Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor (original) (raw)
Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a potential anticancer drug that selectively induces apoptosis in a variety of cancer cells by interacting with death receptors DR4 and DR5. TRAIL can also bind to decoy receptors (DcR1, DcR2, and osteoprotegerin receptor) that cannot induce apoptosis. The occurrence of DR5-responsive tumor cells indicates that a DR5 receptor-specific TRAIL variant will permit tumor-selective therapies. By using the automatic design algorithm FOLD-X, we successfully generated DR5-selective TRAIL variants. These variants do not induce apoptosis in DR4-responsive cell lines but show a large increase in biological activity in DR5-responsive cancer cell lines. Even wild-type TRAIL-insensitive ovarian cancer cell lines could be brought into apoptosis. In addition, our results demonstrate that there is no requirement for antibody-mediated cross-linking or membrane-bound TRAIL to induce apoptosis through DR5.
Keywords: computational protein design, receptor selectivity, biopharmaceutical, death receptor, apoptosis-inducing ligand 2
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) is currently attracting great interest as a potential anticancer therapeutic. TRAIL, in its soluble form, selectively induces apoptosis in tumor cells in vitro and in vivo by a death receptor-mediated process. Unlike other apoptosis-inducing TNF family members, soluble TRAIL appears to be inactive against normal healthy tissue (1). Reports in which TRAIL induces apoptosis in normal cells could be attributed to the specific preparations of TRAIL used (2). TRAIL shows a high degree of promiscuity as it binds to five cognate receptors: DR4 (TRAIL-R1) and DR5 (TRAIL-R2) and the decoy receptors DcR1 (TRAIL-R3), DcR2 (TRAIL-R4), and osteoprotegerin (OPG) (3). Upon binding to TRAIL, DR4 and DR5 receptors recruit Fas-associated death domain, which binds and activates the initiator caspase 8, leading to apoptosis (4–6). DcR1 or DcR2 do not contain a death domain or a truncated death domain, respectively, and therefore could prevent apoptosis by sequestering available TRAIL or by interfering in the formation of a TRAIL–DR4 or –DR5 signaling complex (7).
Use of TRAIL receptor-selective variants could permit better tumor-specific therapies through escape from the decoy receptor-mediated antagonism, resulting in a lower administrated dose with possibly fewer side effects and as alternatives to existing agonistic receptor antibodies (8–10). In experimental anticancer treatments, the receptors DR4 and/or DR5 were shown to be up-regulated after treatment with DNA-damaging chemotherapeutic drugs, and the response to TRAIL-induced apoptosis was significantly increased (3, 11). In addition, irradiation appears to specifically up-regulate DR5 receptor expression, and the combination of irradiation and TRAIL treatment has been demonstrated to have an additive or synergistic effect (12). Thus, we chose to develop DR5 receptor-selective TRAIL variants by using a computational design strategy. Computational design methods have been successfully used to redesign several protein–protein interactions (13–16) but have, as yet, hardly been applied to therapeutic proteins. One exception is the design of dominant negative TNF-α variants that prevent formation of active TNF-α trimers (17). By using the automatic design algorithm FOLD-X (18–20), we were able to redesign TRAIL into exclusively DR5-specific agonistic variants. Because the computational method used in our study is based on general applicable principles and has been successfully tested on a variety of proteins (14, 19, 21–23), our method can be further applied to design other protein therapeutics with reduced promiscuity and improved receptor-binding characteristics.
Results
Modeling of TRAIL–Receptor Complexes.
Monomeric subunits of TRAIL self-associate in bell-shaped homotrimers, the bioactive form of the ligand, like other members of the TNF ligand family (24, 25). A trimer binds three subunits of a cognate receptor, with each receptor subunit bound in the grooves between two adjacent monomer ligand subunits (26, 27). At present, only crystal structures of TRAIL in complex with the DR5 receptor are known (26–28). The sequence alignment of the different TRAIL receptors shows a large overall sequence identity (except for OPG), practically no insertions or deletions, and conservation of all cysteines involved in the formation of internal disulfide bridges (Fig. 5_A_, which is published as supporting information on the PNAS web site). Consequently, good quality homology models of DR4, DcR1, and DcR2, but not of OPG, could be built. The homology models were built by using the what if web interface (29). Afterward, these models were refined by using the protein design options of FOLD-X, removing incorrect side-chain torsion angles, eliminating van der Waals clashes, and accommodating TRAIL and receptor residues to their new interface.
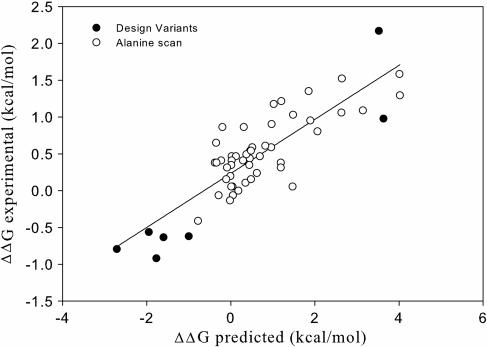
The accuracy of the models and the force field was tested by using the data derived from the alanine scanning of wild-type TRAIL as performed by Hymowitz et al. (30). The predictions of the energy change in the complex formation correlates with the changes in the dissociation constants measured (Fig. 1). The calculated _R_2 factor is 0.6 (_R_2 factors calculated for DR4 and DR5 individually also amount to 0.6). However, several factors involved in accuracy should be taken into account. The methodology used focuses on energy changes in ligand–receptor complex formation. Some mutations to alanine might be predicted not to change receptor-binding affinity but only to produce slight changes in TRAIL stability, thereby affecting the correlation. The prediction error is, on average, within the error of this methodology (0.6–0.7 kcal/mol) (1 kcal = 4.18 kJ). Because many changes in affinity, as measured in the alanine scanning, are within this error, it is not possible to obtain a better correlation. Taken together, these data imply that our method can reliably predict mutations in the receptor-binding interface that will severely affect the complex formation.
