whose transcription requires Ace2p, similar to CTS1 (encoding chitinase), SCW11, YHR143w, and YER124c.
FIG. 5.
ENG1 expression is dependent on Ace2p. Polyadenylated RNA (3 μg) obtained from vegetatively growing wild-type (WT, strain YPA24) cells and the isogenic ace2 (LS61), swi5 (LS64), and ace2 swi5 (LS67) derivatives was denatured, separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized with specific probes for ENG1, CTS1, and ACT1.
Expression of ACF2/ENG2 is increased during the sporulation process.
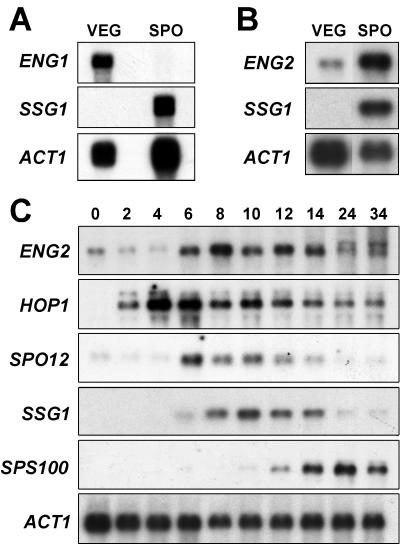
The transcription pattern of the two endo-1,3-β-glucanase-encoding genes during the sporulation process was also analyzed, because during this process extensive synthesis and reorganization of the components of the cell wall occur in order to enclose the four haploid nuclei inside an extremely resistant cell wall (1, 8, 15). ENG1 transcripts were only present in vegetative cells, while a dramatic decrease in transcription was observed during the sporulation process and no expression was detected (Fig. 6A). By contrast, although ACF2/ENG2 was expressed during vegetative growth, its transcription was strongly induced (more than 20-fold) during the sporulation process (Fig. 6B). As a control, the same filters were tested with a specific probe for SSG1, which, as described previously, is only transcribed in sporulating cells and is completely repressed during vegetative growth (37, 54).
FIG. 6.
ENG1 is repressed but ACF2/ENG2 is induced during the sporulation process. (A) Polyadenylated RNA prepared from strain AP1 (a/α) growing vegetatively (VEG) or at 10 h after transfer to sporulation medium (SPO) was denatured, fractionated by electrophoresis (3 μg per lane), and transferred to a nylon membrane. Immobilized RNA was hybridized with radioactively labeled probes specific for ENG1 and SSG1. The ACT1 gene was used as a loading control. (B) Total RNA from strain YPA24 (12 μg per lane) growing vegetatively (VEG) or at 10 h after transfer to sporulation medium (SPO) was hybridized with specific probes for ACF2/ENG2, SSG1, and ACT1. (C) Meiotic time course of ENG2 expression. Total RNA was purified from wild-type cells (strain YPA24) growing vegetatively (time zero) and at the indicated times (in hours) after transfer to sporulation medium. The RNA (12 μg per lane) was hybridized sequentially with the following radioactively labeled gene-specific probes: ACF2/ENG2, HOP1, SPO12, SSG1, and SPS100. The ACT1 gene was used to test for equal loading of RNA in all lanes.
To analyze the ACF2/ENG2 induction kinetics during the sporulation process in greater detail, Northern blot analysis was performed on RNA from the wild-type strain YPA24 during meiosis (Fig. 6C). ACF2/ENG2 transcript levels remained constant during the first 4 h after transfer to sporulation medium, although a sharp rise in the amount of mRNA occurred in the middle period of the sporulation process (6 h), with maximal accumulation at 8 h. The RNA levels remained high until the moment when the first mature asci were observed (12 to 14 h), after which they started to decline slowly.
Several classes of sporulation-specific genes, referred to as early, middle, mid-late, and late genes, are sequentially expressed as the sporulation program proceeds (for reviews, see references 26 and 32). Recently, with whole-genome transcriptional analysis, this classification has been extended to seven different groups, although no induction has been reported for ACF2/ENG2 (7, 47). To determine which of these groups ACF2/ENG2 can be included in, well-known sporulation-specific genes were used as controls, including HOP1, an early gene (22); SPO12, a middle gene (31); SSG1, a mid-late gene (54); and SPS100, a gene belonging the late group (29). The transcription profile of ACF2/ENG2 was very similar to that of SPO12 (Fig. 6C), suggesting that the former is induced with a temporal pattern similar to that of the genes expressed midway through meiosis (32). However, in contrast to the sporulation-specific genes, ACF2/ENG2 transcription was not restricted to the meiotic program.
Eng1p is required for mother-daughter cell separation.
To further elucidate the biological role of ENG1 and ACF2/ENG2, single and double mutants were constructed by replacing the coding region with different selection markers (eng1::URA3 and eng2::kanMX4). Both single and double mutants were viable, and no growth defect in either rich or minimal medium was observed.
