Major Genes Regulating Total Serum Immunoglobulin E Levels in Families with Asthma (original) (raw)
Abstract
Immunoglobulin E (IgE) has a major role in the pathogenesis of allergic disorders and asthma. Previous data from 92 families, each identified through a proband with asthma, showed evidence for two major genes regulating total serum IgE levels. One of these genes mapped to 5q31-33. In the current study, the segregation analysis was extended by the addition of 108 probands and their families, ascertained in the same manner. A mixed recessive model (i.e., major recessive gene and residual genetic effect) was the best-fitting and most-parsimonious one-locus model of the segregation analysis. A mixed two-major-gene model (i.e., two major genes and residual genetic effect) fit the data significantly better than did the mixed recessive one-major-gene model. The second gene modified the effect of the first recessive gene. Individuals with the genotype aaBB (homozygous high-risk allele at the first gene and homozygous low-risk allele at the second locus) had normal IgE levels (mean 23 IU/ml), and only individuals with genotypes aaBb and aabb had high IgE levels (mean 282 IU/ml). A genomewide screening was performed using variance-component analysis. Significant evidence for linkage was found for a novel locus at 7q, with a multipoint LOD score of 3.36 (_P_=.00004). A LOD score of 3.65 (_P_=.00002) was obtained after genotyping additional markers in this region. Evidence for linkage was also found for two previously reported regions, 5q and 12q, with LOD scores of 2.73 (_P_=.0002) and 2.46 (_P_=.0004), respectively. These results suggest that several major genes, plus residual genetic effects, regulate total serum IgE levels.
Immunoglobulin E (IgE) has important functions in the development of allergic disorders and asthma. High total serum IgE levels have been reported to be correlated with the clinical expression of allergy and asthma (Johansson et al. 1972; Burrows et al. 1989; Sears et al. 1991; Halonen et al. 1992). Epidemiologic studies have shown that higher IgE levels are associated with bronchial hyperresponsiveness (BHR), a major component of the asthma phenotype (Hopp et al. 1990; Burrows et al. 1991; Sears et al. 1991). In fact, high total serum levels of IgE predict the development of asthma, independent of other allergic factors. Therefore, an understanding of the genetic mechanisms regulating total serum IgE levels will be important in efforts to dissect the hereditary components of asthma and allergy, complex genetic disorders influenced by the interactions among multiple genes and environmental exposures (Wiesch 1999).
In a study reported elsewhere, our use of two-locus segregation analysis revealed evidence of two major genes and a residual genetic effect regulating total serum IgE levels in the first set of 92 Dutch families ascertained through a parent with asthma (Xu et al. 1995). For the first of these loci, evidence of linkage to chromosome 5q was obtained from both one-locus and two-locus analyses based on a candidate-gene approach (Meyers et al. 1994; Xu et al. 1995). Now that data are available on the total sample of 200 families ascertained through a parent with asthma (all of which originally had been characterized ∼25–35 years earlier), we have performed a genomewide search for genes regulating total serum IgE levels. The purposes of the current study are (1) to determine the relationship between total serum IgE levels and other measures of asthma and allergy, (2) to examine the familial aggregation of total IgE levels by estimating the degree of correlation between relative pairs and by testing the fit of various models by performing one- and two-locus segregation analysis, and (3) to systematically search for the locations of the major genes across the genome, with the use of evenly spaced autosomal markers (∼10 cM apart) and variance-component analyses. High total serum IgE levels, an important phenotype closely associated with asthma and allergic disorders, are an ideal quantitative trait for use with this analytic approach.
Families And Methods
Family Ascertainment
A total of 200 families (1,171 family members) were ascertained through probands who were initially studied during 1962–75 at Beatrixoord Hospital, Haren, the Netherlands, a regional referral center for patients with asthma and other airway obstruction diseases. Patients who had symptomatic asthma without a current asthma exacerbation were referred to this hospital and were admitted for a standardized, complete evaluation. At the time of initial testing, all probands had asthma symptoms, were hyperresponsive to histamine (PC20 forced expiratory volume in 1 s [FEV1] ⩽32 mg of histamine/ml, 30-s method), and were <45 years old. The first 92 probands, together with their spouses, children, children's spouses, and grandchildren >6 years old, were recruited and evaluated in the early 1990s (Panhuysen et al. 1998). To enlarge the sample, 108 probands and their families were collected during 1994–99 by use of the same ascertainment scheme that was used for the first 92 families. This study was approved by the Medical Ethics Committee of the University Hospital Groningen, and all participants signed an informed-consent document. Of the 200 families, 166 consisted of two generations, and 33 consisted of three generations; 1 family consisted of four generations.
Clinical Evaluation
All participants answered a modified British Medical Society respiratory questionnaire, as well as additional questions pertinent to the diagnosis and assessment of asthma and obstructive pulmonary disease. Pulmonary function was tested using standard methods that included spirometry before and after the administration of inhaled salbutamol (800 mg). Testing of bronchial responsiveness to histamine was performed using the method of De Vries et al. (1962), which had been used to assess the initial participants during the period 1962–75. The reactivity-testing protocol consists of having the subject inhale increasing concentrations of histamine, for 30 s of tidal breathing, to a maximum dose of 32 mg of histamine/ml. The test was stopped if FEV1 decreased ⩾20%. Other evaluations included skin tests for responsiveness to 16 common allergens (intracutaneous testing in adults and prick testing in children), a differential blood count (including total eosinophil count), and measures of total serum IgE, as well as IgE specific to house dust and mixed pollens. A positive skin test was defined as the presence of ⩾1 reaction with a wheal diameter ⩾5 mm. Total serum IgE was measured by solid-phase immunoassay (Pharmacia IgE EIA; Pharmacia Diagnostics).
