Nonclassical Action of Retinoic Acid on the Activation of the cAMP Response Element-binding Protein in Normal Human Bronchial Epithelial Cells (original) (raw)
Abstract
Vitamin A (retinol) is essential for normal regulation of cell growth and differentiation. We have shown that the retinol metabolite retinoic acid (RA) induces mucous cell differentiation of normal human tracheobronchial epithelial (NHTBE) cells. However, early biological effects of RA in the differentiation of bronchial epithelia are largely unknown. Here, we showed that RA rapidly activated cAMP response element-binding protein (CREB). However, RA did not use the conventional retinoic acid receptor (RAR)/retinoid X receptor (RXR) to activate CREB. RA activated CREB in NHTBE and H1734 cells in which RARs/RXR were silenced with small interfering RNA (siRNA) targeting RAR/RXR expression or deactivated by antagonist. Inhibition of protein kinase C (PKC) or extracellular regulated kinase (ERK1/2) blocked the RA-mediated activation of CREB. In addition, depletion of p90 ribosomal S6 kinase (RSK) via siRSK1/2 completely abolished the activation, suggesting that PKC, ERK, and RSK are required for the activation. Altogether, this study provides the first evidence that RA rapidly activates CREB transcription factor via PKC, ERK, and RSK in a retinoid receptor-independent manner in normal bronchial epithelial cells. This noncanonical RA signaling pathway may play an important role in mediating early biological effects in the mucociliary differentiation of bronchial epithelia.
INTRODUCTION
Vitamin A (retinol) and its related analogs, collectively called retinoids, play a pivotal role in cell growth and differentiation (for review, see Gudas et al., 1994). In the respiratory system, retinoic acid (RA), the natural metabolite of vitamin A, controls the development and maintenance of differentiated mucociliary epithelial cells. It has been well documented that RA is essential for induction and maintenance of normal mucociliary airway epithelium (McDowell et al., 1984a, 1984b). RA deficiency causes squamous metaplasia, and supplementation with RA restores the normal mucous cell phenotype in cultured primary bronchial epithelial cells (Koo et al., 1999). However, the early effect of RA on mucous cell differentiation of bronchial epithelia is largely unknown.
It has been well documented that the effects of RA at the genomic level are mediated mainly through heterodimerized forms of retinoid receptors, RARs and RXRs, which recognize expression of their target genes through the RA response element (A/G)G(G/T)TCA upon binding of RA and activate gene expression (Giguere, 1994; Mangelsdorf and Evans, 1995; Chambon, 1996). Recent data showed, however, that RA action could be diversified to other cellular signaling pathways, the so-called nongenomic action of RA. For example, RA has been shown to modulate PKC activity by binding directly to PKC. RA binds directly to the C2-domain of PKCα (Ochoa et al., 2002), as determined by structural analysis of the cocrystalized C2-domain of PKC with RA. In addition, RA binding to the phosphatidyl serine binding site of PKCα was determined by photoaffinity labeling assay (Radominska-Pandya et al., 2000). It has also been shown that retinol and its metabolites bind to the cysteine-rich region of the regulatory domain of several PKC isoforms, including α, δ, ζ, and μ (Hoyos et al., 2000; Imam et al., 2001). Therefore, we hypothesized that these diversified actions of RA may play an important role, particularly, in triggering early events of the differentiation of bronchial epithelia, where RA receptors are less abundant to transmit RA action.
The transcription factor CREB plays important roles in controlling cell growth, survival, and cell cycle progression and in determining the fate of many cell types, including vascular smooth muscle cells (Klemm et al., 2001), adipocytes (Reusch et al., 2000), thyrocytes (Nguyen et al., 2000), Sertoli cells (Scobey et al., 2001), and neurons (Sung et al., 2001). Recently, it has been also demonstrated that CREB promotes abnormal proliferation and survival of myeloid cells and was implicated as a proto-oncogene in myeloid transformation (Shankar et al., 2005).
CREB is a member of the closely related CREB/CREM/ATF family of transcription factors, recognize CRE, 5′-TGACGTCA-3′, in the transcription-regulatory regions of its target genes (for review, see Mayr and Montminy, 2001). Recently, genome-wide analysis of the regulatory regions of the CREB revealed numerous novel CREB target genes (Impey et al., 2004). CREB was initially identified as a substrate for protein kinase A (PKA) and has been shown to be activated in response to various extracellular stimuli (e.g., hormones, growth factors, cytokines, and stress signals (for review, see (Johannessen et al., 2004) via diverse signaling molecules, including PKA (Gonzalez et al., 1989; Montminy et al., 1990), PKC (Yamamoto et al., 1988), RSK (Xing et al., 1996), mitogen- and stress-activated protein kinase 1 (Deak et al., 1998), ERK1/2 (Fix et al., 2004), mitogen-activated protein kinase (MAPK)-activated protein 2 kinase (Tan et al., 1996), Akt (Du and Montminy, 1998), and calcium- and calmodulin-dependent protein kinases II (Sun et al., 1994) and IV (Matthews et al., 1994). These kinases phosphorylate CREB at serine 133 residue (Yamamoto et al., 1988; Gonzalez et al., 1989), which is required for the recruitment of CREB-binding protein to induce CREB-mediated transactivation of transcription.
In this study, we sought to elucidate the nonclassical mechanisms of RA activation of the transcription factor CREB in primary human bronchial epithelial cells. We found that RA rapidly activates CREB in a dose- and time-dependent manner and also activates ERK1/2 and RSK, a major upstream kinase of CREB. Rapid and strong activation of ERK1/2 and RSK is required for CREB activation. Our data further demonstrate that RA stimulation of the CREB signaling cascades does not appear to involve nuclear RA receptors.
MATERIALS AND METHODS
Cell Cultures and Chemicals
NHTBE cells (Clonetics, San Diego, CA) were cultured by air-liquid interface method as described previously (Gray et al., 1996; Koo et al., 1999; Kolodziejski et al., 2002). Briefly, the second passage NHTBE cells were seeded at a density of 1 × 105 cells onto 24-mm semipermeable membrane inserts (Transwell-Clear; Corning, Acton, MA) and grown in serum-free growth factor and hormone-supplemented culture medium. Air-liquid interface was started when the cultures were confluent. Human non-small cell lung cancer cell line H1734 was obtained from American Type Culture Collection (Rockville, MD) and grown in RPMI-1640 medium containing 10% fetal bovine serum (FBS). All-trans RA (RA), 9-cis RA, 13-cis RA, retinol, cycloheximide, and actinomycin D (purchased from Sigma-Aldrich, St. Louis, MO), Ro 61-8431 and Ro 26-5405 (kindly provided by Roche Bioscience, Palo Alto, CA) were dissolved in dimethyl sulfoxide (DMSO). Other chemicals including Ro 31-8220, GF 109203X, Rottlerin, SP600125, SB203580, Y27632, U0126, 2′,5′-dideoxyadenosine, and U73122 were purchased from Calbiochem (San Diego, CA) and were dissolved in DMSO.
Western Blot Analysis and Antibodies
Western blot analysis was performed as previously described (Song et al., 2003). Whole-cell extracts were prepared using 2× SDS Laemmli lysis buffer. Cytosolic and nuclear fractions were prepared according to the manufacturer's instructions (Nuclear and Cytoplasmic Extraction Reagent; Pierce, Rockford, IL). Equal amounts of protein (20 μg) were resolved by 10% SDS-PAGE. Antibodies used were mouse monoclonal antibodies against β-actin (clone AC-15; Sigma-Aldrich, St. Louis, MO), rabbit polyclonal antibodies against total CREB and phospho-CREB (Ser-133 phosphorylated; Upstate Biotechnology, Waltham, MA), rabbit polyclonal antibodies against total ERK, phospho-ERK, p90RSK, and phospho-p90RSK (Cell Signaling Technology, Cambridge, MA), and rabbit polyclonal antibodies against RARα, RARβ, RARγ, and RXRα (Santa Cruz Biotechnology, Santa Cruz, CA). Proteins reactive with primary antibody were visualized with an horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence reagents (Amersham Bioscience, Arlington Heights, IL).
Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift assays (EMSA) were performed as described elsewhere (Gray et al., 2001; Aggarwal et al., 2004) with necessary modifications. Briefly, nuclear proteins (8 μg) prepared from RA-treated or untreated cells were incubated with 32P-end-labeled 21-mer double-stranded oligonucleotide containing the consensus binding site for cAMP response element (CRE) with the sequence (5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′) (bold type indicates CRE; Santa Cruz Biotechnology) for 30 min at 37°C, and the DNA-protein complex was resolved in a 6.6% native polyacrylamide gel. For supershift analysis, nuclear extracts were incubated with 100-fold antibodies against the pCREB or CREB, unlabeled probe, or CRE-mutated oligonucleotide probe (5′-AGAGATTGCCTGTGGTCAGAGAGCTAG-3′) (bold, CRE mutant; Santa Cruz Biotechnology) for 30 min at 37°C, and then the complex was analyzed by EMSA. Antibodies against preimmune serum were included as negative controls. The radioactive bands from the dried gels were visualized and quantitated by the PhosphorImager (Molecular Dynamics, Sunnyvale, CA) using ImageQuant software.
Immunofluorescence Analysis
NHTBE cells were fixed in a methanol:acetone mixture (1:1, vol/vol), washed with phosphate-buffered saline (PBS), blocked with 5% preimmune serum for 30 min, and then incubated with pCREB or CREB antibody (1:100 dilution) for 2 h at room temperature. Slides were washed with PBST, incubated with AlexaFluor 488-tagged secondary antibody (Molecular Probes, Eugene, OR) for 1 h at room temperature and counterstained for nuclei with 4′,6-diamidino-2-phenylindole (DAPI) for 30 min. After being washed with PBS, slides were mounted with the SlowFade Antifade kit (Molecular Probes). Staining for pCREB and CREB were visualized with a fluorescence microscope (Axioskop 40; Carl Zeiss, Thornwood, NY), and the images were captured at a magnification of 400× and stored using Axiovision software (Carl Zeiss) as described in the instructions provided by the manufacturer.
Transient Transfection
NHTBE cells were plated in 24-well culture plates at a density of 1 × 104 cells/well. Cells were grown in BEGM without RA. When the cells reached 60-70% confluence, they were subjected to transient transfection with the CRE-promoter-reporter construct (Stratagene, La Jolla, CA) and the β-galactosidase (β-gal) reporter construct (BD Biosciences Clontech, Palo Alto, CA) using a Lipofectamine transfection reagent (Invitrogen, Carlsbad, CA) as recommended by the manufacturer's instructions. Briefly, The cells were treated with a mixture of Lipofectamine reagent and plasmid DNA (0.25 μg/well) for 4 h and, after transfection cells were incubated in 0.5 ml of BEGM without RA for 24 h. Then the cultures were incubated with various concentrations of RA (0.001, 0.01, 0.1, and 1 μM), 1 μM 8-Br-cAMP, or 1 μM Forskolin (positive control; Calbiochem, San Diego, CA) for another 24 h. Cells were also transiently cotransfected with wild-type CREB (CREBwt), mutant CREB (CREB133, S133A), or a dominant-negative CREB (KCREB) construct along with the β-gal reporter construct. After 24 h of transfection, cells were treated with 0.1 μM RA for another 24 h.
Similarly, CRE-promoter activity was measured in H1734 cancer cells using luciferase as a reporter. Briefly, H1734 cells were seeded in 24-well tissue culture plates (2 × 104 cells/well). Cells were grown in RPMI medium with 10% FBS. When the cells reached 60-70% confluence, the cells were transiently cotransfected with CRE reporter construct along with β-gal reporter construct. After 4 h of transfection, the cells were incubated with medium for 24 h and then various log concentrations of RA (0.001, 0.01, 0.1, and 1 μM) were added to the medium for 24 h. Cells were subjected to lysis with 1× reporter lysis buffer (Promega, Madison, WI), and luciferase reporter and β-gal activities were determined by using a luminometer (Lumant LB 9507; EG & Berthold, Germany).
RNA Interference
SMARTpool-sequenced small-interference RNA (siRNA) targeting human RSK1 (Accession no. NM_002953), RSK2 (Accession no. NM_004586), RARα (Accession no. NM_000964), RARβ (Accession no. NM_000965), RARγ (Accession no. NM_000966), or RXRα (Accession no. NM_002957), and nonspecific control pool (siRNA negative control; Dharmacon RNA Technologies, Lafayette, CO) were diluted and stored according to the manufacturer's instructions. The siRNA SMARTpool used in this study contained four pooled RNA duplexes with “UU” 3′-overhangs and 5′-phosphate on the antisense strand. A mixture of several siRNAs ensured effective deletion of the target gene in the cell. H1734 cells at 50% confluence or NHTBE cells at 60-70% confluence were transfected with a final concentration of 100 nM SMARTpool siRNA or nonspecific control pool by using the siIMPORTER siRNA transfection reagent (Upstate Biotechnology) according to the manufacturer's instructions. The cells were treated with RA for 30 min or 24 h after 72 h of transfection, when target protein levels had been reduced by 70-80% as assessed by Western blot analysis.
Reverse Transcription-PCR
Total RNA was extracted using RNeasy mini-kits (Qiagen, Valencia, CA). The reverse transcription (RT) reaction was performed using 1 μg of total RNA that was reverse-transcribed into cDNA using a random hexamer primer (GeneAmp RNA PCR Core kit; Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. PCR conditions were an initial 95°C for 5 min, followed by 30 cycles of 95°C, 55°C, and 68°C for 30 s each. PCR for β-actin (Ambion, Austin, TX) was done as an internal control. Primer sequences were as follows: RARβ sense, AGCCTACGTGCCAAAAAAGG-3′, and antisense, 5′-TCTAGGTGTGGAGGCAAATGG-3′; and CREB sense, 5′-ACCATGGAATCTGGAGCCGAGAAC-3′, and antisense; 5′-CTGTAGGAAGGCCTCCTTGAAAGA-3′. PCR products were then separated in a 2% agarose DNA gel and stained with ethidium bromide.
RESULTS
CREB Is Activated by RA in a Time- and Dose-dependent Manner
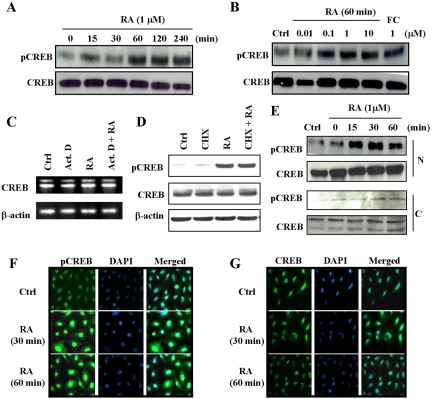
We used Western blot analysis to determine the time and dose responsiveness of RA on the activation of CREB. As shown in Figure 1A, RA induced activation of CREB in a time-dependent manner. CREB activation was detected as early as 15 min after RA treatment and reached a maximum at 1 h; pCREB remained activated for 4 h thereafter. The levels of CREB remained unchanged. It is unequivocally clear that the RA-induced phosphorylation of CREB occurred in a dose-dependent manner (Figure 1B). CREB was activated by as little as 10 nM RA. Forskolin, a known activator of PKA and an upstream activator of CREB, was used as a positive control to monitor the level of comparable activation of CREB. The levels of CREB remained unchanged.