Fig. 1.
Correlation of the predicted changes in binding affinity toward DR4 and DR5 compared with the experimental results of an alanine scanning performed by Hymowitz et al. (30) (open circles) and of the DR5-selective TRAIL variants (closed circles).
Computational Design of the Variants.
For the computational screening, all residues from the TRAIL interface were considered. TRAIL residues interacting with a conserved amino acid environment in all four receptors were disregarded. Amino acids finally considered were as follows: Arg-130, Gly-131, Arg-132, Lys-145, Leu-147, Gly-148, Arg-149, Lys-150, Glu-155, Arg-158, Gly-160, His-161, Tyr-189, Arg-191, Phe-192, Gln-193, Glu-195, Asn-199, Thr-200, Lys-201, Asp-203, Gln-205, Val-207, Gln-208, Tyr-209, Thr-214, Asp-218, Asp-234, Glu-236, His-264, Ile-266, Asp-267, and Asp-269. Tyr-216 was included as a positive control because of its already-known implication in receptor binding (26, 27), and Ser-165, located far away from the receptor-binding interface, was used as a negative control (Fig. 5_B_). At each of the selected positions, FOLD-X placed the 20 natural amino acids while moving the neighboring residues, obtaining a total library of 2,720 models (34 amino acid positions × 20 amino acids × 4 receptors). The energy of interaction was obtained by calculating the sum of the individual energies of the receptor and ligand subunits and subtracting them from the global energy of the complex. In this way, a set of predicted energetic values for the complex formation was obtained and compared with the wild-type TRAIL values. After studying these values together with visual inspection of the mutant models, those variants in which a change in selectivity was predicted were selected for experimental studies (Table 1).
Table 1.
Predicted difference in binding energy (ΔΔ_G_) of DR5-selective variants binding to different receptors when compared with wild-type TRAIL
| Mutations | DR4 | DR5 | DcR1 | DcR2 |
|---|---|---|---|---|
| R130E | 0.75 | −0.2 | 1.76 | 1.52 |
| G160M | −1.11 | −1.52 | −0.18 | −0.65 |
| E195R | 0.11 | −1.11 | 0.2 | −0.79 |
| T214R | 1.85 | −0.17 | 1.94 | 1.89 |
| D269H | 3.52 | −1.6 | 3.78 | 4.43 |
| D269R | 1.95 | −1.95 | 2.45 | 3.28 |
| D269K | 2.43 | −1 | 2.94 | 3.71 |
Prescreen for Selective Receptor Binding.
A fast surface plasmon resonance (SPR)-based receptor-binding prescreen was used to further refine the in silico selection. TRAIL-variant cell extracts were evaluated for binding to DR4-, DR5-, and DcR1-immobilized Ig fusion proteins. The ratios of binding to DR4 and DcR1 receptors with respect to the DR5 receptor were calculated and compared with the ratio obtained for wild-type TRAIL. An increase in the DR5/DR4 binding ratio of ≥25% relative to the ratio of wild-type TRAIL was set as indicative of DR5 selectivity. Several variants comprising a substitution (His, Lys, or Arg) at position Asp-269 and variants with double mutation D269H/E195R and D269H/T214R with reduced binding to the DR4 receptor and increased binding to the DR5 receptor were chosen for further analysis. R191E/D267R, R130E, G160M, I220M, and E195R were also selected, because they also showed an increased DR5/DR4 binding ratio. The effects, however, were smaller than that of the Asp-269 variants (data not shown).
Determination of Receptor Binding.
