Transcriptional Coactivation of Bone-Specific Transcription Factor Cbfa1 by TAZ (original) (raw)
Abstract
Core-binding factor 1 (Cbfa1; also called Runx2) is a transcription factor belonging to the Runt family of transcription factors that binds to an osteoblast-specific _cis_-acting element (OSE2) activating the expression of osteocalcin, an osteoblast-specific gene. Using the yeast two-hybrid system, we identified a transcriptional coactivator, TAZ (transcriptional coactivator with PDZ-binding motif), that binds to Cbfa1. A functional relationship between Cbfa1 and TAZ is demonstrated by the coimmunoprecipitation of TAZ by Cbfa1 and by the fact that TAZ induces a dose-dependent increase in the activity of osteocalcin promoter-luciferase constructs by Cbfa1. A dominant-negative construct of TAZ in which the coactivation domains have been deleted reduces osteocalcin gene expression down to basal levels. NIH 3T3, MC 3T3, and ROS 17/2.8 cells showed the expected nuclear localization of Cbfa1, whereas TAZ was distributed throughout the cytoplasm with some nuclear localization when transfected with either Cbfa1 or TAZ. Upon cotransfection by both Cbfa1 and TAZ, the transfected TAZ shows predominant nuclear localization. The dominant-negative construct of TAZ shows minimal nuclear localization upon cotransfection with Cbfa1. These data indicate that TAZ is a transcription coactivator for Cbfa1 and may be involved in the regulation of osteoblast differentiation.
Osteoblast differentiation is a crucial event during skeletal tissue formation, repair, and maintenance. Osteoblast differentiation depends on the expression of cell-specific transcription factors (15). To date, the only transcription factor known to be specific to osteoblasts is the core-binding factor 1 (Cbfa1), a member of the Runt family of transcription factors (3, 6). Cbfa1, also called Runx2, controls expression of osteoblast-specific genes (e.g., osteocalcin [6, 13, 16]). Deletion of the Cbfa1 gene in mice by homologous recombination leads to the death of homologous mutant mice (Cbfa1−/−) immediately after birth (14, 18). The Cbfa1−/− mutant mice have no osteoblasts or osteoblast progenitors, thus demonstrating that Cbfa1 is necessary for osteoblast differentiation. Ectopic Cbfa1 expression in vivo showed that it is sufficient to induce osteoblast differentiation (20). Using transgenic mice the targeted over- expression of the DNA-binding domain of Cbfa1 to osteoblasts in transgenic mice showed that Cbfa1 is essential not only for bone development but also for the regulation of postnatal osteoblast physiology (7).
The Cbfa1 protein expressed in osteoblasts has 596 amino acids and includes a Runt domain through which it binds DNA. At its N-terminal half is the Q/A region (polyglutamine and polyalanine repeats), which is also part of the activation domain. The C-terminal region of Cbfa1 is rich in proline, serine, and threonine (i.e., the PST region) and contains the nuclear localization signal, along with both an activation and a repressor domain (21). The repressor domain has the sequence VWRPY at its very end. Cbfa1 binds to OSE2, a _cis_-acting element present in the osteocalcin promoter (5), as well as other major genes expressed in osteoblasts such as bone sialoprotein, osteopontin, and α1 type I collagen. The presence of Cbfa1 activity at these promoters that are osteoblast specific but temporally distinct in their regulation suggests that Cbfa1 activity may be modulated by additional transcriptional regulatory proteins. Interestingly, during development a delay is noted between Cbfa1 expression (∼10.5 days postcoitus) and osteoblast differentiation (∼14.5 days postcoitus), suggesting that osteogenesis requires the interactions of Cbfa1 with other proteins to be initiated. The nature of any such an interaction(s) between Cbfa1 and the unknown protein(s) could vary from posttranslational modifications to the binding of other coactivators of transcription.
Transcription factors such as Msx-2, Bapx-1, and Hoxa-2 secreted molecules such as Indian Hedgehog and growth factors such as FGF and FGF-receptor appear to regulate Cbfa1 expression (4). At the functional level, mitogen-activated protein kinase pathway activation by type I collagen signaling via the integrins α2β1 and α1β1 modulates its activity (24). Activation of the cyclic AMP pathway leads to degradation of Cbfa1 through the ubiquitin-proteosome-dependent mechanism (23). The transactivation function of Cbfa1 is further inhibited by the interaction of Cbfa1 at its VWRPY motif with TLE2, a mammalian homologue of Drosophila Groucho (21). At the level of functional regulation, the CCAAT/enhancer-binding proteins (C/EBP) β and δ have been recently shown to synergize with Cbfa1 in the activation of ostsoecalcin gene transcription (9), thus supporting the concept that a combination of factors, rather than a single factor, controls lineage-specific gene expression.
Given the complex nature of osteoblast-specific gene expression and regulation of osteoblast differentiation involving Cbfa1, we sought to identify potential coactivators of transcription that may bind to and regulate the functions of Cbfa1. The data indicate that the transcriptional coactivator TAZ binds to Cbfa1 and modulates Cbfa1 function. In addition, TAZ function as a transcriptional coactivator may be critical in expression of bone-specific genes such as osteocalcin. These data suggest important roles for transcription coactivators such as TAZ in osteoblast differentiation and physiology.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 cells were obtained from R. Juliano (University of North Carolina) and maintained in 100-mm2 dishes in Dulbecco modified Eagle medium (dMEM) with 10% heat-inactivated fetal bovine serum (FBS; Sigma) and antibiotic-antimycotic solution comprising penicillin, streptomycin, and amphotericin B (Sigma). Cells were trypsinized and replated at lower dilution every 3 days. The day before transfection, cells were washed, trypsinized in 0.05% trypsin-0.53 mM EDTA (Gibco-BRL), resuspended in Opti-MEM I (Gibco-BRL) medium with 1% FBS, and replated in the wells (35 mm2) of six-well plates at 1.5 × 104 cells/ml. Jurkat cells were obtained from the cell culture facility at the University of North Carolina and maintained in suspension by using RPMI 1640 medium (Gibco-BRL) containing 10% FBS and antibiotic-antimycotic solution. On the day of transfection, Jurkat cells were washed in phosphate-buffered saline (PBS), resuspended in Opti-MEM I with 1% FBS and placed in the wells of 24-well plates (350 μl/well at 2 × 105 cells/ml). Rat osteosarcoma cells (ROS 17/2.8) were maintained in DMEM-F12 with 10% FBS. MC 3T3 (E1) cells were maintained in α-minimal essential medium with 10% FBS.
Yeast two-hybrid analysis.
The yeast two-hybrid system (Matchmaker GAL4 two-hybrid system; Clontech) was used to identify proteins that bind to mouse Cbfa1. A high-complexity cDNA library (made by using mRNA from 17-day mouse embryos) was purchased from Clontech. This 17-day mouse embryo cDNA library was made by cloning the cDNA into a yeast GAL4 activation domain vector and pretransformed into Saccharomyces cerevisiae host strain Y187. This pretransformed Matchmaker cDNA library served as the prey. The full-length cDNA coding for mouse Cbfa1 was cloned in the vector pCMV5 (pCMV-Cbfa1) and was used as the bait in yeast two-hybrid screening.