Figure 1.
Time- and dose-dependent activation of CREB by RA. NHTBE cells grown in RA-deficient medium for 7 d and then treated with 1 μM RA for the indicated periods of time (A) or with various concentrations of RA for 60 min (B). Vehicle (Ctrl) and forskolin (FC, 1 μM) were used as a negative and a positive control, respectively. Whole-cell extracts were prepared and subjected to Western blot analysis using anti-CREB and anti-pCREB antibodies. To show the effects of actinomycin D and cycloheximide on the expression and activation of CREB, NHTBE cells were preincubated for 2 h with actinomycin D (Act. D) at 0.1 μg/ml or cycloheximide (CHX) at 5 μg/ml and then incubated with 1 μM RA or vehicle control for 4 h. CREB expression was measured using RT-PCR (C) and Western blot analysis (D). β-actin expression was assessed as an internal control in RT-PCR and as a loading control for Western blot analysis (C and D, respectively). The effect of RA on the activation of nuclear CREB. After NHTBE cells were treated with 1 μM RA for the indicated times, nuclear (N) and cytoplasmic (C) extracts were prepared and subjected to Western blot analysis using same antibodies (E). NHTBE cells grown in RA-deficient medium on cover glasses for 7 d were subjected to immunocytofluorescence analysis (F and G). Cells treated with vehicle (control) are shown in the top panels, those treated with 1 μM RA for 30 min in the middle panels, and those treated with 1 μM RA for 60 min in the bottom panels. The cells were fixed and incubated with anti-pCREB (F) or anti-CREB (G) antibodies. The figures shown are representative of three independent experiments.
To determine whether transcriptional activity is involved in the activation of CREB by RA, NHTBE cells grown in RA-deficient medium for 7 d were preincubated with actinomycin D (a general inhibitor of mRNA transcriptional synthesis) for 2 h, followed by incubation with 1 μM RA or vehicle control for 4 h. As shown in Figure 1C, actinomycin D had no effect on RA-induced CREB gene expression. To determine whether new protein synthesis is required for RA action in the CREB pathway, NHTBE cells grown in RA-deficient medium for 7 d were preincubated with cycloheximide (a general inhibitor of protein synthesis) for 2 h, followed by incubation with 1 μM RA or vehicle control for 4 h. As shown in Figure 1D, CREB expression and phosphorylation remained constant when protein synthesis was blocked. Thus, these results showed that the de novo synthesis of mRNA and protein is not required for the activation of CREB by RA, suggesting that RA's action in the CREB pathway is independent of transcriptional or translational regulation.
RA Induces Activation of Nuclear CREB
It has been reported that CREB is translocated after activation by calcium in vascular smooth muscle cells (Stevenson et al., 2001). To determine whether RA-activated CREB translocates from the cytoplasm to the nucleus, we determined the location of CREB and pCREB in nuclear and cytoplasmic fractions isolated from NHTBE cells treated with 1 μM RA for 15, 30, or 60 min. As shown in Figure 1E, the majority of CREB was localized within the nuclear fraction and that RA-activated CREB resided in the nuclear fraction. The activation was maximal within 15 min and continued for another 60 min with no change in the level of CREB in the nuclear fraction (Figure 1E). CREB phosphorylation and CREB levels were much weaker in the cytoplasmic fraction than in the nuclear fraction (Figure 1E). This result is consistent with that of immunocytochemical analysis showing that most of pCREB (Figure 1F) is localized in the nucleus. RA treatment for 30 min activated CREB (Figure 1F, middle panel), and the degree of activation was not changed by prolonged (60 min) treatment with RA (Figure 1F, bottom panel) compared with the untreated control (Figure 1F, top panel). The level of CREB was the same in cells treated for 30 min as in cells treated for 60 min (Figure 1G, middle and bottom panels). Therefore, the results unequivocally prove that the majority of CREB is localized in the nucleus and that RA-induced activation of nuclear CREB.
RA Activates DNA-binding Activity of CREB
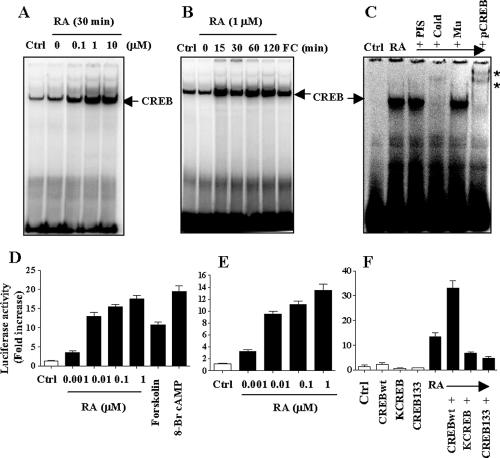
To determine whether RA-activated CREB binds to the specific CRE sequence in DNA, we performed EMSA analysis using NHTBE cells treated with different concentrations of RA for 30 min. As shown in Figure 2A, CREB activated by RA bound to the CRE sequence of a DNA probe in a dose-dependent manner. The maximum binding occurred at an RA concentration of 1 μM. Moreover, CREB activated by RA bound to the CRE sequence of the DNA probe in a time-dependent manner, as shown in Figure 2B. The binding occurred as early as 15 min after treatment and continued for 2 h. Forskolin treatment was used as positive control.
Figure 2.
Induction of DNA-binding activation of CREB and CRE-dependent transactivation by RA. NHTBE cells were then treated with different concentrations of RA as indicated for 30 min (A) or with 1 μM RA for the indicated times (B). Cells used as a control received an equivalent amount of solvent (DMSO). Vehicle (Ctrl) and forskolin (FC) treatment cells were used as negative and positive controls, respectively. After treatment, nuclear extracts were prepared and then assayed for CREB activation by EMSA with a CRE consensus oligonucleotide binding probe. (C) Supershift assay. Nuclear extracts were prepared from untreated cells and cells treated with 1 μM RA A supershift assay was performed by the addition of anti-pCREB antibody, unlabeled CRE oligonucleotide probe (Cold), unlabeled CRE-mutant oligonucleotide probe (Mu), or preimmune serum (PIS). The asterisk (*) indicates shifted bands. (D-F) Transient transfection analysis. (D) NHTBE cells were transiently transfected with a CRE promoter-driven luciferase-containing plasmid. Cells were incubated with different concentrations of RA, forskolin (1 μM), or 8-Br-cAMP (1 μM, as positive control) for 24 h. Cell lysates were assayed for luciferase activity. Vector control is designated Ctrl. (E) H1734 cells were transiently transfected with a CRE-promoter-driven luciferase-containing plasmid. NHTBE cells were transiently cotransfected with a CRE promoter-driven luciferase-containing plasmid and an expression vector for CREBwt, CREB133, or KCREB and a β-gal reporter construct. After transfection, cells were incubated with 1 μM RA or vehicle control (Ctrl) for 24 h. Cell lysates were assayed for luciferase activity (F). The data represent luciferase units normalized to β-gal in the same cell lysate. Data represent the mean ± SE of triplicate experiments, and the figures represent three independent experiments.
Supershift analysis was used to determine whether the retarded band visualized on EMSA in RA-treated cells was indeed due to the binding of CREB to the specific CRE sequence in DNA. As shown in Figure 2C, pCREB and CREB (unpublished data) antibodies shifted the band to a higher molecular mass, and addition of 100-fold excess of unlabeled (cold) CRE consensus oligonucleotide caused complete disappearance of the CREB band, showing that the mutant oligonucleotides could not compete for binding. Preimmune serum (PIS) had no effect. These results suggest that RA activation of CREB indeed results in specific binding to the CRE consensus DNA sequence and that most, if not all, CREB was in a phosphorylated form in the presence of RA, because pCREB almost completely supershifted the retarded band.