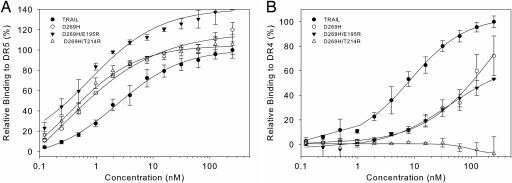
Selected TRAIL variants were purified as described in ref. 22. Analytical size-exclusion chromatography and dynamic light scattering confirmed that the purified TRAIL variants were in a trimeric state and that higher order oligomeric species or aggregates were absent (data not show). Binding of the purified variants to the immobilized DR4-, DR5-, DcR1-, or DcR2-Ig receptor was assessed in real time by using SPR. The TRAIL proteins were initially analyzed at two concentrations (30 and 60 nM). TRAIL variants R191E/D267R and G160M showed stability and folding problems and were therefore discarded. Binding curves of variants showing a significant change in the ratio of DR5/DR4 binding were subsequently recorded for concentrations ranging from 0.1 to 250 nM. The D269H/T214R variant had an improvement comparable with the D269H single mutant variant in DR5-Ig binding, however no detectable binding to DR4-Ig was found (Fig. 2A and B). Apparent K_d values for DR5 binding ranged from 0.6 (D269H/E195R) to 2.5 nM (TRAIL) and from 7.2 (TRAIL) to 244 nM (D269H) for DR4 binding. For D269H/T214R, D269K, and D269R, a proper apparent K_d for DR4 binding could not be determined. Binding of D269H and D269H/E195R toward the decoy DcR1-Ig receptor was >20-fold reduced when compared with wild-type TRAIL. Up to the highest concentration tested (250 nM), D269H/T214R did not show any observable binding to DcR1-Ig (Fig. 6_A, which is published as supporting information on the PNAS web site). D269H and D269H/E195R also showed reduced binding to DcR2-Ig; however, this reduction was much less pronounced than the reduction observed in DcR1 binding. In contrast, D269H/T214R showed a large decrease in binding to DcR2-Ig relative to wild-type TRAIL (Fig. 6_B). Binding to OPG-Ig was also reduced for these three DR5-selective variants, with D269H/E195R showing the largest decrease in binding to this receptor (Fig. 6_C_). A competition ELISA experiment measuring the binding of TRAIL or variants toward immobilized DR5-Ig in the presence of soluble DR4-, DR5-, or DcR1-Ig corroborated the findings of the receptor-binding experiment. Whereas TRAIL binding to immobilized DR5-Ig could be competed by soluble DR4-, DR5-, and DcR1-Ig, binding of the variants could only be antagonized by soluble DR5-Ig (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
Receptor binding of TRAIL and DR5-selective variants toward DR5-Ig as determined by SPR (A) or toward DR4-Ig (B). Receptor binding is calculated relative to the response of TRAIL at 250 nM.
Comparison Between Predictions and Experimentally Obtained Results.
To calculate the correlation between the predicted and experimentally obtained results of our DR5-selective variants, the calculated ΔΔ_G_ values for DR4 and DR5 binding (Table 1) were compared with the ΔΔ_G_ values that stem from the experimentally determined apparent _K_d values (see above). The calculated R_2 factor between these predicted and experimental ΔΔ_G values is 0.9. Adding these values to the alanine scan data set improved the overall calculated _R_2 from 0.6 to 0.7 (Fig. 1).
Biological Activity.
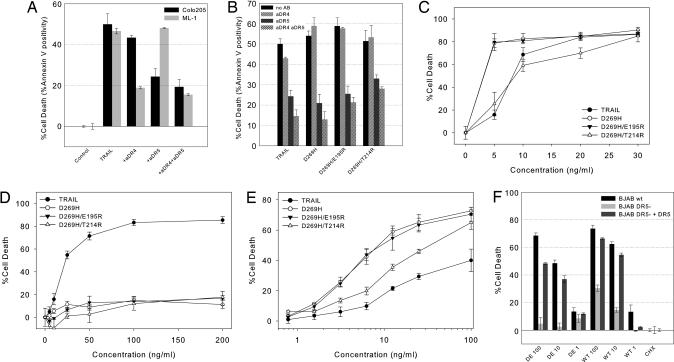
To assess the biological activity related to DR5 binding, various cancer cells were used. Colo205 colon carcinoma cells and ML-1 chronic myeloid leukemia cells express all four TRAIL receptors on the cell surface, as shown by using FACS analysis, (Fig. 8, which is published as supporting information on the PNAS web site), and are sensitive to TRAIL-induced apoptosis. To test the involvement of DR4 versus DR5 in TRAIL-induced cell death, Colo205 cells were treated with neutralizing anti-DR4 or anti-DR5 antibody for 1 h before the addition of TRAIL. Both antibodies reduced TRAIL-mediated cell death and had an additive effect when used in combination (Fig. 3A). However, the DR5-neutralizing antibody was ≈3 times more effective than the DR4-neutralizing antibody, demonstrating that TRAIL-induced apoptosis in Colo205 cells is primarily mediated by DR5. In contrast, the DR4 pathway is the major mediator of TRAIL-induced apoptosis in ML-1 cells (Fig. 3A). To examine whether the DR5-specific TRAIL variants induce cell death in Colo205 cells by way of the DR5 receptor, 1 μg/ml neutralizing anti-DR4 or -DR5 antibodies were administered 1 h before ligand treatment. The presence of the anti-DR4 antibody failed to prevent death induced by the DR5-specific variants. However, 1 μg/ml anti-DR5 antibody significantly reduced the amount cell death (Fig. 3B).
Fig. 3.
Biological activity of TRAIL and DR5-selective variants. (A) Apoptosis-inducing activity of 100 ng/ml TRAIL in the presence of 1 μg/ml DR4 (aDR4), DR5 (aDR5), or DR4 and DR5 (+aDR4+aDR5) receptor-neutralizing antibodies in Colo205 and ML-1 cells. (B) Apoptosis-inducing activity in Colo205 cells of 100 ng/ml TRAIL or DR5-selective variants without the presence of neutralizing DR4 or DR5 antibodies (no AB) or in the presence of neutralizing antibody [aDR4, aDR5, or both (aDR4 aDR5)]. Shown is the cytotoxic potential (% cell death) of TRAIL or DR5-selective variants in Colo205 (C), ML-1 (D), and A2780 (E) and of 1, 10, or 100 ng/ml TRAIL (WT) or D269H/E195R (DE) relative to cycloheximide control (0.33 μg/ml) in BJAB cells responsive to both DR4- and DR5-mediated cell death (BJABwt), BJAB cells deficient for DR5 (BJABDR5 DEF), and BJAB cells deficient for DR5 stably transfected with DR5 (BJABDR5 DEF+DR5) (F) (31).