A large number of mutants that have defects in cell wall construction or remodeling, such as gas1, fks1, and double crh1 crh2 mutants, are hypersensitive to the cell wall-disturbing agents Calcofluor White and Congo Red (46, 50, 51). Additionally, mutations in components of the Pkc1p-Slt2p pathway exhibit a thermosensitive growth phenotype, which can be suppressed by the addition of 1 M sorbitol to the medium (for review, see reference 21). Analysis of eng1, acf2/eng2, and eng1 acf2/eng2 double mutants indicated that they did not display any of these phenotypes, suggesting that no pronounced defect in cell wall architecture would be associated with the loss of these endo-1,3-β-glucanases in S. cerevisiae.
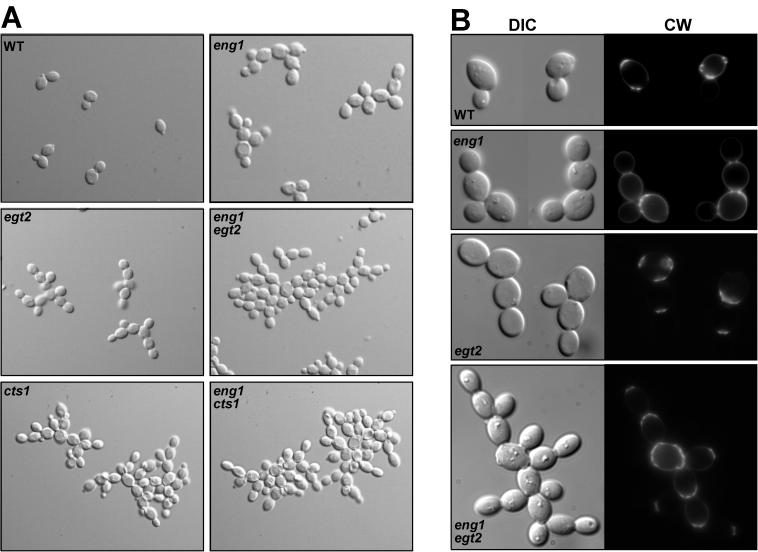
Cell morphology was also analyzed by microscopic observation of wild-type and mutant diploid cells during vegetative growth. Interestingly, exponentially growing eng1 cultures revealed the presence of clusters of cells that had not completed separation (Fig. 7A). When cells were fixed with formaldehyde, only clusters of three or four cells were evident (Fig. 7B), suggesting that the groups of cells were not very tightly associated. These results therefore indicate that eng1 mutants have a defect in cell separation. By contrast, acf2/eng2 cells showed no defect in morphology or chitin distribution (as assessed by Calcofluor White staining) compared with the wild-type strain, and the phenotype of the double eng1 acf2/eng2 mutant was almost identical to that of the eng1 mutant, suggesting that there is no synergistic effect between the two endo-1,3-β-glucanases.
FIG. 7.
eng1 cells have a defect in cell separation. (A) Diploid wild-type (WT, YPA24), eng1 (VB28), egt2 (LS93), eng1 egt2 (LS94), cts1 (LS79), and cts1 eng1 (LE30) cells from exponentially growing cultures were mounted directly on glass slides and photographed with differential interference contrast optics. (B) The same strains grown to mid-log phase were fixed with formaldehyde and stained with Calcofluor White to visualize the chitin. Photographs of differential interference contrast microscopy (DIC, left panels) and fluorescence microscopy (CW, right panels) are shown.
The morphology of eng1 cells clearly resembled the phenotype previously described for egt2 mutants (Fig. 7). Egt2p is required for cell separation, although its role in this process is not clear. It has been proposed that it could be a glucan-degrading enzyme, although no enzymatic activity has been demonstrated, or a protein that regulates certain glucanases (25). Based on the similar phenotype observed for eng1 and egt2 mutants, one intriguing possibility was that Egt2p and Eng1p might act in the same pathway, Egt2p perhaps regulating the activity of the endoglucanase Eng1p. To test this possibility, isogenic single and double mutants were constructed (in a diploid background) and the microscopic appearance of all of them was recorded.
In our strain background, egt2 mutants showed a phenotype similar to that described previously, forming clusters of cells when fresh cultures were observed or groups of three to four cells after fixation. The phenotype of the double eng1 egt2 mutant was more severe than that of any of the single mutants, because cell aggregates much larger than in any of the single mutants were observed, and the cell clusters were tightly associated because they remained together even after fixation (Fig. 7). Similar results were obtained for the haploid strains lacking those genes. This additive phenotype can be taken as an indication that Egt2p does not regulate the activity of Eng1p and that both proteins act in parallel pathways required for completing the mother-daughter separation process.
Chitin distribution in wild-type and mutant cells was analyzed by staining with Calcofluor White, a fluorescent dye that binds this polymer, which is more abundant in the mother bud scars. In wild-type cells, chitin was mainly restricted to the septum that separates mother-daughter cells and to the bud scars present in the mother cell, whereas eng1 mutants showed a more intense staining all over the surface, especially in the mother cell (Fig. 7B). By contrast, staining in egt2 cells was more similar to that in wild-type cells, being restricted to the neck region and bud scars, although the appearance of the scars was more diffuse than in control cells. The double mutant was more similar to egt2 cells, as the fluorescence was preferentially found in the scars and septa.