Molecular Methods
Blood samples were shipped from the Netherlands to the molecular genetics laboratory at the University of Maryland at intervals of ∼2 wk. DNA was isolated by standard protocols using a Puregene kit (Gentra). For the genomewide screen, we used the Weber (version 8) set of markers, which spans the human genome at an average interval of 10 cM and consists of 366 autosomal markers, 86% of which are tri- and tetranucleotide repeats, with an average marker heterozygosity of 76%. We performed multiplex PCR using fluorescently labeled primers, separated the resulting amplified fragments on denaturing polyacrylamide gels, detected the fragments with the use of ABI 377 sequencing machines, and scanned and scored the genotypes, using ABI software. A modified version of the program Linkage Designer (Van Camp 1997) was used to bin the alleles and to check inheritance. The output from Linkage Designer was then analyzed further, for any inconsistencies, by operating the LINKAGE software without disease information. As a final check of the data, we used CRIMAP (Lander and Green 1987) to determine the order and length of the chromosomal map and to detect double recombinants.
Statistical Methods
Total serum IgE levels were logarithm transformed (log10) in order to approximate a normal distribution (all analyses were performed using log10(IgE) levels). Because log levels were higher in male subjects and in younger individuals, the effects of sex and age were included in the genetic analyses. In the variance-component analysis, adjustment of the fixed effects of sex and age was performed simultaneously with fitting of the various models. To estimate the correlation coefficients of IgE for pairs of relatives and for complex segregation analysis, the fixed effects of age and sex were removed by taking the residuals after linear-regression analysis had been performed.
Correlation coefficients of total serum IgE levels among various pairs of relatives were estimated from the sums of squares and from cross-products from the pairs, using the computer program FCOR of S.A.G.E. (Statistical Analysis for Genetic Epidemiology). Three distinct weighting methods were used. In the method of equal weight to pairs, every possible pair has equal weight. The pedigrees with a large number of pairs will contribute more information than will pedigrees that contain a relatively small number of pairs. In the method of equal weight to pedigrees, each pedigree, regardless of its size, contributes equal weight. The data are averaged within pedigrees before they are averaged across pedigrees. In the method of equal weight to nuclear families, only the nuclear families (parents and children) are included.
Complex segregation analyses assuming one- and two-locus models were used to evaluate the transmission of high total IgE levels within the 200 families. An ascertainment correction was not used, for the following reasons: (1) the families were ascertained through a parent (not a child) with asthma, not through probands with specific IgE levels; (2) the probands tend to have high IgE levels but with a large range of values; and (3) we are interested in major genes in the specific population of families with asthma. In the one-locus segregation analysis, various models were evaluated, including a general model and Mendelian major-gene models, an environmental model, a polygenic model, and mixtures of various polygenic models with either a major-gene model or an environmental model. The likelihoods that each of these models fit the observed data were computed using the computer software package Pedigree Analysis Package (PAP), revision 4.0. In the cases of mixed models, the likelihoods were approximated by allowing information from previously analyzed family members to represent information about the entire family (Hasstedt 1982). The parameters in the general models include (1) one allele frequency (q a [corresponding to a high value]) and three genotypic frequencies (_F_AA, F_Aa, and F_aa) that were assumed to occur in Hardy-Weinberg equilibrium; (2) three arbitrary transmission probabilities (τAA, τAa, and τaa), representing the probability that an individual of a given genotype transmitted allele A to the offspring; and (3) three arbitrary genotypic means (μAA, μAa, and μaa), a common variance for all the genotypes, and a residual genetic heritability (h_2), which is partitioned from the variance and represents the additive effects of polygenic loci. In the Mendelian models, the three transmission probabilities were fixed to Mendelian inheritance (τ_AA_=1.0, τ_Aa_=.5, and τ_aa_=0). In the environmental models, the three transmission probabilities were set to be the same (τ_AA_=τ_Aa_=τ_aa), reflecting the independence between the parental genotypes and the transmission probabilities. For the Mendelian-only models and the environmental-only models, h2 is set to 0. For the mixed models, h2 is estimated. Within the Mendelian models, a dominant model is derived by setting μ_Aa_=μ_aa, and a recessive model is derived by setting μ_AA_=μ_Aa.
Two-locus segregation analysis was performed using PAP. Analysis was performed to determine whether two major genes, compared with two environmental factors or one major gene, better modeled the segregation of IgE levels in the families. The parameters in the two-locus segregation analyses included the following: allele frequencies at each locus (_q_a and _q_b), the recombination fraction between the two loci (θ), the means (μ) for the nine possible distributions representing the nine types of individuals (AABB, AABb, AAbb, AaBB, AaBb, Aabb, aaBB, aaBb, and aabb), with a common SD, and h2, which is a measure of residual variance within each type. Mendelian models were derived by setting the nine transmission probabilities (probability that an individual of a given genotype transmits allele A and B to his or her offspring) to Mendelian expectations. The environmental model was derived by setting all nine transmission probabilities to equal values, reflecting the independence between parental genotypes and the transmission probabilities.
Two criteria were used to compare the models. For hierarchical models, the likelihood-ratio test was used. Twice the difference, in likelihoods (–2lnL), between a restricted and an unrestricted model approximately follows a χ2 statistic, with degrees of freedom equal to the difference in the number of parameters used in the two models. The best-fitting model is the one requiring the fewest estimated parameters while giving a log likelihood not significantly smaller than that of the unrestricted model. In the comparison of nonhierarchical models, the Akaike’s (1974) information criterion (AIC) was used, −2lnL + 2_k_, where k is the number of parameters estimated in the models. By this criterion, the most parsimonious model is the one with the smallest AIC score.
