How Clonal Is Staphylococcus aureus? (original) (raw)
Abstract
Staphylococcus aureus is an important human pathogen and represents a growing public health burden owing to the emergence and spread of antibiotic-resistant clones, particularly within the hospital environment. Despite this, basic questions about the evolution and population biology of the species, particularly with regard to the extent and impact of homologous recombination, remain unanswered. We address these issues through an analysis of sequence data obtained from the characterization by multilocus sequence typing (MLST) of 334 isolates of S. aureus, recovered from a well-defined population, over a limited time span. We find no significant differences in the distribution of multilocus genotypes between strains isolated from carriers and those from patients with invasive disease; there is, therefore, no evidence from MLST data, which index variation within the stable “core” genome, for the existence of hypervirulent clones of this pathogen. Examination of the sequence changes at MLST loci during clonal diversification shows that point mutations give rise to new alleles at least 15-fold more frequently than does recombination. This contrasts with the naturally transformable species Neisseria meningitidis and Streptococcus pneumoniae, in which alleles change between 5- and 10-fold more frequently by recombination than by mutation. However, phylogenetic analysis suggests that homologous recombination does contribute toward the evolution of this species over the long term. Finally, we note a striking excess of nonsynonymous substitutions in comparisons between isolates belonging to the same clonal complex compared to isolates belonging to different clonal complexes, suggesting that the removal of deleterious mutations by purifying selection may be relatively slow.
Staphylococcus aureus is a gram-positive pathogen responsible for a wide range of human disease, including septicemia; endocarditis and pneumonia; and wound, bone, and joint infections. Although the vast majority of infections by S. aureus result in asymptomatic carriage, this species nevertheless represents a serious public health burden, particularly in the hospital setting, where clones resistant to methicillin and other classes of antibiotics are endemic and insensitivity to vancomycin is on the increase. Although S. aureus is considered to be an opportunistic pathogen, it is possible that certain clones are more prone to cause invasive disease than are others, due to the presence of virulence factors that increase their chance of gaining access to normally sterile sites. Although many putative virulence factors have been identified in the S. aureus genome (17), the differences in pathogenic potential between naturally occurring isolates remain largely unaddressed.
The extent to which homologous recombination contributes to the emergence and subsequent diversification of clones is also at present unclear, although this question has important implications both for the choice of the most appropriate typing strategy for effective epidemiological surveillance and for vaccine design. The population structure of S. aureus has been studied previously by a variety of techniques, including multilocus enzyme electrophoresis (21, 22), pulsed-field gel electrophoresis (12), and multilocus sequence typing (MLST) (4). These studies have revealed a highly clonal population, consistent with the view that S. aureus, unlike the freely recombining pathogenic species Streptococcus pneumoniae (10), Neisseria meningitidis (9), and Helicobacter pylori (32), is not naturally transformable. However, as discussed elsewhere (11, 30) relatively high rates of recombination within a bacterial population can be masked by sampling bias or the temporary expansion of adaptive genotypes, and detailed analysis of nucleotide sequence data is required to confirm inferences about recombination rates obtained from the apparent extent of linkage disequilibrium in the population.
Here we present an analysis of the sequence data obtained from the characterization by MLST of 334 S. aureus isolates recovered from persons with invasive disease or asymptomatic carriage. These data consist of the nucleotide sequences of ∼450-bp internal fragments of a standard set of seven metabolic housekeeping loci for each isolate. Because it is based on the slowly evolving genomic “core,” MLST provides data that are well suited to studies of global epidemiology and population biology of bacterial pathogens (19). A previous analysis of MLST data from these S. aureus strains proposed that rates of recombination are high in this species, but these data contained errors and this conclusion can no longer be considered valid (3). In this report we used the revised MLST data to reexamine both the distribution of disease-causing strains and carried strains between clonal complexes and the significance of homologous recombination in the diversification of the natural population of S. aureus.
MATERIALS AND METHODS
Bacterial strains.
Three hundred thirty-four isolates of S. aureus were included in this analysis, including 61 isolates from patients with community-acquired invasive disease, 179 isolates from persons with asymptomatic nasal carriage (recovered from healthy blood donors), and 94 isolates from patients with hospital-acquired (nosocomial) invasive disease. This strain collection was recovered from Oxfordshire, United Kingdom, over a 2-year period and is described elsewhere (3).
Of the 155 isolates recovered from patients with cases of invasive disease, 28 were methicillin-resistant S. aureus (MRSA), of which only one was recovered from a patient with a case of community-acquired disease. Twenty-three of the MRSA isolates belonged to a single clone (sequence type 36 [ST36]), three belonged to ST22 (this clone included the single MRSA isolate from the community), and the remaining MRSA isolates corresponded to ST12 and ST38.
MLST.
MLST was carried out with an ABI 3700 capillary sequencer and a standard set of 14 primers as described previously (4). The seven genes included in the S. aureus MLST scheme are arcC, aroE, glpF, gmk, pta, tpi, and yqiL. Information on these loci, the primer sequences, and PCR conditions is available on the MLST website (http://www.mlst.net). Isolates are defined by the alleles present at the seven loci (the allelic profile), and each unique allelic profile is assigned as an ST. Isolates with the same ST, therefore, have identical sequences at all seven MLST loci and are considered to be members of a single clone. The revised data set (3) was carefully checked for errors, particularly the sequences of variant alleles within single-locus variants (SLVs; those strains which differ at only one locus out of seven from an assigned clonal ancestor; see below). All the variant alleles within SLVs were reamplified and resequenced.
Assignment to clonal complexes.
The program BURST was used to divide the 334 isolates into clonal complexes, which are defined as groups of STs in which every ST shares at least five of seven identical alleles with at least one other ST in the group (3). The genotype (ST) that gave rise to each clonal complex (the clonal ancestor) initially will diversify to produce variants that differ at only one of the seven loci (SLVs). Ancestors of clonal complexes were therefore assigned on the basis that they differ at a single locus from the highest number of other genotypes in the clonal complex (i.e., they define the most SLVs). The likely pattern of descent of each ST from the clonal ancestor is then displayed graphically (Fig. 1). (This analysis has previously been carried out on the corrected data set [3].) To assess whether any of the clonal complexes represent hypervirulent lineages (i.e., lineages that contain a disproportionate frequency of disease isolates), we examined the distribution of disease and carriage isolates between clonal complexes by contingency table analysis. The frequencies of isolates from persons with nasal carriage, community-acquired disease, and hospital-acquired disease within all clonal complexes and clones that were represented by more than five isolates were compared, with the remaining singletons arbitrarily placed in a single group. The distribution of isolates of differing epidemiological origins was then tested by the chi-square test.