Plasmid pCMV5-Cbfa1 encoding full-length mouse Cbfa1 cDNA was digested with _Nco_I/_Eco_RI. A 1.2-kb fragment was gel purified and cloned into pGBKT7, resulting in the plasmid pGBKT7-Cbfa1. This plasmid, pGBKT7-Cbfa1, had the Cbfa1 protein as a fusion protein in frame with GAL4. DNA sequencing of pGBKT7-Cbfa1 confirmed the Cbfa1 sequence without any errors. The bait plasmid pGBKT7-Cbfa1 was then transformed into the yeast strain AH109 by using the small-scale transformation protocol (Clontech Matchmaker GAL4 two-hybrid system 3) and plated on an SD/−Trp plate. An overnight culture (concentrated culture) of positive pGBKT7-Cbfa1 bait strain in SD/−Trp medium was used to mate with 1 ml of pretransformed mouse 17-day-old embryo Matchmaker cDNA library in yeast strain Y187 (Clontech) according to the pretransformed Matchmaker library protocol. Then, 100 μl of a 1:10, 1:100, 1:1,000, or 1:10,000 dilution of the resulting mating mixture was spread on 100-mm SD/−Leu, SD/−Trp, or SD/−Leu/−Trp plates to evaluate mating efficiency controls. The remaining mating mixture was plated on 50 SD/−Ade/−His/−Leu/−Trp (QDO) plates (150 mm2, 200 μl per plate). Plates were incubated at 30°C for up to 16 days. Approximately 5 × 105 colonies were screened. Fifty-six positive colonies were picked up and transferred to new SD/−Ade/−His/−Leu/−Trp plates. Of all of the colonies that were replated, three of them grew very rapidly. These three colonies were picked and processed for plasmid preparation according to the Clontech yeast plasmid protocol. The resulting yeast plasmids were electrotransformed into Escherichia coli DH12S. Plasmid DNA was prepared, and the interaction between bait Cbfa1 and prey protein clones was reconfirmed by cotransformation of the bait plasmid pGBKT7-Cbfa1 with each of the three prey plasmids into the yeast strain AH109. The cotransformed yeast was plated on SD/−Ade/−His/−Leu/−Trp/X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates. All of them regrew, and the colonies displayed a blue color after incubation at 30°C for 3 to 5 days. Large-scale preparations of the prey plasmids were made and sequenced. The cDNA sequences were analyzed by using the BLAST program (GenBank).
Plasmid constructs, sequencing, and transfection.
The mouse Cbfa1 cDNA was cloned into the eukaryotic expression vector pCMV5 (pCMV5-Cbfa1). The full-length cDNA for mouse TAZ was amplified by PCR and cloned into the tagged eukaryotic expression vector pcDNA3.1/HisB (Invitrogen; pcDNA-TAZ). A truncated form of mouse TAZ that ends at residue 164 (after the end of the WW domain) was amplified by PCR and also cloned into pcDNA3.1/HisB (pcDNA-N-TAZ). The sequences of all of the constructs were verified by automated DNA sequencing analysis (University of North Carolina). The mouse osteocalcin promoter-luciferase reporter construct (p6OSE2luc) that contains six tandem repeats of the OSE2 element of mouse osteocalcin promoter (5) was used as the reporter gene to study the transcriptional coactivation of Cbfa1. A full-length mouse osteocalcin promoter (∼1 kb) with luciferase as the reporter gene (OG2) was also used in some experiments. To confirm specificity of the effect, we used the other osteoblast-specific element construct (OSE1), also as six tandem repeats (p6OSE1), and a p6OSE2 construct with a mutation in the Cbfa1-binding region (p6OSE2mut) (5). In addition to the osteoblast-specific osteocalcin promoter constructs, other nonspecific promoter constructs were also used to verify the specificity. Promoter-luciferase reporter constructs were obtained for cyclin D1 (2), SMAD-7 (17), acetylcholine receptor (10), and tyrosine aminotransferase (TAT3 [11]) from A. Baldwin, University of North Carolina. Large-scale Plasmid DNA was prepared for each construct by using the EndoFree Maxi plasmid kit from Qiagen. Transient transfection of adherent NIH 3T3 cells, as well as of the Jurkat cells, in suspension was carried out by using Superfect transfection reagent from Qiagen according to the manufacturer's protocol. Cells were transfected with the various constructs singly or in combination, and the total amounts of DNA were always maintained by adding empty vector DNA. At 30 h after the transient transfection, the cells were gently washed in cold PBS and processed for luciferase activity by using the luciferase assay system (Promega). Chemiluminescent detection of luciferase activity was carried out with a Lumat LB 9507 Luminometer (EG&G Berthold).
Immunohistochemistry, RT-PCR, and Northern blot analysis of TAZ expression in bone.
Mandibles and femurs from 7-day-old mice were dissected and fixed in buffered neutral formalin. After brief demineralization in EDTA, the tissues were embedded in paraffin and sections were processed for routine immunohistochemistry. Rabbit polyclonal antibody to mouse TAZ (12) was used to examine the expression of TAZ in osteoblast-like cells of the mandible and femur by using the ABC-Vectastain kit (Vector Labs). Total RNA was isolated from the calvaria, long bones, heart, skin, and brain of 7-day-old mouse pups. One microgram of total RNA from each tissue was reverse transcribed by using oligo(dT) primers and Superscript reverse transcriptase (RT) in a 20-μl reaction. Then, 2 μl of each reverse-transcribed product was amplified by PCR with oligonucleotide primers for mouse TAZ. Primers for mouse beta actin served as controls. In addition, total RNA (30 μg) from calvaria was run on formaldehyde agarose gel and analyzed by Northern blotting with a [32P]dCTP-labeled probe for mouse TAZ.
Immunoprecipitation and immunofluorescent microscopy.
Rat osteosarcoma cells (ROS 17/2.8) were plated in 100-mm2 dishes in DMEM-F12 (Gibco-BRL) with 10% FBS and allowed to adhere and spread for 48 h. The medium was then removed, and the cells were washed gently in cold PBS. The cells were then lysed in modified radioimmunoprecipitation assay buffer containing 50 mM Tris (pH 7.5), 1% NP-40, 0.1% sodium deoxycholate, 150 mM NaCl, 50 mM NaF, 1 mM sodium pyrophosphate, 1 mM sodium vanadate, 0.2 mM calyculin, 1 mM nitrophenylphosphate, 2 mM phenylmethylsulfonyl fluoride, 5 mM benzamidine, and 10 μg of aprotinin/ml. The total cell lysate was cleared by centrifugation at 16,000 × g for 5 min at 4°C. The protein concentration in the lysate was determined by using the bicinchoninic acid assay (Pierce, Rockford, Ill). Cell lysate (200 μg of total protein) was first precleared by using protein G-Sepharose (Pharmacia) and then incubated with polyclonal antibody (4 μl) to Cbfa1 for 2 h at 4°C with gentle shaking. This was followed by the addition of protein G-Sepharose and an additional incubation for 2 h at 4°C. After centrifugation, the protein G-Sepharose beads were washed three times with cold radioimmunoprecipitation assay buffer and then boiled with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer to dissociate the proteins. The immunoprecipitated proteins were separated by SDS-PAGE under reducing conditions, and the proteins were transferred electrophoretically onto polyvinylidene fluoride membrane (Immobilon; Millipore Corp). The membrane was blocked with milk in PBS for 1 h and then incubated with rabbit polyclonal antibody to TAZ (12) for 1 h. The membrane was washed briefly with PBS-Tween (0.1%) and incubated with goat anti-rabbit immunoglobulin G (IgG)-peroxidase conjugates for 1 h. Immunoreactivity was detected on radiographic film by using enhanced chemiluminescence (Amersham Corp).
