Convergence of TOR-Nitrogen and Snf1-Glucose Signaling Pathways onto Gln3 (original) (raw)
Abstract
Carbon and nitrogen are two basic nutrient sources for cellular organisms. They supply precursors for energy metabolism and metabolic biosynthesis. In the yeast Saccharomyces cerevisiae, distinct sensing and signaling pathways have been described that regulate gene expression in response to the quality of carbon and nitrogen sources, respectively. Gln3 is a GATA-type transcription factor of nitrogen catabolite-repressible (NCR) genes. Previous observations indicate that the quality of nitrogen sources controls the phosphorylation and cytoplasmic retention of Gln3 via the target of rapamycin (TOR) protein. In this study, we show that glucose also regulates Gln3 phosphorylation and subcellular localization, which is mediated by Snf1, the yeast homolog of AMP-dependent protein kinase and a cytoplasmic glucose sensor. Our data show that glucose and nitrogen signaling pathways converge onto Gln3, which may be critical for both nutrient sensing and starvation responses.
Carbon and nitrogen are the two most basic nutrient sources for cellular organisms. They are used to produce energy and synthesize a wide range of biomolecules. Energy metabolism and metabolic biosynthesis are carried out by some 500 individual chemical reactions, which are well organized along the centrally placed glycolysis and tricarboxylic acid (TCA) cycle (1). Individual reactions are well calibrated by a feedback regulation, which fine-tunes the flux of metabolites through a particular pathway by temporarily increasing or decreasing the activity of crucial enzymes. In response to the quality of carbon and nitrogen, cells can also regulate the expression of genes involved in different metabolic pathways, particularly those involved in utilization and transport of the available nutrients. Genetic analysis in budding yeast led to the identification of signaling pathways that detect the quality of nutrients and regulate gene expression. In the presence of high-quality nitrogen compounds such as glutamine, the expression of genes necessary for utilization and transport of poor nitrogen sources such as glutamate is repressed. These genes become derepressed in a poor nitrogen source such as glutamate or during nitrogen starvation. This phenomenon is called nitrogen catabolite repression (NCR) (reviewed in references 24 and 33). Likewise, the presence of glucose represses the expression of genes involved in transport and utilization of other carbon sources. This is commonly known as glucose repression (reviewed in references 8 and 21).
A number of NCR transcription factors have now been identified, including the GATA-type transcription factors Gln3 and Gat1/Nil1 (13, 16, 42). Recent studies have shed light on the regulatory events leading to activation of Gln3. Tor1 and Tor2 are the yeast targets of rapamycin proteins (6, 28) and key players of nutrient-mediated signal transduction (recently reviewed in references 17, 29, and 37-39). Both TOR proteins interact with Gln3 and Gat1/Nil1 (3) and control their phosphorylation (2, 3, 9). Nitrogen starvation or inhibition of TOR by rapamycin causes rapid dephosphorylation and nuclear accumulation of Gln3 (2, 3, 9), as well as expression of a wide variety of NCR genes (3, 7, 23, 41). The Tap42-Sit4 phosphatase complex also appears to be involved in the regulation of Gln3. tap42-11, a gain-of-function mutation (18) was found to confer moderate (3) to complete (2) resistance to rapamycin in Gln3 dephosphorylation. Ure2 is a yeast preprion protein that acts as an inhibitor of Gln3 (14, 44). Depletion of Ure2 causes constitutive nuclear accumulation of Gln3 (2, 3, 27). However, the precise roles of Tap42-Sit4 and Ure2 remain to be further defined.
Glucose repression is mediated by the intracellular glucose sensor Snf1, a yeast homolog of AMP-activated protein kinase (AMPK), and becomes activated when the glucose level is low (recently reviewed in references 8, 21, and 22). Snf1 phosphorylates and negatively regulates Mig1, a transcriptional repressor involved in the utilization of alternative carbon sources (34, 43). A second class of glucose sensors is involved in the glucose induction mechanism. Rgt2 and Snf3 are two plasma membrane proteins with strong sequence similarity to hexose transporters (HXT). However, they act as high- and low-glucose sensors, respectively, to activate transcription of HXT genes in response to extracellular glucose levels (reviewed in reference 35). Snf3 and Rgt2 do not appear to be involved in the general glucose repression mechanism (31, 36).
Several studies indicate that not only nitrogen, but also carbon nutrient, controls the expression of some NCR genes (15, 41). However, it is not clear how carbon signaling cross talks with the NCR pathway. In this study, we found that glucose availability regulates Gln3 phosphorylation and subcellular localization via the Snf1 AMPK pathway. Thus, like mitogenic signal transduction pathways, nutrient signaling pathways can closely interact with each other. Such interplay between two key nutrient sensing and signaling pathways may be important for cells to rapidly adjust cellular metabolic activities as well as growth and proliferation in response to changing nutrient conditions.
MATERIALS AND METHODS
Strains and plasmids.