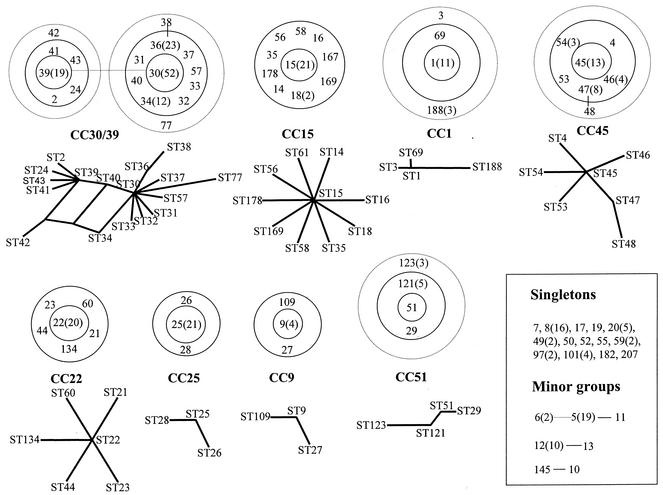
FIG. 1.
Identification of clonal complexes. BURST analysis was used to assign clonal complexes within the Oxford isolate collection. Splits graphs are also shown for each major clonal complex, generated by Splitstree version 3.1 (no splits graphs are shown for the minor groups). For the major clonal complexes, the predicted clonal ancestors are shown in the central ring, SLVs are shown in the middle (solid) ring, and double-locus variants are shown in the outer (dashed) ring. Individual relationships between some STs are shown as a solid line (single-locus difference) or a dashed line (double-locus difference). The values in parentheses are the numbers of isolates of a given ST when more than one example of an ST was observed.
Phylogenetic analysis and tests for recombination.
For each unique ST the sequences of all seven loci were concatenated to produce an in-frame sequence of 3,198 bp. Bayesian maximum likelihood (ML) trees were reconstructed with MrBayes version 2.01 (14), and ML trees made with the use of the HKY85 model of DNA substitution were reconstructed by using PAUP* version 4.0b10 (33). dS/dN ratios were computed by the method of Nei and Gojobori (24) with the Jukes-Cantor correction as implemented in MEGA version 2.1 (S. Kumar, K. Tamura, I. B. Jakobsen, and M. Nei, Arizona State University, Tempe, 2001). Finally, an unweighted pair group method with averages (UPGMA) dendrogram (data not shown) was reconstructed from the pairwise differences in the allelic profiles by using Statistica version 5 (StatSoft Inc., Tulsa, Okla.), and Splits graphs were reconstructed from these data by using Splitstree version 3.1 (15).
Estimates of recombination rates.
BURST was used to identify the most likely (i.e., parsimonious) ancestral ST within each clonal complex and all those strains which have diverged from the predicted clonal ancestor at a single locus but have remained unchanged at the other six (SLVs; Fig. 1). Estimates of the fraction of these SLVs that have arisen by recombination can be made by a method described elsewhere (9, 10). Briefly, the sequences of the alleles that differ between each ancestral ST and its associated SLVs are compared and are assigned as resulting from either a recombinational replacement or a point mutation. Putative point mutations are assigned on the basis of two criteria: firstly, alleles which have arisen by a single point mutation will differ only at a single nucleotide site, and secondly, de novo point mutation will result in an allele that is very likely to be unique within the data set. Variant alleles in SLVs satisfying these two criteria are thus assigned as having arisen by point mutation. Those that differ at multiple nucleotide sites, or which differ at a single site but correspond to alleles found elsewhere in the data set, are assigned as having arisen by recombination.
A comparison between allelic and nucleotide divergence.
If clones diversify predominantly by the stepwise accumulation of point mutations, then two strains which have only very recently diverged will show a high level of similarity both in terms of their respective allelic profiles and in terms of the sequences of their nonidentical alleles. In other words, two very closely related strains will share many identical loci, and furthermore those loci which do differ will do so only at a small number of nucleotide sites. However, if clones diversify predominantly by recombination, then alleles which happen to differ between very closely related strains may do so at a large number of nucleotide sites, owing to the fact that these alleles have been imported from an unrelated lineage. Therefore, under a mutational model, allelic and sequence divergence will show a positive correlation (both parameters will reflect time since the common ancestor), but this relationship will not hold if alleles change predominantly by recombination (when identical alleles are excluded).
We examined the relationship, over all pairwise comparisons, between the number of allelic mismatches and the average number of nucleotide differences per locus (excluding loci that have remained identical). For each pairwise comparison between STs, the number of loci that differed and the mean number of nucleotide differences between nonidentical loci (m) were computed. The average of m was obtained for all pairs of STs that differed at a given number of loci and was plotted against the number of allelic differences. Only a single example of each ST was used in this analysis, since this minimizes the influence of any sampling effects resulting from the overrepresentation of specific clones.
In order to calibrate the results from S. aureus, the approach was also used on MLST data for S. pneumoniae, a naturally transformable species that is known to recombine at a high frequency (10). All 575 unique STs from the S. pneumoniae MLST database (as of August 2002) were analyzed by this approach. A program for implementing this analysis (written by D. A. Robinson) is available on request.
ML analysis of congruence.
Multilocus data sets are ideal for examining the degree to which the phylogenetic signal varies between gene loci. An ML method has been described which scores two trees as either significantly congruent (similar) or no more congruent than two trees of random topology (8). The approach compares the ML scores of gene trees against the 99th percentile of the distribution of scores for 200 trees of random topology given the reference data. The HKY85 model of nucleotide substitution was used for tree reconstruction, with the transition/transversion (Ti/Tv) ratio and the α parameter optimized. (The α parameter describes the extent of nucleotide substitution rate variation between sites assuming a discrete gamma distribution with eight categories.) The likelihood scores for trees of random topology were also computed by reoptimizing these likelihood parameters.
