Initiation Sites Are Distributed at Frequent Intervals in the Chinese Hamster Dihydrofolate Reductase Origin of Replication but Are Used with Very Different Efficiencies (original) (raw)
Abstract
Previous radiolabeling and two-dimensional (2-D) gel studies of the dihydrofolate reductase (DHFR) domain of Chinese hamster cells have suggested that replication can initiate at any one of a very large number of inefficient sites scattered throughout the 55-kb intergenic spacer region, with two broad subregions (ori-β and ori-γ) preferred. However, high-resolution analysis by a PCR-based nascent strand abundance assay of the 12-kb subregion encompassing ori-β has suggested the presence of a relatively small number of fixed, highly efficient initiation sites distributed at infrequent intervals that correspond to genetic replicators. To attempt to reconcile these observations, two different approaches were taken in the present study. In the first, neutral-neutral 2-D gel analysis was used to examine replication intermediates in 31 adjacent and overlapping restriction fragments in the spacer, ranging in size from 1.0 to 18 kb. Thirty of 31 fragments displayed the complete bubble arcs characteristic of centered origins. Taking into account overlapping fragments, these data suggest a minimum of 14 individual start sites in the spacer. In the second approach, a quantitative early labeled fragment hybridization assay was performed in which radioactive origin-containing DNA 300 to 1,000 nucleotides in length was synthesized in the first few minutes of the S period and used to probe 15 clones distributed throughout the intergenic spacer but separated on average by more than 1,000 bp. This small nascent DNA fraction hybridized to 14 of the 15 clones, ranging from just above background to a maximum at the ori-β locus. The only silent region detected was a small fragment lying just upstream from a centered matrix attachment region—the same region that was also negative for initiation by 2-D gel analysis. Results of both approaches suggest a minimum of ∼20 initiation sites in the spacer (two of them being ori-β and ori-γ), with ori-β accounting for a maximum of ∼20% of initiations occurring in the spacer. We believe that the results of all experimental approaches applied to this locus so far can be fitted to a model in which the DHFR origin consists of a 55-kb intergenic zone of potential sites that are used with very different efficiencies and which are separated in many cases by a few kilobases or less.
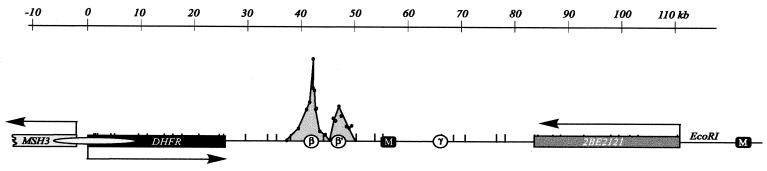
We are interested in the nature of origins of replication in mammalian chromosomes, and have used the dihydrofolate reductase (DHFR) locus of Chinese hamster cells as a model system. Two-dimensional (2-D) gel studies have suggested that replication initiation sites are chosen from a large number of potential sites scattered throughout the 55-kb spacer between the DHFR and 2BE2121 genes, with the central 30- to 35-kb region preferred (see Fig. 1 and references 9 and 12 for reviews). This conclusion derives in part from the observation that several different 4- to 6-kb _Eco_RI and _Xba_I fragments in the intergenic spacer display complete bubble arcs characteristic of centered origins (10, 11, 15, 56). Therefore, we concluded that there must be at least one initiation site near the center of each of these fragments and, by inference, at other locations scattered throughout the spacer that had not yet been analyzed. Furthermore, in neutral-alkaline 2-D gels (44), replication forks have been shown to move in both directions at several positions within the intergenic spacer, suggesting that initiation sites are reasonably frequent and widely scattered (15).
FIG. 1.
Map of the Chinese hamster DHFR locus. An _Eco_RI map of a 120-kb region encompassing the DHFR locus in CHOC 400 cells is shown. The convergently transcribed DHFR and 2BE2121 genes are indicated as rectangular boxes, ori-β, _ori-β_′, and ori-γ are shown as open circles, and a prominent MAR (M) is shown as a black square. The elongated gray oval over the DHFR promoter indicates a diffuse region that appears to be attached to the nuclear matrix as well. The peaks detected at _ori-β, ori-β_′, and ori-γ by the PCR-based small nascent strand abundance assay are based on results shown in reference 31.
The efficiency of initiation at different sites is clearly not uniform throughout the spacer. Relatively low-resolution intrinsic radiolabeling studies reveal two broad subregions within the central core (originally termed ori-β and ori_-γ_ [36]) that incorporate label preferentially at the beginning of S phase. Furthermore, a PCR-based nascent strand abundance assay (55) that focused on the 5-kb sequence encompassing the ori-β region indicated that it represents the predominant initiation site in the DHFR locus (45, 54). A switch in Okazaki fragment template bias was also detected in the ori-β region (4), leading to the proposal that this region represents the primary initiation site in the DHFR locus within a broad zone of very inefficient secondary sites (4, 8).
However, several different _Eco_RI and _Xba_I fragments of about equal size from the central 30- to 35-kb core of the spacer display significant numbers of bubbles on 2-D gels (10, 11, 15). In addition, when the PCR-based nascent strand abundance assay was extended to the 3′ side of ori-β, a second peak of small nascent strands was identified about 5 kb downstream (termed _ori-β_′ [31]). Since the PCR-based assay detected little or no initiation in the valley between ori-β and _ori-β_′ or in the regions immediately surrounding them and since these two sites are situated near the centers of two adjacent _Eco_RI fragments that display complete bubble arcs of similar intensities on 2-D gels (10), ori-β and _ori-β_′ could correspond to highly preferred initiation sites analogous to classic replicators. To accommodate the existing 2-D gel data, additional such sites would have to be situated near the center of the other fragments that displayed complete bubble arcs on 2-D gels (10, 11, 15, 56). By this argument, the DHFR origin would consist of a finite number of fixed initiation sites situated at infrequent intervals, ori-β and _ori-β_′ being among the most active.
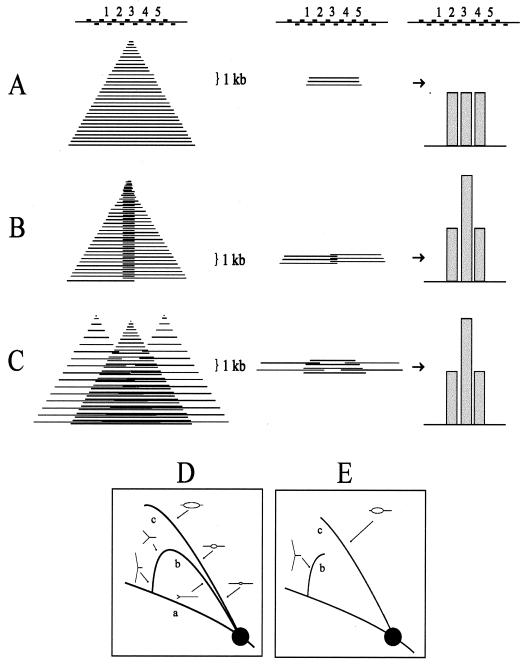
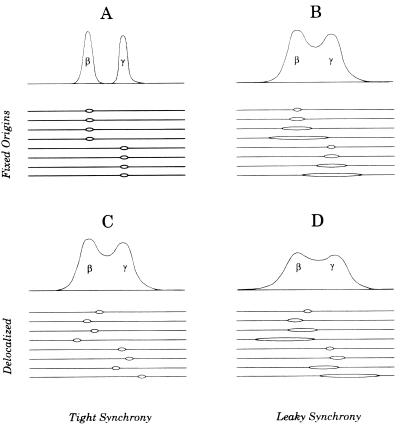
One difficulty with the interpretation that ori-β and _ori-β_′ correspond to fixed initiation sites is illustrated in Fig. 2A. If nascent strands ∼800 nucleotides (nt) in length (in practice, 600 to 1,000 nt) (31) are isolated and their relative abundances are estimated with several closely spaced adjacent primer pairs (e.g., Fig. 2A, sets 2 to 4), then a fixed origin centered at set 3 should measure the same relative abundance of nascent strands as sets 2 and 4 (Fig. 2A, right). Instead, sharp peaks of nascent strands were detected at ori-β and _ori-β_′, with two adjacent primer pairs at the ori-β peak displaying an almost twofold difference in the number of nascent strands detected (see Fig. 1) (31). We suggest that this latter pattern could result, instead, from one of two alternative initiation modes: (i) fixed initiation sites with unequal fork rates in the two directions with neither direction being preferred (shown in Fig. 2B in the extreme, i.e., almost unidirectional) or (ii) a cluster of closely spaced sites, with the greatest abundance occurring in the center of the zone (Fig. 2C).
FIG. 2.