To estimate the effect of a major gene, we used genotypic probability estimators (GPEs) (Elston and Stewart 1971; Hasstedt and Moll 1989), as implemented in PAP. The genotypic probability (P ij; the probability that individual i carries genotype j,) equals the likelihood conditioned on individual i carrying genotype j, divided by the unconditional likelihood and computed with the parameters set at the maximum-likelihood estimates (MLEs). The mean of a trait Y for genotype j is calculated as μ_j_=Σ_P_ ij/n j, where _y_i is the IgE measured on person i, and n j_=Σ_p ij. A _t_-test was used to determine statistical significance.
Variance-component linkage analysis was used to estimate the proportion of the variance attributable to residual genetic effects, random environmental effects, or quantitative-trait loci (QTL). By fitting various models, it is possible to make inferences regarding the localization (the chromosomal regions mapped by the genetic markers, i.e., linkage) and the magnitude of effect sizes of major genes. Analyses were performed using the computer program package Sequential Oligogenic Linkage Analysis Routines (SOLAR) (Almasy and Blangero 1998), which uses the computer programs FISHER and SEARCH (Lange et al. 1988) for likelihood optimization in quantitative-trait analysis. For model fitting, the fixed effects of the covariates sex and age were removed by simultaneously including them in the models. In the most basic model, the expected covariance matrix for a pedigree is written as Ω=2Φσ2_g_+I_σ2_e, where Φ is the kinship matrix, and I is an identity matrix. To test for evidence of major genes, the component of QTL is introduced into the model, and the expected covariance matrix for a pedigree is written as Ω=Πσ2_a_+2Φσ2_g_+I_σ2_e, where Π is the matrix whose elements (Πσ2_m_π_jl_) provide the predicted proportion of a gene that individual j and l share identical by descent (IBD) at a QTL linked to a genetic-marker locus. Marker-specific IBD matrixes (Πm) were generated independently for all 344 markers across the genome. Multipoint IBD matrixes were then generated at 1-cM resolution by incorporating the IBD matrixes at all the neighboring markers and mapping distances between these markers. To test for linkage, the likelihoods of the two models (with the variance due to the QTL estimated or set to zero) are compared. Twice the difference in loge likelihood of these two models yields a test statistic that is asymptotically distributed as a ½:½ mixture of a χ21 variable and a point mass at zero, because the estimated variance due to QTL was fixed to a boundary in the nested model (Self and Kiang 1987). The difference between the two log10 likelihoods produces a LOD score that is equivalent to the classical LOD score of linkage. Tests for linkage and for its effect are repeated throughout the genome.
Results
Characteristics of Patients
Demographic and clinical characteristics of the family members are shown in table 1. There were more male than female probands; the mean age of the probands was 52 years (51 years for spouses and 24 years for children). Although all probands had been hyperresponsive at the time of original testing, 12% were not hyperresponsive at recent testing (30 were not retested because of low lung function [FEV1 ⩽40% predicted level]). A large proportion (82%) of probands were skin-test positive.
Table 1.
Subject Characteristics of Dutch Families with Asthma
| Characteristic | Probands(n = 200) | Spouses(n = 200) | First-Degree Offspring (n = 530) |
|---|---|---|---|
| M:F | 124:76 | 76:124 | 237:293 |
| Mean age (years) [range] | 52.1 [37–76] | 51.1 [33–76] | 24.0 [6–53] |
| PC20 <32 mg/ml (%) [_n_] | 88.2 [170] | 26.1 [199] | 46.5 [525] |
| FEV1, predicted (%) | 69.7 | 98.4 | 93.6 |
| FEV1 <80% predicted (%) | 61.4 | 9.0 | 12.1 |
| Reversibilitya (baseline) | 77.7 | 21.5 | 31.8 |
| Reversibilitya (predicted) | 62.9 | 19.0 | 25.8 |
| Mean IgE (IU/ml) | 92.9 | 26.3 | 64.1 |
| Positive skin test (%) | 81.9 | 30.6 | 54.1 |
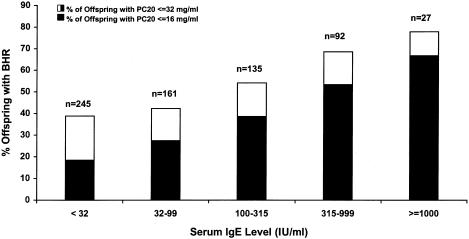
Individuals with BHR had a significantly higher mean IgE level than did individuals without BHR (83 vs. 35 IU/ml; _t_=9.22; P<.0001). The relationship between total serum IgE levels and the presence and degree of BHR is shown in figure 1. In addition, skin-test–positive individuals had significantly higher IgE levels than did individuals with a negative skin test (112 vs. 26 IU/ml; _t_=17.39; P<.0001). Results from multiple-regression analysis, where log10(IgE) was the dependent variable and where BHR, skin-test responsiveness, sex, and age (in years) were independent variables, also suggested that IgE was independently associated with these clinical and demographic variables. The estimated regression coefficients were β=.20 (_P_=.0001) for BHR, β=.59 (_P_=.0001) for skin-test responsiveness, β=-.07 (_P_=.04) for sex (male), and β=-.006 (_P_=.0001) for age.
Figure 1.
