Sequential Roles for Mash1 and Ngn2 in the Generation of Dorsal Spinal Cord Interneurons (original) (raw)
. Author manuscript; available in PMC: 2006 Jan 25.
Published in final edited form as: Development. 2005 May 18;132(12):2709–2719. doi: 10.1242/dev.01859
Summary
The dorsal spinal cord contains a diverse array of neurons that connect sensory input from the periphery to spinal cord motor neurons and brain. During development, six dorsal neuronal populations (dI1-dI6) have been defined by expression of homeodomain factors and position in the dorsoventral axis. The bHLH transcription factors Mash1 and Ngn2 have distinct roles in specification of these neurons. Mash1 is necessary and sufficient for generation of most dI3 and all dI5 neurons. Unexpectedly, dI4 neurons are derived from cells expressing low levels or no Mash1, and this population increases in the Mash1 mutant. Ngn2 is not required for any specific neuronal cell-type but appears to modulate the composition of neurons that form. In the absence of Ngn2 there is an increase in dI3 and dI5 neurons, which is opposite to the activity of Mash1. Mash1 is epistatic to Ngn2, and unlike the relationship between other neural bHLH factors, cross-repression of expression is not detected. Thus, bHLH factors, particularly Mash1 and related family members Math1 and Ngn1, provide a code for generating neuronal diversity in the dorsal spinal cord with Ngn2 serving to modulate the number of neurons in each population formed.
Keywords: spinal cord development, dorsal horn, bHLH, neuronal specification
Introduction
The circuitry of the nervous system depends on complex connections of diverse neuronal cell-types. Thus, defining the mechanisms that regulate neuronal diversity is fundamental for understanding how the nervous system functions. Specification of distinct neuronal sub-types begins as early as the neural tube stage when differences in progenitor cells in the ventricular zone can be detected by differences in gene expression patterns (Briscoe et al., 2000; Caspary and Anderson, 2003; Helms and Johnson, 2003; Jessell, 2000). As the neural tube develops, proliferating cells exit the cell cycle, and the newly born neurons migrate laterally from the ventricular zone and begin to exhibit their specific neuronal identities. There is a complex interplay between extrinsic and intrinsic factors that mediate the precise timing of cell-cycle exit, cell migration, and initiation of cellular properties specific to newborn neurons. It has been demonstrated in the spinal neural tube that the acquisition of identity involves a gradient of extracellular signals that set up a combinational code of transcription factors (Briscoe et al., 2000; Jessell, 2000). In this study, the role of two transcription factors, Mash1 and Ngn2, in the specification of neurons in the dorsal spinal cord is examined.
Neurons in the mouse dorsal spinal cord are largely born between embryonic day 10 to 14 (E10-E14). These neurons provide the network connecting and modulating sensory input from the periphery to the spinal cord and brain. Currently there are 6 early (dI1-dI6) and 2 late (dILA and dILB) dorsal neuron types defined by birthdate, position in the dorsoventral axis, and distinct homeodomain (HD) transcription factor markers (Caspary and Anderson, 2003; Helms and Johnson, 2003). The most dorsal of these, dI1-dI3, are dependent on roof plate signals, whereas dI4-dI6 and dILA/B form independently of these signals and are distinguished by the presence of the HD factor Lbx1 (Gross et al., 2002; Lee et al., 2000; Müller et al., 2002). The combinations of HD factors in early stages of neuronal differentiation have been critical for defining these distinct dorsal interneuron populations. Much less is known about how these neurons contribute to the overall circuitry of the spinal cord (Cheng et al., 2004; Lanuza et al., 2004). Mice mutant for specific HD factors, such as Lbx1 and Tlx1/3, have been used to link the fate of some of these populations to a GABAergic or glutamatergic specific neuronal type at later stages (Cheng et al., 2004; Gross et al., 2002; Müller et al., 2002).
Progenitor populations located in the ventricular zone of the neural tube can also be classified by transcription factor patterns. In ventral regions, the primary determinants of neuronal specification are combinations of HD transcription factors (Briscoe et al., 2000; Ericson et al., 1997; Pierani et al., 2001). In contrast, dorsal progenitor domains have largely been defined by bHLH transcription factors (Gowan et al., 2001). A role for bHLH factors in specification of dorsal neurons was first suggested by expression domains in the dorsoventral axis of Math1 (Atoh1, MGI), Ngn1 (Neurog1, MGI), Mash1 (Ascl1, MGI) (Gowan et al., 2001; Helms and Johnson, 1998; Lo et al., 1991; Ma et al., 1997; Sommer et al., 1996), and more recently, Olig3 (Müller et al., 2005). Each of these factors represents a distinct sub-class of neuronal bHLH (Bertrand et al., 2002). Within the dorsal neural tube, Math1 is in ventricular zone cells adjacent to the roof plate (Helms and Johnson, 1998), Ngn1 is just ventral to the Math1 domain (Gowan et al., 2001; Lee et al., 1998), and Mash1 is ventral to the Ngn1 domain extending almost to the sulcus limitans (Gowan et al., 2001; Guillemot and Joyner, 1993; Lo et al., 1991). There is little if any overlap in Math1, Ngn1 and Mash1 in individual cells (Gowan et al., 2001). The bHLH factors have been shown to have at least two functions during neural development; to induce neuronal differentiation and to specify neuronal sub-types (Cau et al., 2002; Farah et al., 2000; Nakada et al., 2004; Parras et al., 2002; Perez et al., 1999). Furthermore, cross-inhibitory regulation of expression between Math1, Ngn1, and Mash1 has been suggested as a mechanism for refining distinct progenitor domains (Gowan et al., 2001; Parras et al., 2002). In contrast, Ngn2, a bHLH factor most closely related to Ngn1, partially overlaps with Ngn1 and Mash1 (Fig. 1 and unpublished data AWH and JEJ), while Olig3 overlaps with Math1, Ngn1 and dorsal Mash1 (Müller et al., 2005), suggesting different rules and additional complexities for the functions of Ngn2 and Olig3.
Figure 1. Co-localization of Mash1 and Ngn2 in the ventricular zone of the dorsal neural tube.
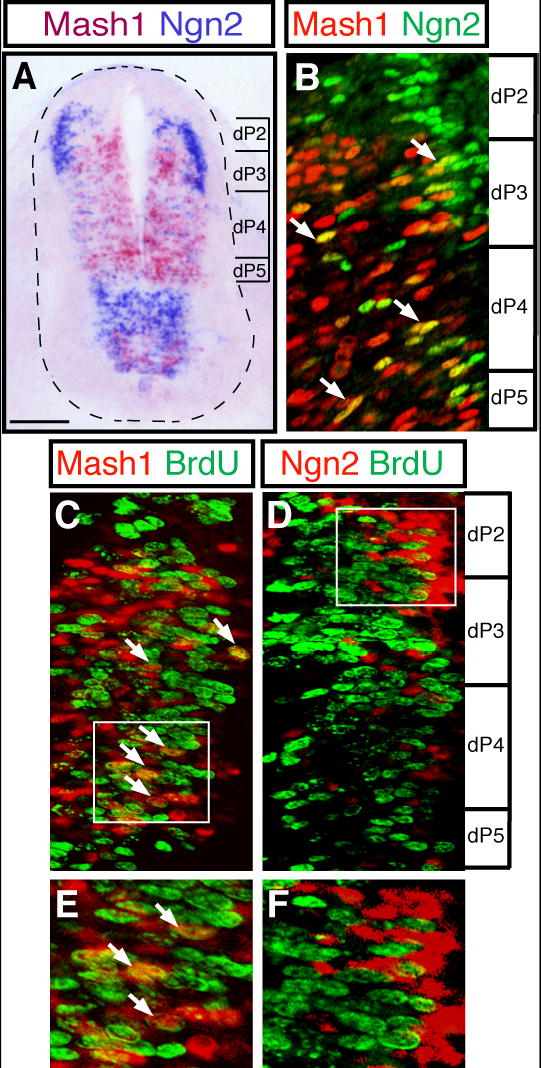
E10.5 embryo transverse sections were processed for mRNA in situ hybridization (A) or immunofluorescence (B-F). (A) Pseudocolored overlay of adjacent sections hybridized with anti-sense Mash1 (pink) or Ngn2 (purple) probes to illustrate expression in the dorsoventral axis and possible overlap. (B) Double-label immunofluorescence shows Mash1 (red) and Ngn2 (green) in ventricular zone cells in the dorsal neural tube (region shown by bracket in (A), ventricular zone is at the left in each panel). Arrows indicate Mash1 and Ngn2 co-expressing cells (yellow). (C-F) Co-labeling with BrdU incorporation was performed to detect cells in S-phase. (C,E) Mash1(red) cells incorporate BrdU (green) (detected as yellow, arrows). (D,F) Ngn2 (red) cells rarely score positive for BrdU incorporation (green). (E,F) are higher magnification images of the regions boxed in (C,D). Scale bar 90 μm in (A) 25 μm in (B-D) and 15 μm in (E,F).