Double cts1 eng1 mutants were also constructed, and the morphology of the cells was analyzed by microscopic inspection. The severe clumpy phenotype observed in cts1 mutants was not exacerbated by eng1 deletion, consistent with the major role played by chitinase in cell separation (Fig. 7A). Interestingly, in all the single and double mutants analyzed, a switch from the typical bipolar budding pattern to a unipolar pattern was observed in a large number of cells. This switch contributes to the formation of linear chains of cells, partly resembling pseudohyphal growth.
Eng1p localizes to the daughter side of the septum.
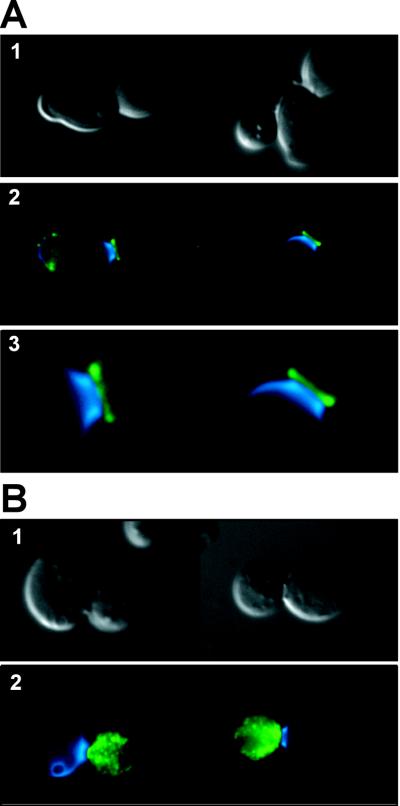
The previous results indicating the participation of Eng1p in cell separation suggested that this endoglucanase must localize to the mother-daughter neck during the time at which the septum is degraded. To confirm this prediction, indirect immunofluorescence with anti-HA monoclonal antibodies was performed in cells carrying the _ENG1_-HA construction (plasmid pVB35), which is fully functional. As expected for a protein presumably involved in septum degradation, the Eng1p endoglucanase mainly localized to the bud neck, although it was distributed asymmetrically in the neck region (Fig. 8A). Thus, when the samples were simultaneously stained with anti-HA antibodies and Calcofluor White and the two fluorescent images were overlaid, the green fluorescence (which corresponds to Eng1p-HA) was always seen at the daughter side of the bud neck, while Calcofluor staining (which corresponds to the primary septum) was concentrated at the opposite side.
FIG. 8.
Eng1p localizes to the daughter side of the septum. Diploid wild-type cells (CEN.PK2 strain) transformed with plasmid pVB35 carrying the ENG1-HA epitope were grown to mid-log phase, fixed, and stained with anti-HA antibodies (HA.11; Babco) to visualize Eng1p by indirect immunofluorescence. Cells were briefly treated with zymolyase 20T and glusulase to partially digest the cell wall (B) or not treated (A) before the primary and secondary antibodies were added. For both panels, images are as follows: 1. differential interference contrast microscopy; 2, overlay of Eng1p-HA fluorescence (green) and Calcofluor White staining of chitin (blue); 3. enlarged view of the mother-daughter neck region (only shown in A).
In newly born cells, a large patch of Eng1-HAp fluorescence remained evident for certain period of time at one of the poles of the cell, which could correspond to the birth scar (data not shown). When immunofluorescence was performed after partially removing the cell wall by mild enzymatic treatment, Eng1p-HA fluorescence was only observed in mother-daughter pairs, exclusively restricted to the newborn cell, confirming that this protein is asymmetrically distributed, accumulating in the daughter cell (Fig. 8B). These results are in good agreement with recently published observations in which chitinase was also found to be localized at the daughter side of the septum (9). In that study, ENG1 was identified as one of the eight genes specifically expressed in daughter cells and for that reason named DSE4.
Thus, all the above observations suggest that the endo-1,3-β-glucanase-encoding gene ENG1 is specifically expressed in daughter cells. The protein is then directed to the septum, where it accumulates in an asymmetrical fashion and helps to degrade the septum from the daughter side to allow cell separation.
DISCUSSION
The yeast cell wall is a complex structure that provides many functions, including maintenance of cell shape, protection from damage by extracellular agents, and protection from osmotic stress. This envelope has often been envisioned as a static structure with a constant composition, although in recent years evidence has accumulated to suggest that it is highly dynamic and changes continuously in response to external and internal cues (8, 43, 57, 58). Whereas 1,3-β-glucan synthase generates long, linear chains of glucan, the 1,3-β-glucan in the cell wall is a branched polymer, indicating that the chains are modified during or after synthesis. The dynamic structural reorganization of the wall architecture during bud emergence, mating, sporulation, and cell separation possibly requires the concerted action of enzymes capable of breaking existing bonds and forming new covalent ones in a process that is highly controlled in both time and space.
