Identification of the SAAT Gene Involved in Strawberry Flavor Biogenesis by Use of DNA Microarrays (original) (raw)
Abstract
Fruit flavor is a result of a complex mixture of numerous compounds. The formation of these compounds is closely correlated with the metabolic changes occurring during fruit maturation. Here, we describe the use of DNA microarrays and appropriate statistical analyses to dissect a complex developmental process. In doing so, we have identified a novel strawberry alcohol acyltransferase (SAAT) gene that plays a crucial role in flavor biogenesis in ripening fruit. Volatile esters are quantitatively and qualitatively the most important compounds providing fruity odors. Biochemical evidence for involvement of the SAAT gene in formation of fruity esters is provided by characterizing the recombinant protein expressed in Escherichia coli. The SAAT enzyme showed maximum activity with aliphatic medium-chain alcohols, whose corresponding esters are major components of strawberry volatiles. The enzyme was capable of utilizing short- and medium-chain, branched, and aromatic acyl-CoA molecules as cosubstrates. The results suggest that the formation of volatile esters in fruit is subject to the availability of acyl-CoA molecules and alcohol substrates and is dictated by the temporal expression pattern of the SAAT gene(s) and substrate specificity of the SAAT enzyme(s).
INTRODUCTION
In addition to their aesthetic qualities, fruits form an important part of our diet—mainly as a source of energy, vitamins, minerals, and antioxidants. Despite the long history of genetic selection of fruit, we still lack valuable information on the molecular, cellular, and physiologic events that control important processes such as flavor formation. Strawberry (Fragaria spp) is unusual because what is called the fruit actually originates from the expansion of the flower base (the receptacle) as a pseudocarp, with the real fruits (achenes) on the epidermal layer. During fruit development and maturation, both physical and morphological changes are often a result of changes in protein concentrations and activities, which may reflect shifts in overall mRNA abundance. The most pronounced changes involve alterations to fruit shape, size, texture, and pigmentation, which coincide with an increase in the soluble solids content and the production of natural aroma and flavor compounds (Perkins-Veazie, 1995).
In recent years, several groups have focused on identifying strawberry genes that are differentially expressed during ripening (Medina Escobar et al., 1997; Manning, 1998; Nam et al., 1999). The advent of molecular tools, such as cDNA microarray analysis, now adds a new dimension to gene expression studies. This type of analysis provides a powerful means for systematically studying the expression profiles of large subsets of genes in given tissues under specific physiologic and environmental conditions. Combining the appropriate biochemical knowledge with gene expression data can provide indirect evidence for the elucidation of gene function. DNA microarray technology has almost exclusively been used to study gene expression in humans and yeast (Schena et al., 1996; DeRisi et al., 1997), with preliminary expression studies reported for the model plant Arabidopsis (Schena et al., 1995; Ruan et al., 1998). However, to our knowledge, none has been reported for a commercial crop.
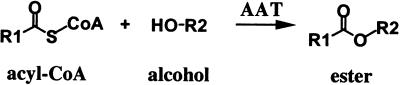
More than 300 compounds have been identified that can contribute to the complex process of aroma biosynthesis in strawberry (reviewed in Maarse, 1991). The major components of strawberry flavor and aroma can be grouped into several chemical classes, including acids, aldehydes, ketones, alcohols, esters, and lactones. Other contributing groups are sulfur compounds, acetals, furans, phenols, epoxides, and hydrocarbons (Zabetakis and Holden, 1997). Esters are one of the most important classes of volatile compounds in fruit flavor; in strawberry alone, more than a hundred different esters have been detected (Honkanen and Hirvi, 1990). Esterification is the result of transacylation from acyl-CoA to an alcohol (Figure 1). The enzyme catalyzing the reaction is termed an alcohol acyltransferase (AAT), a key enzyme in aroma biochemistry. The influence of esters (isoamyl and ethyl acetate) on beer flavor has made AAT one of the most important enzymes in the fermentation process performed by microorganisms. As such, it has been the subject of investigations with both yeast and fungi (Yamakawa et al., 1978; Yoshioka and Hashimoto, 1984; Yamauchi et al., 1989; Malcorps and Dufour, 1992; Fujii et al., 1994, 1996).
Figure 1.
General Scheme for the Esterification Reaction Catalyzed by AAT.
The enzyme AAT catalyzes the transfer of an acyl moiety from acyl-CoA onto the corresponding alcohol, resulting in the formation of an ester.
In plants, AAT activity has been investigated in both flowers and fruit. Volatile esters are constituents of floral scent. Purification of the acetyl-CoA:benzylalcohol acetyltransferase (BEAT) protein from flowers of Clarkia breweri and isolation of the gene encoding it have been reported (Dudareva et al., 1998). BEAT has a high affinity for aromatic alcohols such as benzyl alcohol and cinnamyl alcohol. In melon, banana, and strawberry, AAT proteins have been investigated in crude fruit extracts (Harada et al., 1985; Ueda et al., 1992; Perez et al., 1993, 1996; Olias et al., 1995). Ueda et al. (1992) concluded that the alcohol moieties of the esters produced by crude strawberry fruit extracts reflected the alcohols predominantly synthesized in the fruit and that the acid moieties reflected the acyl-CoA specificity of the AAT enzyme. Olias et al_._ (1995) compared strawberry and banana proteins possessing AAT activity and found clear differences between the alcohol and acyl-CoA specificity of the two enzymes. The strawberry AAT enzyme had high activity with hexanol and with acetyl- or butyl-CoAs. The banana enzyme, on the other hand, had high activity with butanol and acetyl-CoA but showed less activity with butyl-CoA. A clear correlation could be observed between the substrate preference of the enzymes and the volatile esters present in both fruits. In this study, we report the cloning and characterization of a fruit gene encoding an AAT capable of catalyzing the formation of volatile esters in strawberry fruit.
RESULTS
Expression Analysis with cDNA Microarrays
Microarrays were used to examine gene expression quantitatively during strawberry fruit development. In total, 1701 cDNA clones (probes) from strawberry fruit and 480 cDNA clones from petunia corolla were picked at random from cDNA libraries, amplified by polymerase chain reaction (PCR), and arrayed in duplicate on chemically modified microscope slides by using a robotic printing device. The petunia cDNAs were arrayed together with the strawberry cDNAs to assess the specificity of the hybridization assay in the experiments and to identify what genes from strawberry and petunia are highly conserved at the nucleotide level. Three experiments comparing strawberry fruit developmental stages—green with red, white with red, and turning with red—were performed. In each experiment, one mRNA population (target) was labeled with cyanine 3 (Cy3) and the other with Cy5. The labeled targets were then mixed and hybridized simultaneously to a microarray. To exclude artifacts, we performed a reciprocal labeling experiment with each pair of targets, using the same techniques of the first experiment except that the labels were exchanged. After hybridization, the fluorescence pattern of each microarray was recorded for the Cy3 and Cy5 fluorescent dyes, and clones that exhibited differential fluorescence (between the two dyes) were chosen for further analysis.
An image of the microarray after hybridization comparing the green and red stages and demonstrating the dynamic range of expression ratios between the probes arrayed is shown in Figure 2A. The microarray system exhibits high specificity for the hybridized target from strawberry. In a few cases, petunia probes produced a strong signal when the array was hybridized with strawberry targets (Figure 2B); three of these petunia clones (indicated by an intense red signal) were members of the polyubiquitin gene family, which is highly conserved in eukaryotes (∼85% homology among unrelated plant species) (Belknap and Garbarino, 1996).
Figure 2.
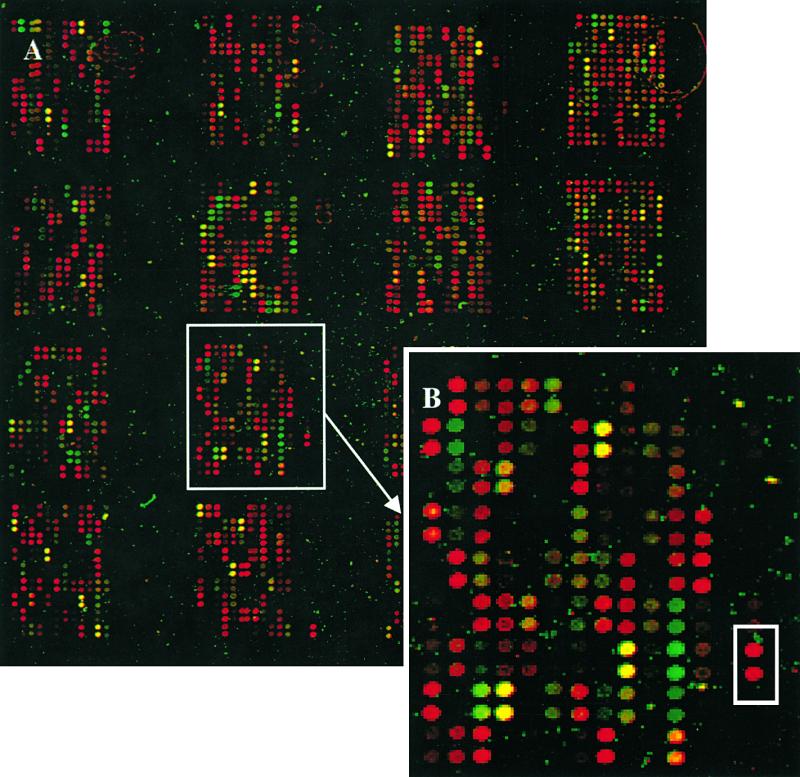
Strawberry and Petunia cDNA Microarrays.