While inroads have been made into identifying important players in the specification of spinal cord neurons, the underlying logic behind a combinatorial code for specification of these neurons is far from complete. Indeed, in the dorsal neural tube a progenitor/neuron relationship has only been defined for Math1+ progenitors with dI1 neurons, Ngn1+ progenitors with dI2 neurons (Gowan et al., 2001), and Olig3+ progenitors with dI1-dI3 (Müller et al., 2005). The generation of dI3, dI4, and dI5 from Mash1+ progenitors has only been inferred by position in the dorsoventral axis (Gross et al., 2002; Müller et al., 2002; Qian et al., 2002). In this study, we show that cells with the highest levels of Mash1 appear to give rise to dI3 and dI5, but not dI4 neurons. Consistent with this lineage relationship, dI5 and most dI3 are lost in the Mash1 mutant, while the dI4 neuronal population appears to increase. In contrast, Ngn2 overlaps with Mash1 and Ngn1 but has later temporal characteristics. Loss of Ngn2 alone, or in combination with Mash1, reveals a function for Ngn2 downstream of Mash1 in modulating the number of Mash1-dependent neurons that form. Mouse mutants where the balance and temporal characteristics of Mash1 and Ngn2 levels have been altered were used to refine a model for how these factors function in generation of the correct composition of neurons in the dorsal spinal cord.
Materials and methods
Bacterial Artificial Chromosome Construct
The Roswell Park Bacterial Artificial Chromosome (BAC) library was screened to identify clones that contained the Mash1 protein coding region. RPCI-428P21 was chosen for further study. It contains a genomic insert of 305 kb with 98 kb 5’ and 206 kb 3’ of the Mash1 translation start codon (Supplemental Fig. 1S). The Mash1 coding region was precisely replaced by EGFP (Clontech) and CRE separated by an IRES using homologous recombination (Yang et al., 1997). The resultant modified BAC named M1-GIC was verified by Southern blot.
Supplemental Figure 1. Generation of the M1-GIC BAC and comparison of Mash1 and GFP.
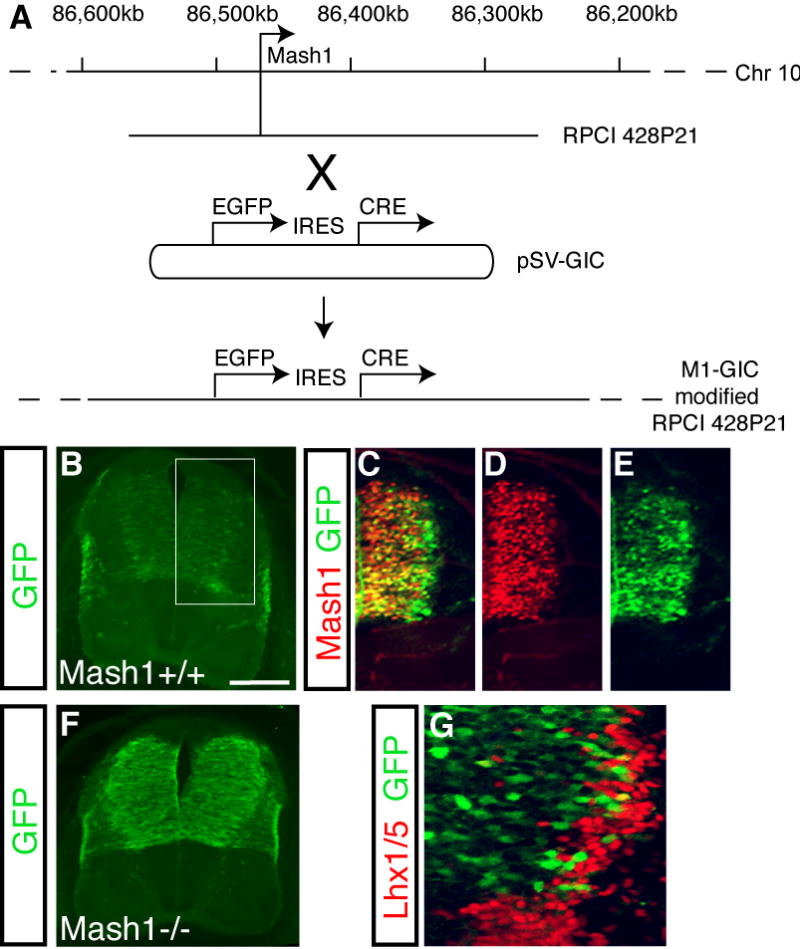
(A) BAC RPCI 428P21 from chromosome 10 containing the Mash1 locus was modified using homologous recombination (Yang et al., 1997). This modification utilized the shuttle vector pSV-GIC that contains a GFP-IRES-CRE cassette flanked by 1kb arms homologous to sequence in the Mash1 locus. This recombination precisely replaces the coding sequence of Mash1 with the GFP-IRES-CRE coding cassette. The resulting modified BAC is called M1-GIC. Transgenic mouse lines were generated with M1-GIC. The GFP reporter is low but it reflects the Mash1 pattern throughout multiple stages and multiple regions including the caudal neural tube (shown here), the telencephalon and diencephalon, sympathetic and enteric nervous systems (data not shown). (B-E) Immunofluorescence on transverse sections of neural tubes from E11.5 M1-GIC embryos with antibodies to GFP (green) and Mash1 (red) show GFP reflects the pattern of Mash1 with boundaries matching precisely in the dorsoventral axis. Persistence of GFP in lateral regions is likely due to the stability of GFP relative to Mash1. (B,F) Immunofluorescence using antibodies to detect GFP demonstrate expression of GFP in the Mash1 null background (F) is dramatically increased over wild type (B). These two images were taken at the identical GAIN to illustrate the difference in levels. This observation is consistent with the negative autoregulation of the Mash1 locus previously reported (Horton et al., 1999; Meredith and Johnson, 2000). (G) Because the persistence of GFP is found continuously throughout the lateral regions adjacent to dI3-dI5, but Mash1 is not required for dI4, we used Lhx1/5, a marker of dI4/6 to examine co-labeling with the GFP. Immunofluorescence using antibodies to detect GFP (green) and Lhx1/5 (red) shows very little overlap of GFP from the M1-GIC line with the dI4/6 population consistent with conclusions drawn from Fig. 4. Scale bar is 175 μm in (B,F) 100 μm in (C-E) and 50 μm in (G).
Transgenic Mouse Generation and Mouse Mutant Strains
Transgenic mice were generated by standard procedures (Hogan et al., 1986) using fertilized eggs from B6D2F1 (C57B1/6 x DBA) crosses. M1-GIC BAC was prepared using a modified Qiagen Midi Prep procedure as directed by manufacturer. The M1-GIC BAC was then injected into the pronucleus of fertilized mouse eggs at 0.5–1ng/μl in 10mM Tris pH7.5, 0.1mM EDTA, 100mM NaCl. Transgenic animals were identified by PCR analysis using tail or yolk sac DNA with primers to CRE: 5’ GGACATGTTCAGGGATCGCCAGGCG 3’ and 5’ GCATAACCAGTGAAACAGCATTGCTG 3’.
The mouse mutant strains used in this study have been previously published: Mash1 (Guillemot et al., 1993), Ngn2 (Fode et al., 1998), Mas_h1_KINgn2 and Ngn2 KIMash1 (Parras et al., 2002) and R26R-YFP (Srinivas et al., 2001). Embryos were staged based on assumed copulation at E0, halfway through the dark cycle. All procedures on animals follow NIH Guidelines and were approved by the UT Southwestern Institutional Animal Care and Use Committee.
Immunofluorescence and mRNA in situ hybridization
Staged embryos were dissected in cold 0.1M Sodium Phosphate buffer pH 7.4, fixed in 4% formaldehyde in 0.1M Sodium Phosphate buffer pH 7.4 for 2 hours at 4°C, sunk in 30% sucrose in 0.1M Sodium Phosphate buffer pH 7.4 overnight at 4°C, embedded in OCT, cryosectioned at 30 μm, and processed for immunofluorescence or mRNA in situ hybridization. All sections shown are from the level of the upper limbs.
For immunofluorescence, slides were incubated in the appropriate dilution of primary antibody in PBS/1% goat serum/0.1% Triton-X-100, followed by either goat anti-rabbit, mouse, or guinea pig IgG conjugated with Alexa Fluor 488, 594, or 647 (Molecular Probes). Primary antibodies used for this study include: rabbit anti-Mash1 (Horton et al., 1999), mouse anti-Ngn2 (Lo et al., 2002), mouse anti-Lhx1/5 (4F2), mouse anti-Islet1/2 (39.4D5), mouse anti-Lmx, (Developmental Studies Hybridoma Bank), rabbit anti-GFP (Molecular Probes), rabbit and guinea pig anti-Brn3a (Fedtsova and Turner, 1997), rabbit, rat, and guinea pig anti-Lbx1 (Gross et al., 2002; Müller et al., 2002), guinea pig anti-Lmx1b (Müller et al., 2002), rabbit anti-Islet1/2 (Tsuchida et al., 1994), rabbit anti-Pax2 (Zymed), and mouse anti-BrdU (Becton Dickinson). For BrdU labeling, pregnant mothers were injected with 200 μg/g of body weight one hour before sacrifice. For double labeling experiments using the anti-BrdU antibody, either Mash1 or Ngn2 antibody staining was carried out in full, followed by treatment with 2N HCl for 20 minutes, 0.1M sodium borate pH 8.5 for 20 minutes and incubation with mouse anti-BrdU antibody as described above. Cell death was detected using TUNEL analysis (Roche) on E10.5 and E11.5 sections. Fluorescence imaging was carried out on a Bio-Rad MRC 1024 confocal microscope. For each experiment multiple sections from at least 3 different embryos were analyzed and counted.
mRNA in situ hybridization was performed essentially as described using a combined protocol from (Birren et al., 1993; Ma et al., 1998). A detailed protocol is available upon request. Mash1, Ngn1, and Ngn2 antisense probes were made from plasmids containing the coding region of each gene (Gowan et al., 2001).