The following yeast strains were used in this study: AH109 (MATa _trp1-901 leu2-3 11 ura3-52 his3-200 gal4_Δ _gal80_Δ LYS2::Gal1UAS-Gal1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 ura3::MEL1UAS-MEL1TATA-lacZ) (Clontech), SZy145 (MATa hisD1 leu2DO met15DO ura3D ure2D::KanMx) (3), SZy158 (MATa ade2::hisG his3D200 leu2 D0 lysD0 met15D0 trp1D63 ura3D0 GLN3-Myc9-TRP1 (3), SZy215 (MATa hisD1 leu2DO met15DO ura3DO) (3), SZy159 (MATa hisD1 leu2DO met15DO ura3DO gln3D::KanMx) (3), SZy686 (MATa hisD1 leu2DO met15DO ura3DO snf1D::KanMx) (this study), SZy687 (MATa hisD1 leu2DO met15DO ura3DO rgt2D::KanMx (this study), SZy688 (MATa hisD1 leu2DO met15DO ura3DO snf3D::KanMx) (this study), SZy812 (MATa hisD1 leu2DO met15DO ura3DO snf4D::KanMx (this study), SZy813 (MATa hisD1 leu2DO met15DO ura3DO snf8D::KanMx (this study), JC423 (MATa ura3-52 ade2-101oc his4-539am snf1D3 SUC2) (10), JC424 (MATa ura3-52 ade2-101 lys2-801 leu2::his3 snf1D Suc2) (10), and JC426 (MATa ura3-52 his3-52 hisD200 ade2-101 lys2-801 trp1-903 tyr1-501 reg1D::Leu2) (20).
The other SZy strains were derived from SZy215 by the single-step PCR-based deletion of the entire open reading frames as described before (5). The origins of other strains are described as above. To analyze Gln3 phosphorylation and localization, pRS315-GLN3-MYC9 and pRS416-GLN3-MYC9, the chromosomal GLN3-MYC9 gene and its natural promoter were generated by PCR from genomic DNA prepared from the GLN3-MYC9 strain (9) and cloned into pRS315 or pRS416. The resulting plasmids were transformed into yeast, shown to express Gln3-MYC9 at a level comparable to that of the chromosomal GLN3-MYC9 gene as judged by Western blotting. Both full-length GLN3 and SNF1 were amplified by PCR and cloned into pACT2 and pAS2-1, respectively (Clontech). The yeast two-hybrid assays were performed with strain AH109 (Clontech), which was tested for growth on Leu− Trp− Ade− and Leu− Trp− His− + 3-AT (2 mM) plates. Full-length Snf1 was cloned into pADH-Leu to generate pADH-SNF1, which was used for complementation studies.
Indirect IF.
pRS315-GLN3-MYC9 and pRS416-GLN3-MYC9 were transformed into the wild-type or mutant strains (SZy159, SZy215, and SZy686). These plasmids expressed GLN3-MYC9 at a level comparable to that in SZy158 (data not shown). In addition, these plasmids fully complemented the ability of Gln3 to regulate GAP1 in the presence and absence of rapamycin (data not shown). The above strains were grown into early log phase (optical density at 600 nm [OD600] = 0.2) at 30°C in synthetic complete (SC) medium and then switched to SC medium minus nitrogen (without ammonium sulfate and amino acids) or SC medium minus glucose for 30 min at 30°C. The cells were fixed and then stained with monoclonal antibody (MAb) 9E10, and indirect immunofluorescence (IF) was analyzed by fluorescence microscopy as described before (3).
Western blotting analysis, GST-pull down, and phosphatase treatment.
Yeast cells were harvested and lysed with glass beads in disruption buffer (DB) (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 50 mM β-glycerophosphate, 10 mM NaF, 100 nM microcystin LR, 1 mM phenylmethylsulfonyl fluoride [PMSF], and a cocktail of protease inhibitors [Roche]) by vortexing. Crude extracts were cleared by centrifugation twice at 20,800 × g for 20 min. For Western blotting analysis, exponential-phase wild-type and mutant yeast cultures (OD600 = 0.4) in SC medium (2% glucose) at 30°C were switched to SC medium minus glucose. Protein samples (10 μg) were used for gel electrophoresis and detected by the ECL enhanced chemiluminescence system (Amersham Life Sciences) with MAb 9E10 for Gln3-MYC9 or 12CA5 and anti-polyhistidine antibodies (Sigma H1029) for HA (hemagglutinin)-Snf1 and Snf1, respectively. In the glutathione _S_-transferase (GST)-pull down experiment, bacterial GST-Gln3 (3) immobilized to glutathione-agarose beads was incubated with cell lysates of the wild-type and _snf1_Δ mutant cells. After extensive washing with DB, the bound proteins were analyzed by Western blotting with an anti-His antibody (Sigma) that recognizes the polyhistidine motif at the N terminus. For the phosphatase treatment, cell extracts expressing Gln3-MYC9 were prepared as described above with a modified DB (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 10 nM calyculin A, 1 mM PMSF, and a cocktail of protease inhibitors [Roche]). The cell extracts were then incubated with calf intestine alkaline phosphatase (CIP) buffer alone, CIP (20 U [Roche]), or CIP plus Na4P2O7 (10 mM) for 10 min at 30°C.
In vitro kinase assay.
To purify active Snf1 from yeast, log-phase _snf1_Δ cells expressing HA-Snf1 or carrying a control vector were shifted to low-glucose conditions (0.05% glucose) for 1 h, lysed by glass beads in extraction buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 50 mM imidazole, 1 mM PMSF, Roche Complete EDTA-free protease inhibitor tablet, 50 mM NaF, 10 mM β-glycerophosphate). After being cleared by centrifugation as described above, Snf1 was incubated with Ni-agarose beads (Qiagen) for 2 h at 4°C and washed 5 times with extraction buffer containing 50 mM imidazole and twice with kinase buffer (50 mM Tris [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol, 20 μM ATP). To assay for Snf1 kinase activity toward Gln3, affinity-purified Snf1 was incubated in the presence of bacterial recombinant GST-Gln3 (1 μg) (3) and [γ-32P]ATP (5 μCi, 10 mCi/ml, 3,000 Ci/mM [Amersham]) in the kinase buffer (25 μl) for 30 min at 25°C. The phosphorylated GST-Gln3 was analyzed by autoradiography.