Colo205 and ML-1 cells were then treated with increasing concentrations of TRAIL or the DR5-specific variants D269H, D269H/E195R, and D269H/T214R, and their cytotoxic potential was measured with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. In Colo205 cells, all TRAIL ligands were biologically active and induced cell death at levels that were either comparable with that of wild-type TRAIL or were up to 5-fold more active than wild-type TRAIL (Fig. 3C and Table 2). Contrary to Colo205 cells, only TRAIL was able to induce cell death in ML-1 cells (Fig. 3D). Similar results were obtained by using EM-2 chronic myeloid leukemia cells expressing only the DR4 receptor and lacking the DR5 receptor and by using the ovarian cancer cell line A2780, which expresses DR5 but lacks DR4 on its surface and is relatively insensitive toward TRAIL-induced cell death (S. de Jong, personal communication). Although EM-2 cells were sensitive to TRAIL-induced cell death (50 ng/ml TRAIL initiating >80% cell death), treatment with any of the DR5 mutants failed to induce significant cell death (Fig. 9, which is published as supporting information on the PNAS web site). In A2780 cells, however, the cytotoxic activity of D269H, D269H/E195R, and D269H/T214R is significantly increased, showing both an increased maximum response and drastically decreased EC50 values when compared with wild-type TRAIL (Fig. 3E and Table 2). An additional experiment using D269H/E195R in wild-type BJAB cells responsive to both DR4- and DR5-mediated cell death (BJABwt), BJAB cells deficient in DR5 (BJABDR5 DEF), and BJAB cells deficient in DR5 and stably transfected with DR5 (BJABDR5 DEF+DR5) (31) confirm our findings. D269H/E195R was able to induce cell death in BJABwt cells but was unable to induce significant cell death in BJABDR5 DEF cells when compared with wild-type TRAIL. In the DR5 transfected BJABDR5 DEF+DR5 cells, however, the cytotoxic potential was restored (Fig. 3F). The cytotoxic effects of these TRAIL variants on noncancerous human umbilical vein endothelial cells was assessed by incubating these cells in the presence of 100 ng/ml TRAIL or TRAIL variants. However, no cytotoxic effects were observed for TRAIL and the receptor-selective TRAIL variants (data not shown). Taken together, the results obtained with the Colo205, ML-1, A2780, and BJAB cell lines show that the biological activity of the D269H, D269H/E195R, and D269H/T214R variants is specifically directed toward the DR5 receptor.
Table 2.
EC50 values of Colo205 and A2780 cells
| Ligand | Colo205 | A2780 | ||
|---|---|---|---|---|
| EC50, ng/ml | Max effect, % cell death | EC50, ng/ml | Max effect, % cell death | |
| TRAIL | 8.6 ± 0.9 | 78 ± 8 | 15.6 ± 3 | 41 ± 3 |
| D269H | 1.8 ± 0.5 | 80 ± 4 | 4.7 ± 0 | 70 ± 5 |
| D269H E195R | 1.5 ± 0.4 | 80 ± 6 | 4.2 ± 1 | 69 ± 2 |
| D269H T214R | 5.1 ± 2.6 | 66 ± 9 | 12.1 ± 4 | 66 ± 11 |
Discussion
Because the DR5 receptor is a good target for TRAIL cancer therapy (see the Introduction), we choose to develop DR5 receptor-selective variants of TRAIL by using a computational design strategy.
Structural Basis for the Changes in Selectivity.
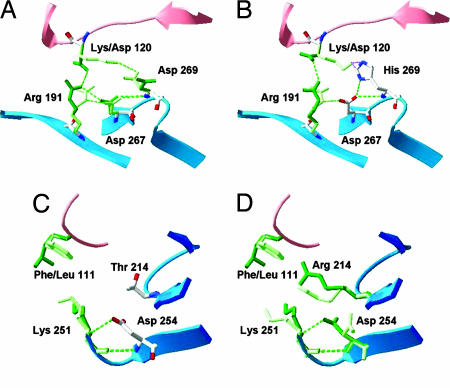
This study shows that residue 269 is one of the most important residues for DR5 selectivity. From the crystal structure of TRAIL in complex with DR5, it can be observed that this amino acid is not interacting directly with the receptor. Studying the models of TRAIL in complex with the other three receptors reveals that Asp-269 from TRAIL is interacting with Lys-120 from the receptor. This lysine residue is conserved among the DR4, DcR1, and DcR2 receptors. In contrast, DR5 has an aspartate at this position (Fig. 4A and B and Fig. 5_A_).
Fig. 4.
Area of interaction of TRAIL and DR4/DR5 receptor around position 269 [TRAIL (A) and D269H variant (B)] and around position 214 [TRAIL(C) and T214R variant(D)]. Red ribbons indicate a receptor, and blue ribbons indicate TRAIL. Residues in DR5 complexes are in dark green, and residues in DR4 complexes are in light green. Arg 191 and Asp 267 are key TRAIL amino acids for DR5 receptor binding in the corresponding binding pocket of the receptor, as observed in the crystal structure of TRAIL in complex with DR5.