In ovo chick electroporation
Fertilized White Leghorn eggs were obtained from the Texas A&M Poultry Department (College Station, TX) and incubated at 37°C. Solutions of supercoiled plasmid DNA (2 μg/ml) in PBS/0.02% Trypan Blue were injected into the lumen of the closed neural tube at stage HH13-14, and embryos electroporated as previously described (Funahashi et al., 1999; Muramatsu et al., 1997; Nakada et al., 2004; Suemori et al., 1990). A GFP expression vector (CMV-eGFP; Clontech) was co-injected as a control to monitor efficiency and extent of electroporation. Embryos were harvested 24 hours later at HH23-24, fixed in 4% formaldehyde for 1 hour, and processed as above for cryosectioning and immunofluorescence. For each experiment multiple sections from at least three electroporated embryos were analyzed and counted. All sections shown were taken between the upper and lower limb regions.
All electroporations utilized the expression vector pMiwIII, which drives expression through a chick β-actin promoter (Matsunaga et al., 2001; Suemori et al., 1990). PCR fragments containing the coding regions of rat Mash1 and mouse Ngn2 were cloned into NcoI and XbaI sites of a modified pMiwIII expression vector. Protein expression was verified by immunofluorescence with antibodies to Mash1 and Ngn2.
Results
Mash1 and Ngn2 partially overlap in the ventricular zone adjacent to dI3, dI4, and dI5 neuronal populations
Mash1 and Ngn2 are present in the ventricular zone of the neural tube at E10.5. Mash1 is in cells adjacent to where dI3, dI4 and dI5 neurons form and in a smaller ventral domain (Gowan et al., 2001; Gross et al., 2002; Müller et al., 2002). In contrast, Ngn2 is present in the ventral and dorsal neural tube, and in the dorsal domain it spans the region where dI2-dI5 are formed (Fig. 1A). As a first step in examining the roles of Mash1 and Ngn2 in dorsal neuron specification, we examined their co-localization in detail at E10.5 using previously characterized rabbit polyclonal antibodies against Mash1 (Horton et al., 1999) and mouse monoclonal antibodies against Ngn2 (Lo et al., 2002). There are many cells where Mash1 and Ngn2 are co-localized, and these are enriched but not restricted to the more lateral ventricular zone (Fig. 1B, arrows). Thus, contrary to the distinct cross-inhibited domains that were observed between Math1, Ngn1, and Mash1, the other bHLH factors present at this time (Gowan et al., 2001), Ngn2 overlaps with Mash1 in a sub-population of cells in the dorsal neural tube.
Mash1 is present throughout the mediolateral extent of the ventricular zone whereas Ngn2 is enriched more laterally (Fig. 1A,B). This spatial pattern suggests these two factors are acting at different times during neuronal development since cells move laterally out of the ventricular zone when they exit the cell cycle and initiate a program of neuronal differentiation. To characterize Mash1 and Ngn2 relative to cell proliferation, we used BrdU incorporation to detect cells in S-phase. In E10.5 embryos exposed to BrdU for 1h before analysis, a subset of Mash1+ cells were detected that incorporate BrdU (Fig. 1C,E, arrows). In contrast, Ngn2+ cells did not score positive for BrdU incorporation (Fig. 1D,F). These results suggest that Mash1 is present at an earlier stage than Ngn2 during neuronal differentiation, and that temporal regulation of these factors may be important for their activities. Alternatively, Mash1 and Ngn2 could be revealing a code for dorsal interneuron specification such that ventricular zone cells containing each bHLH singly, or in combination, give rise to a distinct neuronal population. In this study, we used both loss-of-function and gain-of-function experiments to address the role of Mash1 and Ngn2 in specification of dorsal neuronal sub-types dI2-dI5.
Mash1 is required for the formation of dI3 and dI5 neuronal populations
Mash1 null embryos were analyzed for alterations in the number of dI2-dI6 neurons using multi-label immunofluorescence at E10.5. The neurons are defined by the presence of HD transcription factors, their position in the dorsoventral axis, and a birthdate prior to E11.5 (Table 1). Although Mash1 is in ventricular zone cells adjacent to dI3, dI4, and dI5 neurons, in the Mash1 null there is a striking loss of only two of these populations. There is a 70% loss of dI3 neurons (Isl1) and a complete loss of dI5 neurons (Lmx1b) (Fig. 2, compare E,F,I,J). In contrast, there is a dramatic increase in dI2 (Lhx1/5;Brn3a) and dI4/6 (Pax2) neurons, which appear to fill in at the location of the missing dI3 and dI5 neurons (Fig. 2, compare A,B,I,J). Although in the Mash1 null dI4 and dI6 are not distinguishable, overexpression and lineage analysis (see below) plus the lack of expansion of the ventral ngn1 domain, which marks dI6 progenitors (Fig. 5H), suggest the increase in Pax2 is due to an increase in dI4 neurons. These results demonstrate the requirement for Mash1 in the formation of both dI3 and dI5 neurons, and the repression of dI2 and dI4/6 neurons.
Table 1.
Homeodomain (HD) factors used to define dI1-dI6 dorsal interneuron populations at E10.5*
| Interneuron Population | HD factor |
|---|---|
| dI1 | Lhx2/9, Brn3a |
| dI2 | Lhx1/5, Brn3a |
| dI3 | Islet1, Brn3a |
| dI4 | Lhx1/5, Pax2 |
| dI5 | Lmx1b, Brn3a |
| dI6 | Lhx1/5, Pax2 |
Figure 2. Mash1 is required for dI3 and dI5 neuron formation while Ngn2 represses formation of these populations.
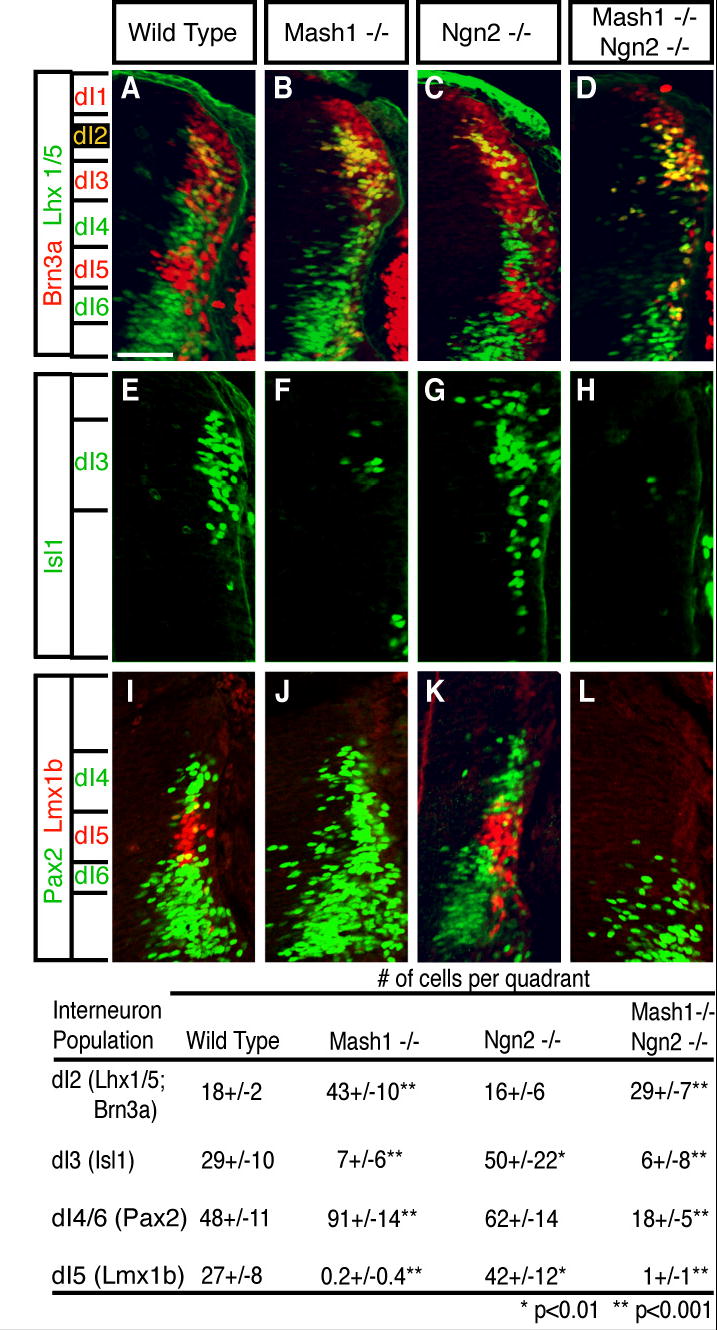
Immunofluorescence on transverse sections of neural tubes in wild type (A,E,I), Mash1_−/_− (B,F,J), Ngn2_−/− (C,G,K) and Mash1_−/−;Ngn2_−/_− (D,H,L) E10.5 mouse embryos. (A-D) Co-labeling with Brn3a and Lhx1/5 (yellow) marks dI2 neurons. There is a significant increase in dI2 neurons in the Mash1_−/− and Mash1_−/−;Ngn2_−/_− embryos but not Ngn2_−/_− relative to wild type. (E-H) Isl1 identifies dI3 neurons (green). In the Mash1_−/− and Mash1_−/−;Ngn2_−/_− mutant embryos, there is a dramatic decrease in dI3 neurons relative to wild type. In contrast, an increase in dI3 neurons in Ngn2_−/_− is detected. (I-L) Pax2 marks dI4 and dI6 neurons (green), and Lmx1b labels dI5 neurons (red). In the Mash1_−/− and Mash1_−/−;Ngn2_−/_− embryos, there is a complete loss of dI5. This is opposite in Ngn2_−/_− embryos where there is an increase in dI5 relative to wild type. dI4/6 neurons increase in the Mash1_−/_−, are unchanged in the Ngn2_−/−, and are dramatically reduced in the Mash1_−/−;Ngn2_−/_− relative to wild type embryos. Individual populations in each mutant were counted on at least three sections from at least three embryos each and cell counts are shown in the table. Because dI4 and dI6 neuronal populations cannot be distinguished in the Mash1 null, all Pax2 cells dorsal to the boundary where the first ventral Pax2 cell was found within the ventricular zone were counted, and as such, they are labeled as dI4/6. Scale bar is 50 μm.