(A) The strawberry and petunia cDNAs were spotted in a 4 × 4 format using a 16-pin print head. In each of the 16 subarrays, the first 12 columns from the left are strawberry probes (total of 1701, in duplicate), and the four columns from the right are petunia probes (total of 480, in duplicate). The array area is 17 × 17 mm. The image is a two-color overlay obtained with green-stage target (fluorescently labeled with Cy5) and red-stage target (fluorescently labeled with Cy3) cohybridized with a single microarray. In the superimposed image, the green-stage target is represented as a green signal and the red-stage target as a red signal. Signal intensities provide an estimate of expression levels, and the green or red spot colors correspond to higher transcript numbers in the green- or red-stage targets, respectively. Genes with no significant difference in expression between the two stages of development show an intermediate yellow or brown color.
(B) Enlarged image of the subarray boxed in (A). Several petunia probes hybridized strongly with targets prepared with mRNA from strawberry at the red fruit stage. One such cDNA clone (boxed) is a member of the highly conserved polyubiquitin gene family.
Detection of Differentially Expressed Genes
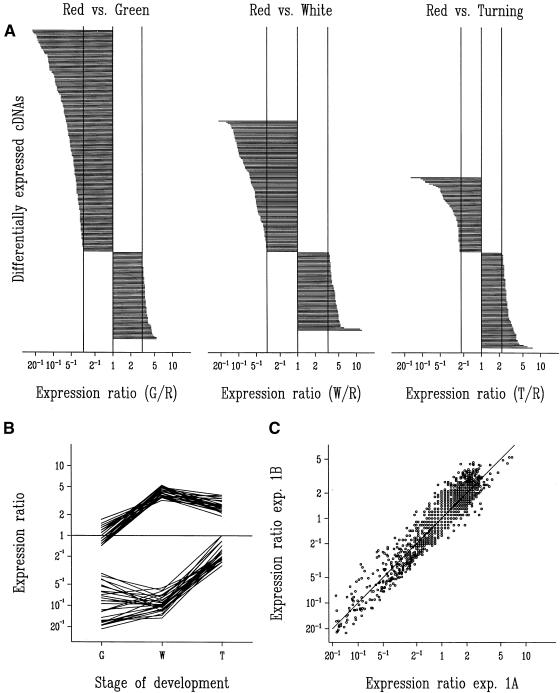
The expression data were transformed onto a log scale and evaluated by statistical analysis of variance models. Of 1701 strawberry cDNA clones, a total of 247, 168, and 137 cDNAs (significant at single test [per probe] P < 0.05; 126, 87, and 76 cDNAs at P < 0.01; 68, 42, and 27 cDNAs at P < 0.001) were differentially expressed during the green, white, and turning stages, respectively, in comparison with the red stage. In total, 401 cDNA clones were identified as being differentially expressed (the same clone can appear at more than one stage). Only expression ratios above a certain value or below the inverse of this value indicate a statistically significant upregulation or downregulation. For the three experiments—green/red, white/red, and turning/red—the threshold ratio for detection was 3.07, 3.32, and 2.24, respectively (at single test with P < 0.05). The maximum observed ratio was 22.5 in the comparison green/red (Figure 3A). The difference in the number of cDNA clones upregulated in the red stage (green/red [177], white/red [105], and turning/red [60]) or downregulated (green/red [70], white/red [63], and green/red [77]) in the three experiments can be attributed to differences in the physiologic states studied and to the source of the probes (red stage).
Figure 3.
Evaluation of Microarray Experiments.
(A) Expression ratios in green- (G), white- (W), and turning- (T) stage targets relative to red- (R) stage target identified a total of 401 probes corresponding to differentially expressed cDNA clones in the three experiments. The ratios are shown on a logarithmic scale and are ordered per experiment. The top series is cDNAs upregulated in the R stage (177, 105, and 60 cDNAs, respectively), and the bottom series is cDNAs downregulated in the R stage (70, 63, and 77 cDNAs, respectively). Vertical lines represent the least significant ratios at P < 0.05 (3.07, 3.32, and 2.24, respectively) and their reciprocals.
(B) Expression ratios for a series of homologous cDNAs in the green, white, and turning stages relative to the red stage (expression ratio marked as 1 for red stage). At the top are 27 cDNAs showing homology to a metallothionein gene (including clone B7); at the bottom, 27 cDNAs show homology to an auxin-induced gene (including clone C58). The GenBank accession numbers (nucleotide sequence) for strawberry cDNA clones B7 and C58 are AI795160 and AI795161, respectively.
(C) Precision of the microarray method is illustrated by the scatter plot of 1701 expression ratios estimated in two partial replicates of the green/red experiment. In the first experiment, green fruit target was labeled with Cy5 and red fruit target was labeled with Cy3 and hybridized with a microarray. A second replicate microarray was then hybridized with the dyes reversed (experiment [exp.] designated 1A). In experiment 1B, the first hybridization of experiment 1A was repeated with a third replicate microarray and was analyzed in a “reversed” manner with the results of the second microarray from experiment 1A.
Sequence alignment of the cDNA clones identified as differentially expressed by microarray analysis revealed that several shared a high degree of sequence similarity. Of 44 redundant cDNA clones (those having at least one other similar cDNA clone represented on the microarray) that were identified as differentially expressed, two were represented 27 times each. One of these clones (C58) was similar to a ripening-induced protein (GenBank accession number AJ001445) and the other (B7) to a putative metallothionein-like protein (GenBank accession number AJ001444). Both of these clones had been previously isolated from the wild strawberry (F. vesca), and their functions have yet to be determined (Nam et al., 1999). The redundancy of the two cDNAs on the microarray allowed us to evaluate the precision of the assay (Figure 3B). Expression ratios in the various developmental stages of both groups of 27 similar cDNA clones correlated well. The two families of cDNAs showed sequence identity in their overlapping regions. However, variability in signal may result from variable probe (tethered nucleic acid) or target (free nucleic acid) length, genetic redundancy (gene family), or some combination of these.
Variability, or interarray differences, was assessed by examining expression ratios of two microarrays hybridized with fluorescent targets used in one of the two green/red labeling experiments. Figure 3C shows a scatter plot of the expression ratios derived from both microarrays for all of the 1701 cDNAs. The fact that the ratios for all 1701 clones fall along the identity line indicates a low variability between the two separate experiments (the variation between the two partial replicates is characterized by a coefficient of variation among the ratios of 21%). After correction for redundancy, 239 unique differentially expressed cDNA clones were classified on the basis of their BLAST (Altschul et al., 1990) search output. We have adapted a systematic way of categorizing expressed sequence tags, as described previously (Tamames et al., 1996). The cDNA clones were divided into different categories: information (DNA, RNA, and protein), energy (primary and secondary metabolism), communication (hormonal regulation, detoxification, signal, stress, defense, and so forth), and unknowns. More than 30% of the cDNA clones were defined as unknowns. These clones had either (1) substantial sequence similarity to genes with unknown function only or (2) no match or only low similarity to other database sequences in the BLAST search. The contribution of clones related to energy pathways was striking. In the green/red experiment, of the 177 cDNA clones identified as being upregulated during the red stage, 53% of these were related to energy, 27% to communication, and 20% to information. A more thorough description of the strawberry microarray experiments (including sequence data) will be documented in a subsequent study.