Northern blotting analysis.
Exponential wild-type and mutant yeast cultures (OD600 = 0.4) in SC medium (2% glucose) at 30°C were switched to SC medium minus glucose. A switch to SC medium containing a low concentration of glucose (0.05%) gave essentially the same results (data not shown). Aliquots of yeast cultures were withdrawn at different times. Total yeast RNA was prepared by the phenol freezing extraction method (40). Total yeast RNA samples (20 μg) were separated on denaturing agarose gels, transferred onto nylon filters, hybridized to 32P-labeled DNA probes, and detected by phosphorimaging as described before (3).
RESULTS
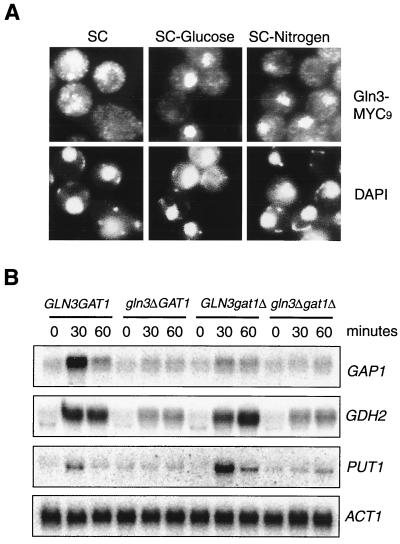
Recent studies indicate that Gln3 localization is controlled by nitrogen availability (2, 3). We found that Gln3 tagged at the C terminus with a nine-MYC epitope (MYC9) primarily localized in the cytoplasm when cells were cultured in SC medium, but accumulated in the nucleus in the absence of nitrogen (Fig. 1A)(2, 3). There was no staining with the nontagged strain (data not shown). For comparison, we also examined the localization of Gln3 in the absence of glucose or both glucose and nitrogen. To our surprise, we also found that glucose starvation led to rapid nuclear accumulation of Gln3 (Fig. 1A). Therefore, although Gln3 is known as a nitrogen sensor in a broad sense, it is also under the control of glucose availability. In essence, Gln3 cytoplasmic localization is a good indicator of the level of preferred nutrients in the environment.
FIG. 1.
Glucose availability regulates subcellular localization of Gln3 and Gln3-dependent NCR genes. (A) Glucose availability regulates subcellular localization of Gln3. Early-log-phase cells (OD600 = 0.2) expressing Gln3-MYC9 in SC medium containing 2% glucose were switched to SC medium, SC medium minus glucose, or SC medium minus nitrogen (SC medium minus ammonium sulfate and amino acids) for 30 min. Gln3-MYC9 localization was then analyzed by indirect IF staining with MAb 9E10. Nuclear DNA was stained with 4",6-diamidino-2-phenylindole. The cell staining images were captured with a SPOT digital camera. (B) Glucose availability regulates Gln3-dependent NCR genes. Exponentially growing wild-type and mutant yeast cells (OD600 = 0.4) in SC medium were switched to SC medium minus glucose. Samples were withdrawn at different times and analyzed for expression of GAP1, GDH2, PUT1, and ACT1 by Northern blot.
Regulation of Gln3 localization by glucose availability suggested a role of glucose in the control of NCR gene expression. To further explore this possibility, we performed Northern blot analysis of several NCR genes. We found that glucose starvation indeed caused an induction of GAP1, GDH2, and PUT1, but not ACT1, the yeast actin gene (Fig. 1B). We next asked whether Gln3 was required for NCR gene expression during glucose limitation. We found that the _gln3_Δ mutation substantially reduced, but did not completely abolish the expression of GAP1, GDH2, and PUT1 (Fig. 1B), which was similar to that during nitrogen starvation (3). We further investigated the role of Gat1 during glucose limitation. We found that Gat1 was also required for the full induction of GAP1. However, it was dispensable for GDH2 induction. In addition, deletion of GAT1 actually enhanced the induction of PUT1. This effect of the _gat1_Δ mutation was dependent on Gln3, since such an effect was blocked by deletion of GLN3 (Fig. 1B). Hence, like Gln3, Gat1 appears to participate in the regulation of NCR genes by glucose. Its role, however, appears to be more complex.