Figure 5. Mash1 inhibits Ngn1 but not Ngn2 expression in the dorsal neural tube.
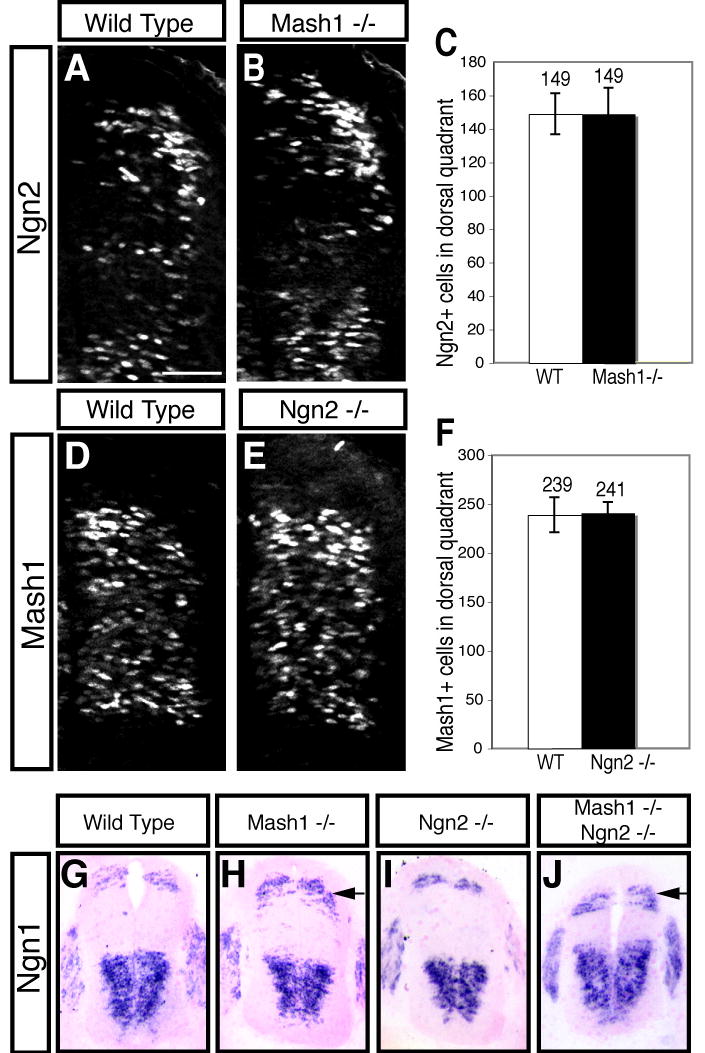
Immunofluorescence (A,B,D,E) and in situ hybridization (G-J) on transverse sections of E10.5 mouse neural tubes. (A-C) There is no change in levels of Ngn2 (green) between wild type (A) and Mash1_−/_− (B) embryos. (D-F) There is no change in expression of Mash1 (red) between wild type (D) and Ngn2_−/_− (E) embryos. (G-J) The dorsal domain of Ngn1 is expanded in Mash1_−/− (H, arrow) and Mash1_−/−;Ngn2_−/_− (J, arrow) relative to wild type (G) embryos. No change was detected in Ngn1 in Ngn2_−/_− (I) embryos. Each panel is representative of the phenotype seen in at least three sections from at least three embryos. Scale bar is 50 μm in (A,B,D,E) and 225 μm in (G-H).
To corroborate the conclusions from the Mash1 loss-of-function analysis, over-expression of Mash1 in the chick neural tube was used to test whether Mash1 is sufficient to promote dI3 and dI5 neuronal fates. A Mash1 expression plasmid (pMiWIII-Mash1) was electroporated into chicken embryos at HH13-14 and its effect on dorsal neuronal markers was analyzed 24 hours later. Excess levels of Mash1 increased the number of dI3 (Isl1) and dI5 (Lmx1b) cells as compared to the control side (Fig. 3C,G, right panel of each pair is the injected side). Furthermore, excess Mash1 decreased the number of dI2 (Lhx1/5;Brn3a) and dI4 (Pax2) neurons (Fig. 3A,E). These results are opposite of that seen in the Mash1 null, and demonstrate that Mash1 is sufficient for the generation of dI3 and dI5 neurons. Interestingly, the increase observed for each marker does not generally extend over the whole dorsoventral length of the neural tube, but rather it is localized around the area of its normal expression domain. This suggests that regional differences along the dorsoventral axis modulate the activity of Mash1, and demonstrate the context dependent nature of its specification function.
Figure 3. Excess Mash1 promotes dI3 and dI5 populations while excess Ngn2 represses them in the chick neural tube.
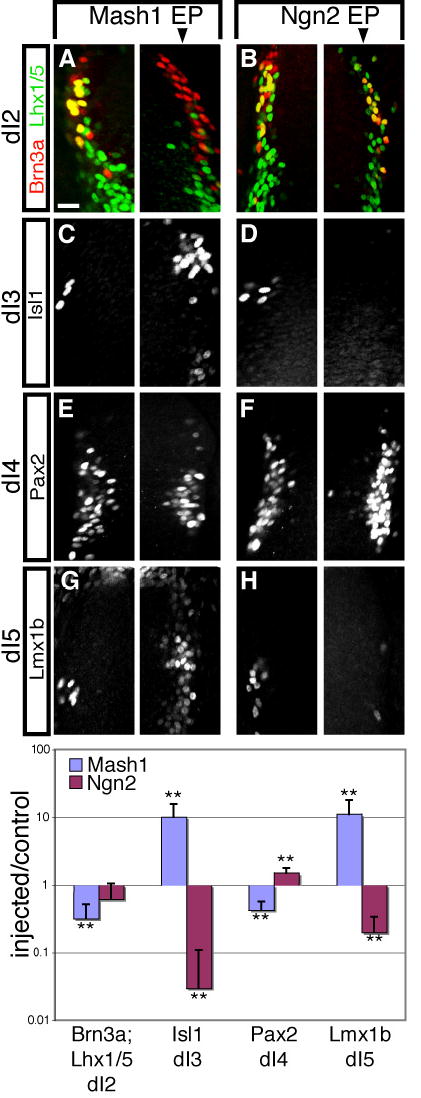
Immunofluorescence on transverse sections of E4 (HH13-14) chick neural tubes electroporated with pMiWIII-Mash1 (A,D,G,J) or pMiWIII-Ngn2 (B,E,H,K) at E3 (HH23-24). The arrow above each row of panels indicates the electroporated side of the neural tube. Neuronal population dI2 was labeled by Brn3a;Lhx1/5 (yellow) (A,B), dI3 by Isl1 (C,D), dI4 by Pax2 (E,F), and dI5 by Lmx1b (G,H). Each panel is representative of the phenotype seen in at least three sections from at least three electroporated embryos. Relative induction or repression is expressed as the number of cells on the injected side divided by the number of cells on the control side and plotted in the graph on a logarithmic scale. ** indicates p<0.001. Scale bar is 25 μm.
Ngn2 acts to limit Mash1 activity in dI3 and dI5 neuron formation
The role of Ngn2 in specification of dorsal neuronal populations was also examined to test the hypothesis that Mash1 and Ngn2 provide a combinatorial code for dorsal neuron identity. In E10.5 embryos null for Ngn2, all neuronal populations, dI1-dI6, were generated (Fig. 2) demonstrating that Ngn2 is not required for any specific dorsal cell-type. However, the composition of neurons that formed was altered with a subtle increase in the number of dI3 (Isl1) and dI5 (Lmx1b) neurons relative to wild type embryos (Fig. 2E,G,I,K), and no significant changes in dI2 (Lhx1/5;Brn3a) and dI4/6 (Pax2) populations were detected (Fig. 2A,C,I,K). Excess levels of Ngn2 in the chick neural tube resulted in a complementary phenotype to the Mash1 experiments in that dI3 (Isl1) and dI5 (Lmx1b) were dramatically decreased but dI4 (Pax2) was slightly increased relative to the non-injected side (Fig. 3B,D,F,H, see graph for cell counts). Together these results demonstrate that Ngn2, although not required for any individual neuronal sub-type, limits the generation of dI3 and dI5 neurons, two populations that require Mash1 activity.
Mash1 is epistatic to Ngn2 in the formation of dI3 and dI5 neurons
The single mutants described above demonstrate that Mash1 and Ngn2 have opposite effects on the number of dI3 and dI5 neurons that form. Furthermore, the expression pattern suggests that Ngn2 may function downstream of Mash1 rather than setting up a combinatorial code for neuronal sub-type specification. If this interpretation of temporal expression is correct, the prediction is that the loss of both Mash1 and Ngn2 should phenocopy the Mash1 null. In support of this hypothesis, embryos null for both Mash1 and Ngn2 have a 70% loss of dI3 and complete loss of dI5 neurons, just as seen in the single Mash1 null (Fig. 2H,L). Furthermore, the increase in dI2 neurons detected in the Mash1 null is also seen in the double mutant (Fig. 2D). These results are consistent with Mash1 functioning upstream of Ngn2 in these populations.