Relationship between total serum IgE levels and BHR, in the offspring of 200 asthmatic parents
Correlation Coefficients of Various Relative Pairs
Correlation coefficients (r) for various pairs of relatives, with equal weights assigned to all pairs, are reported in table 2. The results with equal weights to pedigrees and to nuclear pedigrees were similar (data not shown). There was evidence for familial aggregation of high total serum IgE levels, which was probably the result of a genetic component. The results should be interpreted with caution, since these families were ascertained through an asthmatic parent, rather than being randomly selected from the general population. There was a higher correlation among parent-offspring and sibling pairs (_r_=.24 and _r_=.31, respectively) than among more–distantly related relative pairs (_r_=.12 among grandparent-grandchildren pairs, and r approached 0 for avuncular and first-cousin pairs). As expected for unrelated individuals, there was no evidence of correlation between the spouses (_r_=-.06). The h2 was estimated to be .48 among parent-offspring pairs and .62 among sibling pairs.
Table 2.
Correlation Coefficient of Log IgE among Different Relative Classes (Equal Weights to Pairs)
| Relationship | No. of Pairs | Correlation Coefficient |
|---|---|---|
| First-degree relatives: | ||
| Parent-offspring: | ||
| Mother-daughter | 374 | .28 |
| Mother-son | 315 | .20 |
| Father-daughter | 363 | .25 |
| Father-son | 306 | .23 |
| Overall | 1,358 | .24 |
| Sibling: | ||
| Sister-sister | 195 | .28 |
| Sister-brother | 342 | .32 |
| Brother-brother | 151 | .33 |
| Overall | 688 | .31 |
| Second-degree relatives: | ||
| Grandparent-grandchild | 272 | .12 |
| Avuncular | 559 | .02 |
| Third-degree relatives: | ||
| First cousins | 397 | .03 |
| Spouses | 275 | −.06 |
One-Locus Segregation Analyses
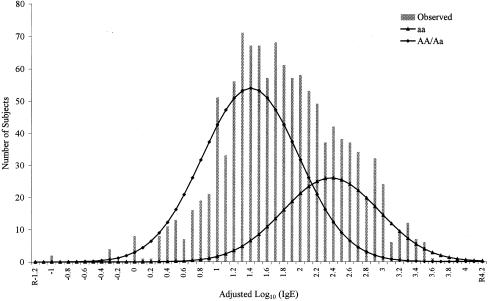
To test whether the familial aggregation of high total IgE levels is due to shared genes or shared environments, complex segregation analyses were performed. In the one-locus segregation analysis, both the sporadic model and the mixed environmental model were rejected (table 3). Although Mendelian-only models were rejected, several mixed Mendelian models were not. These models fit the observed distribution of adjusted log10(IgE) levels and were not significantly different from the general model (_P_>.05). Of the mixed Mendelian models, the mixed recessive model (a major recessive gene and residual genetic effects) was the best-fitting (_P_=.75) and most-parsimonious (smallest AIC) model, although it was only slightly better than the mixed additive model. The MLE of gene frequency _q_a (corresponding to a high IgE level) under the mixed recessive model was .57, which results, under Hardy-Weinberg equilibrium, in genotype frequencies of .68 for AA/Aa and of .32 for genotype aa. When we used GPEs, this recessive gene had a large effect on IgE levels and was responsible for 32.4% of the adjusted log10(IgE) levels in these families. The mean IgE level for genotype aa (μaa) was estimated to be 209 IU/ml, significantly higher than the mean (29 IU/ml) for the other genotypes (μAA, μAa) (_t_-_statistic_=23.89, P = 6.2E–109). This model is plotted in figure 2, illustrating the fit of the model. The residual polygenic component was large, which was evidenced by a high heritability estimate (MLE of _h_2=.49, 95% confidence interval [CI] .35–.63), and a significant improvement in model fitting, compared with a recessive-only model (χ2=40.73; _df_=1; _P_=1.7×10-6).
Table 3.
Segregation Analysis of Adjusted Log10(IgE) under One-Locus Model, in 200 Dutch Families[Note]
| Transmission Probabilityb | Mean | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Modela | _q_A | AA | Aa | aa | AA | Aa | aa | SD | _h_2 | -2lnL | AIC | χ2 | df | P |
| General | .54 | [1] | .47 | [0] | 1.63 | 1.41 | 2.34 | .58 | .57 | 2,356.52 | 2,374.52 | |||
| Env + RGE | .55 | .47 | = AA | = AA | 1.37 | 1.65 | 2.15 | .65 | .65 | 2,365.95 | 2,379.95 | 9.43 | 2 | .009 |
| Add + RGE | .55 | [1] | [.5] | [0] | 1.63 | 1.41 | 2.34 | .58 | .57 | 2,356.69 | 2,368.69 | .17 | 3 | .98 |
| Dom + RGE | .21 | [1] | [.5] | [0] | 1.49 | 2.14 | = Aa | .63 | .47 | 2,365.90 | 2,375.90 | 9.38 | 4 | .052 |
| Rec + RGE | .57 | [1] | [.5] | [0] | 1.45 | = AA | 2.32 | .58 | .49 | 2,358.42 | 2,368.42 | 1.90 | 4 | .754 |
| Add only | .54 | [1] | [.5] | [0] | 1.02 | 1.64 | 2.42 | .5 | [0] | 2,372.41 | 2,382.41 | 25.89 | 4 | .00003 |
| Rec only | .66 | [1] | [.5] | [0] | 1.35 | = AA | 2.22 | .55 | [0] | 2,399.15 | 2,407.15 | 50.63 | 5 | 1.02E–10 |
| RGE | [1] | NA | NA | NA | 1.73 | = AA | = AA | .71 | .55 | 2,366.55 | 2,372.55 | 10.03 | 6 | .04 |
| Sporadic | [1] | NA | NA | NA | 1.74 | = AA | = AA | .7 | [0] | 2,487.04 | 2,491.04 | 130.52 | 7 | 0 |
Figure 2.
Distributions of adjusted log10(IgE) in 200 Dutch families. Vertical bars represent the distribution of the observed adjusted log10(IgE). The two curves represent the distributions of the three genotypes under the best-fitting mixed recessive model.