For this purpose, endo- and exoglucanase activities, as well as glucanosyltransferases located in the fungal cell wall, are likely to play key roles in wall construction during growth, division, and morphogenesis. Most of the 1,3-β-glucanase activity detected in S. cerevisiae was originally attributed to the products encoded by EXG1, EXG2, SSG1, and BGL2 (23, 36, 63). Current knowledge suggests the existence of additional proteins with related activity, indicating that S. cerevisiae contains a complex system of cell wall-remodeling enzymes. Putative genes that might encode these enzymes have been identified by the isolation of soluble cell wall proteins and by sequence comparison with previously characterized glucanases, although no experimental evidence has been offered concerning their hydrolytic activity against glucan polymers (4, 25, 35, 51)
In this report, we have identified two genes (designated ENG1 and ENG2) that code for two new β-glucanases with high specificity for 1,3-β-glycosidic bonds and with an endohydrolytic mode of action. Interestingly, the two proteins have different characteristics, and while Eng1p is a highly glycosylated protein secreted to the cell wall, the protein encoded by ACF2/ENG2 does not contain N-linked carbohydrate and remains inside the cell, suggesting that the two proteins perform different functions in budding S. cerevisiae. Similar proteins were detected in S. pombe, C. albicans, and A. fumigatus, indicating that this family of proteins is widespread in yeast and fungi. Two different forms are present in S. pombe and in C. albicans with characteristics similar to those of the endo-1,3-β-glucanases from S. cerevisiae (Fig. 9A).
FIG. 9.
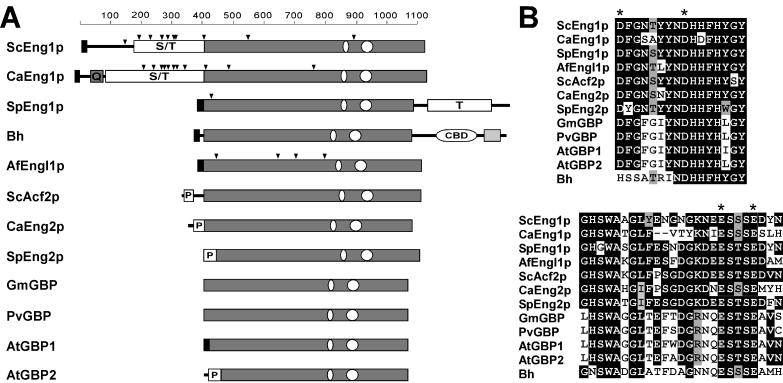
Schematic representation of endo-1,3-β-glucanases and plant β-glucan elicitor binding proteins. (A) The structure of each protein is shown at the same scale (indicated at the top as number of amino acids), with a gray rectangle indicating the region conserved among all the proteins. The abbreviations used to name the proteins are the same as those indicated in Fig. 2. A black box in the N-terminal region indicates the predicted secretory signal sequence, while triangles mark the positions of putative N-glycosylation sites. White boxes represent Ser/Thr-rich regions (indicated by S/T), the Thr-rich domain (marked with a T), Pro-rich regions (P), or a cellulose-binding domain (CBD). A gray box in the sequence of C. albicans Eng1p indicates the presence of a poly-Gln stretch (Q) and a region common to other bacterial xylanases in the sequence of Bh. The oval regions mark the relative positions of the most conserved segments between the proteins whose alignments are shown in panel B, with identities indicated as black boxes and conservative substitutions in gray. The asterisks indicate the conserved Asp and Glu residues that could be part of the catalytic center.
One of the two proteins from each organism (named Eng1p) contained a predicted secretory signal sequence and a Ser/Thr-rich domain, features shared with S. cerevisiae Eng1p. However, the structure of the S. pombe protein is slightly different from that of S. cerevisiae Eng1p, because its Thr-rich domain is located at the C-terminal end of the protein. The pathogenic fungus A. fumigatus also has a protein similar to Eng1p that contains a predicted signal sequence, but no Ser/Thr region could be found. The second protein from S. pombe and C. albicans was more similar to Acf2p/Eng2p (they have accordingly been named SpEng2p and CaEng2p) because a Pro-rich region was present close to the N terminus. Proteins with similarity to S. cerevisiae Eng1p were also present in other yeast and fungal species, including Saccharomyces exiguus, Saccharomyces kluyveri, Kluyveromyces marxianus, Pichia angusta, Debaryomyces hansenii, Pichia sorbitophila, and Candida tropicalis, as found by searching the Génolevures database (60).
A classification of glycosyl hydrolases has been proposed (11; CAZy, http://afmb.cnrs-mrs.fr/≈cazy/CAZY/index.html), and the proteins described here have been grouped in family 81, although no enzymatic activity has previously been reported for any of them. Thus, Eng1p and Acf2p/Eng2p are the first members of this family for which an enzymatic activity has been characterized, supporting their inclusion in the glycosyl hydrolase classification.
Although they have been characterized as a new type of endo-1,3-β-glucanases, it is possible that they also function in vivo as glucanosyltransferases, as has been reported for Bgl2p. This protein was originally described as an exo-1,3-β-glucanase (23) and later as endo-1,3-β-glucanase (36), although further studies have shown that it functions as a β-1,3-glucanosyltransferase, yielding a linear glucan containing a β-1,6 linkage at the transfer site (17). Indeed, glucanosyltransferases can function as hydrolytic enzymes or as transferases, depending on the concentration of the substrate, which serves as both donor and acceptor molecule in this type of reaction. Thus, the possibility that Eng1p (and homologous proteins in other yeasts) might function as a glucanosyltransferase in vivo cannot be ruled out, and a more detailed characterization of its activity will be required to address this possibility.