Surprisingly, embryos null for both Mash1 and Ngn2 have an apparent loss of dI4 neurons (Fig. 2L), a phenotype not predicted from the single mutants. Although dI4 and dI6 cannot be distinguished in the absence of dI5, the position of the Pax2-positive cells in the Mash1/Ngn2 double knockout strongly suggests a complete loss of dI4 neurons, and possibly dI6 neurons as well (Fig. 2L). This loss of dI4 in the double knockout is in contrast to the single knockouts where there was no indication that either Mash1 or Ngn2 are required for dI4 generation. In fact, dI4/6 cells are significantly increased in the Mash1 null (Fig. 2J). Thus, there is an apparent redundant function for Mash1 and Ngn2 in the generation of dI4 neurons.
Mash1-positive cells give rise primarily to dI3 and dI5, but not dI4 neurons
Given the presence of Mash1 in the ventricular zone throughout the dorsoventral domain adjacent to dI3, dI4, and dI5 neurons, it was surprising that dI4 neurons increased in the Mash1 knockout while dI3 and dI5 were lost. This finding suggests either Mash1+ cells become dI4 neurons but do not require Mash1, or that there are distinct low- or non-Mash1+ cells in the ventricular zone that give rise to the dI4 neurons. We used recombination based lineage tracing in vivo to distinguish between these two possibilities. A transgenic mouse was utilized (M1-GIC) that expresses both GFP and Cre in the Mash1 expression pattern from a bacterial artificial chromosome containing 300 kb of genomic sequence surrounding the Mash1 protein coding region (Supplemental Fig. 1S). By crossing the M1-GIC mouse line with a Cre reporter line R26R-YFP, any cell that has expressed the transgene will be permanently labeled with YFP (Srinivas et al., 2001).
Embryos at E11.5 were examined by triple-label immunofluorescence to determine the fate of Mash1+ cells in the dorsal neural tube. An antibody to GFP detects GFP and YFP simultaneously, and thus, these cells are referred to as GFP/YFP-positive cells. GFP/YFP-positive cells in the marginal zone were counted and scored for co-labeling with markers of dI2-dI6 neurons (Fig. 4, Table 2). The majority of GFP/YFP+ cells become dI5 (Lmx1b, 73%) and dI3 (Isl1, 17%), rarely, dI2 (Lhx1/5;Brn3a, 2%) and no dI6 (Lhx1/5, 0%), consistent with the requirement for Mash1 specifically in dI3 and dI5. A notable percentage of GFP/YFP-expressing cells co-label with dI4 (Lhx1/5, 8%) suggesting a Mash1+ cell can become a dI4 neuron. However, this only represents a small percentage of the dI4 cells generated (~3%) and when identified, these co-labeled cells border the dI3 and dI5 domains (Fig. 4E, arrow). We estimate that Cre induced recombination of the reporter gene in the M1-GIC embryos is approximately 50% since essentially all dI5 neurons require Mash1, but we detect only 52% of the dI5 neurons co-labeled with GFP/YFP (44 of 85 total Lmx1b cells). Furthermore, since we detect 25% of the dI3 neurons co-labeled with GFP/YFP (9 of 35 total Isl1 cells), using the efficiency factor we predict ~50% of dI3 neurons are derived from Mash1+ cells, consistent with the partial loss of dI3 detected in the Mash1 null (Fig. 2). Taken together, these results suggest that the function of Mash1 in the generation of dI3 and dI5 neurons is cell-autonomous, and demonstrate that dI2, the majority of dI4, and dI6 neurons develop from cells that contain Mash1 at levels not detected using this transgenic reporter mouse line. Indeed, heterogeneity in endogenous Mash1 levels is detected by immunofluorescence where ventricular zone cells adjacent to dI3 and dI5 have Mash1 at higher levels than cells adjacent to dI4 (Fig. 5D).
Figure 4. Mash1 cells primarily become dI3 and dI5 neurons.
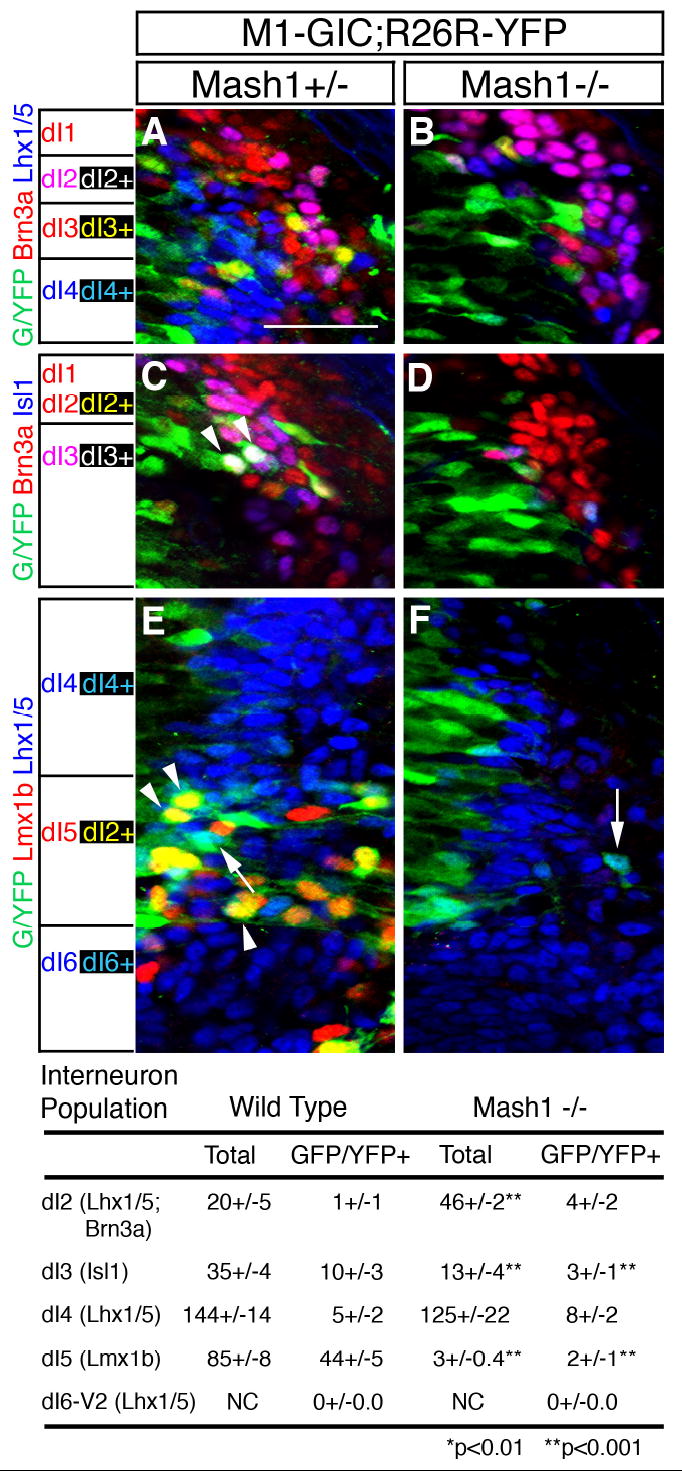
Immunofluorescence on transverse sections of _Mash1+/_− (A,C,E) or Mash1_−/_− (B,D,F) mouse embryos at E11.5 containing the transgene M1-GIC (see Supplementary Fig. 1S) and the Cre reporter allele R26R-YFP. (A-F) anti-GFP antibodies detect both GFP from M1-GIC and YFP when R26R-YFP reports the Cre activity from M1-GIC. These green labeled cells reflect cells with Mash1 or progeny of Mash1+ cells. (A,B) Brn3a;Lhx1/5 (pink) label dI2. These cells would appear white if they co-labeled with antibodies to GFP/YFP. The absence of white cells indicate dI2 do not derive from Mash1+ precursors (A). (C,D) Brn3a;Isl1 (pink) label dI3. White cells indicate co-labeling with antibodies to GFP/YFP and demonstrate Mash1+cells give rise to dI3 neurons (C, arrowheads) and these cells are absent in the null (D). (E,F) Lmx1b (red) labels dI5. The co-labeling with GFP/YFP (yellow) indicates dI5 are derived from Mash1+ cells (E, arrowheads) and these cells are absent in the null (F). Lhx1/5 (blue) labels dI4. Co-labeling with GFP/YFP (turquoise) indicate rare dI4 cells derived from Mash1+ cells (E, arrow), and these cells do not increase in Mash1_−/_− (F). Note that the increase in dI2 and dI4 seen at E10.5 cannot be attributed to fate switching of dI3 and dI5 precursor cells. The color of each neuronal population is shown on the left without and with (black background) co-labeling with the GFP/YFP. The total number of cells for each neuronal sub-type and the number of each that co-labels with GFP/YFP are shown in the table. Individual populations in each mutant were counted on at least three sections from at least three embryos each. Scale bar is 25 μm.
Table 2.