Predictions of the relative abundances of nascent strands with different origin types. As shown in panel A (left), if origin-containing nascent strands in the size range of ∼1,000 nt are analyzed with a series of primer pairs that are positioned within a 1,000-bp stretch, then the three central primer sets (sets 2, 3, and 4) should each detect the same relative abundance of ∼1,000-bp nascent strands (middle and right). The sharp peaks of small nascent strands that were actually measured with closely spaced primer pairs (31) could result from two alternative types of initiation mechanism (unidirectional from a fixed site) (B) or in a cluster of sites within a small region (C). (D) A restriction digest of replication intermediates separated in the first dimension of an agarose gel according to molecular mass and in the second dimension according to both mass and shape (3). After transfer to a membrane and hybridization with a cognate probe for a fragment containing either a single fork or a centered bubble arc (curves b or c) would be obtained. Curve a corresponds to the diagonal of nonreplicating fragments in the genome as a whole. (E) The pattern that would result from a fragment that contains an off-centered origin, which would initially contribute to the bubble arc, but would eventually contribute to the single fork arc when one fork crosses the nearest restriction site.
An important issue to resolve, therefore, is whether ori-β and _ori-β_′, indeed, correspond to fixed initiation sites flanked on either side by silent regions. A second important question is how many initiation sites in addition to ori-β and _ori-β_′ would be uncovered and what their relative activities would be if the entire 55-kb intergenic spacer were examined at much higher resolution. To address these questions, we have taken two different approaches. In the first, we examined replication intermediates in 31 different adjacent and overlapping restriction fragments in the spacer ranging in length from 1 to 18 kb, including 3 small fragments that isolate ori-β, _ori-β_′, and the intervening valley and shoulder of the ori-β peak. In the second, we devised a modified early labeled fragment hybridization (ELFH) assay that allowed us to quantitate the distribution of origin-containing DNA 300 to 1,000 nt in length throughout the spacer region. The results of these two approaches suggest that the DHFR origin comprises a minimum of ∼20 individual start sites that are utilized with very different efficiencies. While the 1-kb region encompassing ori-β is clearly the most efficient region in the spacer, our data suggest that a maximum of ∼20% (and probably far fewer) of starts occur within this region. The only silent region uncovered so far is a short stretch immediately upstream from a prominent, centered, matrix attachment region (MAR). With very few caveats, the results of these analyses are compatible with all of the data that have been obtained in previous studies and provide a unified view of the initiation reaction in the DHFR locus.
(This work fulfills part of the requirements for the Ph.D. degree in biochemistry from the University of Virginia for S.W.)
MATERIALS AND METHODS
Cell culture and synchronization procedures.
CHOC 400 cells were grown in minimal essential medium (MEM) supplemented with nonessential amino acids, 10% Fetal Clone II (HyClone), and 0.1% gentamicin (GIBCO/BRL) and were maintained in an atmosphere of 5% CO2. Cells were arrested in G0 by starvation for isoleucine for 45 h and then were released into complete medium containing either 10 μg of aphidicolin (Sigma)/ml or 400 μM mimosine (Sigma) for 12 h to collect them at or near the G1/S boundary (10, 42). The drug was washed out and replaced with drug-free medium to allow entry into S phase.
2-D gel analysis.
After 12 h in mimosine, CHOC 400 cells were released into the S period by being washed in and returned to prewarmed MEM. Ninety minutes later, which represents the period of maximum initiation in the DHFR origin (10, 11, 15), cells were harvested and replication intermediates were prepared by digesting matrix-attached DNA with the indicated enzymes or enzyme combinations as described previously (14). Replication intermediates were analyzed by neutral-neutral 2-D gel electrophoresis as described previously (3, 14), with the following exceptions: for fragments in the 12- to 15-kb range, the first-dimension gels were 0.3% agarose, run for 60 h at 0.4 V/cm; the second-dimension gels were 0.6% agarose, run for 45 h at 1 V/cm. For fragments in the 1.0- to 2.5-kb range, the first-dimension conditions were the same as previously described (3, 14) and the second-dimension gel was 1.6% agarose, run at 4.5 V/cm for l4 h.
Measuring replication rates.
Rates of DNA synthesis were measured by pulse-labeling cells with medium containing 5 μCi of [3H]thymidine/ml (67 Ci/mmol; ICN) and processing as previously described (42).
In vitro replication and ELFH analysis.
Cells were arrested with either mimosine or aphidicolin for 12 h and were washed with ice-cold MEM to remove the drug and prevent initiation of DNA synthesis. Nuclei were isolated from ∼108 cells by digitonin treatment as previously described (14) and were resuspended in 275 μl of CLB buffer (50 mM KCl, 0.5 mM EDTA, 0.05 mM spermine, 0.125 mM spermidine, 0.5% thiodiglycol, 0.1 mM phenylmethylsulfonyl fluoride, 10 mM Tris-HCl [pH 7.4], 0.1% digitonin) in a 1.5-ml conical tube on ice. To this suspension was added 300 μl pf 2× DNA replication cocktail (4, 57) containing 200 μM (each) unlabeled dCTP, dGTP, and dTTP and 20 μl of [32P]dTTP (3,000 Ci/mmol). Reactions were initiated by incubation at 34°C for the times indicated in the legend to Fig. 3 and 6. Reactions were terminated by the addition of 1.0 ml of ice-cold CLB, and the nuclei were pelleted at 2,000 × g for 5 min in a benchtop centrifuge in the cold, washed, and resuspended in 510 μl of CLB. Aliquots (100 μl) were distributed to five tubes, and nuclei were lysed by the addition of 0.5 ml of 20 mM KCl-70 mM NaCl-20 mM EDTA-20 mM Tris-HCl (pH 7.4), followed by the addition of 0.7 ml of lysis buffer (2% sodium dodecyl sulfate [SDS]-50 mM EDTA-50 mM Tris-HCl [pH 7.4]-1-mg/ml proteinase K [AMRESCO]). After incubation for 3 h at 37°C, 0.4 ml of 5 M NH4 acetate was added, and the DNA was precipitated with 2.5 volumes of cold ethanol at −20°C. DNA was pelleted by centrifugation, redissolved in 2 ml of TEN (10 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, and 10 mM NaCl), reprecipitated in cold ethanol, and finally dissolved in 160 μl of TEN. Denaturation was achieved by the addition of 40 μl of 1 M NaOH and incubation overnight at 24°C.
FIG. 3.
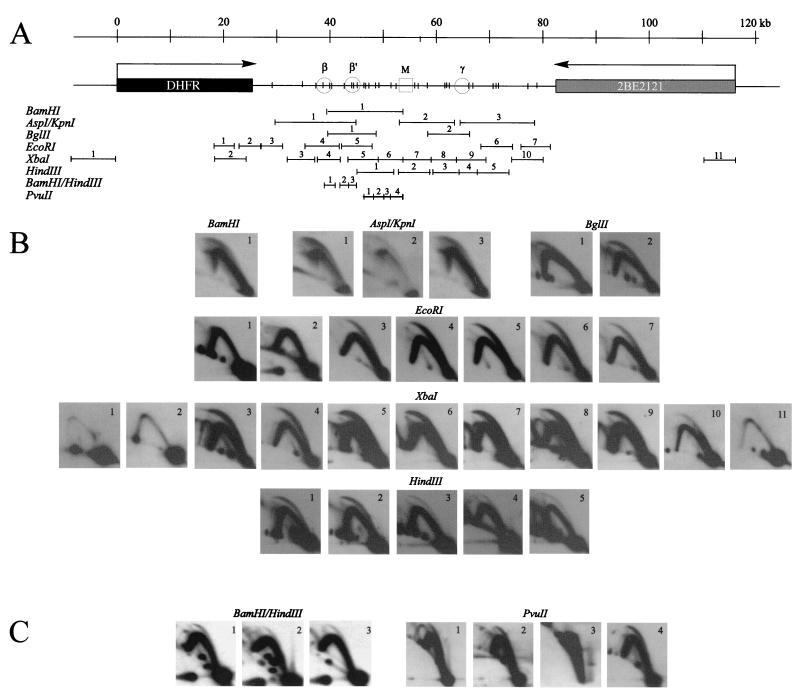
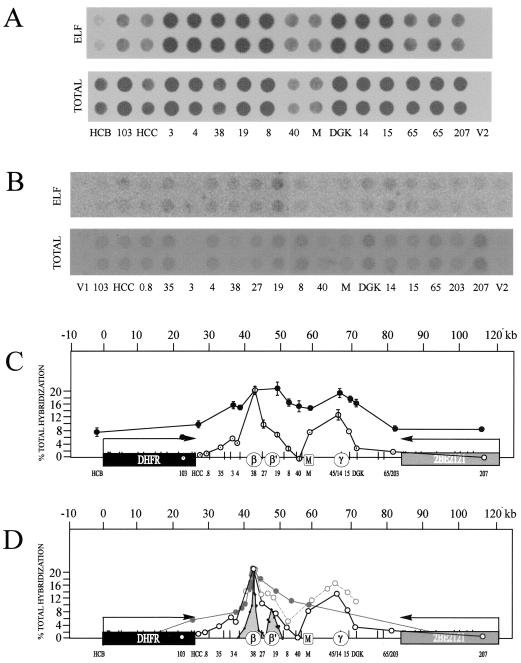
Centered bubble arcs are detected in 30 of 31 adjacent and overlapping restriction fragments in the intergenic region. CHOC 400 cells were arrested in early G1 by isoleucine starvation, followed by release into either 400 μM mimosine or 10-μg/ml aphidicolin for 12 h. At 90 or 30 min after release, respectively, which are the peak initiation periods in this locus after each of these protocols (11, 15), replication intermediates were prepared using one of the enzymes or enzyme combinations shown in panel A. After separation on a neutral-neutral 2-D gel as described in Materials and Methods, digests were blotted onto Hybond-N+ and hybridized as described previously (14) with small cognate single-copy probes for the indicated fragments. (B) Digests yielding fragments in the 3- to 18-kb range. (C) _Bam_HI/_Hin_dIII and _Pvu_II digests yielding a collection of small fragments that together almost reconstitute the 15-kb fragment 1 of _Bam_HI that extends from ori-β to the MAR (A).