Changing this amino acid to another with opposite charge shows two cumulative effects. On one hand, breaking the Asp-269–Lys-120 interaction in the complex between TRAIL and receptors DR4, DcR1, and DcR2 would decrease TRAIL affinity toward them; furthermore, Lys-120 has little space for reaccommodation, and this may even introduce some van der Waals clashes in the area. On the other hand, Asp-120 from the DR5 receptor may interact with the protonated His-269 of TRAIL, improving binding toward this receptor. In summary, this combination of effects explains why a single mutation alone can greatly change the selectivity toward DR5, resulting in better binding to the DR5 receptor and a substantial decrease in binding toward the other receptors. Residue 214 is also important for achieving DR5 selectivity. For the T214R mutation, FOLD-X predicts a decrease in binding affinity for all receptors except DR5 (Table 1). This decrease is due to the presence of a phenylalanine at position 111 in DR4 and a proline in DcR1 and DcR2, which prevent proper accommodation of Arg-214 upon complex formation. As a result, the arginine displaces Asp-254 and breaking intramolecular H bonds. In DR5, a leucine at position 111 allows accommodation of Arg-214 without displacement of Asp-254 (Fig. 4 C and D). An additive effect of mutations toward selectivity can be expected in the cases in which the positions of the mutations are far enough away from each other that they cannot make any unpredictable interaction, e.g., mutations D269H and T214R.
Selective Binding to Different Receptors.
Receptor-binding experiments using SPR and competition ELISA experiments confirmed the modeling predictions. Variants D269H, D269H/E195R, D269K, and D269R are between 70- to 150-fold more selective for the DR5 receptor than for the DR4 receptor when compared with wild-type TRAIL. The D269H/T214R variant showed no binding to the DR4 receptor at the highest concentration used in the assay (250 nM). The dissociation rates of TRAIL and the DR5-selective variants in complex with the DR5 and DR4 receptor were, however, too slow to measure accurately by using SPR, thereby precluding the accurate determination of affinity constants. In the competition ELISA experiment, DR4 was unable to compete with immobilized DR5 for the binding to these designed selective variants, demonstrating that, in the presence of both DR4 and DR5, these variants are markedly more selective toward DR5. The net gain in DR5 selectivity of these variants is the sum of both an increased preference for the DR5 receptor and a reduced preference for the DR4 receptor, exemplifying both positive and negative design principles (15).
Binding of the D269H and D269H/E195R variants to the decoy DcR1 receptor was >20-fold reduced when compared with wild-type TRAIL. The D269H/T214R variant showed no binding to the DcR1 receptor at the highest concentration used in the assay (250 nM). Although binding of the D269H and D269H/E195R variants toward the decoy DcR2 receptor was reduced, the effect was much less pronounced when compared with the reduction in binding as observed with the other receptors. The different environment of Lys-120 in receptor DcR2 when compared with DR4 and DcR1 could explain why the decrease in affinity is smaller in this case in contrast to our predictions. However, the D269H/T214R variant showed an ≈80% decrease in receptor binding to the DcR2 receptor when compared with wild-type TRAIL.
The DR5 Receptor Produces Apoptosis Without Additional Cross-Linking Requirements.
By using several different cancer cell lines, receptor-selective behavior of the DR5-selective variants could also be demonstrated in several in vitro biological assays. In cells with the DR4 receptor as the major mediator of TRAIL-induced apoptosis (ML-1 and EM-2 cells), DR5-selective variants were unable to induce apoptosis even at high concentrations (200 ng/ml). These variants could, however, induce apoptosis in cells with DR5 as the major mediator of TRAIL-induced apoptosis (Colo205), and this induction could be antagonized by using a neutralizing anti-DR5 antibody. The cell death-inducing activity against Colo205 cells was comparable with wild-type TRAIL (EC50 ≈8.6 ng/ml) in the case of D269H/T214R (EC50 ≈5.1 ng/ml) or increased >5-fold in the case of D269H/E195R (EC50 ≈1.5 ng/ml). In the DR5-positive and DR4-negative A2780 cells, the increase in cell death-inducing activity of the DR5-selective variants was even more pronounced. By using the various BJAB cell lines, it was confirmed that D269H/E195R-mediated induction of cell death was dependent on the presence of the DR5 receptor, and it was observed that the presence of only the DR4 receptor was not sufficient to induce cell death for this DR5-selective variant. Taken together, the in vitro biological activity data convincingly demonstrate that differences in receptor selectivity, as measured in the in vitro receptor-binding assay, are both relevant and significant in the in vitro biological context.
Both our results and results recently published in ref. 32 suggest that cross-linking TRAIL or membrane-bound TRAIL is not an absolute prerequisite for DR5-mediated induction of apoptosis, as was concluded by others (33, 34). A 10-fold improvement in DR5-mediated activity of flag-tagged TRAIL upon cross-linking was demonstrated; however, this also resulted in toxicity in normal cynomolgus monkey hepatocytes (32). Our soluble trimeric DR5-selective TRAIL variants are capable of inducing DR5 receptor-mediated apoptosis at lower concentrations than wild-type TRAIL, thus eliminating any requirement for antibody-mediated cross-linking.
Designed Versus Selected Variants.