Identification of the SAAT Gene by cDNA Microarray Technology and Detailed Expression Analysis
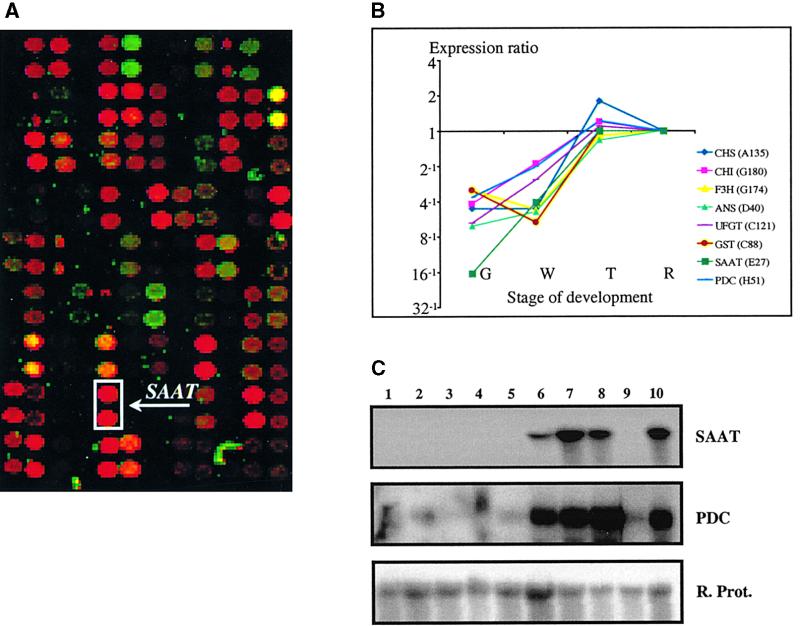
Fruit color is a clear indication of ripening, which in the majority of fruits is ordinarily accompanied by the accumulation of flavor and aroma components (Seymour et al., 1993). We postulated that cDNA clones showing expression profiles similar to those of color genes might be involved in flavor formation. One such cDNA clone, SAAT (for strawberry AAT), showed sequence similarity to genes encoding enzymes with acyltransferase activity. In fruit from several plant species as well as in other organisms such as yeast, biochemical evidence suggests that AAT catalyzes the final step in the synthesis of volatile esters (Ueda and Ogata, 1976; Harada et al., 1985). Quantitative microarray expression analysis revealed that SAAT had 16-fold greater expression during the red stage of fruit development than during the green stage (Figures 4A and 4B). Detailed RNA gel blot analysis (Figure 4C) showed that SAAT is exclusively expressed in the receptacle tissue (fruit without achenes). SAAT expression was first detected during the white stage of fruit development but was greatest between the turning and red stages, as deduced by both microarray and RNA gel blot analyses.
Figure 4.
Analysis of SAAT and PDC Gene Expression in Strawberry.
(A) An overlay image of a subset of a microarray hybridized with samples originating from green and red strawberry fruit mRNA. As given for Figure 2, signal intensities provide an estimate of expression levels, and green or red spot colors correspond to higher transcript levels in the green- or red-stage samples, respectively. The SAAT cDNA appeared as an intense red signal (boxed) and is marked by an arrow (each cDNA was arrayed in duplicate). The physical size of the subarray is ∼4 × 2.5 mm.
(B) Expression profiles of SAAT, PDC, and six strawberry pigmentation-related genes during strawberry development, as detected in the microarray experiments. Expression ratios in green, white, and turning stages are relative to the red stage (expression ratio marked as 1 for the red stage).The strawberry genes and their GenBank accession numbers are as follows: CHS, chalcone synthase (AI795154); CHI, chalcone flavanone isomerase (AI795155); F3H, flavanone-3β-hydroxylase (AI795156); ANS, anthocyanidin synthase (AI795157); UFGT, UDP-glucose:flavonoid-3-_O_-glucosyltransferase (AI795158); GST, glutathione _S_-transferase (AI795159); _SAAT, s_trawberry AAT (AF193789); PDC, pyruvate decarboxylase (AF193791). G, green stage; R, red stage; T, turning stage; W, white stage.
(C) RNA gel blot analysis of the expression of SAAT and PDC in different tissues of strawberry. Lane 1, root; 2, petiole; 3, leaf; 4, flower; 5, green fruit; 6, white fruit; 7, turning fruit; 8, red fruit; 9, achenes; and 10, dark-red fruit. The blot was hybridized with the SAAT probe and then rehybridized with a strawberry cDNA probe showing homology to a gene encoding a ribosomal protein (R. Prot.).
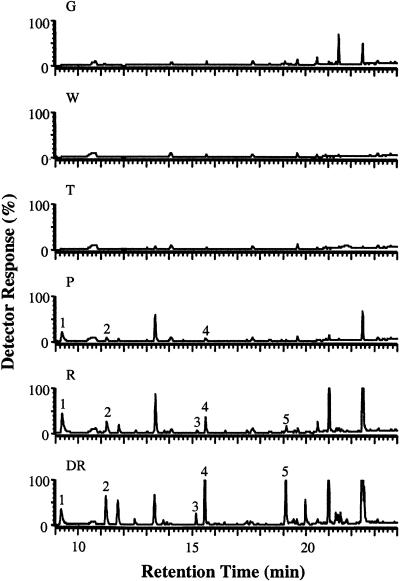
Gas chromatography–mass spectrometry (GC-MS) analysis of different stages of strawberry fruit development (Figure 5) showed that the first detectable sign of release of volatile esters was during the pink stage (between the turning and red stages), the greatest amounts being attained during the dark-red stage. We also investigated the expression profile of a strawberry cDNA showing sequence similarity to an Arabidopsis gene encoding pyruvate decarboxylase (PDC) (GenBank accession number U71122), which may play a role in providing precursors for the formation of ethyl esters (Figures 4B and 4C). The PDC gene expression profile correlated well with that of SAAT during fruit development. However, PDC gene expression was not confined to the receptacle tissue. DNA gel blot analysis suggested that SAAT is not a member of a multigene family (data not shown).
Figure 5.
Volatile Ester Emission during Strawberry Fruit Development.
GC-MS chromatograms (detector response was 100% of 2 × 106 total ion counts) of volatiles in vivo released by strawberry fruits (cv Elsanta) at different stages of development. Developmental stages: DR, dark red; G, green; P, pink; R, red; T, turning; W, white. The five main volatile esters detected are marked with numbers: 1, methyl hexanoate; 2, hexyl acetate; 3, hexyl butanoate; 4, octyl acetate; and 5, octyl butyrate.
Sequence Analysis of SAAT
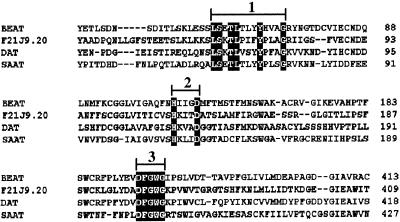
The full-length SAAT cDNA clone is 1618 bp, encoding a polypeptide of 452 amino acid residues with a predicted molecular mass of 50.7 kD. Although a search for sequence homology revealed low sequence identity (29% identical amino acids) between SAAT and its closest sequence homolog (F21J9.20, an Arabidopsis genomic clone; GenBank accession number AC000103), SAAT was found to contain several conserved sequences identifying it as belonging to a plant superfamily of multifunctional acyltransferases (Figure 6). Members of this gene family have a catalytic reaction mechanism related to the ancient chloramphenicol 3-_O-_acetyltransferase (CAT) and dihydrolipoyl _S-_acetyltransferase (DHLAAT) class of enzymes (St. Pierre et al., 1998). Several members of this superfamily with protein sequences showing similarity to SAAT were previously enzymatically characterized and demonstrated to have acyltransferase activity. The Catharanthus roseus gene for deacetylvindoline 4-_O_-acetyl-transferase (DAT; GenBank accession number AF053307) encodes an enzyme catalyzing the biosynthesis of vindoline from acetyl-CoA and deacetylvindoline (St. Pierre et al., 1998). Another gene with some sequence similarity (19.4% identity of amino acids) is BEAT (GenBank accession number AF043464), which encodes the above-mentioned C. breweri enzyme catalyzing the formation of the ester benzyl acetate from benzyl alcohol and acetyl-CoA (Dudareva et al., 1998).
Figure 6.
Protein Sequence Alignment of SAAT and Three Other Related Plant Proteins.
The related protein sequences aligned were F21J9.20 and genomic sequences from Arabidopsis bacterial artificial chromosome F21J9 (GenBank accession number AC000103); BEAT, C. breweri acetyl-CoA:benzylalcohol acetyltransferase _(_GenBank accession number AF043464); and DAT, C. roseus deacetylvindoline 4-_O_-acetyltransferase (GenBank accession number AF053307). The alignments delineate the following conserved motifs: 1, L-S-Xaa-T-L-Xaa-Xaa-Xaa-Y-Xaa-Xaa-Xaa-G; 2, H-Xaa-Xaa-Xaa-D; and 3, DFGWG.
Sequence alignment of the proteins encoded by these genes with the protein sequence of SAAT illustrates the main consensus sequences shared between these proteins (Figure 6). The H-Xaa-Xaa-Xaa-D (Xaa indicates a variable-identity amino acid) motif, corresponding to residues 156 to 160 in SAAT, is the most conserved consensus sequence present even in acyltransferases such as CAT and DHLAAT (Reed and Hackert, 1990) and in carnitine and choline acyltransferases (Brown et al., 1994) from nonplant systems. A second highly conserved motif is DFGWG (corresponding to residues 388 to 392 in SAAT), located near the C terminus. A third consensus sequence, L-S-Xaa-T-L-Xaa-Xaa-Xaa-Y-Xaa-Xaa-Xaa-G, corresponds to residues 66 to 78 in SAAT and is located at the N terminus. This domain is completely identical between the SAAT and BEAT proteins. In addition, the eight residues (LSETLTLY) of the third domain are present in the green alga Chlorella vulgaris acetyl-CoA carboxylase (GenBank accession number BAA57908) carboxyl transferase β subunit. Acetyl-CoA carboxylase is a biotinylated enzyme that catalyzes the ATP-dependent formation of malonyl-CoA from acetyl-CoA and bicarbonate. This function signifies the importance of the domain in reactions using acetyl-CoA as cosubstrate.