FIG. 6.
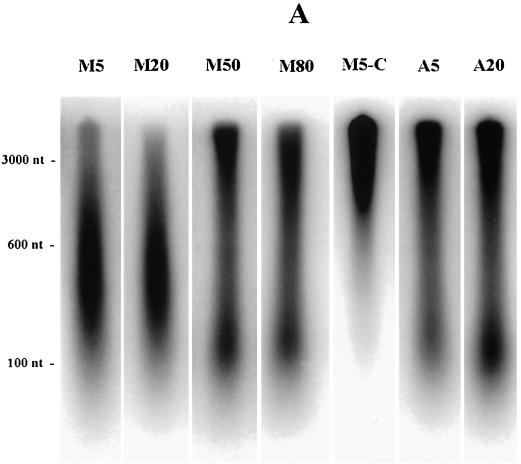
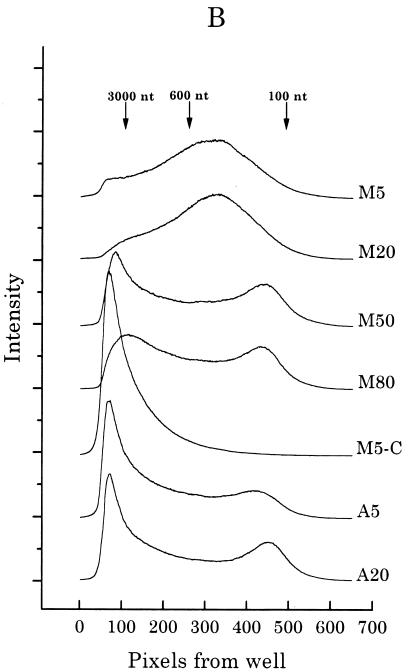
Mimosine-synchronized cells begin synthesizing DNA immediately from sites very close to origins after permeabilization in the presence of dNTPs. (A) CHOC 400 cells were arrested near the G1/S boundary in either 400 μM mimosine or 10-μg/ml aphidicolin, washed with prewarmed drug-free medium, and then incubated in drug-free complete medium (for 5, 20, 50, or 80 min after mimosine removal or for 5 and 20 min after aphidicolin removal) prior to harvesting and isolation of nuclei as described in Materials and Methods. In vitro replication assays were carried out for 1.5 min at 34°C and quenched; one 5-min mimosine-synchronized sample was chased for an additional 30 min in vitro with 100 μM dCTP (lane M5-C). The DNA was then purified as described in Materials and Methods and was separated with an alkaline agarose gel (1.8%) along with a 123-bp ladder (BRL). The gel was transferred to Hybond-N+ and analyzed with the PhosphorImager. (B) Each lane shown in panel A was quantitated as a line graph using an ImageQuant program (the same length and width were scanned in each case), and radioactivity (expressed as relative intensity units) was plotted as a function of distance from the well. The scans are juxtaposed in the figure to allow comparison among samples.
To analyze the sizes of radioactive nascent strands synthesized in vitro (see Fig. 6A), DNA was separated on a 1.5% alkaline agarose gel along with a 123-bp ladder (BRL). For the ELFH experiment (see Fig. 7A), alkali-treated DNA was sonicated to ∼600 nt in length and used directly as a hybridization probe (see below); for another experiment (see Fig. 7B), the DNA preparation was separated on a 1.8% alkaline low-melting agarose gel alongside the 123-bp ladder, and the 300- to 1,000-nt fraction was excised and isolated by repeated freezing and thawing of the agarose plug, followed by centrifugation in the cold. The resulting supernatant fluid was ethanol precipitated and resuspended in 150 μl of TEN buffer. The sonicated labeled DNA and the sized fraction were then used separately as radioactive hybridization probes on Hybond-N+ membranes spotted with 1 μg each of several cloned fragments from the DHFR domain (see Fig. 7). Hybridizations were carried out with 5 ml of Church and Gilbert hybridization solution (5) for 24 h at 65°C. Filters were washed successively with 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M Na citrate]-1% SDS and 0.2× SSC-0.2% SDS at 65°C for 30 min. The resulting signals were collected on a Molecular Dynamics PhosphorImager. In vitro replication reactions were also carried out on NP-40-isolated nuclei as described previously (57) with virtually identical results, as assessed with alkaline agarose gels (S. Wang, unpublished observations).
FIG. 7.
A high-resolution ELFH assay performed with mimosine-synchronized cells suggests that potential initiation sites are distributed at intervals of 1,000 bp or less. (A) Mimosine-synchronized cells were permeabilized and incubated for 1.5 min in vitro as described in Materials and Methods. The DNA was purified, sonicated to ∼600 bp in length, and utilized as a hybridization probe on a series of clones from the DHFR domain (or the pBS and pGem cloning vectors V1 and V2, respectively) that had been dotted onto a nylon filter in a total volume of 5 μl. A duplicate dot blot was hybridized with total CHOC 400 DNA that was sonicated to about 600 bp and labeled with [32P]dCTP as previously described (57). After being hybridized and washed, the filters were exposed to the PhosphorImager. (B) Conditions were the same as described for panel A, except that after purification, the DNA was treated with alkali, separated on an alkaline agarose gel, and the 300- to 1,000-nt fraction was isolated and used to probe a similar series of duplicate clones spotted onto a nylon membrane. (Clone DGK was incorrectly spotted out of linear order on the blots shown in panels A and B relative to its map position and should have been placed between clones 15 and 65.) (C) The percentage of total hybridization specific to each clone was calculated as described in Materials and Methods. The relative radioactivities in duplicate dots were plotted as a function of map position for each clone. Error bars indicate the standard error of the mean of four independent experiments for the total labeled probe shown in panel A (measured by two people independently; filled circles) or for the same experiment shown in panel B (measured by two individuals; open circles). (D) The results from the sized ELFH experiment pictured in panel C and from a previous PCR-based analysis of small nascent strands (31) (grey peaks). Data are also presented from an earlier experiment that measured the intrinsic labeling pattern in the first 30 min of the S period in CHOC 400 cells by an in-gel renaturation procedure (36), indicated by the grey curve with open circles, as well as the data from an earlier low-resolution ELFH experiment with aphidicolin-synchronized cells that lacked indicator clones in the region of ori-γ (19).
The PhosphorImager signals obtained in each experiment were measured by two different individuals. Background contributions and nonspecific vector hybridization were first subtracted from the values obtained for each duplicate control and experimental sample. To correct for differences in fragment length and sequence, the average percentage of total hybridization to duplicate clones was then calculated for both the ELFH and total genomic DNA control samples, and a relative hybridization value was determined by dividing the ELFH value by that for the control. The relative hybridization values were then expressed as the percentage of total relative hybridization (see Fig. 7C).
RESULTS
Neutral-neutral 2-D gel analysis detects centered initiations in 26 different fragments from the intergenic spacer.
With previous neutral-neutral 2-D gel studies with CHOC 400 cells, we analyzed replication intermediates of four _Eco_RI and three _Hin_dIII fragments from the intergenic spacer, which ranged in size from 4.8 to 6.75 kb and which together covered 34 kb (∼60% of the spacer). In these studies, bubble arcs were detected in the early S period in every one of these fragments. Furthermore, each displayed a complete bubble arc that appeared to extend all the way to the 2n position in the mass dimension, which suggests that initiation occurred near the fragment's center (Fig. 2D). Assuming the unlikely event that a single, centered, origin was responsible for the complete bubble arcs in each case and that total coverage of the spacer would detect the same frequency of initiation, we could conclude from these studies that there are at least seven initiation sites or regions within the 55-kb intergenic spacer. An alternative possibility is that each fragment contains several initiation sites, some of which are centered. The latter model would necessarily result in a much higher estimate of potential start sites.
We reasoned that we could discriminate between these two possibilities by digesting genomic DNA containing replication intermediates with several different restriction enzymes. Thus, any fixed origins would be positioned differently with respect to the fragment ends, resulting in some restriction fragments that displayed incomplete bubble arcs and others that displayed only single fork arcs (Fig. 2D); in contrast, if there were a large number of initiation sites at closely spaced intervals, centered bubble arcs would be observed in every fragment tested, regardless of the enzyme used to digest the DNA and regardless of fragment size.