CRE-dependent Transcriptional Activation Is Induced by RA
RA-induced activation of CRE-dependent transcription was determined by transient transfection of NHTBE cells with a CRE-promoter-reporter. As shown in Figure 2D, RA increased CRE promoter activity in a dose-dependent manner, and the increase was 17-fold in cells treated with 1 μM RA. This increase in activity is as high as that induced by treatment with forskolin (11-fold) or 8-Br-cAMP (20-fold), which are known activators of CREB via PKA. The data suggest that RA-activated CREB binds its cognate CRE site in the promoter and induces transcriptional activity of CREB. It has been observed that primary epithelial cells usually have low transfection efficiency. To verify our findings in NHTBE cells, we performed transient transfection analysis with CRE-promoter reporter in lung cancer cell lines, H1734. As shown in Figure 2E, RA increased CRE promoter activity in a dose-dependent manner in the lung cancer cell lines. The increase was 14-fold in H1734 cells at 1 μM RA treatment over that of untreated control; results were consistent with those in NHTBE cells. Further, CRE-promoter activity after RA treatment was 12-fold that in the control (Figure 2F). This increase was further enhanced 19-fold by expression of wild-type CREB, whereas RA-induced CRE promoter activity was markedly suppressed by expression of dominant negative mutant forms of CREB, CREB133, and KCREB in NHTBE cells. These results clearly demonstrate that CRE transactivation by RA is dependent on CREB.
Role of RAR and RXR in Regulating Phosphorylation of CREB and CRE-dependent Transactivation by RA
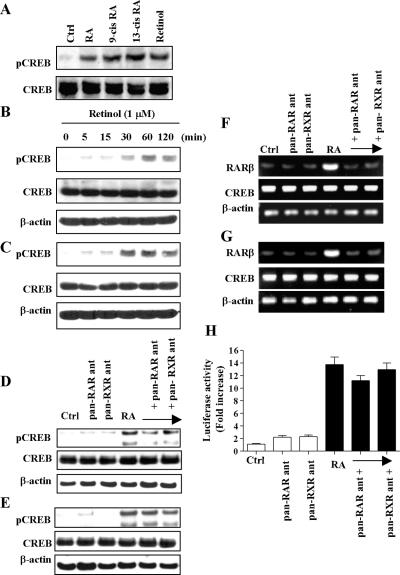
We next determined whether other types of retinoids, such as all-trans RA, 9-cis RA, 13-cis RA, and retinol, also activate CREB in NHTBE cells. CREB was equivalently activated after incubation for 30 min with each of these retinoids (Figure 3A). Of particular interest, retinol also activated CREB as soon as 30 min after treatment (Figure 3, B and C) and was maintained for at least 2 h in NHTBE (Figure 3B) and H1734 (Figure 3C) cells. Thus, it is unequivocally clear from the result that CREB activation by retinoids is not limited to all-trans RA.
Figure 3.
Retinoid receptor-independent activation of CREB by RA. (A) NHTBE cells were treated with 1 μM of all_-trans_ RA, 9-cis RA, 13-cis RA, or retinol for 30 min, and control cells received an equivalent amount of solvent (DMSO). Whole-cell extracts were prepared and subjected to Western blot analysis using anti-pCREB and anti-CREB antibodies. To show the effect of retinol on CREB activation, NHTBE (B) and H1734 (C) cells were treated with 1 μM retinol for the indicated times. Whole-cell extracts were prepared and subjected to Western blot analysis using the same antibodies. To show the effects of pan-RAR antagonist and pan-RXR antagonist on CREB activation induced by RA, NHTBE (D) and H1734 (E) cells were preincubated with pan-RAR or pan-RXR antagonist (10 μM) for 1 h and then incubated with RA (1 μM) for 30 min. Equal amounts of each whole-cell lysate were analyzed by Western blot using same antibodies. To show the effects of pan-RAR antagonist and pan-RXR antagonist on RARβ mRNA expression measured by RT-PCR. NHTBE (F) and H1734 (G) cells were preincubated with 10 μM of pan-RAR antagonist or pan-RXR antagonist for 2 h and then incubated with 1 μM RA or vehicle control for 6 h. Total RNA was used to measure RARβ mRNA level. Expression of β-actin was assessed as internal controls. (H) Effects of pan-RAR antagonist and pan-RXR antagonist on RA-induced CRE-dependent transactivation of the CRE-luciferase reporter. NHTBE cells were transiently transfected with a CRE promoter-driven luciferase-containing plasmid. Cells were incubated with 10 μM pan-RAR antagonist or pan-RXR antagonist for 2 h and then further incubated with 1 μM RA or vehicle control for 24 h. Data represent mean ± SE of triplicate experiments and figures are a representative of three independent experiments.
To determine whether RAR and RXR are involved in the activation of CREB by RA, NHTBE and H1734 cells were pretreated with 10 μM Ro 61-8431 (pan-RAR antagonist) or Ro 26-5405 (pan-RXR antagonist) for 1 h and then were incubated with RA for 30 min. Pretreatment with pan-RAR antagonist or pan-RXR antagonist could not block RA-induced CREB phosphorylation in either NHTBE or H1734 cells (Figure 3, D and E, respectively). These results indicate that the RAR and RXR receptors were not involved in the RA-induced CREB phosphorylation process. The activities of pan-RAR antagonist and RXR antagonist were verified by measuring the mRNA level of the RAR/RXR-mediated RA target gene RARβ. As shown in Figure 3, F and G, the expression of RARβ was almost completely blocked by either antagonist. In contrast, the expression of CREB mRNA was not changed by the antagonist treatment. This result demonstrates the efficiency of the antagonist in inhibiting RAR and RXR activity.
To determine whether RAR/RXR is involved in RA-induced CRE-mediated transcription, transient transfection analysis was performed using CRE-luciferase reporter. As shown in Figure 3H, CRE-luciferase activity was induced by RA and was not affected by the presence of pan-RAR antagonist or pan-RXR antagonist. It is unequivocally clear from these results that the antagonists could not block RA-induced CREB activation or CRE-mediated transcriptional activity, strongly suggesting that RAR and RXR receptors are not involved in RA-induced CRE transactivation.