Functional Expression of the SAAT cDNA in Escherichia coli
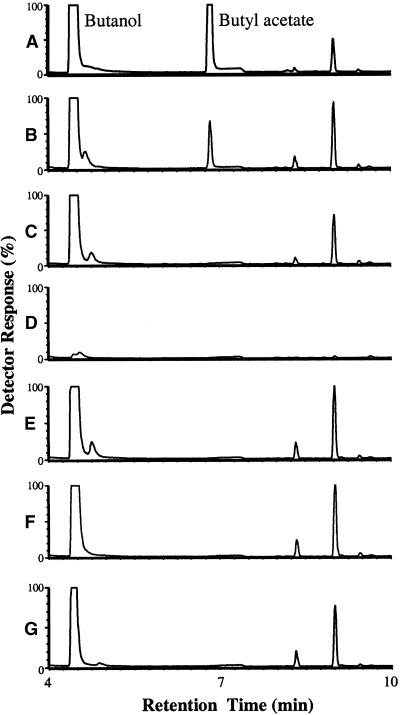
The entire coding region of the SAAT cDNA cloned in frame with a polyhistidine affinity tag (His-Tag; see Methods) was used for heterologous expression studies in E. coli. Protein gel blot analysis revealed a single 58-kD band in the SAAT sample that could not be detected in the control samples (data not shown). The predicted molecular mass of the recombinant SAAT His-Tag protein is 54 kD (50.7 plus 3.8 kD), so the band is within the expected size range. Fractions of the SAAT and control samples eluting from the His-Tag columns were tested for AAT activity. Acetyl-CoA and butanol were used primarily as substrates for the analysis. GC-MS profiles showed an additional peak arising from the SAAT fraction corresponding to the ester butyl acetate that could not be detected in the control fractions (Figure 7). This confirmed that the formation of butyl acetate was a direct result of the activity of the SAAT recombinant protein.
Figure 7.
Verification of Ester Formation by the SAAT Protein by GC-MS.
GC-MS (detector response, 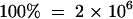 total ion counts) of volatiles produced in the incubation conditions below.
total ion counts) of volatiles produced in the incubation conditions below.
(A) Butanol and butyl acetate standards.
(B) SAAT protein plus butanol plus acetyl-CoA.
(C) As given for (B) but with protein absent.
(D) As given for (B) but with butanol absent.
(E) As given for (B) but with acetyl-CoA absent.
(F) Green fluorescent protein plus butanol plus acetyl-CoA.
(G) Empty pRSET B vector eluate plus butanol plus acetyl-CoA.
Other visible peaks are impurities from the butanol substrate.
Characterization of the SAAT Protein Enzymatic Activity
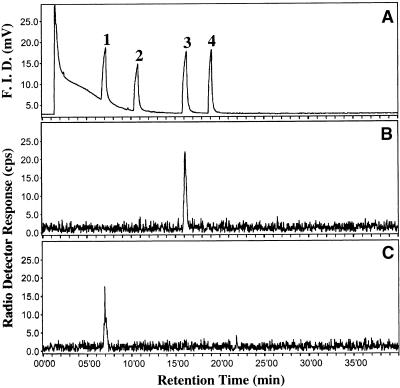
We assessed the substrate specificity of the SAAT recombinant protein in vitro by supplying a range of alcohols and acyl-CoAs and then analyzing the volatiles produced, using both radioactivity detection–gas chromatography (radio-GC) and GC-MS. The enzyme activities with different alcohols and acetyl-CoA as the cosubstrate were assessed by liquid scintillation counting and radio-GC analysis with 14C-acetyl-CoA as cosubstrate (Figure 8 and Table 1). Incubations of SAAT with 14C-acetyl-CoA and 1-octanol produced 14C-1-octylacetate (Figures 8A and 8B), and incubations with 1-hexanol produced 14C-1-hexylacetate (Figures 8A and 8C).
Figure 8.
SAAT-Catalyzed Ester Formation from 14C-Acetyl-CoA.
(A) Flame ionization detector (F.I.D.) signal of unlabeled standards of peak 1, hexylacetate; peak 2, 1-hexanol; peak 3, octylacetate; and peak 4, 1-octanol.
(B) and (C) Radiodetector signal of labeled products formed by the SAAT protein from 0.1 mM 14C-acetyl-CoA and alcohols with (B) 2 mM 1-octanol or (C) 2 mM 1-hexanol.
Table 1.
Substrate Specificity of the SAAT Recombinant Enzyme toward Different Types of Alcohols
| Alcohol | Carbon No. | Ester Formed | Activitya | Reported in Strawberryb |
|---|---|---|---|---|
| Methanol | C1:0 | Methyl acetate | 1.11 ± 0.26 | Yes |
| Ethanol | C2:0 | Ethyl acetate | 0.62 ± 0.10 | Yes |
| 1-Propanol | C3:0 | 1-Propyl acetate | 2.60 ± 0.26 | Yes |
| 2-Propanol | C3:0 | 2-Propyl acetate | 1.30 ± 0.15 | Yes |
| 1-Butanol | C4:0 | 1-Butyl acetate | 2.29 ± 0.25 | Yes |
| 2-Butanol | C4:0 | 2-Butyl acetate | 3.11 ± 0.02 | Yes |
| 3-Methyl-1-butanol (isoamylalcohol) | C5:0 | 3-Methyl-1-butyl acetate (isoamyl acetate) | 3.68 ± 0.25 | Yes |
| 1-Hexanol | C6:0 | 1-Hexyl acetate | 8.44 ± 0.37 | Yes |
| _cis_-2-Hexen-1-ol | C6:1 | _cis_-2-Hexenyl acetate | 6.05 ± 0.55 | No |
| _cis_-3-Hexen-1-ol | C6:1 | _cis_-3-Hexenyl acetate | 4.06 ± 0.14 | Yes |
| _trans_-2-Hexen-1-ol | C6:1 | _trans_-2-Hexenyl acetate | 9.20 ± 0.65 | Yes |
| _trans_-3-Hexen-1-ol | C6:1 | _trans_-3-Hexenyl acetate | 1.25 ± 0.07 | No |
| 1-Heptanol | C7:0 | Heptyl acetate | 14.89 ± 4.12 | No |
| 1-Octanol | C8:0 | 1-Octyl acetate | 16.36 ± 2.69 | Yes |
| 1-Nonanol | C9:0 | 1-Nonyl acetate | 14.00 ± 0.11 | No |
| 1-Decanol | C10:0 | 1-Decyl acetate | 7.79 ± 0.10 | Yes |
| Furfurylalcohol | C5:2 | Furfuryl acetate | 0.72 ± 0.06 | No |
| Benzylalcohol | C7:3 | Benzyl acetate | 0.68 ± 0.04 | Yes |
| 2-Phenylethylalcohol | C8:3 | 2-Phenylethyl acetate | 1.58 ± 0.12 | Yes |
| Linalool | C10:2 | Linalyl acetate | NDc | No |
The enzyme assays were shown to be linear with protein concentration and reaction time for up to 60 min for acetyl-CoA concentrations as low as 0.02 mM (in combination with 20 mM octanol) and hexanol concentrations as low as 2 mM (in combination with 0.1 mM acetyl-CoA). The enzyme exhibited a broad pH range (pH optimum of ∼8.3). With acetyl-CoA, the enzyme accepted a broad range of alcohols as substrate (Table 1). Enzyme activity increased with increasing carbon chain length of the alcohol up to octanol, after which the activity declined. Clear differences in enzyme activity were detected among the four isomers of hexenol tested: _trans_-2-hexenol was a better substrate than _cis_-2-hexenol, whereas _cis_-3-hexenol was a better substrate than _trans_-3-hexenol. The effect of the position of the hydroxy moiety of the alcohol on SAAT activity varied: 1-propanol was a better substrate than 2-propanol, but 2-butanol was a better substrate than 1-butanol. SAAT also accepted the branched primary alcohol isoamyl alcohol. Activity was also detected with aromatic (benzyl and phenylethyl) and cyclic (furfuryl) alcohols, although these activities were much lower than with 1-octanol (4 to 10%). In contrast, no activity could be detected (by GC-MS or liquid scintillation counting) with the terpene alcohol linalool. The ability of SAAT to use different acyl-CoAs as substrates was qualitatively determined by GC-MS. The SAAT recombinant enzyme was capable of using acyl-CoAs up to C10, branched acyl-CoAs, and aromatic acyl-CoAs in combination with 1-propanol or 1-butanol as alcohols (data not shown).