To enrich for replication intermediates in this early firing origin, CHOC 400 cells were arrested in the G1 period by isoleucine deprivation, followed by release into complete medium and collection at the G1/S boundary with the very effective replication inhibitor, mimosine (references 10 and 42 and see below). Cells were then sampled 90 min after release from mimosine, which encompasses a 35- to 40-min lag before DNA synthesis begins in vivo and corresponds to the peak initiation period in this locus (10, 11, 15). In initial experiments, replication intermediates were prepared using either _Bam_HI, AspI, _Bgl_II, _Eco_RI, or _Xba_I to digest the DNA, which allowed us to analyze 26 fragments in the size range of 3.4 to 18 kb (Fig. 3A). Each digest was separated on a neutral-neutral 2-D gel, transferred to a membrane, and hybridized successively with cognate single-copy radioactive probes. The principle of the neutral-neutral 2-D gel replicon mapping method is summarized in Fig. 2D (3), and the resulting autoradiograms are presented in Fig. 4B. A total of 19 fragments from the intergenic region and 5 fragments from the DHFR and 2BE2121 genes were analyzed.
FIG. 4.
Expected patterns of nascent strands after release from a leaky or a nonleaky replication inhibitor with fixed and delocalized origins. To discriminate between two fixed origins at, for example, ori-β and ori-γ versus a delocalized zone with ori-β and ori-γ preferred, it would be necessary to label cells or allow forks to mature for a very short time after release from a very tight G1/S arrest (A and C). If replication forks leak away from origins in the presence of the synchronizing agent, the patterns of labeling would be very similar between the fixed and delocalized initiation modes (B and D).
As shown previously (10, 11, 15), essentially no initiation can be detected in the DHFR gene 90 min after release from mimosine (_Eco_RI fragments 1 and 2 and _Xba_I fragments 1 and 2; the images for these four fragments are overexposed relative to fragments from the spacer on the same blots) (3). Indeed, very few replication forks have yet entered either gene from the spacer region. As predicted, _Eco_RI fragments 4 and 5, which encompass the ori-β and _ori-β_′ regions, respectively, each display a composite pattern consisting of a complete bubble arc and a single fork arc (Fig. 3B) (10, 11, 15, 56). This is the pattern expected of a fragment residing in a broad zone of potential sites, since it will sometimes be replicated from an internal origin (thereby contributing to the bubble arc) and sometimes replicated passively by a fork arising from a neighboring fragment in the same zone (contributing to the simple Y or fork arc) (56).
Superficially, this result is compatible with the PCR data, which suggest that _Eco_RI fragments 4 and 5 contain fixed start sites at ori-β and _ori-β_′ (31). However, both sites would have to be quite inefficient, according to this model, to explain the presence of a large number of single forks that traverse this fragment and that must have initiated from a site or sites located elsewhere in the spacer. In addition, both fragments appear to have complete bubble arcs, even though ori-β is not centered in fragment 4 (see Fig. 1). Furthermore, other enzyme digests position ori-β and _ori-β_′ differently in their respective fragments, yet each of these fragments also displays a complete bubble arc indicative of a centered origin. For example, compare _Bam_HI fragment 1, _Bgl_II fragment 1, _Eco_RI fragment 5, and _Xba_I fragment 5, each of which contains _ori-β_′ (Fig. 3).
Indeed, all 19 adjacent and overlapping intergenic fragments in the range of 3.4 to 18 kb that were examined in this study display the composite pattern consisting of a complete bubble arc and a single fork (Fig. 3B). The _Xba_I digest alone shows unequivocally that there are at least eight initiation sites or zones within the ∼45-kb region of the spacer covered by the _Xba_I digest. To control for the possibility that mimosine induces this polydisperse pattern of initiation, an additional experiment was performed in which cells were collected near the G1/S boundary with aphidicolin and sampled 30 min after release, which represents the peak initiation period after this synchronizing protocol (56). Replication intermediates in this case were digested with _Hin_dIII, which allowed analysis of five nearly contiguous fragments in the 4- to 6-kb range extending from right of _ori-β_′ through ori-γ (Fig. 3A). As shown in Fig. 3B, each of these fragments also displays the composite pattern and centered bubble arc. Based on this digest alone, there are at least five sites in addition to ori-β and _ori-β_′ at which initiation occurs. Thus, the polydisperse initiation pattern displayed by this locus is unlikely to result from the cell synchronization protocol per se.
High-resolution 2-D gel analysis suggests that replication initiation sites can be very closely spaced in the intergenic region.
Given the unlikely circumstance that each of these digests accidentally isolated a fixed initiation site in the center of each fragment examined, we reasoned that several sites might be distributed at frequent intervals within each fragment, some of which are centered. An alternative possibility is that each fragment contains a single fixed origin from which the forks diverge in the two directions at unequal rates, with neither direction preferred. By this scenario, any fragment that displays a bubble arc must contain an initiation site, but it need not be in the center of the fragment. This effect presumably would be most pronounced in larger fragments in which the forks have to travel longer distances.
To obtain a better estimate of the frequency distribution of initiation sites in the intergenic spacer, restriction enzyme digests were chosen that would yield fragments near the size range of the nascent DNA strands analyzed in the PCR-based assay, and conditions were established for adequately separating small fragments on 2-D gels. CHOC 400 cells were synchronized and sampled 90 min after release from mimosine, and replication intermediates were prepared using either a combination of _Bam_HI and _Hin_dIII or _Pvu_II alone to digest the DNA. These two digests yield seven adjacent fragments that range in size from 1.0 to 2.3 kb, encompass ori-β and _ori-β_′, and together nearly reconstitute the 15-kb _Bam_HI fragment 1 (Fig. 3A), which displays a complete bubble arc (Fig. 3B).
The _Bam_HI/_Hin_dIII digest separates ori-β, _ori-β_′, and the region between them into three small fragments (a 1.97-kb _Bam_HI/_Hin_dIII fragment 1, a 1.63-kb fragment 2, and a 1.71-kb _Hin_dIII fragment 3) (Fig. 3A). Thus, we could determine whether these peaks correspond to two fixed origins flanked by silent regions (as in Fig. 4A) or from a collection of start sites within a preferred region (as in Fig. 4C). As shown in Fig. 3C, all three small fragments display bubble arcs, although of varying intensities. This result was expected for fragments 1 and 3, but not for fragment 2, which contains the valley and the very edge of the ori-β peak. Furthermore, the level of initiation detected in fragment 2 is considerable, with the bubble-to-fork arc ratio appearing to be perhaps threefold smaller than that observed for ori-β (fragment 1) and only somewhat smaller than that detected for _ori-β_′ (fragment 3). Any bubbles detected in fragment 2 must have initiated within the fragment itself, which is not predicted if the peaks of nascent strands measured by PCR at ori-β and _ori-β_′ represent fixed initiation sites surrounded by silent regions. We assume that these bubbles arise from the shoulder of the ori-β peak detected by PCR (Fig. 1) and that ori-β corresponds to a collection of start sites (as shown in Fig. 2C and 4C), rather than a single site (as shown in Fig. 2A and 4A). Bubbles were detected in fragment 2 in three additional experiments (P. A. Dijkwel, unpublished observations).
In the _Pvu_II digest, the four adjacent fragments analyzed are 1.5, 1.9, 1.0, and 2.3 kb in length and correspond to the region between _ori-β_′ and the MAR (Fig. 3A). Homologous probes show that fragments 1, 2, and 4 each support substantial amounts of initiation (Fig. 3C). However, _Pvu_II fragment 3 does not display a detectable bubble arc even after the very long film exposure shown here, indicating that it may not be an efficient template for initiation. Together with the results from the _Bam_HI/_Hin_dIII digest, we conclude that the region approximately represented by the 15-kb fragment 1 of _Bam_HI contains at least six individual initiation sites, but at least one silent region lying just upstream from the MAR.
By extrapolation, many of the large fragments analyzed in the experiments shown in Fig. 3B probably also contain multiple internal initiation sites. However, we cannot rule out the possibility that there are also other short silent regions distributed at unknown intervals throughout the spacer, which would only be revealed if the whole region were dissected into very small, adjacent fragments as depicted in Fig. 3C (in practice, it is difficult to analyze more than three or four small fragments in a single gel, owing to an unfortunate distribution of restriction sites).
On the linear map in Fig. 3A, we have placed a vertical bar at the center of each restriction fragment that displayed an apparently centered bubble arc in the gels shown in Fig. 3B and C. Obviously, the bubble arcs in some fragments that have the same approximate centers may arise from the same start site. Nevertheless, we can conclude that there are at least 14 initiation sites situated in nonoverlapping fragments in the spacer: 6 sites displayed by the _Bam_HI/_Hin_dIII and _Pvu_II digests and an additional 8 sites in nonoverlapping _Eco_RI and _Xba_I fragments (Fig. 3A).
In vitro strategy for preparing small, origin-containing nascent strands initiating in the first few minutes of the S period.