Distribution of Total GFP/YFP Expressing Marginal Zone Cells in M1-GIC E11.5 Dorsal Neural Tube
| dI1 | dI2 | dI3 | dI4 | dI5 | dI6 | Total GFP/YFP | |
|---|---|---|---|---|---|---|---|
| # cells | 0 | 1+/−1 | 10+/−3 | 5+/−2 | 44+/−5 | 0 | 60+/−2 |
| % | 0 | 2% | 17% | 8% | 73% | 0 |
Mash1 null cells are stalled in the ventricular zone at E11.5 and do not appear to trans-fate to dI4 neurons
Analysis of the Mash1 knockout revealed the loss of dI3 and dI5 with an increase in dI2 and dI4/6 that appeared to complement the loss (Fig. 2). To determine if the cells that would have become dI3 and dI5 switch their fate to dI2/dI4 in the absence of Mash1, we examined these neuronal populations in M1-GIC;R26R-YFP;Mash1_−/_− embryos at E11.5. The first phenotype noted was that the GFP/YFP level is much higher in the mutant as compared to wild-type (see Supplemental Fig. 1S). This difference in level reflects the negative autoregulation at the Mash1 locus that has previously been reported (Casarosa et al., 1999; Horton et al., 1999; Meredith and Johnson, 2000). The Mash1 null sections showing GFP/YFP (Fig. 4B,D,F) were imaged at much lower GAIN than similar sections from embryos wild-type for Mash1 (Fig. 4A,C,E) (see Supplemental Fig. 1S for images with matched GAIN).
Other than negative autoregulation, the most dramatic phenotype detected using these mouse strains is that the cells that should have expressed Mash1 (GFP/YFP) appear stalled in the ventricular zone with only a few cells detected in the marginal zone (Fig. 4B,D,F). These stalled cells aberrantly located in the ventricular zone have at least partially initiated a differentiation program; they express Lbx1 and Tuj1, markers normally restricted to the marginal zone, and they rarely incorporate BrdU, demonstrating that many have exited the cell-cycle (Supplemental Fig. 2S). TUNEL labeling shows no detectable increase in cell death in the neural tube at E10.5 and E11.5 (data not shown). Taken together, the precursors to dI3 and dI5 are not leaving the ventricular zone in the Mash1 null at E10.5/E11.5, and do not appear to significantly contribute to the increase in dI2 and dI4 since there was no increase in the proportion of GFP/YFP cells co-labeled with dI2 and dI4 markers (Fig. 4B,D,F). Thus, the ectopic dI2 and dI4/6 neurons in the Mash1 null at E10.5 cannot be accounted for by a switch in fate of dI3 and dI5 precursor cells. Furthermore, cells with undetectable levels of Mash1 must give rise to dI2 and dI4/6 neurons, and the number of these cells increase in the Mash1 knockout.
Supplemental Figure 2. Expanded ventricular zone and premature differentiation in the Mash1 null neural tube.
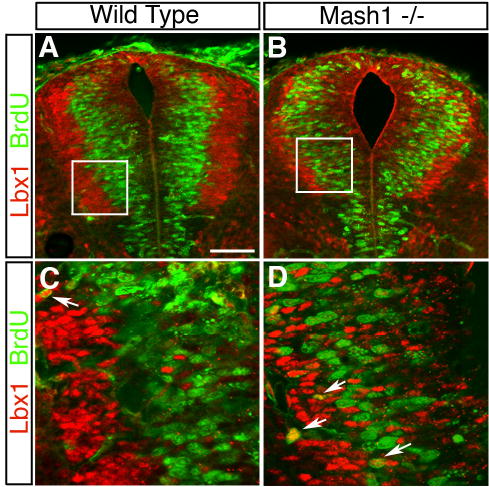
Immunofluorescence of transverse sections of E11.5 wild type (A,C) or Mash1_−/_− (B,D) neural tube showing Lbx1 expression (red) and BrdU incorporation (green). (A,C) In the wild type, Lbx1 expression is in post-mitotic cells that are lateral to the ventricular zone and are almost exclusively BrdU negative (one positive in this section, arrow). (C) is a higher power image of the area boxed in (A). (B,D) In the Mash1 null, the ventricular zone is expanded and appears to bulge. There is no obvious increase in BrdU incorporation but Lbx1+cells are aberrantly located in the ventricular zone suggesting cells are initiating at least some differentiation program but are unable to leave the ventricular zone. Although a few cells incorporate BrdU in lateral regions, rarely have the Lbx1+cells within the ventricular zone incorporated BrdU. Taken together, the coordinated events that normally occur during neuronal differentiation in the dorsal neural tube are disrupted in the Mash1 null. The bulge seen in the dorsal neural tube appears to reflect the inability of cells to completely initiate their program of differentiation and move out of the ventricular zone. Scale bar is 70 μm in (A-B) and 25 μm in (C-D).
Mash1 and Ngn2 levels are independent of each other
Previously it was demonstrated that cross-inhibition between the three bHLH family members, Mash1, Math1, and Ngn1, was used to control cell number and cell-type formed (Gowan et al., 2001; Scardigli et al., 2001). To determine whether this type of regulation is occurring between Mash1 and Ngn2, we examined the levels of Ngn2 in the Mash1 null at E10.5 and vice versa. We observed no significant change in the number of Ngn2+ cells in the dorsal neural tube of Mash1 nulls (Fig. 5A-C) and no significant change in the number of Mash1+ cells in Ngn2 nulls (Fig. 5D-F). These results suggest that Mash1 and Ngn2 do not use cross-inhibition as a mechanism to control the number of dorsal neurons formed, consistent with the co-expression seen with these two bHLH factors. Although difficult to quantify, there may be increased levels of Mash1 in individual cells in the most dorsal region in the Ngn2 null and vice versa in the Mash1 null (Fig. 5, compare A, B, and D,E) and this may account for the increase in dI3 neurons in the Ngn2 null embryos..
We have previously shown that an increase in Ngn1 in the dorsal neural tube leads to an increase in dI2 neurons (Gowan et al., 2001). To test whether an increase in Ngn1 could explain the increase in dI2 neurons in the Mash1 and the Mash1/Ngn2 double knockouts, we examined Ngn1 expression. Indeed, in the Mash1 and the Mash1/Ngn2 double knockouts, we detected an increase in Ngn1 expression in the dorsal neural tube at E10.5 relative to wild type (Fig. 5, compare G with H,J). Consistent with the lack of change of the dI2 population in the Ngn2 null, there was no change in Ngn1 detected (Fig. 5I). Thus, the excess dI2 cells in the Mash1 and Mash1/Ngn2 knockouts is likely due to loss of cross-inhibition of Ngn1 by Mash1 in its dorsal domain of expression (Gowan et al., 2001).
Ngn2 does not directly block Mash1 function in specifying dI3 and dI5 neurons
The preceding data strongly suggest that Ngn2 opposes Mash1 function in generation of dI3 and dI5 neurons, and thus, the levels and timing of Mash1 and Ngn2 determine the number of dI3 and dI5 neurons that form. Possible mechanistic models to explain the phenotypes involve Ngn2 directly opposing Mash1 function by forming a non-functional heterodimer, or by competing with Mash1 on target genes, analogous to interactions between Olig2 and Ngn2 recently reported (Lee et al., 2005). The ability of Ngn2 and Mash1 to form non-functional heterodimers, or to bind similar DNA recognition sites, has been shown in vitro (Gradwohl et al., 1996). To test these models in vivo, we utilized mouse mutant lines that contain replacement mutations where either the Ngn2 protein coding region was swapped into the Mash1 locus (Mash1 KI Ngn2) or the Mash1 protein coding region was swapped into the Ngn2 locus (Ngn2 KI Mash1) (Parras et al., 2002). The heterozygous embryos in each strain shift the balance and temporal relationship of Mash1 and Ngn2. If Ngn2 directly opposes Mash1 function as predicted above, then the Mash1 KI Ngn2/+ would approximate the Mash1 knockout, and the Ngn2 KI Mash1/+ would approximate a Mash1 gain-of-function phenotype. In Ngn2 KI Mash1/+ embryos, we see an increase in dI3 and dI5 reflecting the Mash1 gain-of-function phenotype as predicted from the specification function of Mash1 (Fig. 6F,I,K,N). However, rather than losing dI3 and dI5 in Mash1 KINgn2/+ embryos, dI3 and dI5 are increased (Fig. 6F,G,K,L). Due to variability between these mutant embryos, only the dI3 increase was statistically significant. There is also a significant increase in dI2 neurons in Mash1 KINgn2/+ embryos, possibly reflecting the role of Ngn2 in generation of these neurons (Gowan et al., 2001) (Fig. 6A,B). Although the results confirm the importance of Mash1 in specifying dI3 and dI5 neurons, they contradict the model that Ngn2 directly opposes this activity of Mash1. Rather, these results fit a model that highlights distinct functions for Mash1 and Ngn2 where Mash1 has a major role in neuronal specification and Ngn2 has a role in temporal control of neuronal differentiation.
Figure 6. Distinct functions for Mash1 and Ngn2 in specification of dorsal neurons.