It is also interesting that one group of plant proteins showed a lower level of similarity to the S. cerevisiae endo-1,3-β-glucanases, the β-glucan elicitor binding proteins (Fig. 2A). These proteins are able to bind a β-glucan elicitor molecule that is released from the cell wall of the infecting fungus by the attack of plant 1,3-β-glucanases, and upon binding the β-glucan elicitor molecule, they trigger the defense response that renders the plant more resistant to fungal infections (reviewed in reference 14). The overall identity between the fungal endo-1,3-β-glucanases and plant proteins was rather low (between 20 to 25%), but the similarity was particularly high in two regions of the proteins, amino acids 855 to 870 and 918 to 946 in Eng1p (indicated by ovals in Fig. 9). These two regions, which contained the most conserved sequences of family 81 proteins, partially overlapped the minimal β-glucan elicitor molecule-binding region of G. max GBP, which has been mapped to the 239 to 442 fragment (62). Since one feature common to β-glucan elicitor binding proteins and endoglucanases is their capacity to bind glucan molecules in order to perform their respective functions, the similarity in these regions could be taken as an indication of their importance in the recognition of molecules with similar structure: the glucan elicitors, in the case of β-glucan elicitor binding proteins, and the cell wall glucans, in the case of endoglucanases.
It is remarkable that Eng1p and Eng2p/Acf2p do not show any similarity to the Exg1p, Exg2p, and Ssg1p exo-1,3-β-glucanases and that they lack all of the conserved blocks of amino acids common to these three proteins except for the DHHHY sequence, which is slightly modified in Eng1p and related proteins (DHHFHY; Fig. 9B). The absence of similarities between Eng1p and Eng2p/Acf2p and other glycosyl hydrolase families prevents the identification of the catalytic residues in this family of proteins. However, sequence alignment of fungal proteins revealed that only 11 acidic amino acid residues, which are assumed to act in the cleavage of the O-glycosidic bonds of glucan chains though a general acid catalysis mechanism (56), are conserved in all these proteins. Four of them (marked by asterisks in Fig. 9B) are located in the most conserved regions, and it is tempting to speculate that either the Asp 855 or the Asp 863 residue (referred to the Eng1p sequence) could be involved in the protonation of the glycosidic bond, whereas the glutamic acid present at either position 942 or 946 would act as the nucleophile, mediating the formation of the glycosyl-enzyme intermediate.
Acf2p/Eng2p is an intracellular endo-1,3-β-glucanase.
All the results obtained indicate that Acf2p/Eng2p is not secreted to the exterior of the cell, which is consistent with previous findings that identified this protein as a factor required for actin assembly (30). However, acf2 mutants present normal actin staining, indicating that this protein is not essential for actin assembly in vivo. Those authors reported the isolation of uncharacterized synthetic lethal mutations in combination with acf2 and suggested that there could be other proteins with a redundant function in the cell. Since the double eng1 eng2 deletion mutant is viable and has no defect in actin assembly in vivo (data not shown), the notion that Eng1p might play a redundant role with Acf2p/Eng2p in actin polymerization can be ruled out.
Although there are no previous descriptions of glucanases present in the cytoplasm of budding yeast, reports have been made of the isolation of two proteins, one from Aspergillus nidulans and the other from Pyricularia oryzae, in which a catalytic chitin synthase domain is fused to a myosin motor-like domain. It has been proposed that chitin synthesis in these fungi may be guided by association of the chitin synthases with some cytoskeletal structures (16, 44). Currently, we have no clear explanation for the role that an endo-1,3-β-glucanase might play in the cytoplasm of the cell, but it has been reported that Acf2p can interact in the two-hybrid system with Rvs167p, a protein that belongs to the Abp1p module and that affects actin distribution and the bipolar budding pattern (13, 20). It is very tempting to suggest that this protein could offer a possible link between the cytoskeleton and the cell wall, although further work will be required to address this possibility.
We have also shown that ACF2/ENG2 expression is strongly induced during the sporulation process, suggesting the possible participation of the protein in this process, in which the synthesis and assembly of a specific spore cell wall takes place. However, the sporulation kinetics of acf2/eng2 mutants is similar to that of wild-type strains, and no morphological defect can be observed in the mutants (data not shown), suggesting that the protein is not essential for sporulation to be completed.
Eng1p localizes to the daughter side of the septum and is involved in cell separation.
At cell division in S. cerevisiae, a septum is formed between the mother and daughter cells, composed of three different layers: an internal layer, mainly composed of chitin (the primary septum), surrounded by two layers (the secondary septum) at both sides, which are similar in composition to the cell wall (reviewed in references 2 and 8). To permit cell separation without lysis, a perfect balance between polymer synthesis and partial degradation is required, and this is most clearly seen in the relationship between chitin synthase and chitinase activities. Chitinase, encoded by the CTS1 gene, is required for partial hydrolysis of the primary septum, allowing the separation of the two cells, whereas chitin synthase I (Chs1p) acts as a repair enzyme, replenishing part of the chitin that is lost by chitinase digestion during cytokinesis (3, 27).
In addition to chitin, other cell wall components, such as glucans, must be partially hydrolyzed for cell separation to be completed, and all the data reported here are consistent with the idea that Eng1p is an endo-1,3-β-glucanase involved in cell separation. Thus, for mother-daughter separation to be achieved, a localized degradation of the cell wall components by specific hydrolytic enzymes (chitinases and glucanases) must occur, and this is a complex process which must be tightly controlled both temporally and spatially.