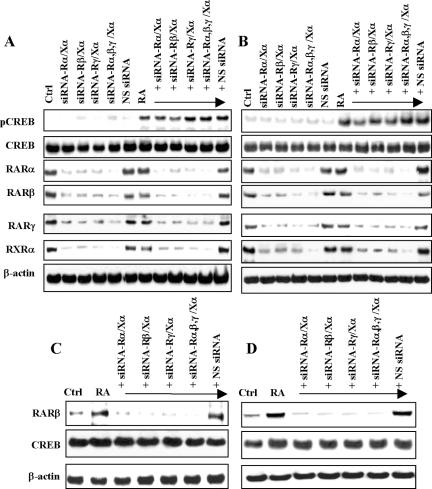
To further confirm the role of the RAR or RXR function in rapid activation of CREB by RA, we used the siRNA approach to deplete expression of RARα, RARβ, RARγ, and RXRα in NHTBE and H1734 cells. We silenced only the α isotype of RXR because it is known that RXRα plays a dominant role in RAR/RXR heterodimers. Cotransfection of SMARTpool-sequenced siRNA targeting human RAR and RXR receptors with a combination of RARα/RXRα, RARβ/RXRα, RARγ/RARα, or RARα, RARβ, RARγ/RXRα, into NHTBE and H1734 cells resulted in maximal silencing of the expression of their target genes 3 d after transfection (Figure 4, A-D). Western blot analysis of RA-induced active CREB levels after transfection by pooled RARα-, RARβ-, RARγ-, and RXRα-specific siRNAs revealed that RA-induced rapid phosphorylation of CREB was not affected by depletion of RARs and RXRα in NHTBE and H1734 cells (Figure 4, A and B, respectively). Silencing of RARs, RXRα, and nonspecific control pool (siRNA negative control) did not affect activation of CREB in the vehicle-treated control. This result suggests that RA-induced rapid CREB activation does not require RARs/RXRα. The specificity of the siRNA targeting each receptor was verified by determining protein levels of each receptor using the cell lysates isolated from the siRNA transfected cells (unpublished data). Although when the silencing of each receptor reached a maximum after 3 d, residual amounts of RARs and RXRα were still detected. To determine the genetic effect of the residual amounts of RAR/RXR, the expression of RARβ protein was measured. As shown in Figure 4C (NHTBE cells) and 4D (H1734 cells), the induction of RARβ expression by 24-h treatment with RA was detected in control cells and in cells transfected with nonspecific siRNA, but this induction was completely inhibited in cells without RAR/RXR heterodimers. These results indicate that even if there are some residual amounts of RARs or RXRα after siRNA transfection, they cannot elicit a biological response such as induction of the expression of the RAR/RXR target gene RARβ and do not affect RA-induced CREB activation. All these results together showed that RA-induced activation of CREB is independent of RAR and RXR.
Figure 4.
Effects of siRNA-mediated silencing of RAR and RXR receptor on RA-induced CREB activation. NHTBE (A) and H1734 (B) cells were transfected with siRNAs of RAR and RXR in the following combinations: siRNA of RARα and RXRα; siRNA-Rα/Xα and siRNA of RARβ and RXRα; siRNA-Rβ/Xα and siRNA of RARγ and RXRα; siRNA-Rγ/Xα and siRNA of RARα, RARβ, RARγ, or RXRα; siRNA-Rαβγ/Xα; or a nonspecific control pool (NS siRNA) alone. Three days after transfection, the cells were incubated with or without RA for 30 min (A and B) or further incubated for 24 h (C and D). After treatment, equal amounts of whole-cell lysates were isolated and subjected to Western blot analysis using the indicated antibodies (A-D). Equal loading was confirmed by stripping the blot and reprobing it for β-actin antibody. All figures are representative of three independent experiments.
Activation of CREB by RA Is Inhibited by PKC and ERK1/2 Inhibitors
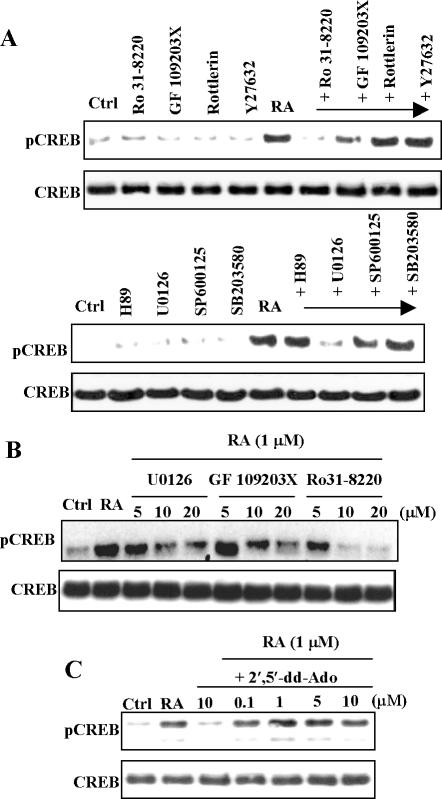
To determine which signaling pathway is involved in RA-induced activation of CREB, we treated NHTBE cells with various signaling inhibitors, such as Ro 31-8220, GF 109203X, and rottlerin (PKC inhibitors); Y27632 (Rock inhibitor); H89 (PKA inhibitor); SP600125 (JNK inhibitor); SB203580 (p38 MAPK inhibitor); and U0126 (MEK1/2 inhibitor). Figure 5, A and B, shows that a 1-h preincubation of these cells with Ro 31-8220, GF 109203X, or U0126 blocked RA induction of CREB activation, whereas other inhibitors did not block RA induction of CREB activation. Inhibitor treatment alone had no effect on CREB activation. We further demonstrated that ERK and PKC inhibitors block RA-induced CREB activation in a dose-dependent manner (Figure 5B). However, the H-89 (a PKA inhibitor) did not have any effect on the RA activation of CREB. To further determine the role of PKA in the activation, NHTBE cells treated with an inhibitor of adenylate cyclase, an upstream activator of PKA, 2′,5′-dideoxyadenosine (2′,5′-dd Ado). As shown in Figure 5C, 2′5′-dd Ado did not have any effect on RA-activation of CREB, suggesting the activation is independent of adenylate cyclase and PKA.
Figure 5.
Effect of signal transduction inhibitors on RA-induced activation of CREB. (A) NHTBE cells were preincubated with various indicated signal transduction inhibitors (10 μM) for 60 min and then treated with 1 μM RA or vehicle control for 30 min. Cells treated as a control received an equivalent amount of solvent (DMSO; Ctrl). (B) NHTBE cells were pre-incubated with various indicated concentrations of signal transduction inhibitors U0126, GF 109203X, or Ro 31-8220 for 1 h and then treated with 1 μM RA for 30 min. (C) NHTBE cells were preincubated with the indicated concentration of adenylate cyclase inhibitor for 1 h and then treated with 1 μM RA for 30 min. Control cells received an equivalent amount of solvent (DMSO; Ctrl). Whole-cell extracts were prepared and subjected to Western blot analysis using anti-pCREB and anti-CREB antibodies. All figures are representative of three independent experiments.
RA Induced Activation of CREB via MAPK Signaling Pathway
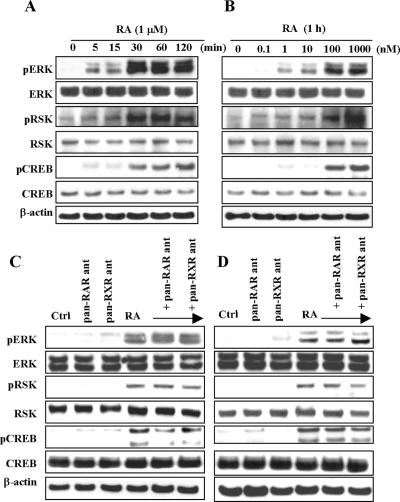
To further determine whether the MAPK pathway is involved in the RA-dependent signal transduction pathways leading to CREB phosphorylation, we determined the activation status of ERK1/2 and RSK. As illustrated in Figure 6, A and B, treatment with 1 μM RA caused strong and rapid increases in the amounts of phosphorylated ERK1/2, phosphorylated RSK, and phosphorylated CREB. This increase was observed as soon as 5 min after incubation with RA, was already maximal at 30 min, and was sustained for as long as 2 h (Figure 6A). A dose of 1 nM RA also caused increases in the amounts of phosphorylated ERK1/2, phosphorylated RSK, and phosphorylated CREB, but the maximal effect occurred at a dose of 1 μM (Figure 6B). In contrast, total levels of ERK1/2, RSK, and CREB as determined with an antibody recognizing the total proteins remained unaltered after RA treatment.
Figure 6.