Two-Locus Segregation Analysis
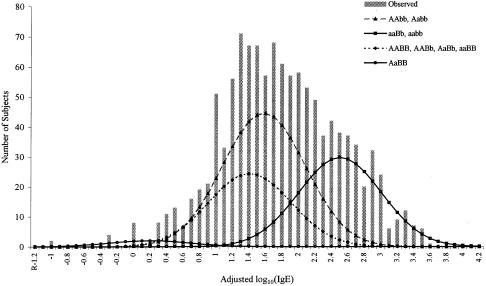
When two-locus segregation analysis was performed, the mixed two-major-gene models (two major genes and residual genetic effects) fitted the data significantly better than did the one-major-gene models (table 4). Of the several mixed two-major-gene models that were tested, a model with a major recessive gene and a dominant modifier gene was the most parsimonious. This model (two-major-gene model 2) fit the data significantly better than did the one-major-recessive-gene model (χ2=27.17; _df_=3; P = 5.42 × 10−6). It also had a much lower AIC value (2,347.25) than did the two-major-environmental-risk-factor model (AIC = 2,371.98). According to this model, the MLE, gene frequency for the first recessive gene, qa, was .55, similar to the gene frequency of the mixed recessive model under one-locus segregation analysis. The MLE of the gene frequency for the second dominant gene, qb, was .8. The second gene modifies the effect of the first recessive gene so that some of the individuals who were homozygous for the high-risk allele at the first gene did not have high IgE levels. Individuals with genotype aaBB (homozygous high-risk allele at the first gene and homozygous low-risk allele at the second locus) had normal IgE levels (mean 23 IU/ml), and only individuals with genotypes aaBb and aabb (29.6% of the total sample) had high IgE levels (mean 282 IU/ml) (fig. 3). The two major genes were responsible for 51.3% of the adjusted log10(IgE) variance; the first gene was responsible for 40.6% of the adjusted log10(IgE) variance, and the second gene was responsible for 9.0% of the adjusted log10(IgE) variance.
Table 4.
Segregation Analysis of Adjusted Log10(IgE) under One- and Two-Locus Models, in 200 Dutch Families
| Mean | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model | _q_a | _q_b | Locus B | AA | Aa | aa | SD | _h_2 | −2lnL | AIC |
| One-locus: | ||||||||||
| Mixed additive | .55 | 1.63 | 1.41 | 2.34 | .58 | .57 | 2356.69 | 2368.69 | ||
| Mixed recessive | .57 | 1.45 | = AA | 2.32 | .58 | .49 | 2358.42 | 2368.42 | ||
| Two-locus: | ||||||||||
| Two-major-gene model 1: | .57 | .83 | .51 | .68 | 2329.19 | 2355.19 | ||||
| Loci A and B additive | BB | 1.51 | .03 | 1.43 | ||||||
| Bb | 1.58 | 1.44 | 2.59 | |||||||
| bb | 1.53 | 1.37 | 2.33 | |||||||
| Two-major-gene model 2: | .55 | .8 | .5 | .52 | 2331.25 | 2347.25 | ||||
| Locus A recessive, locus B dominant | BB | 1.36 | .14 | 1.36 | ||||||
| Bb | 1.36 | 1.36 | 2.45 | |||||||
| bb | 1.53 | 1.53 | 2.45 | |||||||
| Two-major-environmental-risk-factor model | .58 | .83 | BB | 1.37 | .98 | 1.37 | .53 | .9 | 2349.98 | 2371.98 |
| Bb | 1.37 | 1.37 | 2.57 | |||||||
| bb | 1.97 | 1.37 | 2.22 |
Figure 3.
Distributions of adjusted log10(IgE) in 200 Dutch families. Vertical bars represent the distribution of the observed adjusted log10(IgE). The four curves represent the distributions of the nine genotypes under the best-fitting mixed two-major-gene model.
Linkage Analysis Using Variance-Component Analysis
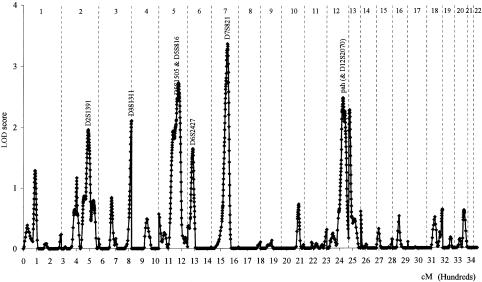
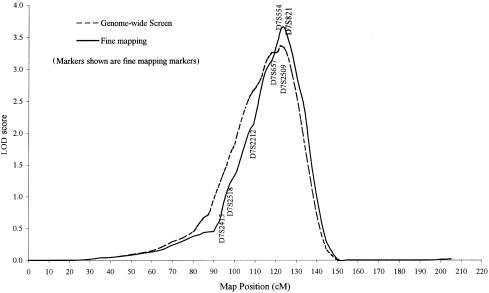
Linkage analysis, using the variance-component approach, was performed to systematically scan the genome for the locations of the major genes regulating total serum IgE levels. Results of the multipoint linkage analysis for the 344 evenly spaced autosomal markers are presented in figure 4. One chromosomal region with a LOD score of 3.36 (P = .00004) was observed at 7q and was flanked by markers D7S820 and D7S821. This locus explained 39% of adjusted log10(IgE) variance, and the residual genetic effect explained 17% of the variance. The evidence for linkage at this region was further supported by two additional analyses. First, the LOD score was strengthened by genotyping more markers in the regions. After seven markers were added to the 30-cM regions, the LOD score increased to 3.65 (_P_=.00002) at the same region (fig. 5). Second, both the first 92 families and the second 108 families provided a positive LOD score at the same region. The LOD score at this region was 2.48 (_P_=.0003) for the first 92 families and 1.88 (_P_=.002) for the second 108 families.