For correct temporal regulation of the genes involved in cell separation, their transcription is strongly regulated during the cell cycle, restricted to the M/G1 transition (61). Two major transcriptional regulators drive the expression of this group of genes, Ace2p and Swi5p. Scrutiny of the expression pattern and regulation of the genes in this group have allowed their classification into several groups whose expression depends on Ace2p, Swi5p, or both (12). Here we have shown that ENG1 expression is dependent only on Ace2p, like that of CTS1, SCW11, YHR143w, and YER124c.
The spatial regulation of hydrolases during cell separation is also important for the process to be successfully completed. Separation in S. cerevisiae is asymmetric and results in the formation of two different marks in the mother and daughter cells. While the birth scar on the daughter side contains no chitin, in the bud scar that remains on the mother side, a chitin ring is still visible after the action of the chitinase. The molecular bases for this asymmetry are now beginning to be understood. It has recently been shown that Ace2p specifically accumulates in the daughter nucleus, where it activates the expression of a group of daughter-specific genes, most of which are involved in cell separation (9). Interestingly, CTS1 and ENG1 are two of the eight genes specifically expressed in daughter cells identified in that screening (CTS1, SCW11, PRY3, CST13, YER124c/DSE1, YHR143w/DSE2, YOR264w/DSE3, and YNR067c/DSE4).
Here, we have demonstrated that ENG1 codes for an endo-1,3-β-glucanase involved in cell separation and have shown by indirect immunofluorescence and Calcofluor White staining that Eng1p localizes asymmetrically to the daughter side of the septum, in good agreement with the proposed daughter-specific expression of ENG1/DSE4. Thus, cell separation requires the participation of at least Cts1p to degrade the chitin-rich primary septum and the endo-1,3-β-glucanase Eng1p (which might be required for partial hydrolysis of the secondary septum). Both of these proteins are located at the daughter side of the septum, contributing to the asymmetric nature of the separation process in budding yeast. The fact that double cts1 eng1 mutants have a defect similar to that of the cts1 single mutant is an indication that dissolution of the chitin present in the neck region (the chitin ring or the primary septum) is more important than glucan degradation for cell separation to be achieved.
To our knowledge, this is the first report of an S. cerevisiae protein required for mother-daughter separation for which β-glucanase activity has been demonstrated and characterized. Although previous reports have proposed that other proteins involved in cell separation could be β-glucanases (such as Egt2p or Sun4p/Scw3p), no experimental confirmation of enzymatic activity was offered (25, 35). In the case of Egt2p, it was suggested that this protein could be a glucan-degrading enzyme or a regulatory protein that activates other glucan-metabolizing enzymes (25). However, no enzymatic activity was reported, and we were unable to detect any increase in hydrolytic activity in cells overexpressing EGT2 under the control of the GAL1 promoter when laminarin (1,3-β) or pustulan (1,6-β) was used as the substrates (data not shown). This suggests that the protein may have a different substrate specificity or, alternatively, that it plays a regulatory role in glucan-degrading enzymes. The additive effect observed for the eng1 and egt2 mutations suggests that these two proteins play independent roles in cell separation, both being required to allow complete digestion of the septum. If Egt2p does indeed play a regulatory role in cell separation, the synergistic effect observed between the eng1 and egt2 mutations would indicate that Eng1p is not the glucanase regulated by Egt2p.
During filamentous growth, a process characterized by persistent cell-cell adhesion, unipolar bud site selection, and apically polarized actin distribution, the CTS1 and EGT2 genes are repressed, contributing to the maintenance of cell-cell adhesion (reviewed in reference 52). The morphology observed in eng1 and eng1 egt2 mutants (defects in cell separation and a preference for unipolar bud site selection) together with the type of regulation similar to that found for chitinase (as regards both transcription and protein localization) suggest that ENG1 could also be downregulated in the induction of pseudohyphal growth. Further experiments in the appropriate strain background will be required to confirm this possibility. Moreover, the fact that a similar protein is present in the pathogenic yeast C. albicans should allow study of the involvement of the endo-1,3-β-glucanase in the yeast-to-hypha transition and its relationship to pathogenicity.
Acknowledgments
We thank Angel Durán, Pilar Pérez, Beatriz Santos, and Jaime Correa for helpful comments and discussions on the manuscript and Nick Skinner for revision of the manuscript.
This research was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (1FD97-1897-C02-02 and BIO2000-1573) and from the European Community (QKL3-2000-01537). V. Baladrón and S. Ufano were recipients of a fellowship from the Ministerio de Educación y Ciencia (Spain), and A. B. Martín was the recipient of a fellowship from the Ministerio de Ciencia y Tecnología (Spain).