Two genes, A and B, are scored as significantly congruent if the difference between the likelihood scores of the trees for gene A and gene B (Δ−lnL) is lower than the Δ−lnL between the 99th percentile of the scores of 200 random trees and the likelihood score for gene A. In all cases, trees are scored against the reference data for gene A. This test therefore examines whether the tree from gene B is a significantly better fit to the data from gene A than are trees of random topology. This method has been used to compare the phylogenetic congruence between MLST genes of a number of different bacterial species (8), including S. aureus (although in this original analysis the results cannot be considered valid; see below). This approach can potentially also be used to compare the effects of recombination on different gene loci believed to be under differing selective pressures. The approach was implemented with PAUP* 4.0b10 (33).
RESULTS
The identification of clonal lineages.
The revised MLST data set for the 334 S. aureus isolates recovered from Oxfordshire, United Kingdom, contains 75 unique genotypes (STs). Forty-nine of these STs were represented by only a single isolate, while 15 were represented by at least five isolates. The largest clone was ST30, which accounts for 52 isolates (15.6% of all isolates). The 75 STs were divided by BURST into eight major clonal complexes, three minor clonal complexes, and 14 singleton STs; these do not belong to any clonal complex, as they differ from every other ST in the data set at three or more of the seven MLST loci (Fig. 1). Of the 14 singleton STs, 6 were represented by more than one strain and hence can be assigned as singleton clones (clones which have no clonal variants). The largest singleton clone in this data set was ST8, which was represented by 16 strains; ST20 was represented by five strains, ST101 was represented by four strains, and ST49, ST59, and ST97 were represented by two strains each. ST8 has been assigned as the predicted ancestor of a large clonal complex when other data sets have been examined (5). The predicted ancestors of the eight major clonal complexes identified in the present data set were ST9, ST15, ST22, ST25, ST30/39, ST45, ST1, and ST51, and these clonal complexes were named on the basis of these assignments, with the prefix “CC” (e.g., CC9). In the case of CC30/39, two closely related ancestors, each defining at least four SLVs, were identified. Ancestral assignments by BURST were consistent with the placing of these STs at internal nodes by splits decomposition analysis (Fig. 1).
Distribution of disease isolates between clonal complexes.
Isolates from persons with nasal carriage, community-acquired invasive disease, and hospital-acquired invasive disease were evenly distributed among the clonal complexes, suggesting no significant differences in their propensity to cause disease (P = 0.24; Table 1). There was a preponderance of isolates from the hospital-acquired disease group in CC30/39, but this was not statistically significant and can be explained by the existence of isolates from the EMRSA-16 clone (ST36) within CC30, all 23 strains of which were from patients with hospital-acquired disease.
TABLE 1.
Distribution by clinical origin of isolates between clonal complexesa
| Clonal complex | No. (%) of isolates | |||
|---|---|---|---|---|
| Nasal carriage | Community- acquired disease | Hospital- acquired disease | Total | |
| CC30 | 46 (26) | 13 (21) | 36 (38) | 95 (28) |
| CC39 | 14 (8) | 4 (7) | 6 (6) | 24 (7) |
| CC15 | 21 (12) | 4 (7) | 6 (6) | 31 (9) |
| CC1 | 6 (3) | 6 (10) | 4 (4) | 16 (5) |
| CC45 | 16 (9) | 6 (10) | 8 (9) | 30 (9) |
| CC22 | 14 (8) | 5 (8) | 6 (6) | 25 (7) |
| CC25 | 8 (4) | 8 (13) | 7 (7) | 23 (7) |
| CC9 | 3 (2) | 0 | 3 (3) | 6 (2) |
| CC51 | 8 (4) | 1 (2) | 1 (1) | 10 (3) |
| CC5 | 13 (7) | 5 (8) | 4 (4) | 22 (7) |
| CC12 | 4 (2) | 3 (5) | 4 (4) | 11 (3) |
| ST8 | 8 (4) | 4 (7) | 4 (4) | 16 (5) |
| Singletons | 18 (10) | 2 (3) | 5 (5) | 25 (7)b |
Direct estimates of recombination within clonal complexes.
Given that it is possible to assign the strains to clonal complexes and to identify the most likely ancestor(s) of each clonal complex, it is possible to estimate whether SLVs have arisen from their respective clonal ancestor by recombination or by mutation. The eight major clonal complexes shown in Fig. 1 provide a total of 35 SLVs for this analysis (Table 2). Twenty-eight of these are within four of the clonal complexes (CC15, CC22, CC45, and CC30/39), while there are only two SLVs in CC25, CC9, and CC51 and only one in CC1.
TABLE 2.
Variant alleles within the SLVs found within eight major clonal complexes
| ST of clonal ancestor | ST of SLV | Variant locus in SLV | Ances- tral allele | SLV allele | No. of nucleotide differences | Other clones containing SLV allele | Amino acid changea |
|---|---|---|---|---|---|---|---|
| 15 | 178 | aroE | 13 | 25 | 1 | None | S-L |
| 15 | 16 | arcC | 13 | 15 | 1 | None | P-S |
| 15 | 18 | aroE | 13 | 15 | 1 | None | Syn |
| 15 | 35 | tpi | 11 | 21 | 1 | None | V-I |
| 15 | 56 | aroE | 13 | 19 | 1 | None | D-E |
| 15 | 58 | aroE | 13 | 30 | 1 | None | Syn |
| 15 | 169 | pta | 12 | 16 | 1 | None | Syn |
| 15 | 14 | arcC | 13 | 11 | 1 | None | Syn |
| 15 | 61 | pta | 12 | 37 | 1 | None | L-F |
| 22 | 21 | glpF | 1 | 13 | 1 | None | Stop |
| 22 | 23 | glpF | 1 | 7 | 1 | None | S-N |
| 22 | 44 | aroE | 6 | 21 | 1 | None | A-P |
| 22 | 60 | gmk | 5 | 7 | 1 | None | S-R |
| 22 | 134 | gmk | 5 | 16 | 1 | None | G-C |
| 25 | 26 | vqiL | 4 | 9 | 1 | None | Syn |
| 25 | 28 | glpF | 4 | 9 | 1 | None | K-N |
| 45 | 4 | aroE | 14 | 10 | 1 | None | Syn |
| 45 | 54 | gmk | 6 | 13 | 1 | None | I-M |
| 30 | 31 | gmk | 2 | 3 | 1 | None | K-I |
| 30 | 32 | aroE | 2 | 12 | 1 | None | P-S |
| 30 | 33 | aroE | 2 | 17 | 1 | None | P-L |
| 30 | 34 | arcC | 2 | 8 | 2 | CC30/39 + 145/10 | D-G + Syn |
| 30 | 37 | pta | 6 | 15 | 1 | None | N-D |
| 30 | 40 | tpi | 3 | 2 | 1 | CC30/39 | A-T |
| 30 | 57 | tpi | 3 | 49 | 1 | None | S-C |
| 30 | 36 | pta | 6 | 3 | 1 | None | A-T |
| 39 | 2 | yqiL | 2 | 26 | 1 | None | Syn |
| 39 | 24 | gmk | 2 | 17 | 1 | None | R-H |
| 39 | 41 | glpF | 2 | 3 | 1 | None | K-I |
| 39 | 43 | yqiL | 2 | 25 | 1 | None | Syn |
| 9 | 27 | tpi | 1 | 22 | 1 | None | V-I |
| 9 | 109 | aroE | 3 | 27 | 1 | None | P-Q |
| 1 | 69 | aroE | 1 | 28 | 1 | None | Syn |
| 51 | 121 | tpi | 7 | 14 | 1 | None | V-A |
| 51 | 29 | glpF | 6 | 17 | 1 | None | G-D |
Thirty-three of the 35 SLVs possessed variant alleles that both are unique within the data set (i.e., found only within the SLV) and differ at only a single nucleotide site from the allele in the putative ancestral ST. These characteristics are consistent with point mutation. In contrast, all the corresponding ancestral alleles were found in more than one ST, thus further supporting the assignments of ancestral and derived genotypes within the clonal complexes. In 28 of 33 cases, the single nucleotide change in the variant allele was a polymorphism that was not present within any of the other alleles in the entire S. aureus MLST data set, which (as of August 2002) contains over 800 strains. These characteristics are consistent with the hypothesis that these alleles have arisen by de novo mutation, rather than by recombination.