The results of 2-D gel analyses and early intrinsic labeling studies are compatible with the suggestion that the DHFR origin consists of a broad zone of very frequently spaced potential initiation sites, with the central ∼30-kb core preferred. Other experimental strategies have suggested that the intergenic region is characterized by a small number of highly preferred, fixed initiation sites (ori-β and _ori-β_′ being two of them) surrounded by very inefficient secondary sites. Indeed, from the results of Okazaki template bias assays, it was originally estimated that >80% of initiations occurring within the 27-kb region examined were located within a 450-bp fragment centered over ori-β (4), while results of the PCR-based nascent strand abundance assay suggested that >90% of initiations in the 12-kb region encompassing ori-β and _ori-β_′ occur at these two sites (31). These models differ in two respects (illustrated diagramatically in Fig. 4, in which _ori-β_′ has been omitted for simplicity): (i) the frequency distribution of sites and (ii) the relative efficiencies of initiation at each site. Clearly, in any quantitative assays designed to distinguish between the two models by analyzing origin-containing DNA, the nascent material analyzed has to be very small (compare Fig. 3A and C to B and D). While the 2-D gel approach described above has allowed us to address the distribution question and even to refine the assay to several fragments in the kilobase range, it is difficult to normalize the signals from one hybridization to another even on a single blot; therefore, the method is relatively insensitive to differences in initiation frequency at different locations.
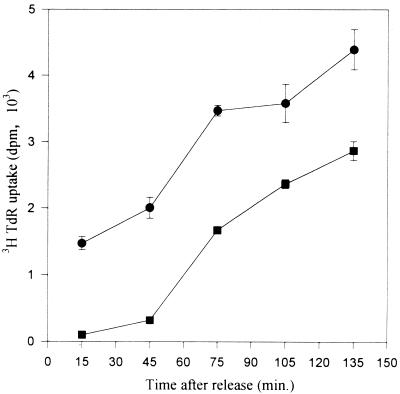
To estimate the relative frequencies at different positions throughout the spacer, we modified the ELFH assay to allow quantitation of origin-containing nascent strands ∼300 to 1,000 nt in length, which approximates the size of the nascent material analyzed in the PCR-based nascent strand abundance assay (31). In initial experiments, we optimized conditions for preparing radiolabeled, origin-containing, nascent DNA in this size range. Cells were arrested in early G1 by isoleucine deprivation and were released into either 400 μM mimosine or 10-μg/ml aphidicolin for 12 h. Drug-containing media were then removed and replaced with drug-free medium, and duplicate wells were pulsed with [3H]thymidine for 30 min beginning at time zero. As shown in Fig. 5, after removal of mimosine, there is a 30- to 40-min lag before a significant amount of label is incorporated into DNA. In contrast, [3H]thymidine incorporation begins immediately after withdrawal of aphidicolin, and the rate extrapolates back to time zero well above the baseline (Fig. 5) (42). Thus, in agreement with earlier studies (37, 42) and unlike aphidicolin, mimosine appears to prevent replication forks from being established or to arrest them very close to their start sites.
FIG. 5.
There is a lag after release from mimosine before CHOC 400 cells begin to incorporate [3H]thymidine into DNA. CHOC 400 cells growing in 24-well dishes were starved for isoleucine for 45 h and were released for 12 h into either 400 μM mimosine (squares) or 10-μg/ml (29.5 μM) aphidicolin (circles). The drugs were washed out with prewarmed drug-free medium and cultures were incubated in complete MEM. At 30-min intervals beginning at time zero, quadruplicate wells were pulse labeled with 5-μCi/ml [3H]thymidine for 30 min. Incorporation was quenched by the addition of citric acid to 0.2 M (43), and specific labeling of DNA was determined as previously described (42). Data are plotted at the midpoint of the pulse period. Bars indicate standard errors of the mean of quadruplicate samples.
To label nascent DNA strands only in the immediate environment of origins of replication, mimosine-synchronized cells were released from the drug-containing medium in vivo and were sampled during the lag period. As a control, another group of cells were synchronized with aphidicolin. Nuclei were isolated in the cold and were exposed to the four deoxynucleoside triphosphates (dNTPs) (one of them radiolabeled) in an in vitro replication cocktail for 1.5 min at 34°C (4). Since mimosine is thought to function in vivo by lowering dNTP pool levels (6, 20, 38, 58), replication should initiate immediately and synchronously when the extracts are supplied with the four dNTPs, conditions facilitating the radiolabeling of very small, origin-containing DNA. To test this proposal, DNA from nuclei subjected to this protocol was purified and separated on an alkaline agarose gel, and radioactivity in each lane was detected with a PhosphorImager. The resulting autoradiogram is shown in Fig. 6A, and densitometric tracings of signal intensities are shown in Fig. 6B.
In nuclei from mimosine-synchronized cells sampled during the lag period (either 5 or 20 min after drug removal), the majority of the DNA synthesized in a 1.5-min incubation in vitro is less than ∼3,000 nt in length in both samples (Fig. 6, lanes M5 and M20). At a probable fork rate of ∼3 kb/min (24), this result indicates a highly synchronous burst of DNA synthesis from positions close to origins at both time points, a result predicted if forks had not significantly diverged away from origins during the lag period. The size distribution suggests that the nascent DNA is composed predominantly of short leading strands, since fragments (∼400 nt) in the center of the major peak are larger than typical Okazaki fragments (50 to 200 nt). However, a component of the labeled material clearly corresponds to Okazaki fragments, which are more visible in the 50- and 80-min samples in which much of the nascent DNA has matured to larger sizes. By 50 or 80 min after mimosine removal (Fig. 6, lanes M50 and M80), a large percentage of the DNA extended and labeled in the 1.5-min in vitro incubation is greater than 3,000 nt in length, in agreement with the observation that DNA synthesis resumes 35 to 40 min after mimosine removal in vivo (Fig. 6) (10, 42). As expected, when the 5-min sample is labeled for 1.5 min in vitro and then chased for 30 min with cold dNTPs, small labeled DNA is chased into high-molecular-weight material (Fig. 6, lanes M5-C).
In contrast, when aphidicolin-synchronized cells are sampled 5 min after drug removal and are then labeled in vitro for 1.5 min, the majority of nascent DNA is much longer than 3,000 nt, some DNA is detected in the Okazaki fragment size range (50 to 200 nt), and a small percentage migrates between 200 and 3,000 nt (Fig. 6). Thus, significant numbers of replication forks have already moved away from their start sites in cells incubated for 12 to 14 h in aphidicolin after release from an early G1 block (42). The pattern is similar in cells sampled 20 min after release from aphidicolin (Fig. 6, lane A20).
Nascent, origin-containing DNA 300 to 1,000 nt in length derives from almost every position tested within the intergenic spacer.
The data shown in Fig. 5 indicated that in mimosine-synchronized cells harvested during the lag period (i.e., 20 min or less after drug removal), the majority of DNA synthesized during a 1.5-min pulse in vitro is ∼5,000 nt or less. Thus, very tight synchrony is achieved, and most of the resulting labeled material must correspond to nascent DNA centered over local origins. Therefore, with an ELFH assay, it should be possible to discriminate between the two different models illustrated in Fig. 4A and C by using this labeled DNA (or a sized fraction thereof) as a hybridization probe on a series of clones scattered throughout the intergenic region.
CHOC 400 cells were synchronized with mimosine, nuclei were isolated 20 min after release, and radiolabeled nascent DNA was synthesized in a 1.5-min incubation in vitro. In the first type of experiment, DNA was isolated, purified, and sonicated to reduce the average size to ∼600 bp. This radiolabeled preparation was hybridized to individual single-copy probes from the intergenic region immobilized on dot blots (see Fig. 7C). The control was total DNA isolated from asynchronous cells sonicated to a similar size and labeled with [32P]dCTP by random priming. Shown in Fig. 7A are the resulting hybridization patterns obtained when either total DNA or the nascent ELFs were used as probes (patterns obtained with the PhosphorImager). The normalized hybridization values (expressed as a percentage of total hybridization to all of the cloned inserts on the blot) are plotted as a function of map position (Fig. 7C).
As can be seen, the nascent strands labeled in vitro hybridized to every probe within the 55-kb intergenic spacer to some degree, although there was clearly a preference for the regions encompassing the _ori-β_-_ori-β_′ and ori-γ regions. Overall, this result was consistent with the 2-D gel results and with previous relatively low-resolution ELFH experiments performed with aphidicolin-synchronized cells performed in our own and other laboratories (19, 57). However, the resolution of this ELFH approach is limited by the size of the largest labeled nascent strands in these preparations (∼5,000 nt) (Fig. 6B) and would average out differences in initiation frequencies at sites spaced more closely than 5 to 10 kb. Furthermore, since the nascent DNA was not sized, it was contaminated with Okazaki fragments that would be distributed throughout the entire domain, probably accounting for significant hybridization to the two genes (Fig. 7C).