REFERENCES
- 1.Byers, B. 1981. Cytology of the yeast life cycle, p. 59-96. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 2.Cabib, E., D. H. Roh, M. Schmidt, L. B. Crotti, and A. Varma. 2001. The yeast cell wall and septum as paradigms of cell growth morphogenesis. J. Biol. Chem. 276**:**19679-19682. [DOI] [PubMed] [Google Scholar]
- 3.Cabib, E., S. J. Silverman, and J. A. Shaw. 1992. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. J. Gen. Microbiol. 138**:**97-102. [DOI] [PubMed] [Google Scholar]
- 4.Cappellaro, C., V. Mrsa, and W. Tanner. 1998. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 180**:**5030-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, M., and D. Botstein. 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell 28**:**145-154. [DOI] [PubMed] [Google Scholar]
- 6.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110**:**119-122. [DOI] [PubMed] [Google Scholar]
- 7.Chu, S., J. DeRisi, J. Mulholland D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282**:**699-705. [DOI] [PubMed] [Google Scholar]
- 8.Cid, V., A. Durán, F. del Rey, M. Snyder, C. Nombela, and M. Sánchez. 1995. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol. Rev. 59**:**345-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colman-Lerner, A., T. E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107**:**739-750. [DOI] [PubMed] [Google Scholar]
- 10.Correa, J., C. R. Vázquez de Aldana, P. San Segundo, and F. del Rey. 1992. Genetic mapping of 1,3-β-glucanase-encoding genes in Saccharomyces cerevisiae. Curr. Genet. 22**:**283-288. [DOI] [PubMed] [Google Scholar]
- 11.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, UK.
- 12.Doolin, M. T., A. L. Johnson, L. H. Johnston, and G. Butler. 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40**:**422-432. [DOI] [PubMed] [Google Scholar]
- 13.Drees, B. L., B. Sundin, E. Brazeau, J. P. Caviston, G. C. Chen, et al. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154**:**549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebel, J. 1998. Oligonucleotide elicitor-mediated activation of plant defense. BioEssays 20**:**569-576. [DOI] [PubMed] [Google Scholar]
- 15.Esposito, R. E., and S. Klapholtz. 1981. Meiosis and ascospore development, p. 211-287. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Fujiwara, M., H. Horiuchi, A. Ohta, and M. Takagi. 1997. A novel fungal gene encoding chitin synthase with a myosin motor-like domain. Biochem. Biophys. Res. Commun. 236**:**75-78. [DOI] [PubMed] [Google Scholar]
- 17.Goldman, R. C., P. A. Sullivan, D. Zakula, and J. O. Capobianco. 1995. Kinetics of β-1,3 glucan interaction at the donor and acceptor sites of the fungal glucosyltransferase encoded by the BGL2 gene. Eur. J. Biochem. 227**:**372-378. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15**:**1541-1553. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein, I. S., G. W. Hay, B. A. Lewis, and F. Smith. 1965. Controlled degradation of polysaccharides by periodate oxidation reduction and hydrolysis, p. 361-370. In R. L. Whistler (ed.), Methods in carbohydrate chemistry, vol. 5. Academic Press, New York, N.Y.
- 20.Goode, B. L., and A. A. Rodal. 2001. Modular complexes that regulate actin assembly in budding yeast. Curr. Opin. Microbiol. 4**:**703-712. [DOI] [PubMed] [Google Scholar]
- 21.Heinisch, J., A. Lorberg, H. P. Schmitz, and J. J. Jacoby. 1999. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 32**:**671-680. [DOI] [PubMed] [Google Scholar]
- 22.Hollingsworth, N. M., L. Goetsch, and B. Byers. 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61**:**73-84. [DOI] [PubMed] [Google Scholar]
- 23.Klebl, F., and W. Tanner. 1989. Molecular cloning of a cell wall exo-β-1,3-glucanase from Saccharomyces cerevisiae. J. Bacteriol. 171**:**6259-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klis, F. M. 1994. Review: cell wall assembly in yeast. Yeast 10**:**851-869. [DOI] [PubMed] [Google Scholar]
- 25.Kovacech, B., K. Nasmyth, and T. Schuster. 1996. EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol. Cell. Biol. 16**:**3264-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Kuranda, M. J., and P. W. Robbins. 1991. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 266**:**19758-19767. [PubMed] [Google Scholar]
- 28.Kuranda, M. J., and P. W. Robbins. 1987. Cloning and heterologous expression of glycosidase genes from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84**:**2585-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law, D. T., and J. Segall. 1988. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol. Cell. Biol. 8**:**912-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lechler, T., and R. Li. 1997. In vitro reconstitution of cortical actin assembly sites in budding yeast. J. Cell Biol. 138**:**95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malavasic, M. J., and R. T. Elder. 1990. Complementary transcripts from two genes necessary for normal meiosis in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 10**:**2809-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58**:**56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell, D. A., T. K. Marshall, and R. J. Deschesnes. 1993. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9**:**715-723. [DOI] [PubMed] [Google Scholar]