For immunofluorescence microscopy, NIH 3T3, MC 3T3, and ROS 17/2.8 cells were plated on clean glass coverslips and allowed to adhere and spread for 24 h in Opti-MEM medium with 1% FBS. Cells were then transfected with pCMV5-Cbfa1, pcDNA-TAZ, or pcDNA-N-TAZ constructs by using Superfect. Double transfection was also carried out by using equal amounts of pCMV5-Cbfa1 and pcDNA-TAZ or of pCMV5-Cbfa1 and pcDNA-N-TAZ. After 30 h, the cells were washed gently three times in PBS. The bound cells were fixed in 3.7% formaldehyde for 10 min and then washed in Tris-buffered saline (TBS; 50 mM Tris-HCl, 150 mM Nacl, 0.1% NaN3 [pH 7.6]) for 10 min. The coverslips, with the cells attached to them, were incubated sequentially in 0.5% Triton for 7 min, TBS for 5 min, primary antibody to Cbfa1 (rabbit polyclonal to Cbfa1) or mouse monoclonal antibody to Xpress Tag (Invitrogen) to stain TAZ and N-TAZ for 60 min at 37°C, TBS for 5 min, secondary antibody (fluorescein isothiocyanate-conjugated goat anti-rabbit from Zymed or Texas red-conjugated goat anti-mouse antibody [Leinco Tech.]), and TBS for 5 min. They were then dipped into water, air dried, and mounted on Mowiol (Calbiochem Corp.). The coverslips were examined under an Olympus fluorescence microscope, and the color images were captured by using the computer-driven SPOT RT system (v3.2.4; Diagnostic Instruments, Inc.). The expression of TAZ proteins was analyzed for intracellular compartments, and cells were grouped accordingly as being predominantly nuclear, predominantly cytoplasmic, or with an equal distribution between the nucleus and the cytoplasm.
Stable transfection and Northern blotting.
Rat osteosarcoma cells (ROS 17/2.8) cells were plated in DMEM-F12 medium with 10% FBS. The eukaryotic expression constructs pcDNA-TAZ and pcDNA-N-TAZ were linearized with the restriction enzyme _Pvu_I and used to transfect ROS 17/2.8 cells by using the reagent Superfect (Qiagen). Cells transfected with the vector pcDNA3.1 and untransfected cells served as controls. Selection of stably transfected cells was carried out by using G418 (Gibco-BRL) at 800 μg/ml. Individual colonies seen after 10 days of culture were isolated and grown further. Total RNA was isolated from clones and blotted onto nylon membranes. The effects of stable transfection of pcDNA-TAZ and pcDNA-N-TAZ on osteocalcin gene expression in ROS17/2.8 cells was analyzed by Northern blotting by using the mouse osteocalcin probe. RNA loading was verified by stripping the membrane and reprobing it with mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe. In addition to the analysis of individual clones, pools of clones were also analyzed by Northern blotting, which was repeated at least three times to confirm the reproducibility of the results.
Deletion mutants of TAZ and Cbfa1 and functional assays.
To determine the role of specific functional domains, several deletion mutants of TAZ and Cbfa1 were assembled in pcDNA vectors. PCR amplification with oligonucleotide primers designed to begin and end at specific domain boundaries was used to clone these deletion mutants. These deletion mutants were used in functional assays in Jurkat cells with p6OSE2luc as the reporter gene.
RESULTS
Cbfa1 binds to TAZ.
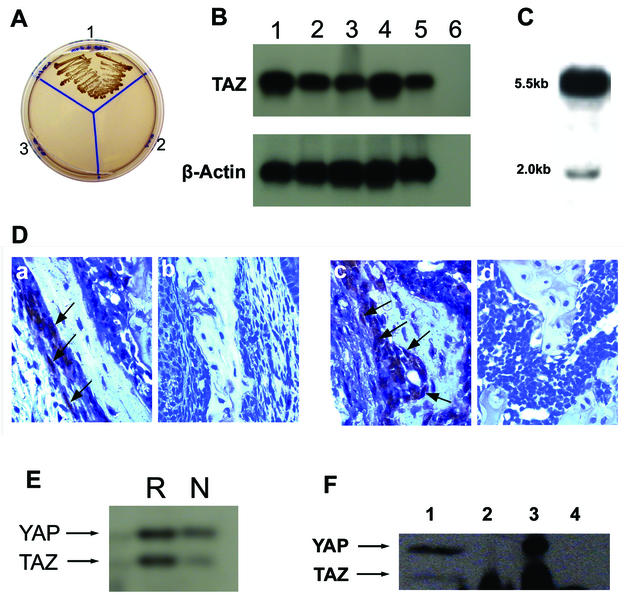
The plasmid pGBKT7-Cbfa1 produces Cbfa1 as a fusion protein in frame with GAL4 and was used as a bait for the yeast two-hybrid analysis. A high-complexity cDNA library made from the mRNA of 17-day mouse embryo (Clontech) was used as the prey. Mating of the yeast strain AH 109 transformed with the bait plasmid pGBKT7-Cbfa1 and the 17-day mouse embryo cDNA library in yeast strain Y187 resulted in 56 positive colonies. High-stringency selection during a second round of screening of these 56 colonies showed that three colonies grew rapidly. These three colonies were picked and processed for plasmid preparation. The interaction between the bait Cbfa1 and the three plasmid clones was verified again by the cotransformation of the bait Cbfa1 plasmid with each of the three prey plasmid clones into the yeast strain AH 109. All of them regrew, and the colonies showed blue color (on X-Gal plates) after incubation at 30°C only for 3 to 5 days. Figure 1A shows the two-hybrid plating results with one of the TAZ clones and the Cbfa1 bait plasmid. Large-scale plasmid preparation and sequence analysis of the three prey clones showed that two of the three clones had complete homology to the recently described mouse transcriptional coactivator TAZ (12). Sequence analysis also showed that we had the full-length cDNA clone for mouse TAZ. The third clone was novel and showed no significant homology to any known gene in the gene bank. Multiple tissue RT-PCR analysis of 7-day-old mouse pups showed TAZ expression in all of the tissues analyzed, although the calvaria and skin showed the most amplification (Fig. 1B). Northern blot analysis of total RNA from 7-day-old mouse calvaria with the TAZ probe showed a 5.5-kb predominant band along with a minor band at ∼2 kb (Fig. 1C). Further evidence of TAZ expression in osteoblasts is provided by the immunohistochemical staining of paraffin-embedded sections of mandible and femurs of 7-day-old mouse pups. As shown in Fig. 1D, the polyclonal rabbit antibody to mouse TAZ shows positive staining in osteoblasts lining the developing bone.
FIG. 1.
(A) Two-hybrid analysis for the specificity of Cbfa1-TAZ interactions. Plasmid DNA from bait Cbfa1 was cotransformed with the prey plasmid containing TAZ cDNA into yeast strain AH 109. Section 1 is the cotransformed mixture, section 2 is the bait alone, and section 3 is the prey alone. Selection was made by growth on SD/−Ade/−His/−Leu/−Trp plates. Note the selective growth only in Section 1. (B) RT-PCR analysis of TAZ and β-actin in 7-day-old mouse pup tissues. Lanes: 1, calvaria; 2, long bones; 3, heart; 4, skin; 5, brain; 6, negative control PCR (i.e., PCR with no RT product). (C) Northern blot of total RNA from 7-day-old mouse calvaria hybridized with cDNA probe for mouse TAZ. Note the prominent 5.5-kb band, along with the less-prominent 2.0-kb band. (D) Immunohistochemistry of developing bone sections from 7-day-old mouse mandible (a and b) and femur (c and d). Paraffin-embedded sections were stained with rabbit antiserum to mouse TAZ (a and c) or normal rabbit serum (b and d). Note the staining of the osteoblast-like cells with the antiserum to TAZ (arrowheads). (E) Western blot of cell extract from ROS 17/2.8 (R) and NIH 3T3 cells (N) analyzed with antibody to mouse TAZ. Note the cross-reactivity to mouse YAP and the weaker expression of TAZ in NIH 3T3 cells compared to ROS 17/2.8 cells. (F) Western blot of immunoprecipitated proteins with antibody to Cbfa1 and Western blotting with antibody to mouse TAZ. Lanes: 1, total lysate from ROS 17/2.8 cells; 2, immunoprecipitation with antibody to Cbfa1; 3, immunoprecipitation with antibody to TAZ; 4, immunoprecipitation with nonspecific rabbit IgG. Note the immunoprecipitation of TAZ by Cbfa1 in lane 2. Interestingly, both TAZ and YAP are immunoprecipitated with antibody to mouse TAZ (lane 3).