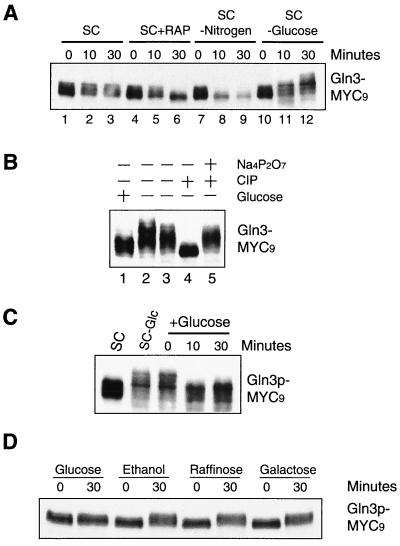
Gln3 is phosphorylated in a TOR-dependent manner in preferred nitrogen, but becomes dephosphorylated under poor nitrogen conditions or during nitrogen starvation (2, 3). We therefore investigated whether glucose starvation affected the level of Gln3 phosphorylation. As shown previously (2, 3), Gln3 appeared as several different phosphorylation forms when grown in SC media, as indicated by its multiple mobility forms on a sodium dodecyl sulfate-polyacrylamide gel (Fig. 2A,lanes 1 through 3). Nitrogen starvation or rapamycin treatment rapidly increased Gln3 gel mobility, eventually to a single, fully dephosphorylated form (Fig. 2A, lanes 6 and 9). In contrast to nitrogen starvation, however, glucose starvation further decreased the gel mobility of Gln3, suggesting that it became additionally phosphorylated (Fig. 2A, lanes 11 and 12). To confirm that this gel mobility change of Gln3 was due to phosphorylation, we treated the lysates of glucose-starved cells with CIP. CIP treatment led to an increase in Gln3 gel mobility (Fig. 2B, lane 4) that comigrated with the fastest form of Gln3 in the unstarved cells (Fig. 2B, lane 1). More importantly, such a gel mobility increase was blocked by Na4P2O7, a potent CIP inhibitor (Fig. 2B, lane 5), indicating that the Gln3 gel mobility increase during glucose starvation was indeed due to phosphorylation. Gln3 phosphorylation by glucose limitation was rapidly reversible once glucose was available (Fig. 2C), suggesting that the phosphorylation or dephosphorylation status of Gln3 can rapidly adapt to different glucose conditions. Gln3 also became phosphorylated when cells were switched from a high glucose concentration (2%) to ethanol (nonfermentable carbon source), raffinose (mimicking a low-glucose condition), and galactose (Fig. 2D) or to a low glucose concentration (0.05%) (data not shown). Thus, Gln3 phosphorylation is controlled by a glucose repression mechanism.
FIG. 2.
Glucose availability regulates phosphorylation of Gln3. (A) Glucose starvation decreases Gln3 gel mobility. Exponentially growing cells in SC medium (OD600 = 0.4) were switched to SC medium, SC medium plus rapamycin (SXC+RAP), SC medium minus nitrogen (SC-Nitrogen), or SC medium minus glucose (SC-glucose) for 0, 10, and 30 min. Gln3 gel mobility was detected by Western blotting. (B) Electrophoretic decrease of Gln3 during glucose limitation is due to phosphorylation. Exponential-phase yeast cells (OD600 = 0.4) in SC medium (lane 1) were switched to SC medium minus glucose for 30 min (lane 2). Lysates of cells under glucose starvation were incubated with a buffer for CIP (lanes 3 and 4) or CIP plus Na4P2O7 (lane 5). Gln3 electrophoretic mobility was detected by Western blotting with MAb 9E10. (C) Phosphorylation of Gln3 during glucose limitation is reversible. Yeast cells cultured in SC medium minus glucose (SC-Glc) were switched to SC medium for 10 and 30 min. Gln3 gel mobility was detected by Western blotting. (D) Glucose limitation causes phosphorylation of Gln3. Exponential cells in SC medium were switched to SC medium plus glucose or SC medium containing one of the following reagents as the sole carbon source for 30 min: ethanol, raffinose, or galactose. Gln3 gel mobility was detected by Western blotting.
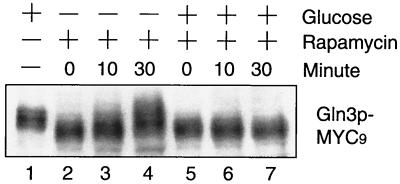
That glucose limitation enhances Gln3 phosphorylation argues against a role of TOR in Gln3 phosphorylation, despite the fact that TOR has been suggested in glucose signaling. To further clarify this, we investigated whether TOR is involved in Gln3 phosphorylation during glucose starvation. In this experiment, we first treated exponentially growing cells in SC medium (2% glucose) with rapamycin for 30 min, which led to inhibition of TOR kinase activity (4) and complete dephosphorylation of Gln3 (Fig. 3, lanes 2 and 5). We then switched the cells to medium containing rapamycin, but with or without 2% glucose. Gln3 remained in the dephosphorylated state under the high-glucose condition (Fig. 3, lanes 5 through 7), but became rapidly phosphorylated in the absence of glucose (Fig. 3, lanes 2 through 4). As expected, the tap42-11 mutation did not have any effect on Gln3 phosphorylation in the presence or absence of glucose (data not shown). Taken together with the data shown in lanes 10 to 12 of Fig. 2A, these results indicate that Gln3 becomes phosphorylated during glucose starvation, regardless of whether rapamycin was present. Therefore, TOR is not required for glucose signaling to regulate Gln3 phosphorylation. Our observation further suggests that the two types of phosphorylations are independent events.
FIG. 3.
TOR is not required for phosphorylation of Gln3 during glucose limitation. Exponentially growing yeast cells in SC medium (OD600 = 0.4) were treated with rapamycin for 30 min to generate completely dephosphorylated Gln3. These cells were then switched to SC medium with or without glucose in the presence of rapamycin for 10 and 30 min. Shown is the gel mobility of Gln3 as detected by Western blotting.