Activation of ERK1/2, RSK, and CREB by RA. NHTBE cells were cultured without RA for 7 d and then incubated for the indicated time periods with 1 μM RA (A) or with the indicated concentration of RA for 1 h (B). Effects of the pan-RAR or pan-RXR antagonist on the activation of ERK1/2, RSK, and CREB were also determined. NHTBE (C) and H1734 (D) cells were preincubated with pan-RAR or pan-RXR antagonist (10 μM) for 1 h and then incubated with RA (1 μM) for 30 min. Equal amounts of whole-cell lysates were subjected to Western blot analysis. Equal loading was confirmed by stripping the blot and reprobing it for β-actin antibody. All figures are representative of three independent experiments.
To further determine whether the activation of ERK1/2 and RSK are dependent or not on RAR/RXR, NHTBE and H1734 cells were pretreated with 10 μM pan-RAR antagonist or 10 μM pan-RXR antagonist for 1 h and then were incubated with RA for 30 min. Pretreatment with pan-RAR antagonist or pan-RXR antagonist could not block RA-induced ERK, RSK, and CREB phosphorylation in either NHTBE or H1734 cells (Figure 6, C and D, respectively). Taken together, these results further confirmed that RA-induced CREB activation is independent of RAR and RXR receptor signaling.
RSK1/2 Are Indispensable for Activation of CREB by RA
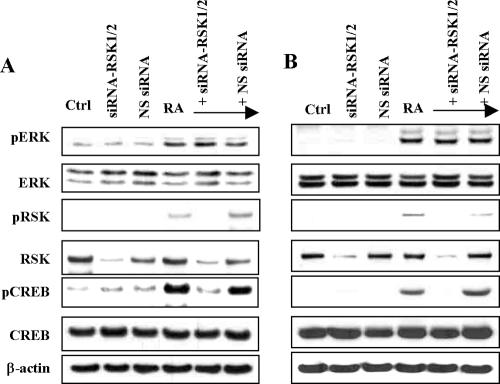
To further determine whether RSK is required for the activation of CREB by RA, we used the siRNA approach to silence expression of RSK1 and RSK2. Cotransfection into NHTBE and H1734 cells of a pool of four SMARTpool siRNAs that target both RSK1 and RSK2 resulted in maximal silencing of protein expression 3 d after transfection (unpublished data). As shown in Figure 7, A and B, depletion of RSK1/2 completely abolished RA-induced activation of CREB. Silencing of RSK1/2, however, did not affect ERK1/2 phosphorylation by RA, because ERK1/2 are upstream kinases of RSK. Transfection of siRSK1/2 or siRNA negative control did not affect CREB activation in control treatment. These results indicate that RSK1/2, an upstream activator of CREB, is indispensable for the activation of CREB by RA.
Figure 7.
Inhibition of RA-induced activation of CREB by siRSK. NHTBE cells (A) and H1734 cells (B) were cotransfected with RSK1 siRNA (siRSK1), RSK2 siRNA (siRSK2) or nonspecific control pool (NS siRNA). Three days after transfection, the cells were incubated with and also without 1 μM RA for 1 h, and then equal amounts of whole-cell lysates were subjected to Western blot using antibodies against using indicated antibodies. Equal loading was confirmed by stripping the blot and reprobing it for β-actin antibody. All figures are representative of three independent experiments.
DISCUSSION
Our findings demonstrate nongenomic receptor-independent RA action resulting in activation of a transcription factor CREB in primary bronchial epithelial cells. This genomic action of RA usually takes hours or even days. However, our work showed that activation of CREB occurred in as little as 15-30 min after RA treatment, implying that the time frame is too short to trigger genetic activation through transcription and translation. In fact, de novo synthesis of mRNA and protein was not required for the activation of CREB by RA, suggesting that RA's action in the CREB pathway is independent of genomic transcriptional or translational regulation. The rapid activation of CREB by RA resulted in increased DNA binding of the active CREB to its cognate CRE binding sequence. This result is in consistent with several studies showing increased DNA binding after CREB activation by various stimuli (Zhang et al., 2004) In contrast, there are few reports that activation of CREB did not lead to increased DNA binding activity in vitro (Hagiwara et al., 1993). Interestingly, most, if not all, of the CREB was present in an active phosphorylated form in the presence of RA, as suggested by the finding that most of the retarded band was supershifted by pCREB antibody.
Our data also suggest that RA activates CREB independently of RAR/RXR, because NHTBE and H1734 cells depleted of RARs/RXRα by means of siRNAs targeting RARα, RARβ, RARγ, and RXRα could still mediate RA-induced activation of CREB. Furthermore, selective pan-RAR and pan-RXR antagonists did not block RA activation of CREB. Moreover, several retinoids (9-_cis_-, 13-_cis_-, and all-trans RA) and even retinol equivalently activated CREB rapidly in as little as 30 min after treatment, suggesting that this action is not limited to all-trans RA. All these results strongly suggest that the action of RA on CREB activation is receptor independent.
The question arises what molecule could mediate this signaling event, if not conventional nuclear retinoid receptors. Our data suggest that PKC is involved in the RA-induced activation of CREB. Several studies provided evidence that RA directly binds to PKC (Radominska-Pandya et al., 2000; Kambhampati et al., 2003; Ochoa et al., 2003), although it is somewhat controversial whether the binding of RA to the PKC activates or inhibits PKC activity. RA activated PKCδ in several cancer cell lines (Kambhampati et al., 2003) or inhibited PKC activity (Radominska-Pandya et al., 2000). It was also reported to be biphasic, depending on the concentration of RA used, being an activator at a low dose and an inhibitor at a high dose (Lopez-Andreo et al., 2005). Thus, it is apparent that the effect of RA on PKC is specific to isotypes, cell types, and the concentration of RA. In our primary bronchial epithelial cells, PKC activity was required for CREB activation. However, further extensive studies are needed to answer the question of what RA target molecule initiates this signaling event in NHTBE cells.
ERKs, which are downstream targets of PKC, are involved in the activation of CREB by RA in NHTBE cells. In accordance with our findings, RA has been shown to activate ERK1/2 (Yen et al., 1998, 1999) and p38 MAPK (Alsayed et al., 2001). It was also recently reported that RA induced activation of CREB via ERK, resulting in expression of the c-fos gene, which does not contain RAREs but contains CRE in its promoter (Canon et al., 2004). However, it is apparent from our data that other MAPK, such as JNK and p38, are not involved in RA activation of CREB. In addition, PKA, one of the well-known CREB kinase, did not participate in RA-mediated CREB phosphorylation.
RA activated RSK is critically required for the activation of CREB, because omitting RSK1/2 by means of siRNAs targeting RSK1/2 completely abolished the activation. It has been shown that RSK is directly phosphorylated by ERK (Sturgill et al., 1988; Frodin and Gammeltoft, 1999). Most of the CREB is located in the nucleus upon stimulation by RA without obvious translocation of CREB from the cytoplasm to the nucleus. Therefore, activated RSKs must translocate to the nucleus and activate CREB at Ser-133 (Ginty et al., 1994; Xing et al., 1996). These findings are in agreement with our recent study that showed that interleukin-1β activated CREB through ERK/RSK1/CREB pathway in human airway epithelial cells (Song et al., 2003). To the best of our knowledge, this is the first report that RSKs are activated by RA in primary bronchial epithelial cells.
Supporting evidence of the activation of transcription factors by RA rather than RAR/RXR has been reported recently. In HL-60, RA increased binding of several transcription factors to their respective consensus sequences (Wang and Yen, 2004). Some of these transcription factors were shown to be involved in the regulation of the RA target gene, Burkitt's lymphoma receptor, cooperatively with RAR/RXR in HL-60 cells. However, it was not known whether RAR/RXR are required for the activation of these transcription factors. In our studies CREB activation by RA in bronchial epithelial cells did not require RAR/RXR.