The two remaining SLVs both belong to CC30/39. The only case of parallel paths in the splits graphs of clonal complexes concerns the relationships of these two SLVs (ST34 and ST40) in CC30/39, which implies that they may have arisen by recombination (Fig. 1). ST34 is the more likely to have arisen by recombination because the variant allele differs at two nucleotide changes from the predicted ancestral allele in ST30, and the variant allele is present in strains outside CC30/39. The origin of ST40 is more difficult to characterize, because it falls midway between the two putative ancestors of this complex (i.e., it is an SLV of both ST30 and ST39, which themselves differ at two loci). ST40 possibly reflects a step in the mutational pathway between these two ancestral clones, and this is supported by the observation that ST40 differs at only a single nucleotide site from ST30 (in tpi) and from ST39 (in pta). This may account for the widespread distribution of the variant alleles in ST40 (at tpi and pta) throughout this clonal complex; thus, in this case the presence of the variant alleles within a number of other genotypes is not convincing evidence for recombination and may instead reflect identity by descent.
If we cautiously assign ST40 and ST34 as having arisen by recombination, then we can estimate that 33 of the SLVs have arisen by point mutation and that only two have arisen by recombination. As a conservative estimate (with respect to the frequency of mutation), it therefore appears that during the initial stages of clonal diversification alleles are at least 15-fold more likely to change by point mutation than by recombination. S. aureus contrasts with N. meningitidis and S. pneumoniae, which are both naturally transformable, where alleles change between 5- and 10-fold more frequently by recombination than by mutation (7).
Of the 33 putative point mutations listed in Table 2, 23 are nonsynonymous, 9 are synonymous, and 1 is a nonsense mutation (resulting in the allele glpF13); the possible significance of this observation is discussed below. The presence of all of these point mutations was verified by reamplifying these gene products and resequencing the alleles on both strands.
This approach has been presented previously with MLST data for S. aureus prior to corrections in the data (8). This original analysis suggested that the impact of recombination on clonal diversification in S. aureus is more significant than point mutation and gave estimates comparable to those for N. meningitidis and S. pneumoniae; this estimate was also noted in a recent review (11; see also reference 31). We wish to emphasize that we believe this original estimate to be erroneous, and that the present analysis of the revised data, as discussed above, most accurately reflects the microevolutionary events occurring within clonal complexes of S. aureus.
The relationship between nucleotide and allelic divergence.
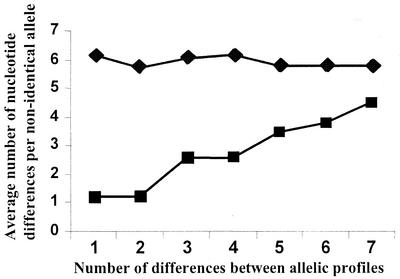
To examine further the validity of the above approach to estimating the contribution of recombination and mutation to clonal diversification, we extended the analysis to all pairwise comparisons of STs, rather than just focusing on the pairs of STs within major clonal complexes that differ at a single locus. The number of allelic mismatches was plotted against the average number of nucleotide changes at nonidentical loci for the S. aureus data set and for the complete S. pneumoniae MLST data set held on the MLST website (Fig. 2). For S. aureus, there was a clear positive trend between the proportion of differing loci and the average number of nucleotide changes per locus, but no such trend was apparent for S. pneumoniae, thus supporting the suggestion of high rates of recombination within the latter species and relatively low rates within the former.
FIG. 2.
Sequence diversity versus allelic diversity. The average number of nucleotide differences per locus (excluding those loci that do not differ) was calculated (y axis) for all pairwise comparisons of the 75 S. aureus STs (squares) and for the 575 S. pneumoniae STs (diamonds). These averages were computed separately for all pairwise comparisons differing at one, two, three, four, five, six, or all seven loci (x axis). There is a clear positive trend between the number of nucleotide differences in nonidentical alleles and the number of allelic differences (i.e., time since divergence) within the S. aureus data, suggesting that recombination has not been sufficiently frequent to overwhelm the stepwise accumulation of diversification through de novo point mutation. However, this trend is not noted within the data from S. pneumoniae.
Phylogenetic validation of the delineation of the clonal complexes.