Figure 4.
Genomewide screen for major genes regulating adjusted log10(IgE) levels in 200 Dutch families, using variance-component analysis. Three hundred forty-four evenly spaced autosomal markers were genotyped. Vertical dotted lines divide genomes into 22 chromosomes.
Figure 5.
Multipoint linkage analysis of log10(IgE) on chromosome 7, using variance-component approach
To examine the impact that outliers had on the results, the linkage data were reanalyzed for all the chromosome 7 markers after deletion of the six outliers with the highest log10(IgE) values. The LOD score changed minimally, from 3.65 to 3.74, suggesting that the results were stable and not driven by the few outliers.
Evidence for linkage at two other regions also was observed. A peak LOD score of 2.73 (_P_=.0002) was found at 5q31, flanked by markers D5S666 and D5S402, a result that was consistent with previous findings (Meyers et al. 1994). This locus explained 37% of adjusted log10(IgE) variance. A peak LOD score of 2.46 (_P_=.0004) was found at 12q, flanked by markers PAH and D12S2070.
There were several other regions with LOD scores of ∼2. Some of these regions (3qter and 13pter) were at the tip of the chromosomes and were supported mainly by one marker. The significance and the interpretation of these regions were uncertain, and genotyping more markers in these regions is necessary. Other regions (2q and 6p) were supported by multiple markers.
Discussion
The family-ascertainment scheme of the current study was appropriate for performing both segregation analysis and variance-component analysis for total serum IgE levels. In this study, all the probands were recruited from a well-defined population sample in the northeastern Netherlands, where little immigration occurs. The probands were recruited because of prior diagnosis of asthma 25–35 years ago and were evaluated using a standardized protocol (Panhuysen et al. 1998). All the children and grandchildren (⩾8 years old) of the probands were studied, regardless of their phenotypic status.
Although many studies have demonstrated heritability of serum IgE levels, the precise mode of genetic control has remained elusive. There have been multiple complex segregation analyses investigating the mode of inheritance of total serum IgE within families. Several previous studies have found evidence for a recessive gene regulating IgE levels, with different estimates of gene frequencies and mean IgE levels (Marsh et al. 1974; Gerrard et al. 1978; Meyers et al. 1987, 1994). Evidence for a codominant mode of inheritance was reported by Martinez et al. (1993), suggesting that homozygotes and heterozygotes have different mean levels although there is clearly overlap between the distributions of total serum IgE levels.
The segregation analysis of the current study represented an extension of the segregation analyses in the first 92 families (Meyers et al. 1994; Xu et al. 1995). In the initial study, evidence was found for a mixed recessive gene (a recessive gene and residual genetic effect) and a mixed model with two major genes (two major genes and residual genetic effect) regulating total IgE levels, for a one-locus model and a two-locus model, respectively. After adding 108 families to our sample, we obtained increased evidence supporting these models. Under a one-locus segregation analysis in the total sample of 200 families, the mixed recessive model fit the data as well as the general model (χ23=1.90; _P_=.59). This could be compared with the relatively poor fit of the mixed recessive model for the first 92 families (χ23=6.90; _P_=.07) (Meyers et al. 1994). In the total sample, the mixed two-major-gene model fit the data significantly better than did the mixed one-major-gene model (χ23=27.17; _P_=5.4×10-6), whereas, in the first 92 families, there was a marginal improvement of the best two-major-gene model over one-major-gene model (χ25=11.9; _P_=.04).
For our previous linkage analyses, we had used a candidate-gene approach and had found evidence for linkage to chromosome 5q in the first 92 families (Meyers et al. 1994; Xu et al. 1995). We have now completed a genomewide search in 200 Dutch families with asthma, using a new analytic method appropriate for investigation of quantitative traits. A region on chromosome 7q reached the criteria for a genomewide significant linkage and provided the strongest evidence for linkage, with a peak LOD score of 3.65 (_P_=.00002) (Lander and Kruglyak 1995). Linkage-analysis results also identified two regions with evidence for linkage at 5q and 12q; both regions have been reported elsewhere and are rich with appropriate candidate genes for immunologic and allergic responses.
There were two linkage studies that systematically searched across the genome for the loci regulating total serum IgE. In a linkage study of 364 subjects in 80 nuclear families subselected from a population sample of 230 families in Busselton in western Australia, Daniels et al. (1996) used 20-cM intervals for their genome screen and, after genotyping 274 autosomal markers, identified four regions that are likely to contain the genes regulating total serum IgE levels; the four regions were 6p, 7p, 11q13, and 16q2. In another genomewide screen linkage study, carried out in Germany in a smaller sample (97 families), Wjst et al. (1999) reported four regions (2p, 6p, 9q2, and 12q) linked to loci regulating total serum IgE.
The possible explanations for the varying results from the various studies include the differences in study populations, ascertainment schemes, sample sizes, and analytical methodologies. The assumptions required for some of the analytical methods oversimplify the complexity of the trait and are probably inappropriate for modeling the genetic regulation of total serum IgE levels. This is especially true for segregation analysis, in which most of the approaches assume only one major factor (a gene). Obviously, this is not realistic for a common trait such as high total serum IgE levels. Alternatively, segregation analysis assuming two major genes, although probably still too simple a model, may significantly improve our ability to model the trait. The effects of two genes and their interactions can be explored under the two-locus model.