Although the biological significance of the receptor-independent action of RA in normal bronchial epithelial cells remained unresolved, particularly its significance in the rapid activation of CREB, we speculate that RA may use CREB (or other transcription factors) to induce early RA response genes that do not have a classical RARE in their regulatory region. Alternatively, RA-activated CREB may induce genes to prepare the cells for initiation of a normal differentiation program in which RA receptors are not sufficient. In fact, we previously observed that expression of RARβ is below detectable level in RA-deficient squamous metaplastic NHTBE cells and that RA increased RARβ expression in these cells 4 h after treatment. RARβ is known to be regulated by CREB through CRE in its promoter (Kruyt et al., 1992). Because RARβ is also regulated by RAR/RXR through RARE in the promoter of the gene, there is an additional possibility that RA-activated CREB synergistically enhances expression of the genes required for induction of normal mucous cell differentiation. Further extensive studies are required to prove these concepts.
Our findings can be extended to explain pathobiologic changes in bronchial epithelia. Our preliminary observation showed that metaplastic squamous bronchial epithelial cells express very low levels of CREB (unpublished data). On the other hand, inappropriate use of this nongenetic RA signaling pathway may give an unfavorable growth advantage to certain malignant cells. Retinoids may sustain CREB activity in premalignant or malignant cells and thus promote malignant cell growth. One recent study showed that CREB acts as a proto-oncogene in leukemogenesis. Again, further studies are needed to test these speculations.
In summary, we demonstrated for the first time that RA activates CREB through activation of the PKC, ERK, and RSK in the absence of the influence of RAR/RXR in normal bronchial epithelial cells. Our findings present strong evidence of a function of RA in a signaling pathway quite distinct from that of the classical retinoic acid receptor paradigm. Receptor-independent nonclassical action of RA in bronchial epithelial cells can expand retinoids' effects on physiological processes, such as normal mucociliary differentiation of bronchial epithelia. Loss or abnormal use of this signaling pathway may cause pathobiologic changes in the epithelia.
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute Grant R01-HL-077556 (to J.S.K.), by National Institute of Environmental Health Sciences Grant K22-ES-000362 (to J.S.K.), and by National Cancer Institute Core Grant CA-16672.
Abbreviations used: CREB, cAMP response element-binding protein; EMSA, electrophoretic mobility shift assay; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; NHTBE, normal human tracheobronchial epithelium; pCREB, phosphorylated cAMP response element-binding protein; PKA, protein kinase A; RA, retinoic acid; RARE, retinoic acid response elements; RAR, retinoic acid receptor; RXR, retinoid X receptor; RSK, p90 ribosomal S6 kinase; siRNA, small interfering RNA.
References
- Aggarwal, S., Takada, Y., Mhashilkar, A. M., Sieger, K., Chada, S., and Aggarwal, B. B. (2004). Melanoma differentiation-associated gene-7/IL-24 gene enhances NF-kappa B activation and suppresses apoptosis induced by TNF. J. Immunol. 173, 4368-4376. [DOI] [PubMed] [Google Scholar]
- Alsayed, Y., Uddin, S., Mahmud, N., Lekmine, F., Kalvakolanu, D. V., Minucci, S., Bokoch, G., and Platanias, L. C. (2001). Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J. Biol. Chem. 276, 4012-4019. [DOI] [PubMed] [Google Scholar]
- Canon, E., Cosgaya, J. M., Scsucova, S., and Aranda, A. (2004). Rapid effects of retinoic acid on CREB and ERK phosphorylation in neuronal cells. Mol. Biol. Cell 15, 5583-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon, P. (1996). A decade of molecular biology of retinoic acid receptors. FASEB J. 10, 940-954. [PubMed] [Google Scholar]
- Deak, M., Clifton, A. D., Lucocq, L. M., and Alessi, D. R. (1998). Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17, 4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, K., and Montminy, M. (1998). CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 273, 32377-32379. [DOI] [PubMed] [Google Scholar]
- Fix, C., Jordan, C., Cano, P., and Walker, W. H. (2004). Testosterone activates mitogen-activated protein kinase and the cAMP response element binding protein transcription factor in Sertoli cells. Proc. Natl. Acad. Sci. USA 101, 10919-10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodin, M., and Gammeltoft, S. (1999). Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol. Cell. Endocrinol. 151, 65-77. [DOI] [PubMed] [Google Scholar]
- Giguere, V. (1994). Retinoic acid receptors and cellular retinoid binding proteins: complex interplay in retinoid signaling. Endocr. Rev. 15, 61-79. [DOI] [PubMed] [Google Scholar]
- Ginty, D. D., Bonni, A., and Greenberg, M. E. (1994). Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell 77, 713-725. [DOI] [PubMed] [Google Scholar]
- Gonzalez, G. A., Yamamoto, K. K., Fischer, W. H., Karr, D., Menzel, P., Biggs, W., 3rd, Vale, W. W., and Montminy, M. R. (1989). A. cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature 337, 749-752. [DOI] [PubMed] [Google Scholar]
- Gray, T., Nettesheim, P., Basbaum, C., and Koo, J. (2001). Regulation of mucin gene expression in human tracheobronchial epithelial cells by thyroid hormone. Biochem J 353, 727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, T. E., Guzman, K., Davis, C. W., Abdullah, L. H., and Nettesheim, P. (1996). Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am. J. Respir. Cell. Mol. Biol. 14, 104-112. [DOI] [PubMed] [Google Scholar]
- Gudas, L. J., Sporn, M. B., and Roberts, A. B. (1994). Cellular Biology and Biochemistry of the Retinoids. In: The Retinoids: Biology, Chemistry, and Medicine, ed. M. B. Sporn, A. B. Roberts, and D. S. Goodman, New York: Raven Press, 443-520.