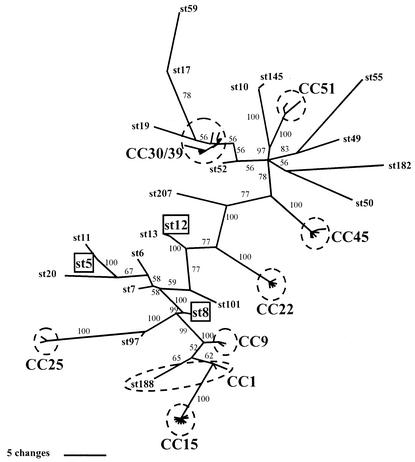
As the biological significance of assigning strains to clonal complexes is of relevance both to the above analyses and to the utility of MLST as a typing scheme, we applied phylogenetic approaches to the concatenated sequences of the seven MLST loci from all 75 unique STs in the S. aureus data set. As this is a large nucleotide sequence data set, ML approaches as implemented in PAUP* are computationally intensive and time-consuming. For this reason, we reconstructed a consensus ML tree by using a Bayesian approach implemented in MrBayes version 2.01. This approach also provides posterior probabilities for the branching order of the resulting consensus tree (14). Six of the eight major clonal complexes identified by BURST are clearly identified as terminal clusters on the unrooted Bayesian tree (Fig. 3) and are supported by posterior probability scores of 100%; the remaining two clonal complexes also formed terminal clusters on the tree but were not so well supported (Fig. 3). With the exception of ST188, all the STs assigned to a specific clonal complex by BURST are also associated with the same cluster in the tree (indicated by the dashed rings in Fig. 3). ST188 is included within CC1 by BURST (because ST188 differs at only two loci from ST1) but is distanced from the main CC1 cluster on the tree. An inspection of the relevant allele sequences in ST188 and ST1 revealed that both varying loci differ at multiple sites, resulting in a total of nine polymorphisms, which on the tree leads to the separation of this ST from the other STs assigned to CC1 by BURST. Presumably these two variant alleles have been imported by recombination.
FIG. 3.
Unrooted Bayesian tree of the concatenated sequences of the 75 different STs of S. aureus. The tree was constructed by using MrBayes version 2.01 (14); six different substitution rates were estimated with site-specific rate variation. Four Markov chains were run for 1,000,000 generations, and the tree was saved (with branch lengths) every 100 generations, resulting in 10,000 trees. These trees were imported into PAUP* 4.0b10 (33), and the first 2,000 trees were discarded as burn-in. A 50% majority rule consensus tree was computed from these trees, and the posterior probability given on each branch is a percentage of these trees supporting each node. All the major clonal complexes are enclosed within dashed rings; singletons or minor groups represented by more than five strains are boxed. With the exception of ST188 the individual STs within each major clonal complex are not labeled.
The relationships between the clonal complexes.
As SLVs appear to arise predominantly by point mutation, we examined the possibility that sufficient phylogenetic signal is present within these data to reconstruct a tree which establishes the relationships between clonal complexes with some confidence. A diverse subsample of 25 STs was chosen which includes the ancestral ST from each of the eight major clonal complexes, one ST from each of the minor clonal complexes, and all the singleton STs (Fig. 1). These 25 STs differ from each other (over all pairwise comparisons) at an average of over 6.3 of 7 loci and are separated from each other by a linkage distance of >0.4 on a UPGMA tree constructed from the pairwise differences in their allelic profiles (tree not shown). The sequences from each of the seven loci from these STs were used in the ML analysis of congruence, and an ML tree was also reconstructed from the concatenated sequences of the seven loci.
Congruence analysis.
By examining the degree of phylogenetic consistency between gene trees, it is possible to gauge the impact of recombination and the feasibility of reconstructing a meaningful intraspecies consensus tree. The results of the congruence analysis for the set of 25 diverse STs are given in Table 3. Of the 42 pairwise comparisons of the seven loci, 23 are significantly congruent. This result contrasts markedly with a previous analysis of congruence on S. aureus MLST data, where only 5 of 42 comparisons were significantly congruent (8); this original estimate is now believed to be unreliable since the analysis was done prior to the corrections in the MLST data set. The significant congruence in 55% of the S. aureus tree comparisons contrasts with that obtained for S. pneumoniae, where all pairwise comparisons were found to be noncongruent (8), and lends further support to the suggestion that recombination is much less frequent in S. aureus than in S. pneumoniae. However, 19 of the pairwise comparisons remain noncongruent, which suggests that recombination has had some impact in S. aureus.
TABLE 3.
ML analysis of congruence
| Locus | Length of MLST allele (bp) | πa | αb | Ti/Tv ratio | −lnL | Δ-lnL (99th)c | Δ−lnL score of gene treed | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| arcC | aroE | glpF | gmk | pta | tpi | yqiL | ||||||
| arcC | 456 | 0.008 | 0.16 | 1.1 | 774 | 92 | 127 | 110 | 128 | 113 | 106 | 102 |
| aroE | 456 | 0.012 | 0.09 | 3.1 | 829 | 118 | 150 | 116 | 112 | 112 | 107 | 115 |
| glpF | 465 | 0.005 | 0.18 | 6 | 727 | 61 | 51 | 40 | 61 | 49 | 40 | 41 |
| gmk | 429 | 0.008 | 0.004 | 7 | 659 | 74 | 123 | 65 | 69 | 63 | 77 | 71 |
| pta | 474 | 0.006 | 0.25 | 1.1 | 766 | 91 | 115 | 95 | 102 | 98 | 76 | 79 |
| tpi | 402 | 0.011 | 0.13 | 1.5 | 735 | 118 | 146 | 122 | 149 | 140 | 109 | 110 |
| yqiL | 516 | 0.008 | 0.007 | 3.8 | 866 | 99 | 114 | 88 | 84 | 93 | 82 | 59 |
Inspection of individual MLST gene trees and evidence for a phylogenetic division within S. aureus.
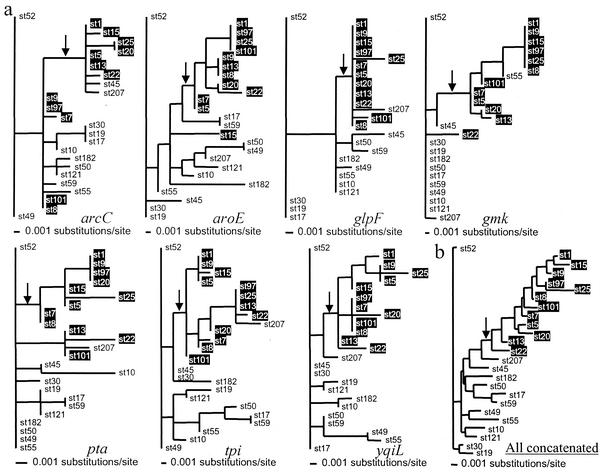
The ML trees for the seven MLST loci from the 25 representative STs are shown in Fig. 4a. Although there are clear inconsistencies between these gene trees, there are also some branches which are well supported by all or most of the trees. In particular, six of seven of the trees support (to various degrees) the division of these diverse STs into two groups; the same division is also supported by the Bayesian tree (Fig. 3), a neighbor-joining tree, and a UPGMA dendrogram of allelic mismatches (not shown). ST207 was the only one of the 25 STs that could not be placed with confidence within either group; it is possible that this reflects a frequent history of recombination in this strain.