TAZ expression was additionally examined in the rat osteosarcoma cell line ROS 17 and in NIH 3T3 cells. Cell lysates prepared from these cells were analyzed by Western blotting with rabbit polyclonal antibody to mouse TAZ. The ROS 17/2.8 cells showed a higher level of TAZ expression compared to the NIH 3T3 cells (Fig. 1E). The binding of TAZ to Cbfa1 was next verified by immunoprecipitation. Lysates from ROS 17/2.8 cells were first immunoprecipitated with polyclonal antibody to Cbfa1, and the precipitated proteins were analyzed by Western blotting with a rabbit polyclonal antibody to mouse TAZ. As shown in Fig. 1F, TAZ coprecipitates with Cbfa1 in lysates prepared from ROS 17/2.8 cells. Immunoprecipitates prepared with nonspecific rabbit serum failed to show any precipitation of TAZ. Interestingly, immunoprecipitation of ROS 17/2.8 cell lysate with antibody to TAZ precipitates both TAZ and YAP (lane 3, Fig. 1F), whereas immunoprecipitation with antibody to Cbfa1 shows only the TAZ band. The rabbit polyclonal antibody to TAZ was made by using a synthetic peptide representing the residues at the carboxy-terminal end of TAZ (12), and since this area shows homology to YAP, the Western blots of total cell lysates and immunoprecipitates with this antibody showed this cross-reactivity with YAP (Fig. 1E and F). Hence, all subsequent experiments involving the intracellular localization of TAZ in cultured cells were carried out by immunofluorescence microscopy with the monoclonal antibody to Xpress tag, which is unique to the transfected constructs of TAZ and N-TAZ.
TAZ functions as transcriptional coactivator for Cbfa1.
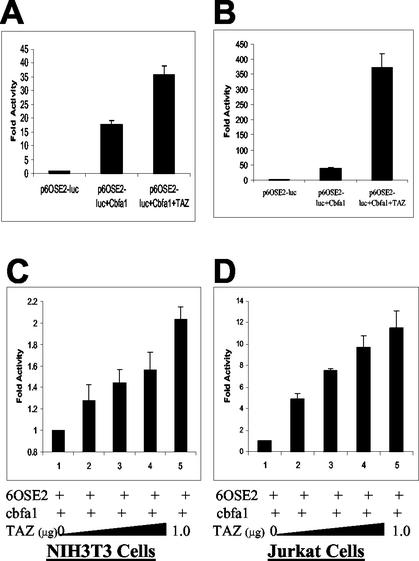
The functional relationship of TAZ-Cbfa1 binding was determined by examination of their independent and mutual effects on a chimeric gene containing six copies of the OSE2 element upstream of the minimum OG2 promoter of the osteocalcin promoter with luciferase construct as the reporter gene (p6OSE2-luc) (4). NIH 3T3 cells were plated in Opti-MEM medium with 1% FBS the day before transfection and assayed for luciferase activity as a measure of transcriptional activation. As shown in Fig. 2A, NIH 3T3 cells transfected with the reporter plasmid p6OSE2-luc alone showed very little basal level activity. Cells transfected with the reporter plasmid and pCMV5-Cbfa1 showed an expected (∼18-fold) increase in transcriptional activity over the basal level. However, cotransfection with pcDNA-TAZ, p6OSE2-luc, and pCMV5-Cbfa1 resulted in a 36-fold increase in the transcriptional activity over basal levels. Since NIH 3T3 cells had shown basal level expression of TAZ, we then tested the effects of TAZ on transcriptional activity of Cbfa1 in Jurkat cells that lack TAZ expression. Jurkat cells transfected with the reporter plasmid p6OSE2-luc showed very little basal level activity, and cotransfection with pCMV5-Cbfa1 showed an ∼39-fold upregulation in reporter gene expression. Cotransfection of the reporter gene with both pCMV-Cbfa1 and pcDNA-TAZ led to a nearly 373-fold increase over the basal level and a nearly 10-fold increase over Cbfa1-alone induced expression of the reporter gene (Fig. 2B). The basal level expression of TAZ in NIH 3T3 cells may be masking some of the transcriptional coactivating effects of transfected TAZ. The coactivation effect of TAZ was further confirmed by the transfection of increasing amounts of TAZ encoding plasmid. As shown in Fig. 2C and D, with constant amounts of p6OSE2-luc and pCMV5-Cbfa1, increasing the amounts of pcDNA-TAZ cotransfection leads to corresponding increases in the transcription of the reporter luciferase gene in both NIH 3T3 and Jurkat cells.
FIG. 2.
(A and B) Transcriptional coactivation assay in NIH 3T3 (A) and Jurkat (B) cells. The results are expressed as fold increases over the control (p6OSE2-luc alone). A total of 1 μg of each construct was used for the NIH 3T3 cells, and 0.5 μg of each construct was used for the Jurkat cells. (C and D) Transcriptional coactivation assay in NIH 3T3 (C) and Jurkat (D) cells demonstrating the dose-responsive effects of TAZ. The results are expressed as the fold increases over p6OSE2-luc+Cbfa1 (positive control). p6OSE2-luc and Cbfa1 were used at 1 μg/well for NIH 3T3 cells and at 0.5 μg/well for Jurkat cells. The data in panels A and B are expressed as fold increases in activity compared to p6OSE2-luc alone, whereas the data in panels C and D are expressed as the fold increases in activity compared to p6OSE2-luc and Cbfa1. The differences in the fold activities expressed between panels A and B and panels C and D are noted.
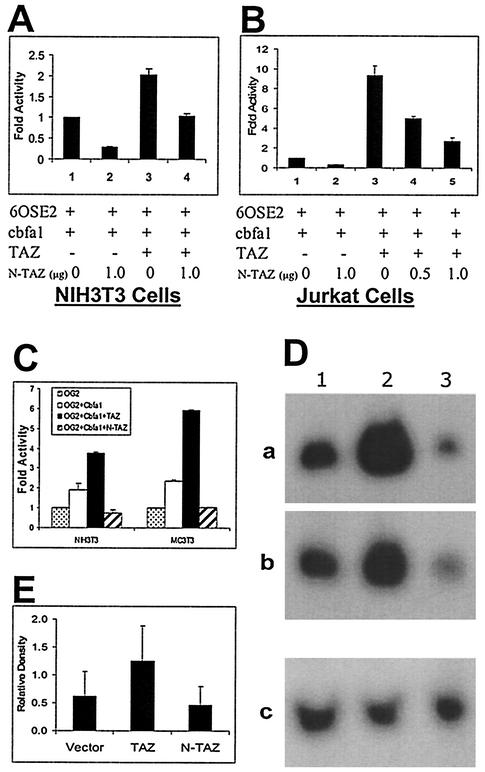
To begin to define the structure-function relationships of this WW domain transcription factor, the functional effect of activation domain deletion was explored. Truncating the TAZ protein at amino acid 164 retains the WW domain (pcDNA-N-TAZ). Cotransfection of NIH 3T3 and Jurkat cells with p6OSE2-luc, Cbfa1, and pcDNA-N-TAZ showed a >50% reduction in the expression of luciferase reporter gene (Fig. 3A and B). Although cotransfection with pcDNA-TAZ led to the expected two- and ninefold increases in luciferase activity in NIH 3T3 and Jurkat cells, respectively, inclusion of pcDNA-N-TAZ along with pcDNA-TAZ reduced the luciferase activity (Fig. 3A and B). The truncated construct pcDNA-N-TAZ may act as a dominant-negative molecule in the transcriptional activation of Cbfa1 by effectively binding Cbfa1 without offering fully effective coactivator function. We further examined the effects of Cbfa1-TAZ cotransfection on the activity of the natural mouse osteocalcin promoter (OG2) in both NIH 3T3 and MC 3T3 (E1) cells. As shown in Fig. 3C, the full-length TAZ upregulated OG2 activity and the dominant-negative construct (N-TAZ) downregulated OG2 activity in both NIH 3T3 and MC 3T3 cells.