Therefore, to increase the resolution of the ELFH assay and to eliminate Okazaki fragment contamination, the in vitro reaction was repeated, the radiolabeled DNA was separated on an alkaline agarose gel, and the 300- to 1,000-nt fraction was excised, purified, and hybridized to a similar collection of single-copy sequences from the intergenic spacer. With this size-selected probe, the distribution of small nascent strands is now seen to be very nonuniform within the spacer and is in relatively good agreement with the distribution of 600- to 1,000-nt nascent strands in the regions of ori-β and _ori-β_′ measured by competitive PCR in an earlier study (Fig. 7D) (31). The superficial difference is the absence of a clear valley between the ori-β and _ori-β_′ positions in the ELFH approach (probe 27), in agreement with the 2-D gel results shown in Fig. 3C (_Bam_HI/_Hin_dIII fragment 2). However, as in the 2-D gel study, probe 27 itself is not directly centered over the valley and is undoubtedly detecting the edge of the ori-β peak. Importantly, the region lying just upstream from the MAR, which was essentially negative for initiation by the criterion of 2-D gel analysis, also appears negative in this high-resolution ELFH analysis (Fig. 7C).
DISCUSSION
For the last several years, there have been opposing views of the nature of the DHFR origin of replication. Combining the results of the more panoramic approaches (e.g., intrinsic radiolabeling [36], low-resolution ELFH assays [19, 57], and neutral-neutral and neutral-alkaline 2-D gel methods [10, 11, 15, 56]), it appeared that nascent strands can initiate at any one of several sites within the intergenic spacer in the early S period but more often near the regions known as ori-β and ori-γ. Thus, the panoramic view led to the proposal that there are multiple start sites with different initiation frequencies distributed randomly at very frequent intervals within the integenic spacer.
In contrast, when the initial focus was on a very narrow subregion of the intergenic spacer with high-resolution methods such as the lagging strand template bias assay (4) or the PCR-based small nascent strand abundance assay (45, 54), it was possible to come away with the view that ori-β is the predominant nascent strand start site in the spacer (7) and thus to consider it a classic replicator. Importantly, however, when the PCR-based assay was subsequently extended beyond the immediate environs of ori-β, a second preferred site or region was discovered (_ori-β_′). We predicted that more such sites would be identified if the analysis were extended even further (particularly to ori-γ) and that aggregate initiations occurring at all such sites in the intergenic spacer would essentially recapitulate the patterns observed with lower-resolution labeling and 2-D gel approaches.
In the present study, we have tested this prediction. We first estimated the minimum number of initiation sites present in the intergenic spacer by performing 2-D gel analysis of 31 adjacent and overlapping restriction fragments and determining how many of these display the complete bubble arcs characteristic of a centered origin. All but one of these fragments displayed complete bubble arcs. These studies suggested a minimum of 14 initiation sites within the 55-kb spacer, a conservative estimate calculated from the number of nonoverlapping fragments displaying this pattern (Fig. 3) (39). Furthermore, in the analysis of seven nonoverlapping fragments in the size range of 1.0 to 2.3 kb that essentially reconstituted a ∼15-kb _Bam_HI fragment in the spacer, all but one of these displayed a complete bubble arc. Thus, by extension, many fragments in the 4- to 18-kb range (Fig. 3B) might be expected to contain several sites firing with different efficiencies, with the result that 20 or more initiation sites reside within the spacer.
We specifically asked whether the dramatic peaks of small nascent strands detected by the PCR approach in the ori-β and _ori-β_′ regions correspond to fixed initiation sites that are surrounded by silent regions. In fact, when the three fragments that isolated ori-β, _ori-β_′, and the region between them were examined with 2-D gels, bubbles were detected in all three fragments, including the one encompassing the valley and a small part of the ori-β peak. Presumably, the replication bubbles we detect in fragment 2 of the _Bam_HI/_Hin_dIII digest arise from the ori-β shoulder. Thus, our data are most consistent with a model in which the ori-β and _ori-β_′ peaks each represent a distribution of initiation sites around a central locus, much like the distribution of nascent strand start sites in Escherichia coli oriC (32) (Fig. 2C and 4C), rather than the very fixed initiation sites identified in simian virus 40 (22) and yeast ARS1 (2) (Fig. 2A and 4A).
We next wanted to determine the relative frequencies of initiation at different sites within the spacer, using a modification of the ELFH assay. To ensure that we could isolate origin-containing fragments in the same size range (∼800 nt) as that analyzed in the PCR-based nascent strand abundance assay, it was necessary to arrest replication forks very close to their start sites. To accomplish this, we took advantage of the interesting properties of mimosine, which, by the criterion of the neutral-neutral 2-D gel approach, is an extremely effective inhibitor of DNA replication when delivered to cells as they approach the G1/S boundary (42). We have shown that the drug can be cross-linked to serine hydroxymethyl transferase, which catalyzes the penultimate step in thymidine biosynthesis (38). Others have shown that mimosine lowers cellular dNTP pool levels (6, 20) and that nuclei from mimosine-synchronized cells have essentially entered the S phase (25) and are capable of synthesizing DNA when supplemented with mammalian S-phase cytosol and the four dNTPs (20).
However, in our view, these observations do not satisfactorily explain why mimosine is so effective at preventing replication fork movement in vivo. One interesting possibility is that, because mimosine depletes cellular dNTP pool levels, it signals a p53-independent G1/S phase checkpoint that was recently identified in CHO cells (35). Thus, in vivo, mimosine may actually prevent replication forks from progressing by some regulatory loop activated by the checkpoint; in vitro, the checkpoint would presumably be nonfunctional and only the addition of deoxyribonucleotides would be required to overcome mimosine arrest. In this regard, mimosine has also been shown to be clastogenic when delivered to cells in S phase (27), and a recent report suggests that it causes single-stranded breaks even when present during G1 (41). Therefore, the proposed single-strand breaks that might result from the level of mimosine utilized here (fewer than one break per 600 kb [41, 59]) could be indirectly responsible for activating a G1/S checkpoint. However, this proposal does not simply explain why hydroxyurea and aphidicolin are much more leaky replication inhibitors (references 37 and 47 and the present study), since hydroxyurea also depletes cellular dNTP pools (52) and aphidicolin has been shown to destabilize replication forks (23, 29, 53). It should also be pointed out that even if mimosine were to elicit single-strand breaks in the region under investigation, we have previously shown that broken bubbles and single forks trace very different patterns on 2-D gels than those detected for bona fide replication bubbles and forks (28). Clearly, there are caveats associated with the use of inhibitors to enrich for S-phase cells in these and other experiments. However, we have demonstrated the presence of complete bubbles arcs in many different fragments from the DHFR intergenic spacer in unsynchronized cells and, as shown here and previously, in cells synchronized with aphidicolin (references 11, 15, and 51; P. A. Dijkwel, unpublished observations). Thus, we consider it likely that the general patterns observed here reflect the in vivo situation.
Regardless of how mimosine prevents DNA replication, the observation that inhibition is overcome in vitro by adding dNTPs (20) suggested a mechanism for studying replication forks from the moment they leave initiation sites. This allowed us to synthesize radiolabeled origin-containing DNA less than ∼5,000 nt in length and from this population to isolate DNA in the size range of 300 to 1,000 nt. This fraction approximates the size of the nascent DNA analyzed by the PCR-based nascent strand abundance assay (31). (The use of aphidicolin as a synchronizing agent here would have allowed many of the small bubbles to mature away from their origins [Fig. 6 and references 37 and 42], thus significantly lowering the amount of small nascent DNA recovered in this approach.)
When this sized nascent DNA was hybridized to a series of probes from the intergenic region, the resulting hybridization pattern afforded a much higher resolution picture than is obtained when the whole (unsized) preparation was used as the probe (Fig. 7B and C) (19, 57). The data also suggested that the sized fraction is not contaminated significantly by Okazaki fragments or other small broken DNA, since no hybridization was detected to fragments in the DHFR gene (compare to unsized labeled DNA) (Fig. 7A). As expected, there are sharp peaks over ori-β and ori-γ, but also a shoulder that accommodates _ori-β_′. Thus, this high-resolution ELFH assay agrees very well with results of the PCR-based nascent strand abundance assay, as well as with the 2-D gel analysis of this region (Fig. 3C).
With the exception of one clone in the series, significant hybridization occurred to every intergenic fragment analyzed in these experiments. It is important to realize that since the labeled nascent DNA is no longer than 1,000 nt and that, with the exception of clones 3 and 4, the fragments tested are more than 1000 bp apart, initiation must occur at each of these locations. Therefore, there must be at least 14 individual start sites within the spacer. This contrasts with the impression received from studying the small nascent strand abundance curve shown in Fig. 1A and 7D (i.e., that ori-β and _ori-β_′ correspond to two fixed sites from which most initiation occurs). In fact, probes 19, 27, and 38 each detect distinct initiation sites or regions, two of which correspond to the ori-β and _ori-β_′ peaks detected by PCR. Thus, there is at least one additional site in the region bracketed by probe 27 that was not uncovered by the PCR-based nascent strand abundance assay. Presumably, an analysis of more, very closely spaced clones or primer pairs would reveal more of the fine structure of this complex origin in the PCR approach and in the ELFH experiments described here. An important conclusion is that only ∼21% of the total hybridization to all clones tested was targeted to probe 38, which contains ori-β. Were it possible to have analyzed many additional clones scattered between those analyzed (Fig. 7C) and had many of these also been positive for initiation, estimates of the percentage occurring at ori-β would be greatly reduced. Therefore, ori-β is a prominent site but does not account for the 80% of initiations suggested to occur at this site, based on the Okazaki template bias assay (4). This result is consistent with earlier studies that determined leading-strand template bias, in which it was not possible to detect a significant bias with probes situated close to the _ori-β_-_ori-β_′ region (21).