There have been difficulties in linkage analysis of quantitative traits in humans. Parametric linkage analysis of quantitative traits has been rarely used, because it requires specifying a genetic model to describe the mode of inheritance, which is usually uncertain. Furthermore, when the SD within the genotype is too large and the distribution of the trait overlaps among the genotypes, the parametric approach is usually uninformative. In some of the earlier studies, nonparametric analysis was performed using the phenotypic information of sib pairs only, either because other relatives were not characterized or because the all-relative-pair approach was not yet available for quantitative traits. Recently, an alternative quantitative-trait linkage analysis, a variance-component method, has been developed (Goldgar 1990; Schork 1993; Amos 1994; Blangero and Almasy 1997). The variance-component method allows for marker-specific effects, residual additive genetic effects, and random environmental effects. The variance-component approach for a linkage study of a quantitative trait has a number of compelling features (Williams et al. 1997; Williams and Blangero 1999). It uses all of the available inheritance information in a pedigree of any size or structure. This not only avoids the violation of true bivariate structure of a sib pair, which can occur in analyses of the phenotypic sib-pair difference, but also uses the available information more efficiently and therefore can achieve greater power to detect linkage. Second, the variance-component approach resolves the problem of independence of sib pairs within a family by maximizing the likelihood of a pedigree that is jointly conditional on all members of the pedigree (Amos et al. 1996). The violation of independence of sib pairs has been a significant problem, because different weighting schemes have tended to produce strikingly different results (Sham et al. 1997).
The results of the present study can be summarized as follows. First, the results confirm the strong association between high total serum IgE levels and BHR and allergy. Second, as observed in previous studies, there is strong aggregation of high total serum IgE levels within families. Third, the segregation analysis provides evidence that major genes with residual genetic effects are responsible for the aggregation of high total serum IgE levels. The presence of at least two major genes, one behaving as a recessive gene and another behaving as a dominant modifier gene, is consistent with the observed distribution of IgE levels in these families. Finally, a genomewide search using variance-component approaches identified several regions that are likely to contain the major genes regulating total serum IgE levels. Regions on chromosomes 5q, 12q, and 6p have been reported elsewhere (Marsh et al. 1994; Meyers et al., 1994; Xu et al. 1995; Barnes et al. 1996; Daniels et al. 1996; Wjst et al. 1999). The novel region on chromosome 7q was confirmed by typing additional markers, and it needs to be replicated in other populations.
Acknowledgments
We thank all of the families whose participation made this project possible. The research was supported in part by the Netherlands Asthma Foundation and by National Institutes of Health grant R01 HL48341. Some of the results reported in this article were obtained by use of the program package S.A.G.E., which is supported by U.S. Public Health Service Resource Grant 1 P41 RR03655 from the National Center for Research Resources.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- LINKAGE, http://linkage.rockefeller.edu/soft/linkage
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for IgE [MIM 147061] and asthma [MIM 600807])
- PAP (Pedigree Analysis Package), ftp://ftp.genetics.utah.edu/pub/software/pap [Google Scholar]
- University of Antwerp DNA Laboratory http://www.uia.ac.be/dnalab/ld/
- S.A.G.E. (Statistical Analysis for Genetic Epidemiology), http://darwin.cwru.edu/pub/sage.html
- SOLAR (Sequential Oligogenic Linkage Analysis Routines), http://www.sfbr.org/sfbr/public/software/solar/index.html
References
- Akaike H (1974) A new look at the statistical model identification. IEEE Trans Automatic Control AC 19:719–723 [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 62:1198–1211 [DOI] [PMC free article] [PubMed]
- Amos CI (1994) Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Zhu DK, Boerwinkle E (1996) Assessing genetic linkage and association with robust components of variance approaches. Ann Hum Genet 60:143–160 [DOI] [PubMed] [Google Scholar]
- Barnes KC, Neely ND, Duffy DL, Freidhoff LR, Breazeale DR, Schou C, Naidu RP, Levett PN, Renault B, Kucherlapti R, Lozzino S, Ehrlich E, Beaty TH, Marsh DG (1996) Linkage of asthma and total serum IgE concentration to markers on chromosome 12q: evidence from Afro-Caribbean and Caucasian populations. Genomics 37:41–50 [DOI] [PubMed] [Google Scholar]
- Blangero J, Almasy L (1997) Multipoint oligogenic linkage analysis of quantitative traits. Genet Epidemiol 14:959–964 [DOI] [PubMed] [Google Scholar]
- Burrows B, Lebowitz MD, Barbee RA, Cline MG (1991) Findings before diagnoses of asthma among the elderly in a longitudinal study of a general population sample. J Allergy Clin Immunol 88:870–877 [DOI] [PubMed] [Google Scholar]
- Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG (1989) Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med 320:271–277 [DOI] [PubMed] [Google Scholar]