FIG. 3.
(A and B) Effects of a dominant-negative construct of TAZ on osteoblast-specific gene expression. Transcriptional coactivation assays in NIH 3T3 (A) and Jurkat (B) cells show the dominant-negative effects of N-TAZ (truncated at the end of WW domain). The results are expressed as fold increases over p6OSE2-luc+Cbfa1 (positive control). p6OSE2-luc, Cbfa1, and TAZ were used at 1 μg/well for NIH 3T3 cells and at 0.5 μg/well for Jurkat cells. (C) Transcriptional coactivation assay in NIH 3T3 and MC 3T3 E1 cells performed with the full-length, natural promoter for osteocalcin (OG2) with luciferase as the reporter gene. The results are expressed as fold increases over the activity with OG2-luc alone. All of the plasmid constructs were used at 1 μg/well. Note the significant upregulation of OG2-luc function in cells cotransfected with Cbfa1 and TAZ and the effects of N-TAZ functioning in a dominant-negative manner. (D) Northern blot analysis of total RNA (30 μg/lane) from ROS 17/2.8 clones that were stably transfected with TAZ or the dominant-negative construct N-TAZ. Lane 1, RNA from stably transfected control clones (pcDNA 3.1); lane 2, RNA from stably transfected TAZ clones; lane 3, RNA from stably transfected N-TAZ clones. Panels a and b represent RNA from separate ROS 17/2.8 clones hybridized with mouse osteocalcin probe. Panel c is the membrane from panel a but stripped and reprobed with GAPDH to confirm loading. Note the significant upregulation of osteocalcin mRNA in TAZ-transfected clones, whereas the clones transfected with the N-TAZ constructs show downregulation of osteocalcin mRNA. (E) Densitometric analysis of bands from Northern blots. Pooled heterogeneous ROS 17/2.8 colonies that were stably transfected with vector alone, TAZ, or the truncated N-TAZ constructs were used to isolate total RNA. Endogenous mRNA levels of osteocalcin were detected by using a probe for rat osteocalcin. The data represent six different Northern blots, and the relative density shown here is the ratio of the signal density of osteocalcin and GAPDH bands.
The effects of transfecting TAZ and the dominant-negative construct of TAZ on endogenous osteocalcin levels in osteoblast cells were additionally evaluated in ROS 17/2.8 cells. Stably transfected cells were analyzed by Northern blotting, and Fig. 3D shows examples of two stably transfected clones each for TAZ and the dominant-negative construct. TAZ significantly upregulated endogenous osteocalcin mRNA levels in these clones, whereas the dominant-negative construct of TAZ showed significant downregulation. In subsequent experiments, clones of transfected ROS 17/2.8 cells were pooled, and total RNA was analyzed by Northern blotting. Repeated analysis on three separate occasions of these pooled cells confirmed the reproducibility of these results (Fig. 3E).
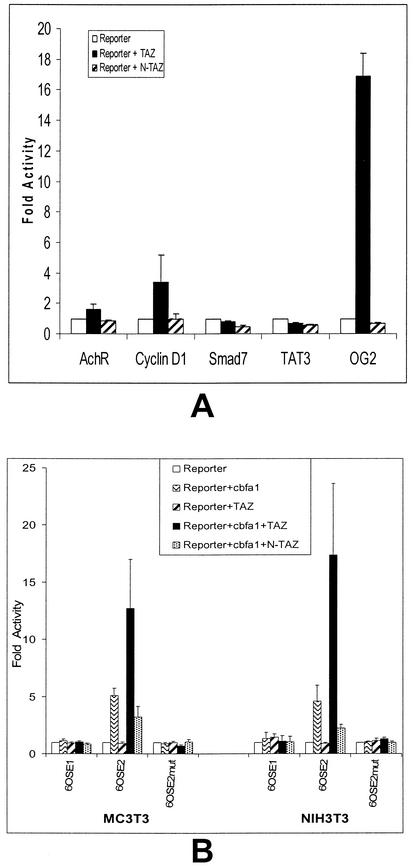
The specificity of TAZ functioning as a transcriptional coactivator for Cbfa1 was examined by plating Jurkat cells in Opti-MEM medium with 10% FBS for 24 h to allow the expression of other transcriptional factors. The cells were then cotransfected with pcDNA-TAZ and four other promoter-luc constructs: cyclin D, SMAD-7, acetylcholine receptor (AchR), and TAT3 with osteocalcin-luc (OG2) as the positive control. All of these promoter constructs were tested individually for transcriptional coactivation by TAZ. As shown in Fig. 4A, TAZ induced upregulation of osteocalcin gene expression (∼16-fold over basal levels) and showed minor upregulation (∼3-fold over basal levels) of only the cyclin D gene. The other promoter constructs showed no significant activity, suggesting that TAZ may possess some selectivity in its function as a transcriptional coactivator. The specificity of effect was also evaluated by testing the effects of TAZ on the second osteoblast specific element (OSE1) within the osteocalcin promoter. Transcriptional assay with p6OSE1-luc constructs showed no effects of Cbfa1, TAZ, or combinations of Cbfa1-TAZ. Similarly, a mutation in the active promoter construct-p6OSE2 in the region of Cbfa1 binding (5) also showed no effect of TAZ. These results were reproducible in both NIH 3T3 and MC 3T3 cells (Fig. 4B). Compared to Jurkat cells, TAZ alone showed no effect in NIH 3T3 or MC 3T3 cells on the transcription of osteocalcin gene.
FIG. 4.
(A) Transcriptional coactivation assay of other nonosteogenic promoters by TAZ (0.5 μg) in Jurkat cells. The promoters with luciferase as reporter gene used were AchR, cyclin D1, SMAD-7, TAT3, and the natural mouse osteocalcin promoter (OG2). Note the significant transcriptional activation of OG2-luc and to some extent cyclin D1, along with the lack of effect in the other three constructs. (B) The specificity of TAZ effect on the osteocalcin gene was further analyzed by using the mouse OSE1 promoter construct and a mouse OSE2 promoter construct with a mutation in the Cbfa1 binding region. Note the lack of effect of TAZ in the transcriptional coactivation assay with these OSE1 and mutant OSE2 constructs.