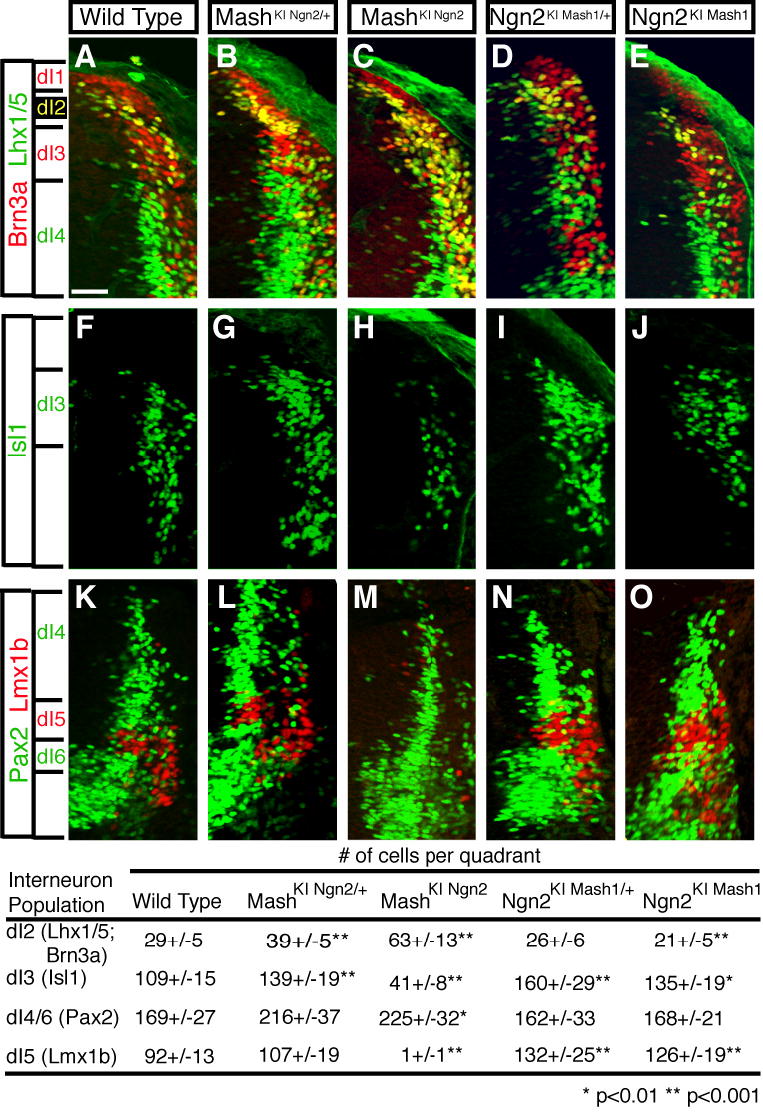
Immunofluorescence on cross sections of E11.5 mouse neural tubes where Mash1 and Ngn2 have been swapped into the Ngn2 and Mash1 locus, respectively: wild type (A,F,K), Mash1 KINgn2/+ (B,G,L), Mash1 KINgn2 (C,H,M), Ngn2 KIMash1/+ (D,I,N), and Ngn2 KIMash1(E,J,O). (A-E) Co-localization of Brn3a (red) and Lhx1/5 (green) detects dI2 neurons (yellow). (F-J) Isl1 (green) detects dI3 neurons. (K-O) Pax2 (green) and Lmx1b (red) detect dI4 and dI5 neurons, respectively. Cell counts for each marker on at least three sections from at least three embryos of each genotype are shown in the table. Because dI4 and dI6 populations cannot be distinguished in the Mash1 null, all Pax2 counts were completed by drawing a boundary line at the first Pax2 expressing cell found nearest the ventricular zone, and as such, they are labeled as dI4/6. ** indicates p<0.001 and * indicates p<0.01. Scale bar is 50 μm.
The Mash1 and Ngn2 knock-in strains have also been used to address whether these related bHLH factors are able to compensate for each other’s function. It has been reported that Ngn2 and Mash1 share the ability to induce neuronal differentiation, but are distinct in specifying neuronal identity (Parras et al., 2002). The functions of Mash1 and Ngn2 in the dorsal neural tube are similar to those attributed to them in other regions of the nervous system. Ngn2 is unable to compensate for Mash1 function in specification as shown by its inability to rescue the generation of dI3 and dI5 neurons (Fig. 6F,H,K,M), and its inability to suppress the ectopic formation of dI2 and dI4/6 neurons (Fig. 6A,C,K,M). Similarly, Mash1 does not compensate for Ngn2 since in both Ngn2_−/_− and Ngn2 KI Mash1/+ embryos the number of dI3 and dI5 neurons is elevated (Fig. 6F,J,K,O), reflecting the specification function of Mash1 distinct from Ngn2. The number of dI2 neurons was decreased in this mutant line (Fig. 6A,E), suggesting that ectopic Mash1 in dI2 precursors is inhibitory for dI2 generation as is predicted from Mash1 repression of Ngn1 expression (Gowan et al., 2001). Together, these results confirm that Mash1 function is critical for the specification of dorsal neuronal sub-types dI3 and dI5, and Ngn2 cannot replace this function. In addition, although no specification function can be ascribed to Ngn2, it functions in controlling the number of different neuronal sub-types that form.
Discussion
A transcription factor code for specification of dorsal interneurons
Formation of the neuronal network in the dorsal spinal cord requires the generation of correct numbers of neurons with specified identities. Regulation of this process starts early in nervous system development as cells in the ventricular zone of the neural tube have distinct identities that are determined by transcription factors of the HD and bHLH families. Here we demonstrate that Mash1 is in precursors to, and required for dI3 and dI5 neurons. Furthermore, the number of dI3 and dI5 neurons that form depends on the activity of Ngn2. Thus, the balance of Mash1 and Ngn2 activity plays a central role in setting up the correct composition of neurons found in the dorsal spinal cord.
Surprisingly, dI4 neurons, which appear to arise from the domain containing Mash1, do not require Mash1 or Ngn2 alone. Rather, dI4 neurons appear to arise from cells with low levels of Mash1 or none at all. Thus, the control of dI4/6 cell number implies a non-autonomous mechanism with respect to Mash1. Indeed, the heterogeneous Mash1 levels may reflect function of the Notch-signaling pathway in the neural tube at this time (Lindsell et al., 1996; Ma et al., 1997). In addition, we recently identified another bHLH factor, PTF1a (Krapp et al., 1996), which is required for dI4, and is present in the dI4 precursor domain where Mash1 levels are low (S. Glasgow and J.E.J., unpublished). We have shown that Mash1 levels are heterogeneous in the ventricular zone; thus, the cells with the highest levels of Mash1 preferentially go on to become dI3 and dI5 neurons, and the cells with distinctly lower levels of Mash1, either remain as progenitor cells or become dI4 neurons. These data combined with previous reports demonstrating the requirement of Math1 for dI1 neurons (Bermingham et al., 2001; Helms and Johnson, 1998) and Ngn1/2 for dI2 neurons (Gowan et al., 2001), demonstrate an emerging bHLH transcription factor code for specification of the early born dorsal neuronal populations.
Additional refinements of the code are required to explain what determines whether a Mash1 precursor will develop into a dI3 or a dI5 neuron. Over-expression of Mash1 in the chick neural tube resulted in an increase in both neuronal cell-types, however, the normal position of the ectopic neurons in the dorsoventral axis was essentially maintained. This result suggests that Mash1 interacts with other factors to specify dI3 versus dI5, and that these factors are likely restricted to defined domains in the dorsoventral axis of the neural tube (Fig. 3C,G). The identity of these interacting factors is suggested in recent studies. Olig3, a bHLH factor present in precursors to dI1-dI3, when combined with Mash1, induces the dI3 phenotype but not dI5 (Müller et al., 2005). In contrast, Lbx1, a HD class transcription factor expressed just as the cells become post-mitotic, is required for specification of dI4-dI6 (Gross et al., 2002; Müller et al., 2002). When Lbx1 is co-electroporated with Mash1 into the chick neural tube, there is an increase in dI5 and decrease in dI3 (AWH, YN, JEJ, unpublished data). Furthermore, in PTF1a deficient embryos, the loss of PTF1a leaves cells with Mash1 and Lbx1, and this combination results in a fate switch from dI4 to dI5 (S. Glasgow and J.E.J., unpublished). Thus, additional refinements to the transcription factor code suggest that combinations of factors such as Mash1 plus Olig3 specifies dI3 (Müller et al., 2005), Mash1 followed by Lbx1 specifies dI5, and Mash1low plus PTF1a followed by Lbx1 specifies dI4 (S. Glasgow and J.E.J., unpublished). In addition, upstream factors controlling expression of the bHLH genes, such as seen with the function of the HD factor Gsh2 in suppressing Ngn1 and inducing Mash1 (Kriks et al., 2005), are also critical for generating the correct composition of neuronal subtypes. Identification of transcriptional targets for the bHLH and HD factors, as has been reported in ventral spinal cord development (Lee et al., 2005; Lee and Pfaff, 2003), will be required to determine the mechanisms controlling the specification of these dorsal neurons.
Sequential actions of Mash1 and Ngn2 in specifying dorsal horn neurons
In the dorsal telencephalon, loss of Ngn2 results in an increase in Mash1+ cells and a subsequent increase in GABAergic neurons presumably derived from these ectopic Mash1-expressing cells (Fode et al., 2000). Thus, one function of Ngn2 in forebrain development is to suppress levels of Mash1. This interpretation is similar to the cross-repression seen between Math1, Ngn1 and Mash1 in the dorsal spinal neural tube (Gowan et al., 2001). However, in the dorsal neural tube, Ngn2 overlaps with both Mash1 (Fig. 1) and Ngn1 (AWH and JEJ, unpublished data). This co-localization is found in cells that do not incorporate BrdU suggesting that Ngn2 is somewhat temporally delayed relative to Mash1 and Ngn1. Regardless, the overlap suggests there is little if any transcriptional cross-repression mechanism regulating the expression of Mash1 and Ngn2 in the dorsal neural tube. Indeed, in this domain, the number of Mash1+ cells is not increased in the Ngn2 null, nor is the number of Ngn2+ cells increased in the Mash1 null (Fig. 5). Thus, rather than cross-inhibition, these results support a model where Mash1 appears to be temporally upstream of Ngn2.
Although Mash1 and Ngn2 do not appear to cross-repress each other’s expression, it is clear that Ngn2 limits the apparent activity of Mash1 in inducing dI3 and dI5 neurons. This is evident in the Ngn2 null where there is an increase in the Mash1-dependent dI3 and dI5 populations, and in over-expression of Ngn2 in chick where there is a decrease in the number of dI3 and dI5 neurons. However, the hypothesis that Ngn2 directly blocks Mash1 activity, either by forming inactive complexes or by blocking shared DNA binding sites, as has been reported for Olig2 and Ngn2 (Lee et al., 2005), appears incorrect since increasing Ngn2 and decreasing Mash1 in a cell, as occurs in Mash1 KINgn2/+, does not decrease dI3 and dI5 as would be predicted.