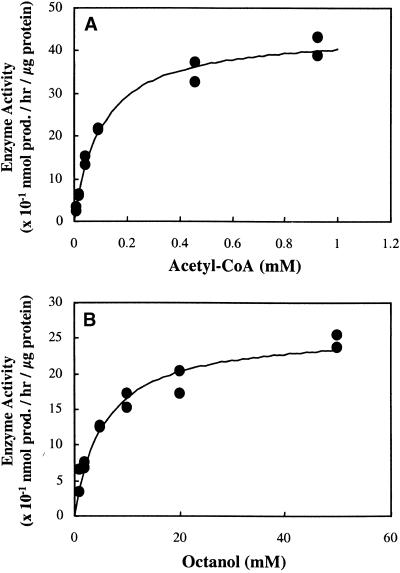
The kinetic properties of SAAT were determined for acetyl-CoA and five different alcohols by liquid scintillation counting. The substrate–activity relationships showed the typical saturation curve (Figure 9). The apparent _K_m and _V_max for acetyl-CoA were 104.2 μM and 4.5 nmol of product hr–1 μg–1 protein, respectively (Figure 9A). The apparent values of _K_m (in millimoles per liter) and _V_max (in nanomoles of product per hour per microgram of protein) for 1-butanol were 46.1 and 0.4, for 1-hexanol 8.9 and 0.6, for 1-octanol 5.7 and 2.6 (Figure 9B), for _trans_-2-hexenol 16.8 and 1.1, and for _cis_-2-hexenol 17.9 and 0.6, respectively. The SAAT enzyme characteristics support the data presented in Table 1, with the enzyme activity increasing with carbon chain length from butanol through hexanol to octanol (with a concomitant increase in _V_max and a decrease in _K_m). The difference in activity with the hexenol isomers appears to result from differences in _V_max values rather than in affinity.
Figure 9.
AAT Activity of SAAT as a Function of Substrate Concentration for the Formation of Octyl Acetate.
(A) For acetyl-CoA (in the presence of 20 mM 1-octanol).
(B) For 1-octanol (in the presence of 0.1 mM acetyl-CoA).
Equations for fitted curves are (A) 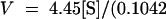 +
+ 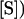 ,
, 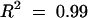 and (B)
and (B) 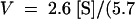 +
+ 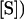 ,
, 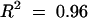 . Data were obtained by liquid scintillation counting. prod., product.
. Data were obtained by liquid scintillation counting. prod., product.
DISCUSSION
Accessing Genetic Information by Using cDNA Microarrays
In this study, we have reported the successful application of DNA microarray technology to help identify a novel flavor gene with subsequent verification of function. The capability of the technology to act as a powerful tool to link gene to function will no doubt aid future research into plant functional genomics. Statistical analysis of experimental data confirmed the reproducibility of our microarray system. The system allows for a 95% reliable detection of four- to fivefold differences in mRNA expression levels, with a lower limit of two- to threefold differences, as detected in other nonplant systems (Schena et al., 1996; DeRisi et al., 1997).
The use of simultaneous, two-color hybridization schemes increases the precision of the differential measurements by eliminating artifacts associated with comparing separate microarrays (Schena et al., 1995). However, because ratios obtained with a single microarray are nonetheless generated with two separate scans, artifacts may still arise as a result of differences in the two channels that are not related to altered mRNA quantities. The use of reciprocal labeling schemes for the two targets in independent experiments helps correct for dye-related differences, sample contaminants, dust, different laser settings, nonlinearities of photomultiplier tubes, and other artifacts that can erroneously affect the fluorescent readout of the two channels. Our microarray assays exhibited high specificity to the strawberry targets, as demonstrated by the lack of hybridization to the petunia clones (except for the highly homologous polyubiquitin cDNA clones, which produced a strong signal). Such homologous genes offer the possibility for future analyses of syntenic expression between plant species.
The presence of different types of volatile esters in ripe fruit of many species raises several questions concerning the mechanisms of ester formation and the genetic controls governing the entire process. A strawberry gene (SAAT) isolated from the ripe red fruit was shown in the course of this study to encode an enzyme with AAT activity. The evidence for the identification of SAAT as a putative AAT originated from microarray expression profiling data, coupled with biochemical data concerning volatile ester release during fruit development. SAAT gene expression is fruit specific, commencing at the white stage of fruit ripening, before any detectable formation of volatile esters in strawberry (cv Elsanta), and reaching maximal levels between turning and red stages. This result correlates with AAT activities reported for several commercial strawberry cultivars during fruit development (Perez et al., 1996). The expression pattern supports the proposition that the absence of ester formation in immature stages of fruit ripening results from a lack of ester-producing enzymes (Yamashita et al., 1977). Pyruvate decarboxylase has been proposed to supply precursors for the formation of esters (Zhou et al., 1995), and the fact that its expression profile is similar to that of SAAT suggests that both may be under the same genetic regulation.
Biochemical Evidence for Involvement of SAAT in the Formation of Fruity Esters
Extensive in vitro enzyme assays revealed that a comprehensive array of acyl-CoA molecules and alcohols (both short and long chains with even and uneven numbers of carbons; aromatic, cyclic, and aliphatic; branched and unbranched; saturated and unsaturated) may serve as substrates for the SAAT enzyme in vivo. Esters identified by our analyses (1-butyl acetate, 1-hexyl acetate, and 1-octyl acetate) as the major volatiles emitted in the latter stages of fruit ripening were demonstrated to be genuine products of the recombinant SAAT enzyme. Perez et al. (1993)(1996) partially purified an AAT protein from strawberry (cv Chandler) fruits and tested AAT activity in several strawberry cultivars. The enzyme described by Perez et al. showed broad substrate specificity, with a preference for the medium-chain alcohol hexanol and a lower activity with shorter chain alcohols (its activity with ethanol was 25% of that with hexanol). We also included longer chain alcohols in our enzyme assays and found even greater activity with 1-heptanol and 1-octanol, but less with 1-nonanol and 1-decanol than with 1-hexanol. Perez et al. (1996) reported only 14% activity with 3-hexen-1-ol relative to 1-hexanol; however, it is unclear whether they used the _cis_- or _trans_-isomer of 3-hexenol. The activity of the SAAT enzyme with _cis_-3-hexen-1-ol and with _trans_-3-hexen-1-ol was 46 and 14%, respectively, of that with 1-hexanol (Table 1). Thus, the relative activity of AAT with 3-hexenol reported by Perez et al. agrees closely with the relative activity we found for _trans_-3-hexenol.
The SAAT enzyme was also capable of using the C10 decanoyl-CoA as a cosubstrate. This ability may account for the presence of long-chain esters of this type in strawberry (Honkanen and Hirvi, 1990). Although the acyclic monoterpene alcohol linalool has been identified as a component of volatiles emitted by the strawberry cultivar Elsanta (Ulrich et al., 1997), linalool was not used as substrate by the recombinant SAAT enzyme. This is in agreement with the absence of linalool esters in strawberry volatile profiles, including that of the Elsanta cultivar (Honkanen and Hirvi, 1990).
The _K_m value of SAAT for acetyl-CoA (104 μM) was somewhat higher than that reported by Perez et al. (1996) for a partially purified strawberry AAT (65 μM) and by Harada et al. (1985) for a banana AAT (50 μM). The calculated _K_m value of SAAT for some alcohols (especially that for the best substrate, 1-octanol, or 5.7 mM) are comparable with the data for butanol (3 mM) reported by Perez et al. (1996). However, the values are higher than the 0.4 mM for isoamyl alcohol reported by Harada et al. (1985) for banana AAT. Although our _K_m data are comparable with the data for AATs reported in the literature, they are ∼1000-fold higher than the _K_m values reported for enzymes with a high substrate specificity, such as amorpha-4,11-diene synthase (_K_m of 0.6 μM; Bouwmeester et al., 1999b) and (–)-α-gurjunene synthase (_K_m of 5.5 μM; Schmidt et al., 1999). Even cytochrome P-450 enzymes that catalyze the hydroxylation of (+)- and (−)-limonene to _trans_-carveol show _K_m values between 10 and 20 μM (Karp et al., 1990; Bouwmeester et al., 1999a). All of these terpene biosynthetic enzymes with low _K_m values show high substrate specificity. Further research is required to find out whether the relatively high _K_m values of AATs can be correlated with the relatively low substrate specificity of these enzymes.
The wide range of alcohols and acyl-CoAs accepted by the SAAT enzyme cannot completely explain the differences in ester composition between the developmental stages and different strawberry cultivars. An important factor is the availability of substrates, which may depend on the activity of various catabolic pathways (e.g., lipid breakdown). Another factor that may influence the ester composition is the activity of the esterases that perform the reverse reaction from AAT, which has been described for several fruits, including strawberry (Ueda and Ogata, 1976). Surprisingly, the molecular mass of the partially purified AAT enzyme from strawberry (Perez et al., 1996) was 70 kD, in contrast to the 54 kD estimated for the SAAT recombinant enzyme. Other plant AAT enzymes, such as those from banana and C. breweri (Harada et al., 1985; Dudareva et al., 1998), were also estimated to have a lower molecular mass (40 and 58 kD, respectively) than that estimated for the partially purified strawberry enzyme by Perez et al. (1996). In addition, the different _K_m value found for 1-butanol (46.1 versus the 3 mM reported by Perez et al., 1996) could indicate that the enzyme purified by Perez et al. is a different AAT from the one described here.