FIG. 4.
ML trees. Trees are shown for each MLST gene (a) and for the concatenated sequences of all seven genes (b), with a sample of 25 diverse STs. Each tree was computed by using the HKY85 model of nucleotide substitution, with the α parameter (describing the shape of the gamma distribution of rate variation with eight discrete categories) and the Ti/Tv ratio optimized during tree construction. Despite the presence of multiple inconsistencies between these trees, there is evidence for a conserved division, which is marked with an arrow in each tree. STs belonging to one side of this division (group 1) are given in white on a black background. There are some exceptions to this division, particularly at the arcC locus (see text). The branching order of the concatenated tree (b) is very similar to Fig. 3, which was computed by a Bayesian approach, the main difference being the positions of ST17 and ST59.
Although the significance of this observation is unclear, the placing of strains within one or the other of these two groups provides a rough approximation of how closely a given gene fits the consensus tree or how much recombination has occurred in any given strain. For example, the arcC locus was found to be the most atypical in congruence analysis; of the 19 noncongruent comparisons, 11 involve this gene (Table 3), and an inspection of the arcC tree reveals that the two major groups are very poorly supported (Fig. 4a).
The relationships between STs within each major group are often inconsistent between gene loci. This is compatible with a history of recombination but may also reflect an insufficient number of phylogenetically informative sites to allow a robust reconstruction of the branching order. To investigate further the relationships between the 25 diverse STs, an unrooted ML tree was reconstructed from the concatenated sequences of the seven loci by using the same likelihood model that was used to reconstruct trees for individual gene loci, and this tree also supports the conserved node evident in the individual gene trees (Fig. 4b).
Synonymous and nonsynonymous substitutions.
The suggestion that recombination has distorted the relationships between clonal lineages seems at odds with the evidence that clones diversify predominantly by point mutation rather than by recombination. A possible way to reconcile these conflicting lines of evidence is suggested by the observation that 23 of the point mutations within SLVs were nonsynonymous, 9 were synonymous, and 1 resulted in the generation of a stop codon in glpF (Table 1). The genes used for MLST were chosen on the basis that they are ubiquitous “core” housekeeping genes, subject to stabilizing selection; it might therefore be expected that synonymous mutations, which are far more likely to be neutral, should outweigh nonsynonymous substitutions. In order to compare the ratio of synonymous to nonsynonymous substitutions within clonal complexes to that between clonal complexes, the average dS/dN ratios were computed over all pairwise comparisons for each locus from the diverse set of 25 STs. On average over all loci, the dS/dN ratio was far higher for comparisons between the diverse genotypes (8.6) than for comparisons between SLVs and their clonal ancestors (1.3) (Table 4). In comparing SLVs with their ancestral sequences, we found 729.76 synonymous sites and 2,468.24 nonsynonymous sites (these values are averages for the ancestral sequences of each SLV). Nine synonymous substitutions, and 23 nonsynonymous substitutions within SLVs (Table 2) correspond to a dS of 0.012, a dN of 0.009, and a dS/dN ratio of 1.32. This implies that many of the nonsynonymous point mutations observed within the SLVs will in time be lost from the population by purifying selection. The relative impact of point mutation in the long-term evolution of this species may therefore be inflated when only the initial stages of clonal diversification are examined.
TABLE 4.
dS/dN ratios for 25 diverse genotypes
| Locus | dS/dN ratio |
|---|---|
| arcC | 7.64 |
| aroE | 6.74 |
| glpF | 10.47 |
| gmk | 11.8 |
| pta | 6.54 |
| tpi | 7.38 |
| yqiL | 9.39 |
| Avg | 8.57 |
DISCUSSION
Delineation of clonal groups and a comparison of isolates from persons with disease and with asymptomatic carriage.
The MLST data analyzed in this report confirm previous suggestions, originally by Musser et al. using multilocus enzyme electrophoresis (22), that the population structure of S. aureus is highly clonal. We have explored the clonal structure of a well-defined “snapshot” of a localized population from Oxfordshire, United Kingdom, and present evidence that the clonal complexes within this population tend to be separated from each other by large gaps in sequence space (Fig. 3) and result from the star-like diversification of the clonal ancestors (Fig. 1). In the absence of comparable samples from disease patients and carriers from different geographical locations, it is unclear at present how much of the global diversity of this species is represented within this localized sample.
Disease isolates are equally represented in all the clonal complexes, suggesting that there is no link between MLST genotype and the propensity to cause disease. This finding appears to be in contrast to a recent study by Booth et al., who detected differences in the frequencies of specific lineages (as defined by pulsed-field gel electrophoresis) when comparing samples of clinical and carried isolates (1). A possible explanation is that the present comparison is based upon clinical and carried isolates drawn from a single well-defined population, thus minimizing any differences between the samples that reflect geographical or temporal structuring and are unrelated to virulence.
There is strong evidence in some pathogens for marked differences in the population structures of isolates recovered from persons with disease and carriage. For example, population studies of the gram-positive pathogen S. pneumoniae have demonstrated that isolates from carriers are more diverse than those from disease patients (20, 28, 34), and a recent study suggests that different clones (as defined by MLST) and serotypes show differing potential to cause invasive disease (2). The carriage population structure of the gram-negative pathogen N. meningitidis has also been shown to be more diverse than samples associated with invasive disease (16), and there is some experimental evidence that different clones of this species may differ in their ability to cause invasive disease (35).
The explanation as to why the subpopulations of invasive and asymptomatically carried S. aureus appear to be identical by MLST in the present study is unclear but may in part reflect the fact that “invasive disease” encompasses a very wide range of disease symptoms caused by this species and the associated plethora of putative virulence determinants so far identified (17). Furthermore, despite this finding we do not argue that all S. aureus isolates are equally virulent. The influx and loss of virulence determinants carried on mobile elements will play a large part in determining the virulence of an isolate. The movement of these genes may occur so rapidly that their presence or absence is only weakly linked to the relatively stable clonal background defined by MLST.