- Daniels SE, Bhattacharrya S, James A, Leaves NI, Young A, Hill MR, Faux JA, Ryan GF, LeSouef PN, Lathrop GM, Musk AW, Cookson WOC (1996) A genome-wide search for quantitative loci underlying asthma. Nature 383:247–250 [DOI] [PubMed] [Google Scholar]
- De Vries K, Goei JT, Booy-Noord H, Orie NGM (1962) Changes during 24 hours in the lung function and histamine hyperreactivity of the bronchial tree in asthmatic and bronchitic patients. Int Arch Allergy 20:93–101 [DOI] [PubMed] [Google Scholar]
- Elston RC, Stewart J (1971) A general model for the genetic analysis of pedigree data. Hum Hered 21:523–542 [DOI] [PubMed] [Google Scholar]
- Gerrard JW, Rao DC, Morton NE (1978) A genetic study of immunoglobulin E. Am J Hum Genet 30:46–58 [PMC free article] [PubMed] [Google Scholar]
- Goldgar DE (1990) Multipoint analysis of human quantitative genetic variation. Am J Hum Genet 47: 957–967 [PMC free article] [PubMed] [Google Scholar]
- Halonen M, Stern D, Taussig LM, Wright A, Ray CG, Martinez FD (1992), The predictive relationship between serum IgE levels at birth and subsequent incidences of lower respiratory illnesses and eczema in infants. Am Rev Respir Dis 146:866–870 [DOI] [PubMed] [Google Scholar]
- Johansson SG, Bennich HH, Berg T (1972) The clinical significance of IgE. Prog Clin Immunol 1:157–181 [PubMed] [Google Scholar]
- Hasstedt SJ (1982) A mixed-model likelihood approximation on large pedigrees. Comput Biomed Res 15:295–307 [DOI] [PubMed] [Google Scholar]
- Hasstedt SJ, Moll PP (1989) Estimation of genetic model parameters: variables correlated with a quantitative phenotype exhibiting major locus inheritance. Genet Epidemiol 6:319–332 [DOI] [PubMed] [Google Scholar]
- Hopp RJ, Bewtra AK, Nair NM, Townley RG (1984) Specificity and sensitivity of methacholine inhalation challenge in normal and asthmatic children. J Allergy Clin Immunol 74:154–158 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lange K, Weeks D, Boehnke M (1988) Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol 5:471–472 [DOI] [PubMed] [Google Scholar]
- Marsh DG, Bias WB, Ishizaka K (1974) Genetic control of basal serum immunoglobulin E level and its effect on specific reaginic sensitivity. Proc Natl Acad Sci USA 71:3588–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh DG, Neely JD, Breazeale DR, Ghosh B, Friedhoff LR, Ehrlich-Kautzy E, Schou C, Krishnaswamy G, Beaty TH (1994) Linkage analysis of IL-4 and other chromosome 5q31.1 markers and total serum IgE concentrations. Science 264:1152–1156 [DOI] [PubMed] [Google Scholar]
- Martinez FD, Holberg CJ, Halonen M, Morgan WJ, Wright AL, Taussig LM (1994) Evidence for Mendelian inheritance of serum IgE levels in Hispanic and non-Hispanic white families. Am J Hum Genet 55:555-565 [PMC free article] [PubMed] [Google Scholar]
- Meyers DA, Beaty TH, Freidhoff LR, Marsh DG (1987) Inheritance of total serum IgE (basal levels) in man. Am J Hum Genet 41:51–62 [PMC free article] [PubMed] [Google Scholar]
- Meyers DA, Postma DS, Panhuysen CIM, Xu J, Amelung PJ, Levitt RC, Bleecker ER (1994) Evidence for a locus regulating total serum IgE levels mapping to chromosome 5. Genomics 23:464–470 [DOI] [PubMed] [Google Scholar]
- Panhuysen CI, Bleecker ER, Koeter GH, Meyers DA, Postma DS (1998) Characterization of obstructive airway disease in family members of probands with asthma: an algorithm for the diagnosis of asthma. Am J Respir Crit Care Med 157:1734–1742 [DOI] [PubMed] [Google Scholar]
- Schork NJ (1993) Extended multipoint identity-by-descent analysis of human quantitative traits: efficiency, power, and modeling considerations. Am J Hum Genet 53:1306–1319 [PMC free article] [PubMed] [Google Scholar]
- Sears M, Burrows B, Flannery EM, Herbison GP, Hewitt CJ, Holdaway MD (1991) Relation between airway responsiveness and serum IgE in children with asthma and in apparently normal children. N Engl J Med 325:1067–1071 [DOI] [PubMed] [Google Scholar]
- Self SG, Kiang KY (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under non-standard conditions. J Am Stat Assoc 82:605–610 [Google Scholar]
- Sham PC, Zhao JH, Curtis D (1997) Optimal weighting scheme for affected sib-pair analysis of sibship data. Ann Hum Genet 61:61–69 [DOI] [PubMed]
- Van Camp G, Balemans W, Wellems PJ (1997) Linkage Designer and Linkage Reporter software for automated gene localization studies. Trends Genet 13:82 [Google Scholar]
- Wiesch DG, Meyers DA, Bleecker ER (1999) Genetics of asthma. J Allergy Clin Immunol 104:895–901 [DOI] [PubMed] [Google Scholar]
- Williams JT, Blangero J (1999) Comparison of variance components and sibpair-based approaches to quantitative trait linkage analysis in unselected samples. Genet Epidemiol 16:113–134 [DOI] [PubMed] [Google Scholar]
- Williams JT, Duggirala R, Blangero J (1997) Statistical properties of a variance components method for quantitative trait linkage analysis in nuclear families and extended pedigrees. Genet Epidemiol 14:1065–1070 [DOI] [PubMed] [Google Scholar]
- Wjst M, Fischer G, Immervoll T, Jung M, Saar K, Rueschendorf F, Reis A, Ulbrecht M, Gomolka M, Weiss EH, Jaeger L, Nickel R, Richter K, Kjellman NI, Griese M, von Berg A, Gappa M, Riedel F, Boehle M, van Koningsbruggen S, Schoberth P, Szczepanski R, Dorsch W, Silbermann M, Wichmann HE (1999) A genome-wide search for linkage to asthma. German Asthma Genetics Group. Genomics 58:1–8 [DOI] [PubMed] [Google Scholar]
- Xu J, Levitt RC, Panhuysen CI, Postma DS, Taylor EW, Amelung PJ, Holroyd KJ, Bleecker ER, Meyers DA (1995) Evidence for two unlinked loci regulating total serum IgE levels. Am J Hum Genet 57:425–430 [PMC free article] [PubMed] [Google Scholar]