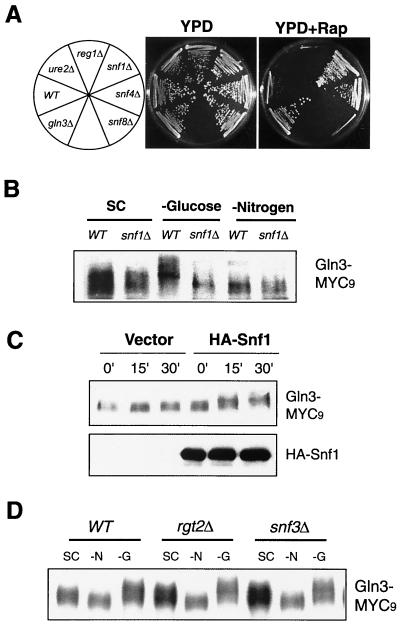
We have recently conducted a chemical genomics screen for genes that, when deleted, confer resistance or sensitivity to rapamycin (12). We have recently completed screening the entire yeast genome. Like GLN3 and URE2, deletion of several genes in the Snf1 pathway conferred significant differential rapamycin sensitivity. Deletion of SNF1, the yeast AMPK and SNF4, an activator of Snf1, rendered rapamycin resistance (Fig. 4A).In contrast, deletion of REG1, a negative regulator of Snf1, caused rapamycin hypersensitivity (Fig. 4A). These results suggest that Snf1 negatively participates in a cellular process that also involves TOR. We therefore examined whether Snf1 was required for increased Gln3 phosphorylation during glucose limitation. We found that the _snf1_Δ mutation did not affect normal phosphorylation of Gln3 in SC medium or dephosphorylation of Gln3 during nitrogen starvation (Fig. 4B) or rapamycin treatment (data not shown). However, the _snf1_Δ mutation completely abolished the ability of Gln3 to become phosphorylated in the absence of glucose (Fig. 4B). These results demonstrate that Snf1 is required for glucose, but not nitrogen, signaling to Gln3. To confirm this, we further examined Gln3 phosphorylation during glucose limitation in the _snf1_Δ strain that carried a plasmid-borne SNF1 or a vector control. The plasmid-borne SNF1, but not the vector control, was able to restore Gln3 phosphorylation upon glucose limitation (Fig. 4C). In contrast, deletion of RGT2 and SNF3 did not affect Gln3 phosphorylation under different nutrient conditions (Fig. 4D). Therefore, the cytoplasmic, but not the cell surface, glucose sensors regulate Gln3 phosphorylation or dephosphorylation, further supporting the hypothesis that Gln3 regulation is mediated by the glucose repression mechanism.
FIG. 4.
The role of glucose sensors in the in vivo phosphorylation of Gln3. (A) Mutations in the Snf1 pathway confer differential rapamycin sensitivity. The wild-type (WT) and mutant yeast strains were streaked onto YPD and YPD plus rapamycin (YPD+RAP) (25 nM) plates and incubated at 30°C for 2 (YPD) and 4 (YPD plus rapamycin) days, respectively. (B) Snf1 is required for Gln3 phosphorylation during glucose starvation, but not during nitrogen starvation. Exponentially growing wild-type and _snf1_Δ cells expressing Gln3-MYC9 in SC medium (OD600 = 0.4) were switched to SC medium or SC medium minus glucose or nitrogen for 30 min. (C) Recombinant Snf1 restores the deficiency of Gln3 phosphorylation during glucose limitation in the _snf1_Δ strain. The _snf1_Δ strains carrying a plasmid expressing recombinant Snf1 or a vector control were examined for Gln3-MYC9 phosphorylation at different times after glucose limitation. (Upper panel) Western blot for Gln3-MYC9. (Lower panel) Western blot for Snf1. (D) Rgt2 and Snf3 are not required for Gln3 phosphorylation during glucose limitation. Exponentially growing wild-type and mutant cells expressing Gln3-MYC9 in SC medium (OD600 = 0.4) were switched to SC medium or SC medium minus nitrogen or glucose for 30 min. Gln3 phosphorylation was examined by Western blotting.
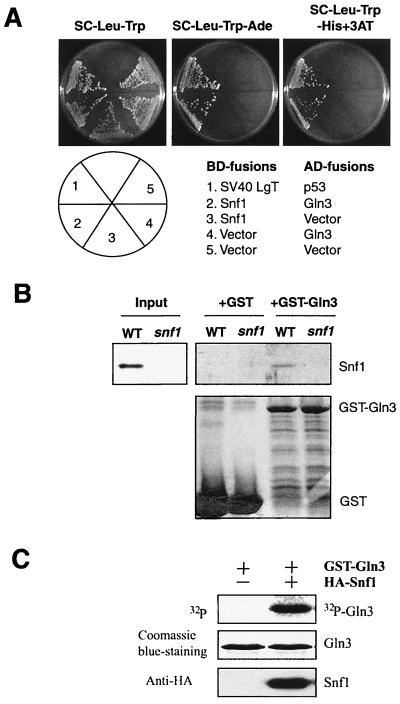
Snf1 is known to interact with several transcriptional regulators, including Mig1 (34). Hence, we investigated a possible interaction between Snf1 and Gln3 by using the yeast two-hybrid system. We found that Gal4 DNA-binding domain (BD)-Snf1 specifically interacted with Gal4 activation domain (AD)-Gln3, but not with Gal4 AD alone (Fig. 5A).Conversely, Gal4(AD)-Gln3 interacted with Gal4(BD)-Snf1, but not Gal4(BD) alone (Fig. 5A). To further confirm the interaction between Snf1 and Gln3, we tested the ability of Snf1 to bind to Gln3 in an in vitro GST-pull down assay (Fig. 5B). In this experiment, we incubated lysates from the wild-type and _snf1_Δ strains with bacterially produced GST-Gln3 or GST. We found that Snf1 specifically bound to GST-Gln3, but not to GST alone (Fig. 5B). These results demonstrate that Snf1 and Gln3 are capable of binding to each other. In addition, Snf1 significantly phosphorylated a bacterial recombinant Gln3 in vitro (Fig. 5C), suggesting that Snf1 may be directly responsible for Gln3 phosphorylation in vivo .