Other DR5 receptor-selective TRAIL variants were recently isolated by using phage display (32). These variants were selected from saturation mutagenesis libraries that were constructed on the basis of a previously performed alanine scan (30). Remarkably, the best DR5-selective mutant (DR5–8) contained six amino acid substitutions. The mutations we found (e.g., D269H, E195R, and T214R) to induce DR5 selectivity were not identified by the phage-display approach. In a partial dissection to determine the role of each mutation in selectivity, Kelley et al. (32) could not eliminate any of the mutations without losing selectivity and/or biological activity. It was concluded that, to achieve receptor selectivity, multiple amino acid substitutions were required. However, our results clearly demonstrate that, in case of the D269H/T214R variant, only two amino acid substitutions are required to obtain complete receptor selectivity. Having fewer mutations relative to the wild-type sequence appears favorable in view of a potential use of the DR5-selective variants as anticancer therapeutics, because fewer mutations are likely to reduce the risk of an immunogenic response.
Conclusion
This study shows that computational redesign of the receptor-binding interface of TRAIL to obtain DR5-selective variants is achievable. In vitro analysis demonstrates that our DR5-selective mutants have increased affinity for DR5, whereas they do not bind to DR4. Our DR5-selective variants show high activity toward DR5-responsive cancer cells without the need for additional cross-linking. Consequently, these variants are of interest for development as a potential anticancer therapeutic. Previously, we designed TRAIL variants with improved thermal stability by using a computational redesign strategy (22). Computational protein redesign methods are therefore a valuable addition to other protein engineering methodologies, such as directed evolution or experimental high-throughput approaches, as a tool for the improvement of protein properties. Combining computational and experimental screening methods is a powerful approach in protein engineering; a preliminary computational screening of proteins helps to identify the most important positions involved in protein–protein interactions and therefore decreases the number of variants to screen.
Methods
All reagents were of analytical grade unless specified otherwise. Recombinant TRAIL Ig receptor fusion proteins were ordered from R & D Systems. PBS (pH 7.4) and RPMI medium 1640 were obtained from Invitrogen. All other chemicals were from Sigma. All buffers used in SPR, ELISA, and biological activity assays were of physiological pH and ionic strength.
Computational Design of the Variants.
Homology models of DR4, DcR1, and DcR2 were built by using the what if (29) web interface based on human TRAIL in complex with the DR5 ectodomain (26). Afterward, these models were refined by using the protein design options of FOLD-X, removing incorrect torsion angles, eliminating van der Waals clashes, and accommodating TRAIL and receptor residues to their new interface and to build up the putative interactions between TRAIL and the three noncrystallized receptors through rotamer substitution. The crystal complex structure of TRAIL with the DR5 receptor was also refined this way (see Supporting Methods, which is published as supporting information on the PNAS web site). A detailed description of the empirical force field FOLD-X is available in ref. 18 and at http://fold-x.embl-heidelberg.de.
In addition, the modified version of FOLD-X used in this work (20) is able to perform amino acid mutations, accommodating this new residue and its surrounding amino acids in the following way: It first mutates the selected position to alanine and annotates the side-chain energies of the neighbor residues. Then it mutates this alanine to the selected amino acid and recalculates the side-chain energies of the same neighboring residues. Those that exhibit an energy difference are then mutated to themselves to see whether another rotamer will be more favorable. This feature allows for proceeding through the whole computational design process by using just a single force field. The method does not guarantee a global minimum, but we have found that it is able to find the wild-type side-chain conformations when doing side-chain reconstruction from a polyAla backbone (F. Stricher and L. Serrano, personal communication).
Side-Directed Mutagenesis, Expression, and Purification of Selectivity Mutants.
cDNA corresponding to human soluble TRAIL (amino acids 114–281) was cloned in pET15B (Novagen) by using NcoI and BamHI restriction sites. Mutants were constructed by PCR as described in ref. 22. Homotrimeric TRAIL proteins were purified by using a three-step purification process as described in ref. 22.
SPR Receptor-Binding Assay.
Binding experiments were performed by using a SPR-based biosensor, Biacore 3000. Immobilization of the DR4- and DR5-Ig receptors on the sensor surface of a Biacore CM5 sensor chip was performed by following a standard amine-coupling procedure according to the manufacturer’s instructions. Receptors were coated at a level of ≈600–800 resonance units. Eighty microliters of TRAIL and variants were injected 3-fold at concentrations ranging from 250 to 0.1 nM at 70 μl/min and at 37°C by using PBS (pH 7.4) supplemented with 0.005% vol/vol P20 (Biacore) as running and sample buffer. Binding of ligands to the receptors was monitored in real time. Due to the very slow dissociation of the TRAIL–receptor complex, only pre-steady state binding data could be obtained. Furthermore, a fast initial dissociation was observed directly after the end of injection, pointing at some heterogeneity in complex formation. To obtain data that represent proper high-affinity complex formation, the response at each concentration was recorded 30 s after the end of the injections (contact time, 30 s). The response data as a function of TRAIL concentration were fitted by using a four-parameter equation to give an apparent affinity constant. Between injections, the receptor/sensor surface was regenerated by using 3 M sodium acetate (pH 5.2) injections. DcR1-, DcR2-, and OPG-Ig were captured by using a protein A-modified (Sigma) CM5 sensor chip, and the protein A sensor surface was regenerated by using 0.5 M glycine (pH 2). For the prescreening assay, 1:50 diluted clarified Escherichia coli BL21 extracts were injected at 50 μl/min (see Supporting Methods).