Ancestry of SAAT
The presence of several consensus sequences in the SAAT protein sequence assigns it to a superfamily of multifunctional proteins responsible for CoA-dependent acyl transfer (St. Pierre et al., 1998). The H-Xaa-Xaa-Xaa-D motif was initially detected in mammalian systems (Reed and Hackert, 1990; Shaw and Leslie, 1991), and site-directed mutagenesis experiments with the rat carnitine palmitoyltransferase suggested that both the conserved histidine and the conserved aspartate residues are part of the protein catalytic site that mediates acyl transfer and cleavage of free CoA (Brown et al., 1994). The AAT enzyme from yeast (encoded by ATF1; GenBank accession number D63449) is capable of reacting with acetyl-CoA and with various kinds of short-chain alcohols (Yoshioka and Hashimoto, 1984). Despite the similarity in catalytic activity between SAAT and ATF1, they show very low sequence identity (only 4% at the amino acid level). This suggests a lack of direct evolutionary relationships between the two enzymes. However, we identified two regions with sequence conservation. One is an H-Xaa-Xaa-Xaa-D motif (residues 191 to 195 in ATF1 and residues 57 to 61 in SAAT); the other is a stretch of 12 amino acids located between two tryptophan residues (W-Xaa-Xaa-F-F-X-P-L-Xaa-F-Xaa-W; residues 330 to 341 in ATF1 and residues 380 to 391 in SAAT). The latter conserved sequence overlaps the D-F-G-W-G motif (previously described).
The isolation of other fruit AATs will allow us to study the significant factors fundamental to the diversity of volatile ester profiles within and between fruit species and will pave the way for natural flavor production. Furthermore, it should provide the means for improving flavor characteristics of commercial fruits lost through successive years of breeding and selection.
METHODS
RNA Isolation, cDNA Library Construction, Mass Excision, and Sequence Analysis
Total RNA was isolated from strawberry (Fragaria × ananassa) cultivar Elsanta and from petunia (Petunia hybrida) variety W115, as described previously (van Tunen et al., 1989; Schultz et al., 1994). Red, ripe fruit tissue from strawberry, including the achenes_,_ and open corolla tissue from petunia were used to construct cDNA libraries, both in the UNI–XR vector (Stratagene, La Jolla, CA), and the inserts were directionally cloned. In total, 20 × 103 plaque-forming units from the nonamplified strawberry and petunia libraries were excised (ExAssist/SOLR system; Stratagene). High-quality plasmid DNA from 1701 strawberry and 480 petunia colonies picked randomly was extracted with the BioROBOT 9600 (Qiagen, Chatsworth, CA). We partially sequenced 1100 strawberry (out of 1701) and 480 petunia cDNAs from the 5′ end before performing microarray experiments. Sequencing was performed by using the Applied Biosystems (Foster City, CA) dye terminator cycle sequencing Ready Reaction kit and the Applied Biosystems 373 and 370A DNA sequencers. Comparison analysis of the sequences was conducted by using the advanced basic local alignment search tool, BLAST X server (Altschul et al., 1990), and the National Center for Biotechnological Information (www.ncbi.nlm.nih gov) nonredundant protein database. Software used for DNA and protein analysis were the Geneworks program (IntelliGenetics, Oxford, UK) and the DNASTAR program (DNASTAR, Madison, WI).
Fluorescent Targets and Probes
Total RNA (as much as 1.5 mg) was isolated from strawberry fruits at four selected stages of development: green fruit (medium-size green berries), white fruit (no sign of pigment), turning (half of each berry is colored red), and red (firm, fully red berries). This RNA was used to prepare mRNA with an mRNA purification kit (Pharmacia Biotechnology). First-strand cDNA was prepared as follows: oligo(dT) 21mer (4 μg) was annealed to 5 μg of mRNA by heating the reaction to 65°C for 3 min and then transferring to 25°C for 10 min. To the reaction mixture (50 μL total), the following components were added (final concentrations or amounts): 1 × first strand buffer (Gibco); 1 × DTT (0.1 M); 0.6 units/μL ribonuclease block; 500 μM each dATP, dGTP, and dTTP; 40 μM dCTP; 40 μM cyanine 3 (Cy3)–dCTP or Cy5–dCTP; and 6 units/μL Superscript II RNase H (Gibco) reverse transcriptase. Reverse transcription was performed at 37°C for 2 hr, after which the samples were precipitated with ethanol and resuspended in 10 μL of Tris-EDTA buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0). cDNA/mRNA hybrids were denatured by boiling for 3 min, and the samples were chilled on ice. The RNA was degraded by adding 0.25 μL of 1 M NaOH (10 min at 37°C). Samples were neutralized by adding 2.5 μL of 1 M Tris-Cl, pH 6.8, and 2.0 μL of 1 M HCl and then precipitated with ethanol. The cDNA, pelleted by centrifugation, was washed with 80% ethanol, dried, and dissolved in 6.5 μL of double-distilled H2O. After adding 2.5 μL of 20 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 1 μL of 2% SDS, the final concentration of the target was ∼0.5 μg/μL.
A total of 1701 strawberry and 480 petunia cDNAs were amplified by polymerase chain reaction (PCR) with use of the T3 and T7 universal primers and the GeneAmp PCR system 9600 (Perkin-Elmer, Foster City, CA). Redundant clones were not eliminated (∼30%). The primers contained a C6 amino modification (Isogen Bioscience BV, Maarssen, The Netherlands) at the 5′ end. The PCR reaction (in 100 μL total volume) was prepared in a 96-well PCR plate (MicroAmp; Perkin-Elmer) by mixing the following components: 10 × PCR buffer (1 × PCR buffer is 1.5 mM MgCl2), 0.2 mM for each deoxynucleotide triosphosphate, 1 μM for each T3 and T7 modified primers, 2.5 units of Taq DNA polymerase (Qiagen), and 10 ng of plasmid DNA template. The PCR program used 30 cycles of 30 sec at 94°C, 30 sec at 55°C, and 60 sec at 72°C. PCR products were purified with the QIAquick PCR purification kit (Qiagen) and eluted in 100 μL of 0.1 × Tris-EDTA buffer, pH 8.0. Samples were completely dried, resuspended in 7.5 μL of 5 × SSC (∼1 mg/mL), and transferred to a 384-format plate to be used later for application to microscope slides.
Arraying and Slide Processing
Amplified cDNA sequences were applied onto silylated microscope slides (CEL Associates, Houston, TX) by using a 16-pin printhead and a custom-built arraying robot. Strawberry (1701) and petunia (480) cDNA clones were arrayed in duplicate (4362 total probes per array) within a 1.7-cm2 area. Each probe contained 0.5 to 10.0 ng of PCR product. After arraying, the slides were left overnight to dry. For slide processing before hybridization, slides were soaked twice in 0.2% SDS and twice in double-distilled H2O successively for 2 min at room temperature with gentle agitation and then transferred to 95 to 100°C for 2 min (DNA denaturation). After the slides had dried for 5 min at room temperature, they were transferred into a sodium borohydrate solution (1 g of NaBH4 dissolved in 300 mL of PBS and 100 mL of 100% ethanol) for 5 min, rinsed three times in 0.2% SDS for 1 min and in double-distilled H2O for 1 min, and allowed to dry.
Hybridization, Scanning, and Data Acquisition
Portions (2.5 μL) of each fluorescent target were mixed together, and a total of 5 μL was applied to the microarray under a 22 × 22-mm cover slip. The microarray was placed in a custom-made hybridization chamber and incubated for 6 hr at 62°C in dry, hot air. After hybridization, the array was washed by agitation successively in 1 × SSC containing 0.1% SDS and in 0.1 × SSC containing 0.1% SDS for 5 min each at room temperature. The arrays were dried and scanned for fluorescence emission. Arrays were scanned with the ScanArray 3000 (General Scanning, Watertown, MA). A separate scan was conducted for each of the two fluorescent dyes used for hybridization. Using AIS software (Imaging Research, St. Catherines, Ontario, Canada), the integrated optical density of each individual probe on the array was measured. Extraction of the data for each probe was performed by using a grid to place a defined circle fitting the size of the DNA spots and measuring the integrated absorbance inside each circle.
Statistical Analysis of Reciprocal Two-Color Microarray Experiments
For statistical modeling, an appropriate scale of analysis was chosen in which factors under study act linearly and uncontrolled error has a gaussian (normal) distribution with constant variance. Preliminary analysis indicated that the fluorescence measurements should be natural logarithm–transformed for proper statistical analysis of variance. The transformed data were averaged over the two neighboring replicates and then were regressed on experimental factors (namely, dye [Cy3 or Cy5], mRNA source [e.g., green or red], and PCR amplicon [1701 cDNA clones]) and on their two-way and three-way interactions, by using standard software for analysis of variance (Genstat 5, Rothamsted, UK).