- Hagiwara, M., Brindle, P., Harootunian, A., Armstrong, R., Rivier, J., Vale, W., Tsien, R., and Montminy, M. R. (1993). Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol. Cell. Biol. 13, 4852-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos, B., Imam, A., Chua, R., Swenson, C., Tong, G. X., Levi, E., Noy, N., and Hammerling, U. (2000). The cysteine-rich regions of the regulatory domains of Raf and PKC as retinoid receptors. J Exp Med 192, 835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam, A., Hoyos, B., Swenson, C., Levi, E., Chua, R., Viriya, E., and Hammerling, U. (2001). Retinoids as ligands and coactivators of PKC alpha. FASEB J. 15, 28-30. [DOI] [PubMed] [Google Scholar]
- Impey, S., McCorkle, S. R., Cha-Molstad, H., Dwyer, J. M., Yochum, G. S., Boss, J. M., McWeeney, S., Dunn, J. J., Mandel, G., and Goodman, R. H. (2004). Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119, 1041-1054. [DOI] [PubMed] [Google Scholar]
- Johannessen, M., Delghandi, M. P., and Moens, U. (2004). What turns CREB on? Cell Signal. 16, 1211-1227. [DOI] [PubMed] [Google Scholar]
- Kambhampati, S. et al. (2003). Activation of PKC delta by all-trans-retinoic acid. J. Biol. Chem. 278, 32544-32551. [DOI] [PubMed] [Google Scholar]
- Klemm, D. J., Watson, P. A., Frid, M. G., Dempsey, E. C., Schaack, J., Colton, L. A., Nesterova, A., Stenmark, K. R., and Reusch, J. E. (2001). cAMP response element-binding protein content is a molecular determinant of smooth muscle cell proliferation and migration. J. Biol. Chem. 276, 46132-46141. [DOI] [PubMed] [Google Scholar]
- Kolodziejski, P. J., Musial, A., Koo, J. S., and Eissa, N. T. (2002). Ubiquitination of inducible nitric oxide synthase is required for its degradation. Proc. Natl. Acad. Sci. USA 99, 12315-12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, J. S., Yoon, J. H., Gray, T., Norford, D., Jetten, A. M., and Nettesheim, P. (1999). Restoration of the mucous phenotype by retinoic acid in retinoid-deficient human bronchial cell cultures: changes in mucin gene expression. Am. J. Respir. Cell. Mol. Biol. 20, 43-52. [DOI] [PubMed] [Google Scholar]
- Kruyt, F. A., Folkers, G., van den Brink, C. E., and van der Saag, P. T. (1992). A cyclic AMP response element is involved in retinoic acid-dependent RAR beta 2 promoter activation. Nucleic Acids Res. 20, 6393-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Andreo, M. J., Torrecillas, A., Conesa-Zamora, P., Corbalan-Garcia, S., and Gomez-Fernandez, J. C. (2005). Retinoic acid as a modulator of the activity of protein kinase calpha. Biochemistry 44, 11353-11360. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf, D. J., and Evans, R. M. (1995). The RXR heterodimers and orphan receptors 83, 841-850. [DOI] [PubMed] [Google Scholar]
- Matthews, R. P., Guthrie, C. R., Wailes, L. M., Zhao, X., Means, A. R., and McKnight, G. S. (1994). Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Mol. Cell. Biol. 14, 6107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr, B., and Montminy, M. (2001). Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2, 599-609. [DOI] [PubMed] [Google Scholar]
- McDowell, E. M., Keenan, K. P., and Huang, M. (1984a). Effects of vitamin A-deprivation on hamster tracheal epithelium. A quantitative morphologic study. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 45, 197-219. [DOI] [PubMed] [Google Scholar]
- McDowell, E. M., Keenan, K. P., and Huang, M. (1984b). Restoration of mucociliary tracheal epithelium following deprivation of vitamin A. A quantitative morphologic study. Virchows Arch. B Cell Pathol. Incl. Mol. Pat. 45, 221-240. [DOI] [PubMed] [Google Scholar]
- Montminy, M. R., Gonzalez, G. A., and Yamamoto, K. K. (1990). Characteristics of the cAMP response unit. Metabolism 39, 6-12. [DOI] [PubMed] [Google Scholar]
- Nguyen, L. Q., Kopp, P., Martinson, F., Stanfield, K., Roth, S. I., and Jameson, J. L. (2000). A dominant negative CREB (cAMP response element-binding protein) isoform inhibits thyrocyte growth, thyroid-specific gene expression, differentiation, and function. Mol. Endocrinol. 14, 1448-1461. [DOI] [PubMed] [Google Scholar]
- Ochoa, W. F., Corbalan-Garcia, S., Eritja, R., Rodriguez-Alfaro, J. A., Gomez-Fernandez, J. C., Fita, I., and Verdaguer, N. (2002). Additional binding sites for anionic phospholipids and calcium ions in the crystal structures of complexes of the C2 domain of protein kinase calpha. J. Mol. Biol. 320, 277-291. [DOI] [PubMed] [Google Scholar]
- Ochoa, W. F., Torrecillas, A., Fita, I., Verdaguer, N., Corbalan-Garcia, S., and Gomez-Fernandez, J. C. (2003). Retinoic acid binds to the C2-domain of PKC(alpha). Biochemistry 42, 8774-8779. [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya, A., Chen, G., Czernik, P. J., Little, J. M., Samokyszyn, V. M., Carter, C. A., and Nowak, G. (2000). Direct interaction of all-transretinoic acid with PKC. Implications for PKC signaling and cancer therapy. J. Biol. Chem. 275, 22324-22330. [DOI] [PubMed] [Google Scholar]
- Reusch, J. E., Colton, L. A., and Klemm, D. J. (2000). CREB activation induces adipogenesis in 3T3-L1 cells. Mol. Cell. Biol. 20, 1008-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey, M., Bertera, S., Somers, J., Watkins, S., Zeleznik, A., and Walker, W. (2001). Delivery of a cyclic adenosine 3′,5′-monophosphate response element-binding protein (creb) mutant to seminiferous tubules results in impaired spermatogenesis. Endocrinology 142, 948-954. [DOI] [PubMed] [Google Scholar]
- Shankar, D. B., Cheng, J. C., Kinjo, K., Federman, N., Moore, T. B., Gill, A., Rao, N. P., Landaw, E. M., and Sakamoto, K. M. (2005). The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell 7, 351-362. [DOI] [PubMed] [Google Scholar]
- Song, K. S., Seong, J. K., Chung, K. C., Lee, W. J., Kim, C. H., Cho, K. N., Kang, C. D., Koo, J. S., and Yoon, J. H. (2003). Induction of MUC8 gene expression by interleukin-1 beta is mediated by a sequential ERK MAPK/RSK1/CREB cascade pathway in human airway epithelial cells. J. Biol. Chem. 278, 34890-34896. [DOI] [PubMed] [Google Scholar]
- Stevenson, A. S., Cartin, L., Wellman, T. L., Dick, M. H., Nelson, M. T., and Lounsbury, K. M. (2001). Membrane depolarization mediates phosphorylation and nuclear translocation of CREB in vascular smooth muscle cells. Exp. Cell Res. 263, 118-130. [DOI] [PubMed] [Google Scholar]
- Sturgill, T. W., Ray, L. B., Erikson, E., and Maller, J. L. (1988). Insulin-stimulated MAP-2 kinase phosphorylates and activates ribosomal protein S6 kinase II. Nature 334, 715-718. [DOI] [PubMed] [Google Scholar]
- Sun, P., Enslen, H., Myung, P. S., and Maurer, R. A. (1994). Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 8, 2527-2539. [DOI] [PubMed] [Google Scholar]
- Sung, J. Y., Shin, S. W., Ahn, Y. S., and Chung, K. C. (2001). Basic fibroblast growth factor-induced activation of novel CREB kinase during the differentiation of immortalized hippocampal cells. J. Biol. Chem. 276, 13858-13866. [DOI] [PubMed] [Google Scholar]
- Tan, Y., Rouse, J., Zhang, A., Cariati, S., Cohen, P., and Comb, M. J. (1996). FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15, 4629-4642. [PMC free article] [PubMed] [Google Scholar]
- Wang, J., and Yen, A. (2004). A novel retinoic acid-responsive element regulates retinoic acid-induced BLR1 expression. Mol. Cell. Biol. 24, 2423-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, J., Ginty, D. D., and Greenberg, M. E. (1996). Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273, 959-963. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K. K., Gonzalez, G. A., Biggs, W. H., 3rd, and Montminy, M. R. (1988). Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature 334, 494-498. [DOI] [PubMed] [Google Scholar]
- Yen, A., Roberson, M. S., and Varvayanis, S. (1999). Retinoic acid selectively activates the ERK2 but not JNK/SAPK or p38 MAP kinases when inducing myeloid differentiation. In Vitro Cell Dev. Biol. Anim. 35, 527-532. [DOI] [PubMed] [Google Scholar]
- Yen, A., Roberson, M. S., Varvayanis, S., and Lee, A. T. (1998). Retinoic acid induced mitogen-activated protein (MAP)/extracellular signal-regulated kinase (ERK) kinase-dependent MAP kinase activation needed to elicit HL-60 cell differentiation and growth arrest. Cancer Res. 58, 3163-3172. [PubMed] [Google Scholar]
- Zhang, B., Liu, S., Perpetua, M. D., Walker, W. H., and Harbrecht, B. G. (2004). Cytokines increase CRE binding but decrease CRE-mediated reporter activity in rat hepatocytes by increasing c-Jun. Hepatology 39, 1343-1352. [DOI] [PubMed] [Google Scholar]