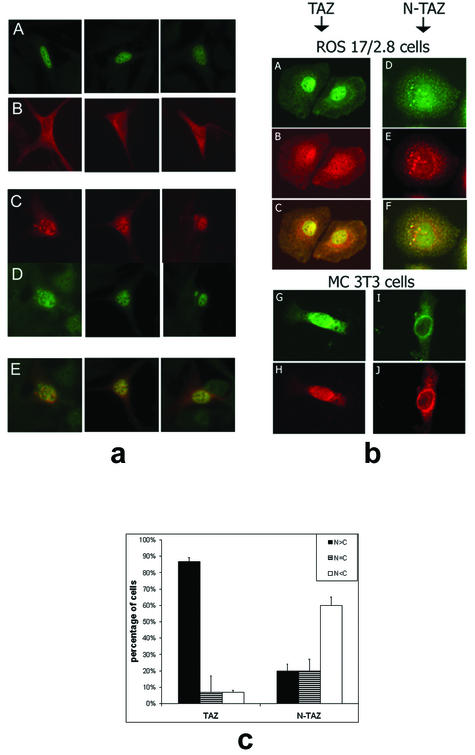
An initial evaluation of the interaction of TAZ with Cbfa1 was attained by the cellular localization of TAZ in transfected NIH 3T3 and MC 3T3 E1 cells. Because both of these cell lines show basal level expression of endogenous TAZ, cellular localization of TAZ was evaluated by using a monoclonal antibody to the Xpress tag present in the transfected pcDNA-TAZ construct. The transfection of NIH 3T3 cells by Cbfa1 alone revealed expression confined to the nucleus (Fig. 5a), and transfection by pcDNA-TAZ showed localization of TAZ throughout the cell but with tendency toward a perinuclear distribution (Fig. 5a). The cotransfection of Cbfa1 and TAZ resulted in the nuclear localization of both TAZ and endogenous Cbfa1 (Fig. 5a). Computer-generated microphotograph overlays confirm the nuclear translocation of TAZ in cells cotransfected with Cbfa1 (Fig. 5a).
FIG. 5.
(a) Immunocytochemistry of NIH 3T3 cells transfected with either Cbfa1 alone (A), TAZ alone (B), or both Cbfa1 and TAZ (C to E). The cells were fixed 24 h after transfection and analyzed by immunofluorescence (rabbit P/C antibody to Cbfa1-fluorescein isothiocyanate-conjugated secondary antibody and/or mouse M/C antibody to Xpress tag-rhodamine-conjugated goat anti-mouse IgG). Note the expected nuclear localization of Cbfa1 in panel A and the mostly cytoplasmic or perinuclear location of tagged TAZ in panel B. Upon cotransfection with Cbfa1, the staining for tagged TAZ is mostly in the nucleus (see panel C). Panel D shows Cbfa1 expression in the nucleus of TAZ-Cbfa1-transfected cells. Panel E shows computer-generated overlays of individual cells stained for both Cbfa1 and TAZ, giving a yellow color where there is double staining. These data clearly suggest nuclear translocation of TAZ upon cotransfection with Cbfa1. (b) Immunohistochemistry of ROS 17/2.8 cells stably transfected with full-length TAZ (A to C) or the dominant-negative construct N-TAZ (D to F). Also shown is immunohistochemistry of MC 3T3 cells transiently transfected with full-length TAZ and Cbfa1 (G and H) or the dominant-negative N-TAZ and Cbfa1 (I and J). The cells in panels A, D, G, and I were stained with antibody to Cbfa1, and the cells in panels B, E, H, and J were stained with antibody to Xpress tag fused to TAZ constructs. Note the predominantly nuclear location of Cbfa1 in panels A and G, whereas in panels D and I the Cbfa1 is located in the cytoplasmic or perinuclear area. Similarly, panels B and H show predominantly nuclear localization of TAZ, whereas panels E and J show the perinuclear presence of N-TAZ. Panels C and F are computer-generated overlays of panels A and B and panels D and E, respectively. (c) Percentage of stably transfected ROS 17/2.8 cells showing the location of TAZ and N-TAZ in the various cellular compartments.
How TAZ may function was further explored by examining the localization of the dominant-negative mutant (N-TAZ) after stable transfection of ROS 17/2.8 cells and transient transfection of MC3T3 cells. In both osteoblast cell lines, the dominant-negative construct was present predominantly in a perinuclear location (Fig. 5b). Consistent with a role for TAZ in binding Cbfa1, colocalization of Cbfa1 with N-TAZ showed Cbfa1 to also reside in a perinuclear location. This is in marked contrast to its localization in the nucleus upon cotransfection with full-length TAZ or in untransfected cells (Fig. 5c).
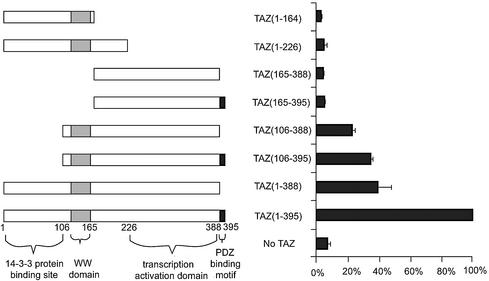
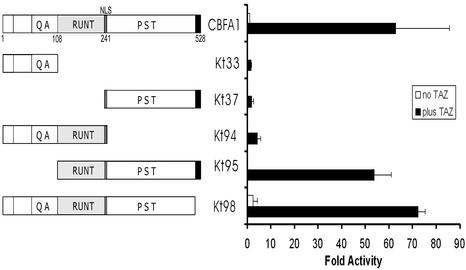
Finally, several deletion mutants of TAZ and Cbfa1 were used to test the interactions of specific domains. Initially, seven deletion mutants of TAZ were tested in functional assays upon cotransfection with full-length Cbfa1 and p6OSE2-luc reporter constructs in Jurkat cells. As shown in Fig. 6, deletion of the C-terminal PDZ-binding motif alone in TAZ reduced the activity of p6OSE2-luc to ca. 40% of the positive control TAZ. Similarly, deleting the 14-3-3 protein binding domain of TAZ also reduced the activity to <40%. The most dramatic effect was seen when the WW domain and the transcriptional activation domains were deleted. In a reverse experiment, full-length TAZ was tested in functional assays with several deletion mutants of Cbfa1. As shown in Fig. 7, deletion of the Runt and PST domains eliminated the effects of TAZ. Interestingly, eliminating the N-terminal portion up to the beginning of the Runt domain had increased activity by nearly 50-fold when TAZ is included compared to the control, wild-type Cbfa1 alone. Also, as expected, eliminating the C-terminal repressor region of Cbfa1 appeared to further increase the activity over that seen with the positive control Cbfa1 and TAZ.
FIG. 6.
Transcriptional coactivation assay in Jurkat cells with 6OSE2-luc as reporter construct to show the effects of deleting specific domains of TAZ on the transcriptional activity of full-length Cbfa1. Cbfa1 and 6OSE2-luc were transfected at 0.5 μg/well in each of the experiments. The various mutant constructs of TAZ (shown on the left) were used at 0.5 μg/well and assayed 30 h after transfection. Data are expressed as percentages of the positive control (full-length TAZ and Cbfa1 activity as 100%). Note the virtual elimination of activity upon deleting the WW and transcriptional activation domains.
FIG. 7.
Transcriptional coactivation assay in Jurkat cells with 6OSE2-luc as a reporter construct to show the effects of deleting specific domains of Cbfa1 on the transcriptional coactivation function of full-length TAZ. TAZ and 6OSE2-luc were transfected at 0.5 μg/well in each of the experiments. The various mutant constructs of Cbfa1 (shown on the left) were used at 0.5 μg/well and assayed 30 h after transfection. Data are expressed as fold increases in activity over that of the control (with full-length Cbfa1 activity being considered equal to “1”).
DISCUSSION
TAZ was identified here as a transcriptional coactivator that binds to the osteoblast-specific transcriptional factor Cbfa1. When Cbfa1 was used as bait to identify binding proteins from a 17-day mouse embryo library in the yeast two-hybrid system, only three clones that showed rapid growth, indicating the strong binding of Cbfa1were identified. Two of these three clones were identical copies of TAZ, and subsequent analysis reconfirmed the functional binding of TAZ to Cbfa1. TAZ is known to function as a coactivator for transcription factors with PPXY motifs, and Cbfa1 does have a PPPY motif. The present series of functional studies established the transcriptional coactivation of Cbfa1 as an important function of TAZ.