FIG. 5.
Snf1 interacts with Gln3. (A) Snf1 interacts with Gln3 in a yeast two-hybrid assay. Gal4 DNA binding domain (BD)-Snf1 specifically interacts with Gal4 activation domain (AD)-Gln3, but not with AD alone. The two-hybrid reporter strain (AH109) carrying different BD and AD fusion plasmids was assayed for growth on an adenine dropout plate or a histidine dropout plate containing 3-AT (2 mM). LgT, large T antigen. (B) Snf1 binds to GST-Gln3. Extracts of wild-type (WT) and _snf1_Δ yeast strains were assayed for their ability to bind to immobilized, bacterially produced GST or GST-Gln3. The bound materials were detected by Western blotting with an anti-polyhistidine antibody that recognizes Snf1. (C) Snf1 phosphorylates Gln3 in vitro. HA-Snf1 was immunoprecipitated and assayed for its ability to phosphorylate bacterial recombinant GST-Gln3 in the presence of [γ-32P]ATP.
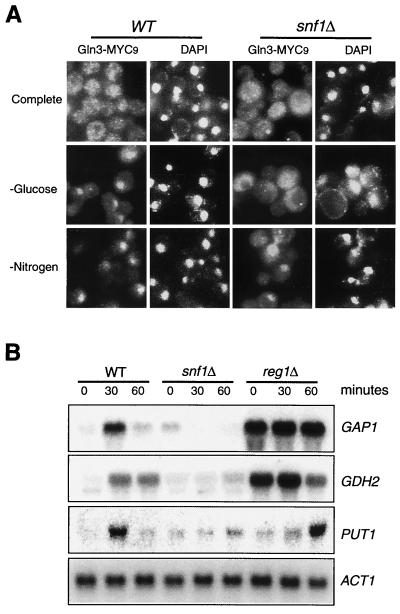
To further confirm the role of Snf1 in glucose signaling to Gln3, we performed indirect IF staining of Gln3-MYC9 in the wild-type and _snf1_Δ cells. Gln3 primarily localized in the cytoplasm in both cell types in SC medium and accumulated in the nucleus in the absence of nitrogen or glucose in the wild-type strain (Fig. 6A).However, Gln3 remained in the cytoplasm even in the absence of glucose in the _snf1_Δ strain (Fig. 6A). In contrast, deletion of SNF1 did not affect the nuclear accumulation of Gln3 during nitrogen starvation. Therefore, Snf1 is required for nuclear accumulation during glucose, but not nitrogen starvation. We next investigated whether Snf1 was involved in the regulation of NCR genes by Northern blot analysis of GAP1, GDH2, and PUT1. As seen earlier (Fig. 1B), glucose limitation caused an induction of these genes in the wild-type yeast (Fig. 6B). This induction, however, was virtually absent in the _snf1_Δ mutant strain (Fig. 6B). Reg1 is a negative regulator of Snf1, and deletion of REG1 causes constitutive activation of Snf1 and derepression of many Snf1-regulated genes (32). We found that the _reg1_Δ mutation caused constitutive expression of GAP1 and GDH2 even in the presence of 2% glucose, further supporting the notion that Snf1 is a critical regulator of NCR-sensitive genes via Gln3 and Gat1 (Fig. 6B). Interestingly, the _reg1_Δ mutation only slightly delayed PUT1 induction, suggesting that additional factors in the Snf1 pathway are required for Snf1-regulated PUT1 expression.
FIG. 6.
Snf1 is required for Gln3 nuclear accumulation and induction of NCR genes during glucose limitation. (A) Early-log-phase (OD600 = 0.2) wild-type (WT) or _snf1_Δ yeast cells expressing Gln3-MYC9 in SC medium were switched to SC medium, SC medium minus glucose (−glucose), or SC medium minus nitrogen (−Nitrogen) for 30 min. Gln3 localization was detected by indirect IF with the 9E10 MAb. DAPI, 4",6-diamidino-2-phenylindole. (B) Snf1 is required for expression of NCR genes during glucose limitation. Exponential wild-type or mutant yeast cells (OD600 = 0.4) in SC medium were shifted to SC medium minus glucose. Aliquots of culture were withdrawn at different times (as shown) and analyzed for expression of selected NCR genes by Northern blotting.