Importantly, the only fragment that did not display a detectable bubble arc on 2-D gels was the 1.0-kb _Pvu_II fragment, which was also the only clone that failed to hybridize significantly to radiolabeled origin-containing DNA in the size range of 300 to 1,000 nt in the modified ELFH assay. This result suggests that when both methods are adjusted to afford approximately the same resolution (a few kilobases or less), the results of the two approaches are very similar.
In the results shown in Fig. 7D, we incorporated the data from four different approaches into one graph for the purposes of comparison: (i) an early intrinsic labeling study (36), (ii) an early ELFH assay using aphidicolin-synchronized cells, (iii) the PCR-based nascent strand abundance assay (31), and (iv) the high-resolution ELFH experiment described here. Each curve has been arbitrarily expanded so that the signals at ori-β are equal. In our opinion, all of this data is very consistent. It is not difficult to appreciate why the lack of probes in the ori-γ region in the earlier ELFH study (19) would give the impression that most initiations occur at the ori-β locus. However, when the resolution is increased and a more extensive part of the spacer is examined, as in the higher-resolution ELFH study presented here, a more comprehensive picture of initiation in the region is obtained. Remarkably, the shape of the curve obtained with the 300- to 1,000-nt probe in the ELFH assay (Fig. 7D) is very similar to data obtained almost 12 years ago by intrinsically labeling replicating DNA as cells entered the S period and analyzing restriction fragments in the 1- to 2.5-kb range in the intergenic spacer by a modified in-gel renaturation method (36). With the exception of regions in which fragments were not examined in the older study (e.g., near probe 40 and to the left of probe 38) (36), the patterns are remarkably similar. Thus, when the size of the nascent DNA analyzed is reduced to a few kilobases or less, two things are apparent: (i) replication initiates at many positions throughout the spacer, and (ii) regions (sites?) differ markedly in their efficiency of utilization.
We believe that all of the data that has been collected on this locus can now be reconciled with a general model in which potential sites with different efficiencies are distributed at intervals of a few kilobases or less throughout the intergenic region. This does not exclude the possibility that individual sites such as ori-β and _ori-β_′ each contain a genetic element recognized by sequence-specific _trans_-acting proteins that facilitate initiation in their immediate environments (i.e., such as simian virus 40 T antigen [51] or E. coli dnaA protein [33]). Indeed, a recent genetic assay analyzed the results of selected mutations on the ability of a small _ori-β_-containing fragment to serve as a replicator for that fragment when inserted at ectopic chromosomal positions (1). It was found that some small deletions do, in fact, down-regulate initiation by as much as 50% in the immediate environment of ori-β by the criterion of the nascent strand abundance assay.
However, it is important to point out that ori-β does not contain sequences that are required for initiation at sites distant from itself in the intergenic spacer: when a 4.5-kb fragment containing ori-β was selectively deleted in loco from the spacer by homologous recombination, initiation was basically unchanged in the remainder of the intergenic region, and all copies of the locus replicated at approximately the same time during the S period as in the wild-type control (30). If ori-β corresponds to a local replicator and if such replicators are distributed as frequently throughout the intergenic spacer as preferred nascent strand start sites appear to be, then there must be enormous numbers of such sites in the mammalian genome. So far, there are no obvious sequence motifs or combinations thereof shared by ori-β, _ori-β_′, and ori-γ that could correspond to specific protein binding sites (see reference 16 for analysis of other candidate origins; P. J. Mosca, H.-B. Lin, and L. D. Mesner, unpublished observations). Perhaps future sequence analysis of many such preferred sites will reveal relaxed motifs that attract critical initiation proteins.
Whether or not some of these sites correspond to classic genetic replicators is almost a semantic question at this point, since each appears to control initiation only in its immediate environment and then only very inefficiently, rendering it difficult to define the boundaries of any given replicon. Efficiencies in some cases could result from interference by neighboring active start sites, as has been shown for closely spaced yeast ARS elements (3). It does seem likely, however, that whether or not individual start sites are called replicators, the base composition and/or sequence of some regions (e.g., ori-β, _ori-β_′, and ori-γ) will attract initiation complexes more efficiently than other regions and that changes in these sequences will affect their abilities to load these complexes. The chromatin environment will also undoubtedly play a role in the efficiency of initiation site usage, as was pointed out in a recent review (18, 46). In this regard, it is interesting that the center of the regions that we previously termed ori-β and ori-γ are almost precisely centered between the 3′ ends of the two genes and the silent region lying upstream from the MAR. Thus, it is conceivable that usage of inchoate sites will be determined not only by sequence but also by local transcriptional activity and/or chromatin architecture.
Will this polydispersed initiation mode be found in other higher eukaryotes? Broad replication initiation zones have also been identified in the ribosomal DNA locus in Xenopus (26), humans (40), and Chinese hamsters (34); the rhodopsin locus in Chinese hamsters (13); and the _a_-polymerase and histone loci in Drosophila melanogaster (48-50). The only system that appears to contain a very fixed, solo initiation site is the lamin B2 origin in human cells, which was defined by the PCR-based nascent strand abundance assay (17) and by replication initiation point mapping (1). This situation is unusual, however, since the intergenic region between the two local transcription units is only ∼500 bp in length. Thus, if preferred sites or regions are distributed at kilobase intervals or less on average and if active genes are not used as templates for initiation, then only one such site would be isolated in the spacer between the two genes in the lamin B2 locus. The same situation may apply to oriGNAI3, which has been localized within a 1.7-kb region in the Chinese hamster genome (53a). In all of the cases in which initiation is distributed throughout broad zones, it will be extremely interesting to understand how the homologues of yeast initiation proteins, which recognize very fixed sites in the yeast genome, are distributed in mammalian genomes.
Acknowledgments
We thank David Gilbert (SUNY, Syracuse) for providing us with probe DGK and Howard Cedar (Hebrew University, Jerusalem) for providing probe HCC. We are indebted to Carlton White and Kevin Cox for expert technical assistance and to the other members of our laboratory for very useful discussions. We especially thank two of the reviewers for insightful comments on a previous version of the manuscript that caused us to think about these experiments more critically.
This work was supported by NIH grant RO1 GM26108 to J.L.H.