The combinations Cy3–red and Cy5–green were scored on microarray 1 and Cy5–red and Cy3–green on microarray 2, and the fluorescence values were obtained in four separate laser scans (i.e., statistically, the factors dye, tissue, and microarray are partially confounded). yijk denotes the natural logarithm of the fluorescence measurement for dye i (with i 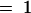 , 2 for the dyes Cy3 and Cy5), target j (with j
, 2 for the dyes Cy3 and Cy5), target j (with j 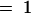 , 2 for green and red), and probe k (k
, 2 for green and red), and probe k (k 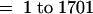 ), already averaged over the two neighboring replicates. In the following text, dots denote averages, for example, y_·1_k denotes the average over the two dyes. We used similar definitions for y_·1_k, _y_·1, and _y_·2. On the two microarrays, y_11_k_−_y_22_k and y_21_k_−_y_12_k are then measures of the upregulation of probe k in target green relative to target red. The average difference,
), already averaged over the two neighboring replicates. In the following text, dots denote averages, for example, y_·1_k denotes the average over the two dyes. We used similar definitions for y_·1_k, _y_·1, and _y_·2. On the two microarrays, y_11_k_−_y_22_k and y_21_k_−_y_12_k are then measures of the upregulation of probe k in target green relative to target red. The average difference,
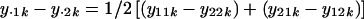
is not influenced by either dye or microarray-related variability. It can, however, be influenced by differences introduced by experimental artifacts specific to each of the two targets. The average expression difference between the two targets, _y_·1−_y_·2, originated partly from upregulation and downregulation of subsets of the 1701 cDNAs. This difference between the targets was also due to various experimental artifacts. We searched for probe-specific relative upregulation or downregulation differences averaged over the dye conditions. In statistical terms, the two-way target-by-probe interactions are tested against the lowest stratum error, the mean square of residuals mean squares, estimated from the three-way dye-by-target-by-probe variation. Differences (_Diff_k) were tested per probe (k) by using a standard two-sided t test on
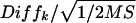
where the factor 1/2 arises from the implicit averaging over two fluorescence dyes, at P = 0.05 (used in the analysis), P = 0.01, and P = 0.001 single-test (per probe) significance levels. The exponent of the estimated probe-specific differences on the natural logarithmic scale provided the probe-specific cDNA ratios at the original scale. In the case of a genuine four- to fivefold difference, the t test is significant with a probability of 95% (power of the test).
RNA Gel Blot Analyses
For RNA gel blot analysis, total RNA was isolated from different strawberry tissues, as described by Schultz et al_._ (1994). Ten micrograms of glyoxal (1.5 M)-denaturated total RNA was electrophoresed and transferred to a Hybond N+ membrane (Amersham). After fixation (2 hr at 80°C), blots were hybridized as described by Angenent et al. (1992). The hybridization probes were made by random labeling oligonucleotide priming (Feinberg and Vogelstein, 1984) of the entire cDNAs. Blots were washed two times for half an hour each in 0.1 × SSC and 0.1% SDS at 65°C.
Analyses of Volatiles Released by Ripening Strawberry Fruits
Volatiles released by strawberry fruits in vivo were sampled by solid-phase microextraction (SPME). Intact fruits of approximately the same size at different stages of ripening were placed under a glass funnel (diameter of 8 cm) that was closed with aluminum foil, thus creating a headspace. The needle of the SPME device (Supelco Inc., Bellefonte, PA) was inserted into the funnel through the foil, and volatiles were trapped by exposing a fused silica fiber coated with 100 μM polydimethylsiloxane to the strawberry headspace for 30 min. The SPME-trapped strawberry volatiles were analyzed by gas chromatography–mass spectrometry (GC-MS), as described by Verhoeven et al. (1997). Volatile compounds were identified by screening the National Institute of Standards and Technology library for comparable mass spectra and by comparison with authentic reference compounds (Aldrich).
Expression of SAAT in Escherichia coli
The expression vector pRSET B (Invitrogen, Carlsbad, CA) was used for the expression of SAAT (for strawberry alcohol acyltransferase [AAT]) in E. coli (Stratagene; BL21 Gold DE3 strain). The BamHI and HindIII restriction sites of the original pRSET B were primarily used for cloning the gene encoding the green fluorescent protein (GFP); this construct as well as the empty pRSET B vector served as controls for the experiments. Cloning the GFP gene to the pRSET B vector inserted an additional SalI restriction site at the 3′ and, together with the BamHI site located at the 5′ end of the GFP gene, served as sites for cloning the SAAT cDNA. The SAAT coding region was amplified by using the forward primer 5′-CGGATCCGGAGAAAATTGAGGTCAG-3′ and the reverse primer 5′-CGTCGACCATTGCAC-GAGCCACATAATC-3′. The primers added BamHI and SalI restriction sites at both ends of the PCR fragments for cloning to the pRSET B vector. The forward primer eliminated the native methionine ATG codon of SAAT and formed a fusion protein at the N terminus with a peptide that included an ATG translation initiation codon, a series of six histidine residues (His-Tag), and an anti-Xpress (Invitrogen) epitope.
For small-scale purifications (50 mL of bacterial culture), the His-Tag protein purification was performed by using Ni-NTA spin columns according to the manufacturer's instructions (Qiagen). Larger scale purifications (250 mL of bacterial culture) were conducted with B-PER 6XHis columns (Pierce, Rockford, IL). The soluble fraction of the lysate and the eluate from the columns were analyzed by SDS-PAGE (10%, stained with Coomassie Brilliant Blue R 250) and protein gel blotting (the first antibody was anti-Xpress [Invitrogen], and the second antibody was GAR-AP [Boehringer]). The first eluates (200 μL) from the His-Tag columns were used for enzyme activity assays.
Enzymatic Characterization of the Recombinant SAAT
The AAT activity of the _SAAT_-encoded protein was established in two ways. For determining activity with different alcohols and acetyl-CoA, 14C-acetyl-CoA was used as substrate, and the formation of radiolabeled products was analyzed by radioactivity detection–gas chromatography (radio-GC) and quantified by liquid scintillation counting. Radio-GC was performed essentially as described previously (Bouwmeester et al., 1999c). Temperature programming was as follows: 70°C for 10 min, increased at 10°C/min to 270°C, and held at 270°C for 10 min. To quantify the enzyme activity, we diluted 1.4 μg of SAAT protein to 100 μL with 50 mM Tris buffer, pH 8.3, to which 14C-acetyl-CoA (0.1 mM at 0.1 Ci/mol in routine assays) and alcohol (20 mM in routine assays, 2 μL of hexane stock) were then added. After incubation for 30 min at 30°C, the assays were left on ice for 15 min, and 700 μL of hexane was added. Assays were vortex-mixed and centrifuged, and a 600-μL portion of the hexane phase was removed for liquid scintillation counting. The ratio of hexane-soluble radioactivity (esters) to the total radioactivity added as acetyl-CoA was used to calculate product formation.
To determine the utility of different acyl-CoA molecules, we stirred 325 μL of buffer (50 mM Tris-HCl, pH 8.0, containing 1 mM DTT), 25 μL of protein eluate, and 25 μL of acyl-CoA (4 mM in buffer) in a glass vial. The enzyme reaction was started by adding 25 μL of alcohol (160 mM in buffer). After a 15-min incubation with continuous stirring (35°C), solid CaCl2 was added (final concentration of 5 M). Volatiles released into the vial headspace (at 35°C with stirring) were subsequently trapped by SPME for 15 min and analyzed by GC-MS, as described above.
Acknowledgments
We thank Maayan Aharoni for her contribution to data analysis; Nick Mace and Ken Smith for scanning the microarrays in the first experiments; and Robert Hall, Michel Ebskamp, and especially Oscar Vorst and Andy Pereira for helpful discussions and comments on the manuscript. We thank Dr. Twan America for his help with bacterial expression and Gerie van der Heijden for his help with image analysis.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215**,** 403–410. [DOI] [PubMed] [Google Scholar]
- Angenent, G.C., Busscher, M., Franken, J., Mol, J.N., and van Tunen, A.J. (1992). Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4**,** 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap, W.R., and Garbarino, J.E. (1996). The role of ubiquitin in plant senescence and stress responses. Trends Plant Sci. 1**,** 331–335. [Google Scholar]
- Bouwmeester, H.J., Konings, M.C.J.M., Gershenzon, J., Karp, F., and Croteau, R. (1999. a). Cytochrome P-450 dependent (+)-limonene hydroxylation in fruits of caraway. Phytochemistry 50**,** 243–248. [Google Scholar]
- Bouwmeester, H.J., Wallaart, T.E., Janssen, M.H.A., van Loo, B., Jansen, B.J.M., Posthumus, M.A., Schmidt, C.O., de Kraker, J.-W., König, W.A., and Franssen, M.C.R. (1999. b). Partial purification and characterization of amorpha-4,11-diene synthase. The sesquiterpene synthase catalyzing the first probable step in the biosynthesis of artemisinin. Phytochemistry 52**,** 843–854. [DOI] [PubMed] [Google Scholar]
- Bouwmeester, H.J., Verstappen, F.W.A., Posthumus, M.A., and Dicke, M. (1999. c). Spider mite–induced (3_S_)-(E)-nerolidol synthase activity in cucumber and lima bean. The first dedicated step in acyclic C11-homoterpene biosynthesis. Plant Physiol. 121**,** 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, N.F., Anderson, R.C., Caplan, S.L., Foster, D.W., and McGarry, J.D. (1994). Catalytically important domains of rat carnitine palmitoyltransferase II as determined by site-directed mutagenesis and chemical modification. Evidence for a critical histidine residue. J. Biol. Chem. 269**,** 19157–19162. [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278**,** 680–686. [DOI] [PubMed] [Google Scholar]
- Dudareva, N., D'Auria, J.C., Nam, K., Raguso, R.A., Pichersky, E., and Nam, K.H. (1998). Acetyl-CoA:benzylalcohol acetyltransferase—An enzyme involved in floral scent production in Clarkia breweri. Plant J. 14**,** 297–304. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1984). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Addendum. Anal. Biochem. 137**,** 266–267. [DOI] [PubMed] [Google Scholar]
- Fujii, T., Nagasawa, N., Iwamatsu, A., Bogaki, T., Tamai, Y., and Hamachi, M. (1994). Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl. Environ. Microbiol. 60**,** 2786–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, T., Yoshimoto, H., Nagasawa, N., Bogaki, T., Tamai, Y., and Hamachi, M. (1996). Nucleotide sequences of alcohol acetyltransferase genes from lager brewing yeast, Saccharomyces carlsbergensis. Yeast 12**,** 593–598. [DOI] [PubMed] [Google Scholar]
- Harada, M., Ueda, Y., and Iwata, T. (1985). Purification and some properties of alcohol acetyltransferase from banana fruit. Plant Cell Physiol. 26**,** 1067–1074. [Google Scholar]
- Honkanen, E., and Hirvi, T. (1990). The flavour of berries. In Food Flavours, I.D. Morton and A.J. Macleod, eds (Amsterdam: Elsevier Scientific Publications), pp. 125–193.