DISCUSSION
There are two major glucose sensing and signaling mechanisms in yeast: the one mediated by the cell surface glucose sensors Rgt2 and Snf3, which belong to the glucose transporter family, also called the glucose induction mechanism (recently reviewed in reference 35); and the one mediated by the cytoplasmic glucose sensor Snf1, also called the glucose repression mechanism (recently reviewed in references 8, 21, and 22). Similarly, nitrogen sensing and signaling also involve two distinct sensors. The cell surface amino acid sensor Ssy1 is related to amino acid permeases and acts analogously to Rgt2 and Snf3 (19, 25, 26). On the other hand, Tor1 and Tor2 are the cytoplasmic nitrogen sensors. The role of Gln3 in nitrogen sensing and signaling is dependent on TOR proteins, but not Ssy1 (P. G. Bertram and X. F. S. Zheng, unpublished observation). In this study, we show that Gln3 is regulated by Snf1 (this study), but not Snf3 and Rgt2 (Fig. 4D), the two cell surface glucose sensors (31, 36). Therefore, regulation of Gln3 by both nitrogen and glucose utilizes very similar strategies (for a model, see Fig. 7).In addition to regulating Gln3 nuclear accumulation, phosphorylation may also modulate the ability of Gln3 to transactivate NCR genes, since there is a quantitative difference in GAP1 expression levels under different conditions. The GAP1 induction level was much higher during nitrogen starvation (∼100-fold) (3) than during glucose starvation (∼11.5-fold) (Fig. 1B).
FIG. 7.
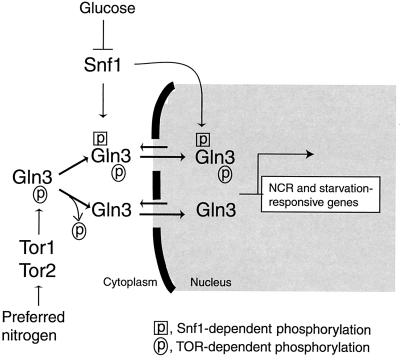
Model for regulation of Gln3 by both nitrogen and glucose. TOR and Snf1 independently mediate nitrogen and glucose signaling pathways, respectively, to regulate Gln3 nuclear localization and specificity for its target genes. Our results suggest that phosphorylation controls the subcellular localization of Gln3. TOR-dependent phosphorylation appears to keep Gln3 in the cytoplasm, while Snf1-dependent phosphorylation leads to nuclear accumulation of Gln3 by promoting nuclear import and/or inhibiting nuclear export.
Both TOR and Snf1 are cytoplasmic kinases that control the phosphorylation of Gln3 in response to different nutrients. However, phosphorylation by these two kinases has opposite effects. TOR-dependent phosphorylation is consistent with its role in keeping Gln3 in the cytoplasm (2, 3). In contrast, Snf1-dependent phosphorylation correlates well with increased nuclear accumulation (this study). The opposing effects of the two different phosphorylations also suggest that TOR-dependent and Snf1-dependent phosphorylation events occur at distinct sites. Two additional observations further support this hypothesis. Inhibition of TOR by rapamycin treatment did not affect glucose limitation-dependent phosphorylation. In contrast, deletion of Snf1 had little effect on the phosphorylation or dephosphorylation events under different nitrogen conditions. Snf1-dependent phosphorylation may promote Gln3 nuclear import and/or inhibit Gln3 export (Fig. 7). The TOR- and Snf1-dependent phosphorylation sites need to be identified in order to clearly define their roles in the regulation of Gln3 localization and functions.
Why does the cell need to up-regulate NCR-sensitive genes during glucose limitation? Both carbon- and nitrogen-containing compounds can be ultimately utilized for energy metabolism as well as metabolic biosyntheses. For example, while glycolysis and the TCA cycle use glucose to generate NADH leading to ATP synthesis, many of the intermediate products are also precursors for biosynthesis of amino acids, nucleotides, and lipids (1). These genes are induced by rapamycin or nitrogen starvation (23). Conversely, amino acids can be used as precursors for biosynthesis of other amino acids, nucleotides, and lipids as well as for energy metabolism. For example, GDH2 encodes glutamate dehydrogenase that catalyzes the conversion of glutamate and NAD to α-ketoglutarate, NH4+, and NADH. While NADH is directly used to generate ATP, α-ketoglutarate can be directly fed into the TCA cycle. During glucose limitation, the cell may need to mobilize other available nutrients as alternative carbon sources, including amino acids. In addition to energy metabolism and metabolic biosyntheses, nitrogen and glucose starvation results in many other common cell physiological changes, such as exit from the cell cycle, accumulation of glycogen, reduced protein synthesis, and increased autophagy. For example, limitation of both nitrogen and glucose leads to increased expression of several genes required for autophagy, including APG and vacuolar protease genes (11, 41), as well as those involved in ribosomal biogenesis (30). Therefore, in addition to regulation of NCR genes, Gln3 appears to be important for general starvation responses as well.
Acknowledgments
We thank D. Dean for use of his fluorescence microscope and M. Johnston for strains and plasmids.
This work was supported by grants from the NIH (R01CA77668) and the Howard Hughes Medical Institute (X.F.Z.).
REFERENCES
- 1.Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. Watson. 1994. Molecular biology of the cell. Garland Publishing, Inc., New York, N.Y.