A recent study by Peacock et al. (27), of the same bacterial strain collection on which the present study is based, noted that the presence or absence of seven putative virulence factors is significantly correlated with the epidemiological origin of the strain (i.e., from disease or asymptomatic carriage). This study demonstrated that bacterial factors do contribute toward the ability of S. aureus to cause disease, whereas the MLST data indicate that these differences are generally not reflected in the “core” genome. Put another way, isolates of the same ST differ in their content of virulence genes and may therefore differ in their ability to cause disease. Unlike earlier studies of the impact of recombination (8), the study by Peacock et al. was carried out with reference to the corrected S. aureus MLST data set, and their conclusions are therefore not compromised by the original errors in the data.
The MLST data also provide no evidence that strains responsible for nosocomial disease represent a distinct subpopulation from strains causing community-acquired disease or strains recovered from asymptomatic carriers. Although the acquisition of genes conferring drug resistance within certain clones confers a strong selective advantage in the hospital environment, MRSA and vancomycin-insensitive S. aureus clones being the most important examples, the MLST data reveal that these clones have evolved from genotypes which were already common in the population (5).
Evidence concerning recombination rates. (i) Clonal diversification.
The analysis of diversification within clonal complexes suggests that alleles, and individual nucleotide sites, are at least 15-fold more likely to change by point mutation than by recombination. This estimate is supported by a similar analysis of 117 isolates recovered from Nottingham, United Kingdom, in which the same clonal complexes are present (13). These data add a further 13 SLVs to the clonal complexes, 12 of which appear to have arisen by point mutation and 1 of which appears to have arisen by recombination (data not shown).
Extending the analysis to include all pairwise comparisons between different genotypes reveals a trend of increasing nucleotide divergence with increasing allelic divergence. This result is also consistent with a predominantly mutational mode of evolution. The power of this approach is limited by the number of loci used for MLST; in the S. aureus data set over 50% of all pairwise comparisons differ at all seven loci. This analysis reduces all of these comparisons, which will include the vast majority of comparisons between clonal complexes, to a single data point.
The results of both of these analyses contrast strikingly with those obtained with MLST data from S. pneumoniae and N. meningitidis and highlight the fact that the initial stages of clonal diversification in S. aureus appear to be predominantly driven by point mutation, rather than recombination.
(ii) Phylogenetic relationships between clonal lineages.
Although there is limited evidence for recombination over the short term (within clonal complexes), or the short to medium term (as suggested by the relationship between allelic and nucleotide diversity), there is evidence that recombination has contributed to the evolution of the S. aureus population over the longer term. Many of the phylogenetic relationships between the closely related clonal complexes are poorly supported and inconsistent between individual gene trees (Fig. 4a), although this may in part reflect factors other than recombination, such as a paucity of phylogenetically informative sites. Despite these inconsistencies, there is also evidence for a conserved node dividing the subsample of 25 diverse STs approximately into halves. The significance of this observation within the context of the evolutionary history of this population is unclear; nevertheless, it serves well to illustrate the middle ground occupied by the significance of homologous recombination on the core genome of this species over the long term. On the one hand, and in contrast to S. pneumoniae and N. meningitidis, recombination has not been so frequent as to completely eliminate the intraspecies phylogenetic signal in this species. On the other hand, certain alleles at specific loci appear to have been horizontally transferred between the two major phylogenetic groups, and statistical tests of congruence between loci identified 45% of tree comparisons as being not significantly congruent.
Interestingly, over half of these noncongruent comparisons involved the arcC locus, which encodes carbamate kinase. An inspection of the genome sequence in the vicinity of arcC reveals the presence of a putative virulence factor, clumping factor B (clfB), approximately 1 kb downstream of arcC, and two further putative virulence factors, aureolysin (aur) (29) and isaB, approximately 6 kb upstream of this locus. clfB is known to be associated with the cell wall (25, 26), and isaB is known to elicit an immune response (18); both of these genes probably encode proteins that are exposed to the host immune response and hence are likely to be subject to diversifying selection. Recombinational replacements within these genes, selected as they introduce genetic diversity, will frequently extend into flanking genes and may influence the sequence evolution of arcC. Although this explanation needs to be examined more closely, such a hitchhiking effect has also been noted within an MLST gene (ddl) of S. pneumoniae (6).
The analysis of congruence described here can thus be used to identify gene loci in which recombination has had a particularly high impact on the phylogenetic signal; this has two implications. Firstly, genes such as arcC, which appear to be behaving atypically, could subsequently be removed from the analysis in order to reconstruct the most meaningful phylogeny for a given group of strains. Alternatively, the approach may be employed to investigate differences between loci where there is some a priori reason that they may exhibit various degrees of congruence; thus, the likely effect of diversifying selection on genes encoding proteins exposed to the host immune response can be systematically examined.
The short-term survival of nonsynonymous point mutations.
The evidence discussed above suggests an inflation of mutation, relative to recombination, over very short-term evolution. The ratio of nonsynonymous to synonymous substitutions occurring within clonal complexes approaches parity, whereas for pairwise comparisons of diverse STs it is >8 (Table 4). This suggests that de novo nonsynonymous mutations, though mostly deleterious, are rarely lethal, and most will survive long enough to be sampled. It is particularly striking that one of these point mutations has resulted in a stop codon in the glpF gene. However, the action of purifying selection will mean that few of these deleterious mutations will survive over the longer term.
A precedent is set by the study of Nachman et al. (23), who compared the ratios of synonymous and nonsynonymous substitutions within the mitochondrial gene NADH dehydrogenase subunit 3 (ND3) of humans and chimpanzees. They found a higher ratio of nonsynonymous substitutions when comparing sequences within a species than when comparing sequences from different species. Their conclusion was that most of the intraspecies protein polymorphisms are slightly deleterious and are lost from the population before becoming fixed in the different species. For S. aureus, it is possible that most of the polymorphisms within clonal complexes are also slightly deleterious and will mostly have been eliminated in those rare adaptive genotypes that occasionally give rise to new clonal complexes.
Concluding remarks.