REFERENCES
- 1.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287**:**2023-2026. [DOI] [PubMed] [Google Scholar]
- 1a.Altman, A. L., and E. Fanning. 2001. The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21**:**1098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279**:**95-98. [DOI] [PubMed] [Google Scholar]
- 3.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51**:**463-471. [DOI] [PubMed] [Google Scholar]
- 4.Burhans, W. C., L. T. Vassilev, M. S. Caddle, N. H. Heintz, and M. L. DePamphilis. 1990. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell 62**:**955-965. [DOI] [PubMed] [Google Scholar]
- 5.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81**:**1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai, Y., B. Gold, J. K. Vishwanatha, and S. L. Rhode. 1994. Mimosine inhibits viral DNA synthesis through ribonucleotide reductase. Virology 205**:**210-216. [DOI] [PubMed] [Google Scholar]
- 7.DePamphilis, M. L. 1993. Eukaryotic DNA replication: anatomy of an origin. Annu. Rev. Biochem. 62**:**29-63. [DOI] [PubMed] [Google Scholar]
- 8.DePamphilis, M. L. 1993. Origins of DNA replication that function in eukaryotic cells. Curr. Opin. Cell Biol. 5**:**434-441. [DOI] [PubMed] [Google Scholar]
- 9.DePamphilis, M. L. 1997. The search for origins of DNA replication. Methods 13**:**211-219. [DOI] [PubMed] [Google Scholar]
- 10.Dijkwel, P. A., and J. L. Hamlin. 1992. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol. Cell. Biol. 12**:**3715-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkwel, P. A., and J. L. Hamlin. 1995. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell. Biol. 15**:**3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkwel, P. A., and J. L. Hamlin. 1996. Sequence and context effects on origin function in mammalian cells. J. Cell. Biochem. 62**:**210-222. [DOI] [PubMed] [Google Scholar]
- 13.Dijkwel, P. A., L. D. Mesner, V. V. Levenson, J. d'Anna, and J. L. Hamlin. 2000. Dispersive initiation of replication in the Chinese hamster rhodopsin locus. Exp. Cell Res. 256**:**150-157. [DOI] [PubMed] [Google Scholar]
- 14.Dijkwel, P. A., J. P. Vaughn, and J. L. Hamlin. 1991. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol. Cell. Biol. 11**:**3850-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dijkwel, P. A., J. P. Vaughn, and J. L. Hamlin. 1994. Replication initiation sites are distributed widely in the amplified CHO dihydrofolate reductase domain. Nucleic Acids Res. 22**:**4989-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobbs, D. L., W. L. Shaiu, and R. M. Benbow. 1994. Modular sequence elements associated with origin regions in eukaryotic chromosomal DNA. Nucleic Acids Res. 22**:**2479-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giacca, M., L. Zentilin, P. Norio, S. Diviacco, D. Dimitrova, G. Contreas, G. Biamonti, G. Perini, F. Weighardt, and S. Riva. 1994. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. USA 91**:**7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294**:**96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, D. M., H. Miyazawa, and M. L. DePamphilis. 1995. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol. Cell. Biol. 15**:**2942-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert, D. M., A. Neilson, H. Miyazawa, M. L. DePamphilis, and W. C. Burhans. 1995. Mimosine arrests DNA synthesis at replication forks by inhibiting deoxyribonucleotide metabolism. J. Biol. Chem. 270**:**9597-9606. [DOI] [PubMed] [Google Scholar]
- 21.Handeli, S., A. Klar, M. Meuth, and H. Cedar. 1989. Mapping replication units in animal cells. Cell 57**:**909-920. [DOI] [PubMed] [Google Scholar]
- 22.Hay, R. T., and M. L. DePamphilis. 1982. Initiation of SV40 DNA replication in vivo: location and structure of 5′ ends of DNA synthesized in the ori region. Cell 28**:**767-779. [DOI] [PubMed] [Google Scholar]
- 23.Huberman, J. A. 1981. New views of the biochemistry of eucaryotic DNA replication revealed by aphidicolin, an unusual inhibitor of DNA polymerase alpha. Cell 23**:**647-648. [DOI] [PubMed] [Google Scholar]
- 24.Huberman, J. A., and A. D. Riggs. 1966. Autoradiography of chromosomal DNA fibers from Chinese hamster cells. Proc. Natl. Acad. Sci. USA 55**:**599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes, T. A., and P. R. Cook. 1996. Mimosine arrests the cell cycle after cells enter S-phase. Exp. Cell Res. 222**:**275-280. [DOI] [PubMed] [Google Scholar]
- 26.Hyrien, O., and M. Mechali. 1993. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 12**:**4511-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jha, A. N., P. M. Hande, L. H. Mullenders, and A. T. Natarajan. 1995. Mimosine is a potent clastogen in primary and transformed hamster fibroblasts but not in primary or transformed human lymphocytes. Mutagenesis 10**:**385-391. [DOI] [PubMed] [Google Scholar]
- 28.Kalejta, R. F., and J. L. Hamlin. 1996. Composite patterns in neutral/neutral two-dimensional gels demonstrate inefficient replication origin usage. Mol. Cell. Biol. 16**:**4915-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalejta, R. F., and J. L. Hamlin. 1997. The dual effect of mimosine on DNA replication. Exp. Cell Res. 231**:**173-183. [DOI] [PubMed] [Google Scholar]
- 30.Kalejta, R. F., X. Li, L. D. Mesner, P. A. Dijkwel, H. B. Lin, and J. L. Hamlin. 1998. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2**:**797-806. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi, T., T. Rein, and M. L. DePamphilis. 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18**:**3266-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohara, Y., N. Tohdoh, X. W. Jiang, and T. Okazaki. 1985. The distribution and properties of RNA primed initiation sites of DNA synthesis at the replication origin of Escherichia coli chromosome. Nucleic Acids Res. 13**:**6847-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornberg, A., and T. A. Baker. 1992. DNA Replication. W. H. Freeman, New York, N.Y.
- 34.Larner, J. M., H. Lee, R. D. Little, P. A. Dijkwel, C. L. Schildkraut, and J. L. Hamlin. 1999. Radiation down-regulates replication origin activity throughout the S phase in mammalian cells. Nucleic Acids Res. 27**:**803-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, H., J. M. Larner, and J. L. Hamlin. 1997. A p53-independent damage-sensing mechanism that functions as a checkpoint at the G1/S transition in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA 94**:**526-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leu, T. H., and J. L. Hamlin. 1989. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol. Cell. Biol. 9**:**523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levenson, V., and J. L. Hamlin. 1993. A general protocol for evaluating the specific effects of DNA replication inhibitors. Nucleic Acids Res. 21**:**3997-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, H. B., R. Falchetto, P. J. Mosca, J. Shabanowitz, D. F. Hunt, and J. L. Hamlin. 1996. Mimosine targets serine hydroxymethyltransferase. J. Biol. Chem. 271**:**2548-2556. [DOI] [PubMed] [Google Scholar]
- 39.Linskens, M. H., and J. A. Huberman. 1990. Ambiguities in results obtained with 2D gel replicon mapping techniques. Nucleic Acids Res. 18**:**647-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Little, R. D., T. H. Platt, and C. L. Schildkraut. 1993. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 13**:**6600-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikhailov, I., G. Russev, and B. Anachkova. 2000. Treatment of mammalian cells with mimosine generates DNA breaks. Mutat. Res. 459**:**299-306. [DOI] [PubMed] [Google Scholar]
- 42.Mosca, P. J., P. A. Dijkwel, and J. L. Hamlin. 1992. The plant amino acid mimosine may inhibit initiation at origins of replication in Chinese hamster cells. Mol. Cell. Biol. 12**:**4375-4383. (Erratum **13:**1981, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moscovitis, G., and A. B. Pardee. 1980. Citric acid arrest and stabilization of nucleoside incorporation into cultured cells. Anal. Biochem. 101**:**221-224. [DOI] [PubMed] [Google Scholar]
- 44.Nawotka, K. A., and J. A. Huberman. 1988. Two-dimensional gel electrophoretic method for mapping DNA replicons. Mol. Cell. Biol. 8**:**1408-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelizon, C., S. Diviacco, A. Falaschi, and M. Giacca. 1996. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol. 16**:**5358-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pemov, A., S. Bavykin, and J. L. Hamlin. 1998. Attachment to the nuclear matrix mediates specific alterations in chromatin structure. Proc. Natl. Acad. Sci. USA 95**:**14757-14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reichard, P. 1993. From RNA to DNA, why so many ribonucleotide reductases? Science 260**:**1773-1777. [DOI] [PubMed] [Google Scholar]
- 48.Shinomiya, T., and S. Ina. 1991. Analysis of chromosomal replicons in early embryos of Drosophila melanogaster by two-dimensional gel electrophoresis. Nucleic Acids Res. 19**:**3935-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinomiya, T., and S. Ina. 1993. DNA replication of histone gene repeats in Drosophila melanogaster tissue culture cells: multiple initiation sites and replication pause sites. Mol. Cell. Biol. 13**:**4098-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinomiya, T., and S. Ina. 1994. Mapping an initiation region of DNA replication at a single-copy chromosomal locus in Drosophila melanogaster cells by two-dimensional gel methods and PCR-mediated nascent-strand analysis: multiple replication origins in a broad zone. Mol. Cell. Biol. 14**:**7394-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simmons, D. T. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 55**:**75-134. [DOI] [PubMed] [Google Scholar]
- 52.Skoog, L., and B. Nordenskjold. 1971. Effects of hydroxyurea and 1-beta-d-arabinofuranosyl-cytosine on deoxyribonucleotide pools in mouse embryo cells. Eur. J. Biochem. 19**:**81-89. [DOI] [PubMed] [Google Scholar]
- 53.Snapka, R. M., C. G. Shin, P. A. Permana, and J. Strayer. 1991. Aphidicolin-induced topological and recombinational events in simian virus 40. Nucleic Acids Res. 19**:**5065-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Toledo, F., B. Baron, M. A. Fernandez, A. M. Lachages, V. Mayau, G. Buttin, and M. Debatisse. 1998. oriGNAI3: a narrow zone of preferential replication initiation in mammalian cells identified by 2D gel and competitive PCR replicon mapping techniques. Nucleic Acids Res. 26**:**2313-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vassilev, L. T., W. C. Burhans, and M. L. DePamphilis. 1990. Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol. Cell. Biol. 10**:**4685-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vassilev, L. T., and E. M. Johnson. 1989. Mapping initiation sites of DNA replication in vivo using polymerase chain reaction amplification of nascent strand segments. Nucleic Acids Res. 17**:**7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaughn, J. P., P. A. Dijkwel, and J. L. Hamlin. 1990. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell 61**:**1075-1087. [DOI] [PubMed] [Google Scholar]
- 57.Wang, S., P. A. Dijkwel, and J. L. Hamlin. 1998. Lagging-strand, early labelling, and two-dimensional gel assays suggest multiple potential initiation sites in the Chinese hamster dihydrofolate reductase origin. Mol. Cell. Biol. 18**:**39-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Y., J. Zhao, J. Clapper, L. D. Martin, C. Du, E. R. DeVore, K. Harkins, D. L. Dobbs, and R. M. Benbow. 1995. Mimosine differentially inhibits DNA replication and cell cycle progression in somatic cells compared to embryonic cells of Xenopus laevis. Exp. Cell Res. 217**:**84-91. [DOI] [PubMed] [Google Scholar]
- 59.Ward, J. F. 1990. The yield of DNA double-strand breaks produced intracellularly by ionizing radiation: a review. Int. J. Radiat. Biol. 57**:**1141-1150. [DOI] [PubMed] [Google Scholar]