- 2.Beck, T., and M. N. Hall. 1999. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcriptional factors. Nature 402**:**689-692. [DOI] [PubMed] [Google Scholar]
- 3.Bertram, P. G., J. Choi, J. Carvalho, W. D. Ai, C. B. Zeng, T. F. Chan, and X. F. S. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p and phosphatases. J. Biol. Chem. 275**:**35727-35733. [DOI] [PubMed] [Google Scholar]
- 4.Bertram, P. G., C. Zeng, J. Thorson, A. S. Shaw, and X. F. Zheng. 1998. The 14-3-3 proteins positively regulate rapamycin-sensitive signaling. Curr. Biol. 8**:**1259-1267. [DOI] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14**:**115-132. [DOI] [PubMed] [Google Scholar]
- 6.Cafferkey, R., M. M. McLaughlin, P. R. Young, R. K. Johnson, and G. P. Livi. 1994. Yeast TOR (DRR) proteins: amino-acid sequence alignment and identification of structural motifs. Gene 141**:**133-136. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas, M., N. Cutler, M. Lorenz, C. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13**:**3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2**:**202-207. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho, J., P. G. Bertram, S. R. Wente, and X. F. S. Zheng. 2001. Phosphorylation regulates the interaction between Gln3p and the nuclear import factor Srp1p. J. Biol. Chem. 276**:**25359-25365. [DOI] [PubMed] [Google Scholar]
- 10.Celenza, J. L., and M. Carlson. 1986. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233**:**1175-1180. [DOI] [PubMed] [Google Scholar]
- 11.Chan, T.-F., P. G. Bertram, W. Ai, and X. F. S. Zheng. 2001. Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J. Biol. Chem. 276**:**6463-6467. [DOI] [PubMed] [Google Scholar]
- 12.Chan, T. F., J. Carvalho, L. Riles, and X. F. S. Zheng. 2000. A chemical genomics approach toward understanding the global functions of TOR. Proc. Natl. Acad. Sci. USA 97**:**13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffman, J. A., R. Rai, T. Cunningham, V. Svetlov, and T. G. Cooper. 1996. Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen-catabolic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 16**:**847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coschigano, P. W., and B. Magasanik. 1991. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione _S_-transferases. Mol. Cell. Biol. 11**:**822-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coschigano, P. W., S. M. Miller, and B. Magasanik. 1991. Physiological and genetic analysis of the carbon regulation of the NAD-dependent glutamate dehydrogenase of Saccharomyces cerevisiae. Mol. Cell. Biol. 11**:**4455-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courchesne, W. E., and B. Magasanik. 1988. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J. Bacteriol. 170**:**708-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis, P. B., S. Fumagalli, and G. Thomas. 1999. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 9**:**49-54. [DOI] [PubMed] [Google Scholar]
- 18.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10**:**1904-1916. [DOI] [PubMed] [Google Scholar]
- 19.Didion, T., B. Regenberg, M. U. Jorgensen, M. C. Kielland-Brandt, and H. A. Andersen. 1998. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27**:**643-650. [DOI] [PubMed] [Google Scholar]
- 20.Flick, J. S., and M. Johnston. 1990. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 10**:**4757-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62**:**334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67**:**821-855. [DOI] [PubMed] [Google Scholar]
- 23.Hardwick, J. S., F. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96**:**14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofman-Bang, J. 1999. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol. 12**:**35-73. [DOI] [PubMed] [Google Scholar]
- 25.Iraqui, I., S. Vissers, F. Bernard, J. O. de Craene, E. Boles, A. Urrestarazu, and B. André. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19**:**989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19**:**5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulkarni, A. A., A. T. Abul-Hamd, R. Rai, H. El Berry, and T. G. Cooper. 2001. Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J. Biol. Chem. 276**:**32136-32144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunz, J., R. Henriquez, U. Schneider, M. Deuter-Reinhard, N. R. Movva, and M. N. Hall. 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73**:**585-596. [DOI] [PubMed] [Google Scholar]
- 29.Kuruvilla, F., and S. L. Schreiber. 1999. The PIK-related kinases intercept conventional signaling pathways. Chem. Biol. 6**:**R129-R136. [DOI] [PubMed] [Google Scholar]
- 30.Kuruvilla, F. G., A. F. Shamji, and S. L. Schreiber. 2001. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc. Natl. Acad. Sci. USA 98**:**7283-7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, H., and R. F. Gaber. 1996. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol. Biol. Cell 7**:**1953-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludin, K., R. Jiang, and M. Carlson. 1998. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95**:**6245-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magasanik, B. 1992. The molecular and cellular biology of yeast Saccharomyces, vol. 2., p. 283-318. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Ostling, J., and H. Ronne. 1998. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur. J. Biochem. 252**:**162-168. [DOI] [PubMed] [Google Scholar]
- 35.Özcan, S., and M. Johnston. 1999. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63**:**554-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Özcan, S., and M. Johnston. 1996. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol. Cell. Biol. 16**:**5536-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raught, B., A.-C. Gingras, and N. Sonenberg. 2001. The target of rapamycin (TOR) protein. Proc. Natl. Acad. Sci. USA 98**:**7037-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohde, J., J. Heitman, and M. Cardenas. 2001. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276**:**7027-7036. [DOI] [PubMed] [Google Scholar]
- 39.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103**:**253-262. [DOI] [PubMed] [Google Scholar]
- 40.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18**:**3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shamji, A., F. Kuruvilla, and S. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10**:**1574-1581. [DOI] [PubMed] [Google Scholar]
- 42.Stanbrough, M., D. W. Rowen, and B. Magasanik. 1995. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc. Natl. Acad. Sci. USA 92**:**9450-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18**:**6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickner, R. B. 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264**:**566-569. [DOI] [PubMed] [Google Scholar]