TAZ (transcriptional coactivator with PDZ-binding motif) is a recently described transcriptional coactivator factor with binding sites for 14-3-3 proteins, PDZ proteins and a WW domain (12). TAZ is known to bind to 14-3-3, a conserved family of ∼30-kDa proteins that bind to a large number of intracellular phosphoproteins involved in signal transduction to regulate differentiation, cell cycle progression, and apoptosis (1, 8, 12). TAZ has 395 amino acid residues, shares homology with YAP (Yes-associated protein), and contains a single WW domain. It functions as a transcriptional coactivator by binding to the PPXY motif present in targeted transcription factors (12). The C terminus of TAZ has the highly conserved PDZ-binding motif that helps localization of TAZ into discrete nuclear foci and is essential for TAZ-stimulated gene transcription. PDZ domains are present in a large number of proteins and proteins with PDZ domains may be membrane associated or in some instances end up in the nucleus (19, 22). The binding of TAZ to 14-3-3 proteins can inhibit nuclear translocation of TAZ and thus inhibit the transcriptional coactivation function (12).
The function of WW domain is clear in that it binds to the PPXY motifs in transcription factors and may thus be facilitating the interaction between the transactivation domains of TAZ and the activation domains of transcription factors. However, the role of the PDZ domain-binding region at the C-terminal end of TAZ is unclear. The PDZ domain-binding motif at the C-terminal end of TAZ may serve to localize the protein into punctate nuclear foci, as well as plasma membrane-associated complexes (12). The high-level expression of TAZ mRNA has been detected in the mouse heart, lung, liver, and kidney (12). However, the tissue distribution pattern for TAZ mRNA is slightly different in humans, where it was additionally found in the skeletal muscle, spleen, small intestine, and placenta (12). The present data on the expression of TAZ protein in osteoblasts lining the developing bone and the presence of TAZ mRNA in 7-day-old mouse calvaria that is enriched in osteoblasts demonstrates that TAZ is expressed in differentiating osteoblasts. More importantly, since TAZ mRNA is also found in nonmineralizing tissues such as the heart, lung, liver, and muscle, TAZ may be either functional as a transcriptional coactivator on a more general level or it may be functioning at different capacities in these various tissues. Cotransfecting TAZ with four different promoter constructs (cyclin D1, TAT3, AchR, and Smad7) revealed that TAZ function was largely restricted to enhancement of transcription of Cbfa1-transactivating promoters with minimal modulation of transcription at the other promoters tested. The current data derived from Jurkat cells grown in the presence of 10% FBS further indicates the possible restricted activity of TAZ. Since the PPXY motif is found in many other transcription factors, such as c-Jun, c/EBPα, and AP2 (12), Jurkat cell expression of the other four transcription factors might have been sensitive to TAZ transfection but was not. A preferential interaction of TAZ with the Runt family of transcription factors is an interesting possibility.
The present findings demonstrate that TAZ functions as a transcriptional coactivator for the osteoblast-specific transcription factor Cbfa1. It is interesting that although both YAP and TAZ are expressed in ROS 17/2.8 cells, a polyclonal antibody to Cbfa1 pulled down detectable levels of endogenous TAZ, whereas YAP could not be detected on Western blots. The polyclonal antibody to TAZ used here cross-reacts with YAP and, although it detected YAP in the lysate of ROS 17/2.8 cells immunoprecipitated with antibody to TAZ, it could not detect any YAP in Cbfa1 antibody precipitates. In fact, ROS 17/2.8 cells had higher levels of endogenous YAP in their lysates compared to TAZ. This finding is difficult to explain given the previous findings by other investigators (25) that YAP may bind to PEBP2α. It is possible that interactions of endogenous Cbfa1/TAZ and Cbfa1/YAP may be of a different nature and involve other proteins and thus could vary between cell types. Our initial experiments with NIH 3T3 cells showed only a twofold increase in transcription of osteocalcin promoter constructs. This minor upregulation of osteocalcin promoter activity in transfected NIH 3T3 cells could be attributed to the basal level expression of TAZ in these cells since NIH 3T3 cells showed some basal level. When TAZ was cotransfected with Cbfa1 and osteocalcin promoter construct in Jurkat cells that do not have any expression of TAZ (12), a nearly ninefold increase in the transcription of the luciferase reporter gene was noted. In addition, a clear dose responsiveness could be established for the transcriptional coactivation effects of TAZ in the reporter system used in both NIH 3T3 and Jurkat cells. Furthermore, a truncated construct of TAZ that includes only the first 164 amino acids with the PPXY motif-binding WW domain intact showed a dramatic reduction in the coactivation of Cbfa1 transcriptional activity. Perhaps the most convincing data were from the Northern blot analysis of the effects of TAZ and a dominant-negative construct of TAZ on endogenous osteocalcin mRNA expression in stably transfected rat osteosarcoma cells (ROS 17/2.8). These findings clearly indicate that TAZ may be serving as a transcriptional coactivator for the osteoblast-specific transcription factor Cbfa1. In addition, TAZ failed to activate the other OSE1 in the osteocalcin gene, as well as the OSE2 construct with a mutation in the Cbfa1-binding region. In a recent study, Gutierrez et al. (9) examined the possible coregulation of Cbfa1 by C/EBP_β_ and -δ. Interestingly, C/EBP and Cbfa1 factors interact together in a synergistic manner to enhance osteocalcin transcription in the cell culture model (ca. 40-fold). Hence, it appears that TAZ and C/EBP factors may be serving as coactivators of Cbfa1 at different sites on the osteocalcin promoter.
The transcriptional coactivation of Cbfa1 by TAZ was accompanied by the nuclear translocation of TAZ. NIH 3T3 cells transfected with Cbfa1 alone showed the expected nulcear localization of the transcription factor. TAZ, on the other hand, when transfected alone into NIH 3T3 cells and visualized by using a monoclonal antibody to the Xpress tag showed a more generalized pattern of expression in the cytoplasm and nucleus, with a tendency toward perinuclear accumulation. However, upon cotransfection with Cbfa1, most of the TAZ was found to localize into punctate nuclear bodies. When the truncated form of TAZ (N-TAZ) was also cotransfected with Cbfa1 and visualized by double fluorescence microscopy, the truncated N-TAZ was also found as punctate bodies but in a perinuclear location. Interestingly, stably transfected ROS 17/2.8 and MC 3T3 cells both showed significant cytoplasmic retention of Cbfa1 upon cotransfection with N-TAZ. These findings suggest that Cbfa1-TAZ binding interactions may be occurring in the cytoplasm itself. In fact, the dominant-negative effect seen with the use of N-TAZ in functional assays may be due to the retention of significant amounts of Cbfa1 in the cytoplasm bound to N-TAZ and unable to translocate into the cytoplasm. The finding of perinuclear retention of the truncated, dominant-negative construct of TAZ is in agreement with the findings by Kanai et al. (12), suggesting that the PDZ-binding motif may be important in the nuclear localization of TAZ. It is interesting that even with the full-length Cbfa1, the removal of the PDZ-binding motif of TAZ significantly reduces the transcriptional coactivation function. Although binding to Cbfa1 may be one of the mechanisms by which TAZ would translocate into the nucleus, the role of PDZ proteins also appears to be important. As far as the functional interactions between TAZ and Cbfa1 domains are concerned, data from analyses of deletion mutants suggest that all of the domains of TAZ are important in enhancing the transcriptional functions of Cbfa1. However, in the case of Cbfa1, deletion of the N-terminal end (up to the Runt domain) and the C-terminal repressor region enhances 6OSE2-luc activity by >50-fold when cotransfected with TAZ, suggesting that with Runt as a DNA-binding region and the PST region for its PPXY motif, the transcriptional coactivation function of TAZ is significant.