An alternative model is that Ngn2 increases the probability that a cell will permanently exit the cell-cycle. Even subtle perturbations in the probability of cell-cycle exit could modulate the number of cells of a specific cell-type that are formed. This function for Ngn2 in controlling the timing of differentiation is similar to that attributed to Ngn2 in motor neuron formation, where the balance of Olig2 and Ngn2 control the number of cells that will undergo neuronal differentiation (Lee et al., 2005). In Mash1 KINgn2/+ embryos, Ngn2 is present earlier than in wild type, and Mash1 levels are decreased, but an increase in dI3 and dI5 is still detected. This increase in dI3 and dI5 could reflect Ngn2 inducing premature differentiation. In contrast, in the Ngn2 null, the increase in dI3 and dI5 populations could result from extra divisions of the dI3/dI5 precursor cells due to a subtle shift in timing of cell-cycle exit. What is clear is that there is a fundamental difference in how Ngn2 functions relative to the other bHLH factors such as Mash1, Math1, and Ngn1. These latter factors function in specifying neuronal identity in the dorsal spinal cord. In contrast, Ngn2 is not required for any specific cell-type but is required to get a normal composition of neurons formed. Further support for this difference in how Ngn2 functions relative to Mash1 is seen in the ventral neural tube where Ngn2 but not Mash1 can synergize with the HD factors Lhx3 and Isl1 in post-mitotic cells to generate motor neurons (Lee and Pfaff, 2003).
Mash1 is required for lateral movement of differentiating cells from the ventricular zone
The generation of transgenic mice expressing GFP and Cre from a Mash1 locus containing BAC allows a unique look at the behavior of the cells in the absence of Mash1 function. The GFP reporter in M1-GIC;Mash1_−/_− mice indicates that in the absence of Mash1, the cells do not switch fate and contribute to the increased dI4 population, but rather these cells primarily remain in the ventricular zone (Fig. 1S, compare E and F; Fig. 2S, compare A and B). Although stalled in the ventricular zone, the cells continue some aspects of the neuronal differentiation process as illustrated by ectopic Lbx1 and Tuj1 within the ventricular zone of Mash1 nulls (Fig. 2S; data not shown). Furthermore, these aberrant Lbx1+ cells rarely incorporate BrdU suggesting many have left the cell-cycle (Fig. 2S). Thus, in the absence of Mash1, the cells apparently undergo multiple aspects of differentiation but they do not have the characteristic lateral movement to the marginal zone.
Conclusions
The data reported here, combined with previous studies (Bermingham et al., 2001; Gowan et al., 2001; Helms and Johnson, 1998; Müller et al., 2005), demonstrate that bHLH factors are required for generating the correct number and types of neurons in the dorsal neural tube. The precise mechanisms for how these factors interact to result in this neuronal diversity is still not clear and will require identification of downstream targets to define where these pathways intersect. Furthermore, identification of additional bHLH family members and co-factors such as HD proteins, and a more detailed understanding of non-cell autonomous mechanisms involving Notch/delta signaling, will be required to fully understand the formation of the neuronal network in the dorsal spinal cord.
Acknowledgments
These studies could not be performed without the generosity of multiple laboratories in providing cell-type specific antibodies including S. Morton and T. Jessell for guinea pig anti-Lmx1b and rabbit anti-Isl1/2, E. Turner for rabbit and guinea pig anti-Brn3a, T. Müller and C. Birchmeier for guinea pig and rabbit anti-Lbx1, M. Goulding for rabbit and rat anti-Lbx1, L.-C. Lo and D. Anderson for mouse anti-Ngn2, and the Developmental Studies Hybridoma Bank for mouse anti-Lhx1/5, mouse anti-Isl1, and mouse anti-Lmx. We thank M. Goulding, T. Müller and C. Birchmeier for sharing data prior to publication. These studies also benefited from technical assistance provided by Ms P. Parab and Ms C. Weldon. This work was supported by NIH RO1 HD37932 and RO1 NS32817 to JEJ.
References
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on MATH1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neuroscience. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Birren SJ, Lo L, Anderson DJ. Sympathetic neuroblasts undergo a developmental switch in trophic dependence. Development. 1993;119:597–610. doi: 10.1242/dev.119.3.597. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Alessandra P, Jessell TM, Ericson J. A homeodomain protein code specifies progeitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Caspary T, Anderson KV. Patterning cell types in the dorsal spinal cord: what the mouse mutants say. Nat Rev Neurosci. 2003;4:289–97. doi: 10.1038/nrn1073. [DOI] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–7. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–80. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Fedtsova N, Turner EE. Inhibitory effects of ventral signals on the development of Brn-3.0-expressing neurons in the dorsal spinal cord. Dev Biol. 1997;190:18–31. doi: 10.1006/dbio.1997.8691. [DOI] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN2 is a detemination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;120:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Funahashi J, Okafuji T, Ohuchi H, Noji S, Tanaka H, Nakamura H. Role of Pax-5 in the regulation of a mid-hindbrain organizer's activity. Dev Growth Differ. 1999;41:59–72. doi: 10.1046/j.1440-169x.1999.00401.x. [DOI] [PubMed] [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Fode C, Guillemot F. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev Biol. 1996;180:227–241. doi: 10.1006/dbio.1996.0297. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Joyner A. Expression of murine Achaete-Scute and Notch homologues in the developing central nervous system. Mech Dev. 1993;42:171–185. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–76. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–925. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Curr op in Neurobiology. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Hogan, B., Costantini, F. and Lacy, E. (1986). Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press.
- Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol Cell Neurosci. 1999;14:355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genetics. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagen-buchle O, Wellauer PK. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Kriks, S., Mizuguchi, R., Lanuza, G. M., Nakafuku, M. and Goulding, M. (2005). Genetic interactions involving the repression of Ngn1 by Gsh2 specify dorsal interneuron fate in the spinal cord. Developmentin press [DOI] [PubMed]
- Lanuza GM, Gosgnach S, Pierani A, Jessell TM, Goulding M. Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron. 2004;42:375–86. doi: 10.1016/s0896-6273(04)00249-1. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Dietrich P, Jessell TM. Genetic ablation reveals that the roof plate is essential for dorsal interneuron specification. Nature. 2000;403:734–740. doi: 10.1038/35001507. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Mendelsohn M, Jessell TM. Neuronal patterning by BMPs: a requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev. 1998;12:3394–3407. doi: 10.1101/gad.12.21.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes & Development. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cellular Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Lo LC, Johnson JE, Wuenschell CW, Saito T, Anderson DJ. Mammalian achaete-scute homolog 1 is transiently expressed by spatially-restricted subsets of early neuroepithelial and neural crest cells. Genes Dev. 1991;5:1524–1537. doi: 10.1101/gad.5.9.1524. [DOI] [PubMed] [Google Scholar]
- Lo LC, Dormand E, Greenwood A, Anderson DJ. Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homologs in cultured neural progenitor cells. Development. 2002;129:1553–1567. doi: 10.1242/dev.129.7.1553. [DOI] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;120:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga E, Araki I, Nakamura H. Role of Pax3/7 in the tectum regionalization. Development. 2001;128:4069–77. doi: 10.1242/dev.128.20.4069. [DOI] [PubMed] [Google Scholar]
- Meredith A, Johnson JE. Negative regulation of Mash1 expression in CNS development. Dev Biol. 2000;222:336–346. doi: 10.1006/dbio.2000.9697. [DOI] [PubMed] [Google Scholar]
- Müller T, Anlag K, Wildner H, Britsch S, Treier M, Birchmeier C. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes & Development. 2005;19:733–743. doi: 10.1101/gad.326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor Lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Muramatsu T, Mizutani Y, Ohmori Y, Okumura J. Comparison of three nonviral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochem Biophys Res Commun. 1997;230:376–380. doi: 10.1006/bbrc.1996.5882. [DOI] [PubMed] [Google Scholar]
- Nakada Y, Hunsaker TL, Henke RM, Johnson JE. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes & Development. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez SE, Rebelo S, Anderson DJ. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development. 1999;126:1715–28. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- Pierani A, Moran-Rivard L, Sunshine MJ, Littman DR, Goulding M, Jessell TM. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron. 2001;29:367–384. doi: 10.1016/s0896-6273(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Qian Y, Shirasawa S, Chen C, Cheng L, Ma Q. Proper development of relay somatic sensory neurons and D2/D4 interneurons requires homeobox genes Rnx/Tlx3 and Tlx1. Genes & Development. 2002;16:1220–1233. doi: 10.1101/gad.982802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R, Schuurmans C, Gradwohl G, Guillemot F. Crossregulation between neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Sommer L, Ma Q, Anderson DJ. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogenity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suemori H, Kadodawa Y, Goto K, Araki I, Kondoh H, Nakatsuji N. A mouse embryonic stem cell line showing pluripotency of differentiation in early embryos and ubiquitous beta-galactosidase expression. Cell Differ Dev. 1990;29:181–6. doi: 10.1016/0922-3371(90)90120-l. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, SL P. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
References
- Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol Cell Neurosci. 1999;14:355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- Meredith A, Johnson JE. Negative regulation of Mash1 expression in CNS development. Dev Biol. 2000;222:336–346. doi: 10.1006/dbio.2000.9697. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]