- 34.Mithöfer, A., J. Fliegmann, G. Neuhaus-Url, H. Schwarz, and J. Ebel. 2000. The hepta-β-glucoside elicitor-binding proteins from legumes represent a putative receptor family. Biol. Chem. 381**:**705-713. [DOI] [PubMed] [Google Scholar]
- 35.Mouassite, M., N. Camougrand E. Schwob, G. Demaison, M. Laclau, and M. Guerin. 2000. The ′SUN′ family: yeast SUN4/SCW3 is involved in cell septation. Yeast 16**:**905-919. [DOI] [PubMed] [Google Scholar]
- 36.Mrsa, V., F. Klebl, and W. Tanner. 1993. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-β-glucanase. J. Bacteriol. 175**:**2102-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muthukumar, G., S. H. Suhng, P. T. Magee, R. D. Jewell, and D. A. Primerano. 1993. The Saccharomyces cerevisiae SPR1 gene encodes a sporulation-specific exo-1,3-β-glucanase which contributes to ascospore thermoresistance. J. Bacteriol. 175**:**386-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nebreda, A. R., C. R. Vázquez, T. G. Villa, J. R. Villanueva, and F. del Rey. 1987. Heterogeneous glycosylation of the EXG1 gene product accounts for the two extracellular exo-β-glucanases of Saccharomyces cerevisiae. FEBS Lett. 220**:**27-30. [DOI] [PubMed] [Google Scholar]
- 39.Nebreda, A. R., T. G. Villa, J. R. Villanueva, and F. del Rey. 1986. Cloning of genes related to exo-β-glucanase production by Saccharomyces cerevisiae: characterization of an exo-β-glucanase structural gene. Gene 47**:**245-259. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, M. J. 1957. Colorimetric analysis of sugars. Methods Enzymol. 3**:**85-86. [Google Scholar]
- 41.Ng, R., and J. Abelson. 1980. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 77**:**3912-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nombela, C., M. Molina, R. Cenamor, and M. Sánchez. 1988. Yeast β-glucanases, a complex system of secreted enzymes. Microbiol. Sci. 5**:**328-332. [PubMed] [Google Scholar]
- 43.Orlean, P. 1997. Biogenesis of the yeast wall and surface components, p. 229-362. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Park, I. C., H. Horiuchi, C. W. Hwang, W. H. Yeh, A. Ohta, J. C. Ryu, and M. Takagi. 1999. Isolation of csm1 encoding a class V chitin synthase with a myosin motor-like domain from the rice blast fungus Pyricularia oryzae. FEMS Microbiol. Lett. 170**:**131-139. [DOI] [PubMed] [Google Scholar]
- 45.Peterson, G. L. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83**:**346-356. [DOI] [PubMed] [Google Scholar]
- 46.Popolo, L., D. Gilardelli, P. Bonfante, and M. Vai. 1997. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J. Bacteriol. 179**:**463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Dawis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeast. Nat. Genet. 26**:**415-423. [DOI] [PubMed] [Google Scholar]
- 48.Pringle, J., A. E. M. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast., p. 565-601. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 49.Pringle, J. R. 1991. Staining of bud scars and other cell wall chitin with Calcofluor. Methods Enzymol. 194**:**732-735. [DOI] [PubMed] [Google Scholar]
- 50.Ram, A. F., A. Wolters, R. Ten Hoopen, and F. M. Klis. 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to Calcofluor White. Yeast 10**:**1019-1030. [DOI] [PubMed] [Google Scholar]
- 51.Rodríguez Peña, J. M., V. Cid, J. Arroyo, and C. Nombela. 2000. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 20**:**3245-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rua, D., B. T. Tobe, and S. J. Kron. 2001. Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol. 4**:**720-727. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.San Segundo, P., J. Correa, C. R. Vázquez de Aldana, and F. del Rey. 1993. SSG1, a gene encoding a sporulation-specific 1,3-β-glucanase in Saccharomyces cerevisiae. J. Bacteriol. 175**:**3823-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Sinnott, M. 1990. Catalytic mechanisms of enzymic glycosyl transfer. Chem. Rev. 90**:**1171-1202. [Google Scholar]
- 57.Smits, G. J., J. Kapteyn, H. van den Ende, and F. M. Klis. 1999. Cell wall dynamics in yeast. Curr. Opin. Microbiol. 2**:**348-352. [DOI] [PubMed] [Google Scholar]
- 58.Smits, G. J., H. van den Ende, and F. M. Klis. 2001. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147**:**781-794. [DOI] [PubMed] [Google Scholar]
- 59.Somogyi, M. 1952. Notes on sugar determination. J. Biol. Chem. 195**:**19-23. [PubMed] [Google Scholar]
- 60.Souciet, J. L., M. Aigle, F. Artiguenave, G. Blandin, M. Bolotin-Fukuhara, et al. 2000. Genomic exploration of the hemiascomycetous yeasts. 1. A set of yeast species for molecular evolution studies. FEBS Lett. 487**:**3-12. [DOI] [PubMed] [Google Scholar]
- 61.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9**:**3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Umemoto, N., M. Kakitani, A. Iwamatsu, M. Yoshikawa, N. Yamaoka, and I. Ishida. 1997. The structure and function of a soybean β-glucan-elicitor-binding protein. Proc. Natl. Acad. Sci. USA 94**:**1029-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vázquez de Aldana, C. R., J. Correa, P. San Segundo, A. Bueno, A. R. Nebreda, E. Méndez, and F. del Rey. 1991. Nucleotide sequence of the exo-1,3-β-glucanase gene, EXG1, of the yeast Saccharomyces cerevisiae. Gene 97**:**173-182. [DOI] [PubMed] [Google Scholar]
- 64.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast 12**:**259-265. [DOI] [PubMed] [Google Scholar]