- Karp, F., Mihaliak, C.A., Harris, J.L., and Croteau, R. (1990). Monoterpene biosynthesis: Specificity of the hydroxylations of (−)-limonene by enzyme preparations from peppermint (Mentha piperita), spearmint (Mentha spicata), and perilla (Perilla frutescens) leaves. Arch. Biochem. Biophys. 276**,** 219–226. [DOI] [PubMed] [Google Scholar]
- Maarse, H. (1991). Volatile Compounds in Foods and Beverages. (New York: Marcel Dekker).
- Malcorps, P., and Dufour, J.P. (1992). Short-chain and medium-chain aliphatic-ester synthesis in Saccharomyces cerevisiae. Eur. J. Biochem. 210**,** 1015–1022. [DOI] [PubMed] [Google Scholar]
- Manning, K. (1998). Isolation of a set of ripening-related genes from strawberry: Their identification and possible relationship to fruit quality traits. Planta 205**,** 622–631. [DOI] [PubMed] [Google Scholar]
- Medina Escobar, N., Cardenas, J., Valpuesta, V., Munoz Blanco, J., and Caballero, J.L. (1997). Cloning and characterization of cDNAs from genes differentially expressed during the strawberry fruit ripening process by a MAST-PCR-SBDS method. Anal. Biochem. 248**,** 288–296. [DOI] [PubMed] [Google Scholar]
- Nam, Y.W., Tichit, L., Leperlier, M., Cuerq, B., Marty, I., and Lelievre, J.M. (1999). Isolation and characterization of mRNAs differentially expressed during ripening of wild strawberry (Fragaria vesca L_._) fruits. Plant Mol. Biol. 39**,** 629–636. [DOI] [PubMed] [Google Scholar]
- Olias, J.M., Sanz, C., Rios, J.J., and Perez, A.G. (1995). Substrate specificity of alcohol acyltransferase from strawberry and banana fruits. In Fruit Flavors: Biogenesis, Characterization and Authentication, R.L. Rouseff and M.M. Leahy, eds (Washington, DC: American Chemical Society), pp. 134–141.
- Perez, A.G., Sanz, C., and Olias, J.M. (1993). Partial purification and some properties of alcohol acyltransferase from strawberry fruits. J. Agric. Food Chem. 41**,** 1462–1466. [Google Scholar]
- Perez, A.G., Sanz, C., Olias, R., Rios, J.J., and Olias, J.M. (1996). Evolution of strawberry alcohol acyltransferase activity during fruit development and storage. J. Agric. Food Chem. 44**,** 3286–3290. [Google Scholar]
- Perkins-Veazie, P. (1995). Growth and ripening of strawberry fruit. Hort. Rev. 17**,** 267–297. [Google Scholar]
- Reed, L.J., and Hackert, M.L. (1990). Structure–function relationships in dihydrolipoamide acyltransferases. J. Biol. Chem. 265**,** 8971–8974. [PubMed] [Google Scholar]
- Ruan, Y., Gilmore, J., and Conner, T. (1998). Towards Arabidopsis genome analysis: Monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 15**,** 821–833. [DOI] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270**,** 467–470. [DOI] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Heller, R., Chai, A., Brown, P.O., and Davis, R.W. (1996). Parallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93**,** 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, C.O., Bouwmeester, H.J., Bülow, N., and König, W.A. (1999). Isolation, characterization and mechanistic studies of (−)-α-gurjunene synthase from _Solidago canadensi_s. Arch. Biochem. Biophys. 364**,** 167–177. [DOI] [PubMed] [Google Scholar]
- Schultz, D.J., Craig, R., Cox-Foster, D.L., Mumma, R.O., and Medford, J.I. (1994). RNA isolation from recalcitrant plant tissue. Plant Mol. Biol. Rep. 12**,** 310–316. [Google Scholar]
- Seymour, G.B., Taylor, J.E., and Tucker, G.A. (1993). Biochemistry of Fruit Ripening. (London: Chapman and Hall).
- Shaw, W.V., and Leslie, A.G. (1991). Chloramphenicol acetyltransferase. Annu. Rev. Biophys. Biophys. Chem. 20**,** 363–386. [DOI] [PubMed] [Google Scholar]
- St. Pierre, B., Laflamme, P., Alarco, A.M., and De Luca, V. (1998). The terminal _O_-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A–dependent acyl transfer. Plant J. 14**,** 703–713. [DOI] [PubMed] [Google Scholar]
- Tamames, J., Ouzounis, C., Sander, C., and Valencia, A. (1996). Genomes with distinct function composition. FEBS Lett. 389**,** 96–101. [DOI] [PubMed] [Google Scholar]
- Ueda, Y., and Ogata, K. (1976). Studies of biosynthesis of esters in fruit volatiles. II. Esterification of added alcohol in separated cells from banana, strawberry and melon. J. Food Sci. Technol. Tokyo 23**,** 288–294. [Google Scholar]
- Ueda, Y., Tsuda, A., Bai, J.H., Fujishita, N., and Chachin, K. (1992). Characteristic pattern of aroma ester formation from banana, melon, and strawberry with reference to the substrate specificity of ester synthetase and alcohol contents in pulp. J. Jpn. Soc. Food Sci. Technol. 39**,** 183–187. [Google Scholar]
- Ulrich, D., Hoberg, E., Rapp, A., and Kecke, S. (1997). Analysis of strawberry flavour—Discrimination of aroma types by quantification of volatile compounds. Z. Lebensm. Unters. Forsch. 205**,** 218–223. [Google Scholar]
- van Tunen, A.J., Hartman, S.A., Mur, L.A., and Mol, J.N.M. (1989). Regulation of chalcone flavanone isomerase (CHI) gene expression in Petunia hybrida: The use of alternative promoters in corolla, anthers and pollen. Plant Mol. Biol. 12**,** 539–551. [DOI] [PubMed] [Google Scholar]
- Verhoeven, H., Beuerle, T., and Schwab, W. (1997). Solid-phase microextraction: Artifact formation and its avoidance. Chromatographia 46**,** 63–66. [Google Scholar]
- Yamakawa, Y., Goto, S., and Yokotsuka, I. (1978). Fractionation and some properties of acetic-ester synthesizing enzyme from Cladosporium cladosporioides No. 9. Agric. Biol. Chem. 42**,** 269–274. [Google Scholar]
- Yamashita, I., Iino, K., Nemoto, Y., and Yoshikawa, S. (1977). Studies on flavor development in strawberries. IV. Biosynthesis of volatile alcohol and esters from aldehyde during ripening. J. Agric. Food Chem. 25**,** 1165–1168. [Google Scholar]
- Yamauchi, H., Hasuo, T., Amachi, T., Akita, O., Hara, S., and Yoshizawa, K. (1989). Purification and characterization of acyl coenzyme A: Alcohol acyltransferase of Neurospora sp. Agric. Biol. Chem. 53**,** 1551–1556. [Google Scholar]
- Yoshioka, K., and Hashimoto, N. (1984). Ester formation by brewers' yeast during sugar fermentation. Agric. Biol. Chem. 45**,** 333–340. [Google Scholar]
- Zabetakis, I., and Holden, M.A. (1997). Strawberry flavour: Analysis and biosynthesis. J. Sci. Food Agric. 74**,** 421–434. [Google Scholar]
- Zhou, L., Ke, D., Kader, A.A., Zhou, L.L., and Ke, D.Y. (1995). Mode of controlled atmosphere action on ester biosynthesis of strawberries. Acta Agric. Univ. Pekinensis 21**,** 180–186. [Google Scholar]