The results discussed above present a complex picture of the influence of recombination on the evolution and population structure of S. aureus. Firstly, the striking clonal structure of the population is, with the caveats outlined in the introduction, an indication that recombination has had negligible impact on the diversification of the core genome of this species. Such a view is consistent with an examination of intraclonal diversity, which suggests that the vast majority of clonal variants arise by point mutation, rather than recombination. Going further back in the tree, and hence considering longer time scales, phylogenetic approaches suggest that at least some recombination has occurred. This may be explained, at least in part, by purifying selection resulting in the extinction of many de novo point mutations over time. Finally, the atypical phylogenetic signal within arcC demonstrates that recombination has had more influence on some gene loci than others, despite the fact that MLST genes were chosen on the basis that they are likely to represent the stable “core” of the genome. Thus, the degree to which the perceived importance and stability of essential metabolic genes equate with their phylogenetic consistency remains an open question.
Acknowledgments
E.J.F. and J.E.C. are funded by an MRC Research Career Development award (no. G120/614). N.P.J.D. and S.J.P. are funded by The Wellcome Trust. B.G.S. is a Wellcome Trust Principal Research Fellow. M.C.E. is a Royal Society University Research Fellow.
We are very grateful to Eddie Holmes and Laurence Hurst for useful discussions and critical comments on the manuscript and to Paul Wilkinson for technical assistance.
REFERENCES
- 1.Booth, M. C., L. M. Pence, P. Mahasreshti, M. C. Callegan, and M. S. Gilmore. 2001. Clonal associations among Staphylococcus aureus isolates from various sites of infection. Infect. Immun. 69**:**345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. Clonal relationships between invasive and carriage Streptococcus pneumoniae, and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis., in press. [DOI] [PubMed]
- 3.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science **292:**114-116. (Retraction, **295:**971, 2002.) [DOI] [PubMed]
- 4.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38**:**1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99**:**7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., and B. G. Spratt. 1999. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol. Biol. Evol. 16**:**1687-1695. [DOI] [PubMed] [Google Scholar]
- 7.Feil, E. J., M. C. Enright, and B. G. Spratt. 2000. Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res. Microbiol. 151**:**465-469. [DOI] [PubMed] [Google Scholar]
- 8.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98**:**182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16**:**1496-1502. [DOI] [PubMed] [Google Scholar]
- 10.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154**:**1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55**:**561-590. [DOI] [PubMed] [Google Scholar]
- 12.Grundmann, H., S. Hori, M. C. Enright, C. Webster, A. Tami, E. J. Feil, and T. Pitt. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 40**:**4544-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundmann, H., A. Tami, S. Hori, M. Halwani, and R. Slack. 2002. Nottingham Staphylococcus aureus population study: prevalence of MRSA among elderly people in the community. Br. Med. J. 324**:**1365-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17**:**754-755. [DOI] [PubMed] [Google Scholar]
- 15.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14**:**68-73. [DOI] [PubMed] [Google Scholar]
- 16.Jolley, K. A., J. Kalmusova, E. J. Feil, S. Gupta, M. Musilek, P. Kriz, and M. C. Maiden. 2000. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 38**:**4492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357**:**1225-1240. [DOI] [PubMed] [Google Scholar]
- 18.Lorenz, U., K. Ohlsen, H. Karch, M. Hecker, A. Thiede, and J. Hacker. 2000. Human antibody response during sepsis against targets expressed by methicillin resistant Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 29**:**145-153. [DOI] [PubMed] [Google Scholar]
- 19.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95**:**3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller-Graf, C. D., A. M. Whatmore, S. J. King, K. Trzcinski, A. P. Pickerill, N. Doherty, J. Paul, D. Griffiths, D. Crook, and C. G. Dowson. 1999. Population biology of Streptococcus pneumoniae isolated from oropharyngeal carriage and invasive disease. Microbiology 145**:**3283-3293. [DOI] [PubMed] [Google Scholar]
- 21.Musser, J. M., and V. Kapur. 1992. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J. Clin. Microbiol. 30**:**2058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musser, J. M., P. M. Schlievert, A. W. Chow, P. Ewan, B. N. Kreiswirth, V. T. Rosdahl, A. S. Naidu, W. Witte, and R. K. Selander. 1990. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc. Natl. Acad. Sci. USA 87**:**225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nachman, M. W., W. M. Brown, M. Stoneking, and C. F. Aquadro. 1996. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics 142**:**953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3**:**418-426. [DOI] [PubMed] [Google Scholar]
- 25.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30**:**245-257. [DOI] [PubMed] [Google Scholar]
- 26.Peacock, S. J., I. de Silva, and F. D. Lowy. 2001. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 9**:**605-610. [DOI] [PubMed] [Google Scholar]
- 27.Peacock, S. J., C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O'Neill, and N. P. Day. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70**:**4987-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson, D. A., K. M. Edwards, K. B. Waites, D. E. Briles, M. J. Crain, and S. K. Hollingshead. 2001. Clones of Streptococcus pneumoniae isolated from nasopharyngeal carriage and invasive disease in young children in central Tennessee. J. Infect. Dis. 183**:**1501-1507. [DOI] [PubMed] [Google Scholar]
- 29.Sabat, A., K. Kosowska, K. Poulsen, A. Kasprowicz, A. Sekowska, B. van Den Burg, J. Travis, and J. Potempa. 2000. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect. Immun. 68**:**973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90**:**4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spratt, B. G., W. P. Hanage, and E. J. Feil. 2001. The relative contributions of recombination and point mutation to the diversification of bacterial clones. Curr. Opin. Microbiol. 4**:**602-606. [DOI] [PubMed] [Google Scholar]
- 32.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95**:**12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swofford, D. L. 2000. PAUP and other methods. Phylogenetic analysis using parsimony, 4th ed. Sinauer Associates, Sunderland, Mass.
- 34.Takala, A. K., J. Vuopio-Varkila, E. Tarkka, M. Leinonen, and J. M. Musser. 1996. Subtyping of common pediatric pneumococcal serotypes from invasive disease and pharyngeal carriage in Finland. J. Infect. Dis. 173**:**128-135. [DOI] [PubMed] [Google Scholar]
- 35.Townsend, R., L. Goodwin, T. M. Stevanin, P. B. Silcocks, A. Parker, M. C. Maiden, and R. C. Read. 2002. Invasion by Neisseria meningitidis varies widely between clones and among nasopharyngeal mucosae derived from adult human hosts. Microbiology 148**:**1467-1474. [DOI] [PubMed] [Google Scholar]