Biological Activity.
Cell lines and treatment.
Colo205 colon cancer cells, A2780 ovarium cancer cells, ML-1 myeloid leukemia cells, and the BJAB cell lines were maintained in RPMI medium 1640, 10% FCS/1% penicillin/1% streptomycin, in a humidified incubator at 37°C in a 5% CO2 environment. In the medium of BJABDR5 DEF+DR5 cells, puromycin (Sigma) was added to a final concentration of 1 μg/ml. TRAIL receptor inhibitors (neutralizing antibodies) were always added 1 h before TRAIL addition.
Annexin V staining.
The Colo205 and ML-1 cells were seeded the day before the experiment at 105 cells per ml in 24-well plates (1 ml per well), and were treated with 1 μg/ml anti-DR4- and/or anti-DR5-neutralizing antibodies for 1 h. Wild-type TRAIL, D269H, D269H/E195R, or D269H/T214R (100 ng/ml) was added to the cells and incubated for 2 h and 30 min. After treatment, the cells were harvested by scraping them gently off the wells and then spinning them down. Control or treated Colo205 and ML-1 cells were harvested and collected by centrifugation, washed once in Annexin V incubation buffer, and resuspended in 400 μl of fresh incubation buffer. One microliter of Annexin V was added to the samples, incubated at room temperature for 10 min, and immediately measured on a FACSCalibur flow cytometer (Becton Dickinson). Results were expressed as a percent of Annexin V-positive cells.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay was performed as described in ref. 22. BJAB cell lines were incubated with 1, 10, or 100 ng/ml TRAIL or D269H/E195R in the presence of 0.33 μg/ml cycloheximide (Sigma). For the EC50 determination, Colo205 cells were treated with serial dilutions (0–25 ng/ml) of TRAIL or mutants, and cytotoxicity was determined as described in ref. 22. EC50 values were calculated by using a four-parameter fit.
Supplementary Material
Supporting Information
Acknowledgments
We thank Dr. Andrew Thorburn (University of Colorado Health Sciences Center, Aurora) for kindly providing the BJAB cell lines; Dr. Steven de Jong and Derk-Jan de Groot (both at University Medical Center, Groningen, The Netherlands) for providing the A2780 cell line and characterizing the BJAB cell lines; and Johanna Vrielink, Dr. Rob van Weeghel, Ron Suk, and Cátia Rodrigues for helpful discussions and technical assistance. This research was partly funded by European Union Fifth Framework Program Grant QLK3-CT-2001-00498. R.H.C. was supported by the European Community Initiative Interreg IIIA.
Abbreviations
OPG
osteoprotegerin
SPR
surface plasmon resonance
TNF
tumor necrosis factor
TRAIL
TNF-related apoptosis-inducing ligand.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ashkenazi A., Pai R. C., Fong S., Leung S., Lawrence D. A., Marsters S. A., Blackie C., Chang L., McMurtrey A. E., Hebert A., et al. J. Clin. Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence D., Shahrokh Z., Marsters S., Achilles K., Shih D., Mounho B., Hillan K., Totpal K., Deforge L., Schow P., et al. Nat. Med. 2001;7:383–385. doi: 10.1038/86397. [DOI] [PubMed] [Google Scholar]
- 3.LeBlanc H. N., Ashkenazi A. Cell Death Differ. 2003;10:66–75. doi: 10.1038/sj.cdd.4401187. [DOI] [PubMed] [Google Scholar]
- 4.Kischkel F. C., Lawrence D. A., Chuntharapai A., Schow P., Kim K. J., Ashkenazi A. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 5.Sprick M. R., Weigand M. A., Rieser E., Rauch C. T., Juo P., Blenis J., Krammer P. H., Walczak H. Immunity. 2000;12:599–609. doi: 10.1016/s1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 6.Bodmer J. L., Holler N., Reynard S., Vinciguerra P., Schneider P., Juo P., Blenis J., Tschopp J. Nat. Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 7.Kimberley F. C., Screaton G. R. Cell Res. 2004;14:359–372. doi: 10.1038/sj.cr.7290236. [DOI] [PubMed] [Google Scholar]
- 8.Griffith T. S., Rauch C. T., Smolak P. J., Waugh J. Y., Boiani N., Lynch D. H., Smith C. A., Goodwin R. G., Kubin M. Z. J. Immunol. 1999;162:2597–2605. [PubMed] [Google Scholar]
- 9.Ichikawa K., Liu W., Zhao L., Wang Z., Liu D., Ohtsuka T., Zhang H., Mountz J. D., Koopman W. J., Kimberly R. P., et al. Nat. Med. 2001;7:954–960. doi: 10.1038/91000. [DOI] [PubMed] [Google Scholar]