In conclusion, the present findings suggest that TAZ may be a specific transcriptional coactivator of Cbfa1. The transcription-activating function of TAZ may be specific with respect to Cbfa1 involving the PPPY motif in the PST region. All of the domains of TAZ appear to be necessary for the transcriptional activation of Cbfa1. Conversely, deletion of the QA region and repressor domain in Cbfa1 significantly enhances the transcription coactivating function of TAZ. The binding of TAZ to Cbfa1 appears to occur in the cytoplasm since the translocation of TAZ to the nucleus is predominant only when cotransfected with Cbfa1. More importantly, other proteins, such as the PDZ proteins, may be necessary in the translocation of Cbfa1-TAZ complexes to the nucleus. This is shown by the predominant perinuclear and/or cytoplasmic retention of the truncated TAZ (N-TAZ) and Cbfa1 in cotransfected osteoblast cell lines. Future studies should further investigate these TAZ-Cbfa1 complexes and the kinetics of their activities in osteoblast and nonosteoblast cell lines. The findings presented here suggest that TAZ is a coactivator of Cbfa1 and that coactivating factors such as TAZ and C/EBP_β_ and -δ may be important regulators of Cbfa1 function.
Acknowledgments
This research was supported by grants from the National Institutes of Health.
We thank, M. B. Yaffe at the Massachusetts Institute of Technology for providing the antibody to mouse TAZ.
REFERENCES
- 1.Aitken, A. 1996. 14-3-3 and its possible role in co-ordinating multiple signaling pathways. Trends Cell Biol. 6**:**341-347. [DOI] [PubMed] [Google Scholar]
- 2.Albanese, C., J. Johnson, G. Watanabe, D. N. Eklund, Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21 mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270**:**23589-23597. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, C., L. R. McCabe, J.-Y. Choi, S. W. Hiebert, J. L. Stein, G. S. Stein, and J. B. Lian. 1997. _Runt_-homology domain proteins in osteoblast differentiation: AML-3/Cbfa1 is a major component of a bone specific complex. J. Cell. Biochem. 66**:**1-8. [DOI] [PubMed] [Google Scholar]
- 4.Ducy, P. 2000. Cbfa1: a molecular switch in osteoblast biology. Dev. Dynam. 219**:**461-471. [DOI] [PubMed] [Google Scholar]
- 5.Ducy, P., and G. Karsenty. 1995. Two distinct osteoblast-specific _cis_-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 15**:**1858-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ducy, P., R. Zhang, V. Geoffrey, A. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcription activator of osteoblast differentiation. Cell 89**:**747-754. [DOI] [PubMed] [Google Scholar]
- 7.Ducy, P., M. Starbuck, M. Priemel, J. Shen, G. Pinero, V. Geoffry, A. Amling, and G. Karsenty. 1999. A Cbaf1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 13**:**1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu, H., R. R. Surbamaniam, and S. C. Masters. 2000. 14-3-3 proteins: structure, functions and regulation. Annu. Rev. Pharmacol. Toxicol. 40**:**617-647. [DOI] [PubMed] [Google Scholar]
- 9.Gutierrez, S., A. Javed, D. K. Tennant, M. van Rees, M. Montecino, G. Stein, J. Stein, and J. B. Lian. 2002. CCAAT/Enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277**:**1316-1323. [DOI] [PubMed] [Google Scholar]
- 10.Guttridge, D. C., M. W. Mayo, L. V. Madrid, C. Y. Wang, and A. B. Baldwin, Jr. 2000. NF-κB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science 289**:**2363-2366 [DOI] [PubMed] [Google Scholar]
- 11.Iniguez-Lluhf, J. A., D. Y. Lou, and K. R. Yamamoto. 1997. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J. Biol. Chem. 272**:**4149-4156. [DOI] [PubMed] [Google Scholar]
- 12.Kanai, F., P. A. Marignani, D. Sarbassove, R. Yagi, R. A. Hall, M. Donowitz, A. Hisaminato, T. Fujiwara, Y. Ito, L. C. Cantly, and M. B. Yaffe. 2000. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 24**:**6778-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karsenty, G. 2000. Bone formation and factors affecting this process. Matrix Biol. 19**:**85-89. [DOI] [PubMed] [Google Scholar]
- 14.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimuzu, R. T. Bronson, Y. H. Gao, M. Inada, M. Sato, R. Okamato, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89**:**755-764. [DOI] [PubMed] [Google Scholar]
- 15.Lian, J. B., G. S. Stein, J. L. Stein, and A. J. van Wijnen. 1998. Transcriptional control of osteoblast differentiation. Biochem. Soc. Trans. 26**:**14-21. [DOI] [PubMed] [Google Scholar]
- 16.Lian, J. B., G. S. Stein, J. L. Stein, and A. J. van Wijnen. 1998. Osteocalcin gene promoter: unlocking the secrets for regulation of osteoblast growth and differentiation. J. Cell. Biochem. Suppl. 20**/31:**62-72. [DOI] [PubMed] [Google Scholar]
- 17.Nagarajan, R. P., J. Zhang, W. Li, and Y. Chen. 1994. Regulation of Smad7 promoter by direct association with smad3 and smad4. J. Biol. Chem. 274**:**33412-33418. [DOI] [PubMed] [Google Scholar]
- 18.Otto, F., A. P. Thornwell, T. Crompton, A. Denzel, K. C. Gilmour, I. R. Rosewell, G. W. Stamp, R. S. Beddington, S. Mundlos, B. R. Olsen, P. Selby, and M. J. Owen. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89**:**765-771. [DOI] [PubMed] [Google Scholar]
- 19.Poulat, F., P. S. Barbara, M. Desclozeaux, S. Soullier, B. Moniot, N. Bonneaud, B. Boizet, and P. Berta. 1997. The human testis determining factor SRY binds a nuclear factor containing PDZ protein domains. J. Biol. Chem. ••**:**7167-7172. [DOI] [PubMed]
- 20.Takeda, S., J. P. Bonnamy, M. J. Owen, P. Ducy, and G. Karsenty. 2001. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 15**:**467-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thirunavakkarasu, K., M. Mahajan, K. W. McLarren, S. Stifani, and G. Karsenty. 1998. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transcriptional function and its inability to heterodimerize with Cbfβ. Mol. Cell. Biol. 18**:**4197-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas, M. K., K. M. Yao, M. S. Tenser, G. G. Wong, and J. F. Habener. 1999. Bridge-1, a novel PDZ domain coactivator of E2A-mediated regulation of insulin gene transcription. Mol. Cell. Biol. 19**:**8492-8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tintut, Y., F. Parhami, V. Le, G. Karsenty, and L. L. Dmeer. 1999. Inhibition of osteoblast-specific transcription factor by the cAMP pathway in osteoblastic cells. J. Biol. Chem. 274**:**28875-28879. [DOI] [PubMed] [Google Scholar]
- 24.Xiao, G., D. Jiang, P. Thomas, M. D. Benson, K. Guan, G. Karsenty, and R. T. Franceschi. 2000. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor Cbfa1. J. Biol. Chem. 275**:**4453-4459. [DOI] [PubMed] [Google Scholar]
- 25.Yagi, R., L. F. Chen, K. Shigesada, Y. Murakami, and Y. Ito. 1999. A WW domain-containing Yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18**:**2551-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]