- 10.Chuntharapai A., Dodge K., Grimmer K., Schroeder K., Marsters S. A., Koeppen H., Ashkenazi A., Kim K. J. J. Immunol. 2001;166:4891–4898. doi: 10.4049/jimmunol.166.8.4891. [DOI] [PubMed] [Google Scholar]
- 11.Wen J., Ramadevi N., Nguyen D., Perkins C., Worthington E., Bhalla K. Blood. 2000;96:3900–3906. [PubMed] [Google Scholar]
- 12.Chinnaiyan A. M., Prasad U., Shankar S., Hamstra D. A., Shanaiah M., Chenevert T. L., Ross B. D., Rehemtulla A. Proc. Natl. Acad. Sci. USA. 2000;97:1754–1759. doi: 10.1073/pnas.030545097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shifman J. M., Mayo S. L. J. Mol. Biol. 2002;323:417–423. doi: 10.1016/s0022-2836(02)00881-1. [DOI] [PubMed] [Google Scholar]
- 14.Reina J., Lacroix E., Hobson S. D., Fernandez-Ballester G., Rybin V., Schwab M. S., Serrano L., Gonzalez C. Nat. Struct. Biol. 2002;9:621–627. doi: 10.1038/nsb815. [DOI] [PubMed] [Google Scholar]
- 15.Havranek J. J., Harbury P. B. Nat. Struct. Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 16.Kortemme T., Joachimiak L. A., Bullock A. N., Schuler A. D., Stoddard B. L., Baker D. Nat. Struct. Mol. Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 17.Steed P. M., Tansey M. G., Zalevsky J., Zhukovsky E. A., Desjarlais J. R., Szymkowski D. E., Abbott C., Carmichael D., Chan C., Cherry L., et al. Science. 2003;301:1895–1898. doi: 10.1126/science.1081297. [DOI] [PubMed] [Google Scholar]
- 18.Guerois R., Nielsen J. E., Serrano L. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 19.Kiel C., Serrano L., Herrmann C. J. Mol. Biol. 2004;340:1039–1058. doi: 10.1016/j.jmb.2004.05.050. [DOI] [PubMed] [Google Scholar]
- 20.Schymkowitz J. W., Rousseau F., Martins I. C., Ferkinghoff-Borg J., Stricher F., Serrano L. Proc. Natl. Acad. Sci. USA. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiel C., Wohlgemuth S., Rousseau F., Schymkowitz J., Ferkinghoff-Borg J., Wittinghofer F., Serrano L. J. Mol. Biol. 2005;348:759–775. doi: 10.1016/j.jmb.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 22.van der Sloot A. M., Mullally M. M., Fernandez-Ballester G., Serrano L., Quax W. J. Protein Eng. Des. Sel. 2004;17:673–680. doi: 10.1093/protein/gzh079. [DOI] [PubMed] [Google Scholar]
- 23.Kempkens Ö., Médina E., Fernandez-Ballester G., Özüyaman S., Le Bivic A., Serrano L., Knust E. Eur. J. Cell Biol. 2006 doi: 10.1016/j.ejcb.2006.03.003. in press. [DOI] [PubMed] [Google Scholar]
- 24.Locksley R. M., Killeen N., Lenardo M. J. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 25.Bodmer J. L., Schneider P., Tschopp J. Trends Biochem. Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 26.Mongkolsapaya J., Grimes J. M., Chen N., Xu X. N., Stuart D. I., Jones E. Y., Screaton G. R. Nat. Struct. Biol. 1999;6:1048–1053. doi: 10.1038/14935. [DOI] [PubMed] [Google Scholar]
- 27.Hymowitz S. G., Christinger H. W., Fuh G., Ultsch M., O’Connell M., Kelley R. F., Ashkenazi A., de Vos A. M. Mol. Cell. 1999;4:563–571. doi: 10.1016/s1097-2765(00)80207-5. [DOI] [PubMed] [Google Scholar]
- 28.Cha S. S., Sung B. J., Kim Y. A., Song Y. L., Kim H. J., Kim S., Lee M. S., Oh B. H. J. Biol. Chem. 2000;275:31171–31177. doi: 10.1074/jbc.M004414200. [DOI] [PubMed] [Google Scholar]
- 29.Vriend G. J. Mol. Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]
- 30.Hymowitz S. G., O’Connell M. P., Ultsch M. H., Hurst A., Totpal K., Ashkenazi A., de Vos A. M., Kelley R. F. Biochemistry. 2000;39:633–640. doi: 10.1021/bi992242l. [DOI] [PubMed] [Google Scholar]
- 31.Thomas L. R., Johnson R. L., Reed J. C., Thorburn A. J. Biol. Chem. 2004;279:52479–52486. doi: 10.1074/jbc.M409578200. [DOI] [PubMed] [Google Scholar]
- 32.Kelley R. F., Totpal K., Lindstrom S. H., Mathieu M., Billeci K., Deforge L., Pai R., Hymowitz S. G., Ashkenazi A. J. Biol. Chem. 2005;280:2205–2212. doi: 10.1074/jbc.M410660200. [DOI] [PubMed] [Google Scholar]
- 33.Muhlenbeck F., Schneider P., Bodmer J. L., Schwenzer R., Hauser A., Schubert G., Scheurich P., Moosmayer D., Tschopp J., Wajant H. J. Biol. Chem. 2000;275:32208–32213. doi: 10.1074/jbc.M000482200. [DOI] [PubMed] [Google Scholar]
- 34.Wajant H., Moosmayer D., Wuest T., Bartke T., Gerlach E., Schonherr U., Peters N., Scheurich P., Pfizenmaier K. Oncogene. 2001;20:4101–4106. doi: 10.1038/sj.onc.1204558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information