Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II (original) (raw)
Abstract
In budding yeast, the mitogen-activated protein kinase (MAPK) Hog1 coordinates the transcriptional program required for cell survival upon osmostress. The Hot1 transcription factor acts downstream of the MAPK and regulates a subset of Hog1-responsive genes. In response to high osmolarity, Hot1 targets Hog1 to specific osmostress-responsive promoters. Here, we show that assembly of the general transcription machinery at Hot1-dependent promoters depends on the presence of Hot1 and active Hog1 MAPK. Unexpectedly, recruitment of RNA polymerase (Pol) II complex to target promoters does not depend on the phosphorylation of the Hot1 activator by the MAPK. Hog1 interacts with the RNA Pol II and with general components of the transcription machinery. More over, when tethered to a promoter as a LexA fusion protein, Hog1 activates transcription in a stress- regulated manner. Thus, anchoring of active Hog1 to promoters by the Hot1 activator is essential for recruitment and activation of RNA Pol II. The mammalian p38 also interacts with the RNA Pol II, which might suggest a conserved mechanism for regulation of gene expression by SAPKs among eukaryotic cells.
Keywords: Hog1/Hot1/osmostress/RNA polymerase II/SAPK
Introduction
Mitogen-activated protein kinase (MAPK) cascades are common signaling modules found in both higher and lower eukaryotic cells (Robinson and Cobb, 1997). Budding yeast has several MAPK cascades, one of which contains a relative of the p38 family of stress-activated MAPKs. This kinase, Hog1, coordinates cellular responses to increases in external osmolarity by inducing diverse osmoadaptive responses (Hohmann, 2002). Recent genome-wide transcriptional studies have revealed that a large number of genes are regulated by osmotic stress in a _HOG1_-dependent manner, suggesting a key role for the MAPK in stress-induced gene expression (Posas et al., 2000; Rep et al., 2000). There is not a uniform mechanism by which stress-activated MAPKs (SAPKs), and MAPKs in general, modulate gene expression. It has been reported that SAPKs can modify gene regulation by direct phosphorylation of transcription factors, both activators and repressors. In addition, it has been reported that in response to stress, and through intermediate kinases (MSKs), they can induce the phosphorylation of com ponents involved in chromatin remodeling and histones themselves (Kyriakis and Avruch, 2001; de Nadal et al., 2002).
In yeast, several transcription factors have been proposed to act under the control of the Hog1 MAPK (i.e. Msn1, Msn2, Msn4, Hot1, Sko1 and Smp1). Each factor seems to be controlling a small subset of the osmoresponsive genes, and thus deletion of a particular transcription factor has a very limited effect on general osmostress gene expression. Although little is known about how Hog1 regulates the activity of downstream factors, two different mechanisms by which it can regulate the activity of a transcription factor have recently been proposed. One mechanism involves the MEF2-like transcription factor Smp1. Smp1 activator is directly phosphorylated by Hog1 on several residues within its transactivation domain, and this phosphorylation by the MAPK is essential for Smp1-mediated gene expression (de Nadal et al., 2003). Similarly, other members of the MEF2 family of transcription factors are regulated by SAPKs (McKinsey et al., 2002). A second mechanism involves the Sko1 transcription factor. Sko1 is an ATF/CREB-related factor (Nehlin et al., 1992; Vincent and Struhl, 1992) which inhibits transcription of several genes that are inducible by osmotic stress (Proft and Serrano, 1999; Garcia-Gimeno and Struhl, 2000). Sko1 represses gene expression by recruiting the general corepressor complex Ssn6–Tup1. Release from Ssn6–Tup1 repression in response to osmotic stress requires direct phosphorylation by the Hog1 MAPK (Proft et al., 2001). Interestingly, Hog1 phosphorylation switches Sko1 activity from a repressing to an activating state, which involves recruiting of SWI/SNF and SAGA complexes (Proft and Struhl, 2002).
Here, we analyze the mechanism by which the Hot1 activator controls Hog1-mediated osmostress gene expression. Hot1 is a transcription factor related to Msn1. It controls a small subset of genes involved in the production of osmolyte (Rep et al., 1999). Hot1 interacts with Hog1 (Rep et al., 1999), and this interaction is critical for recruitment of the MAPK to Hot1-dependent promoters and essential for their transcriptional induction upon stress (Alepuz et al., 2001). Thus, apart from the role of Hog1 in the modification of transcription factor activity, its specific association with stress-responsive promoters suggests a new direct involvement of a signaling kinase in complex recruitment and activity.
In this work we present evidence for such a model showing that recruitment of the Hog1 MAPK by the Hot1 activator is critical for gene expression. We show that phosphorylation of Hot1 is not required for gene expression or binding of the MAPK to the promoters. Instead the critical step to induce gene expression is the Hog1 directed recruitment of the RNA polymerase (Pol) II complex to the promoter. Our results indicate that Hot1 mediates transcription by the anchoring of a complex that includes active Hog1 MAPK and RNA Pol II holoenzyme. Interestingly, SAPK p38, the mammalian homolog of Hog1, interacts with the core of the RNA Pol II in HeLa cells. These data suggest that a novel conserved mechanism for regulation of gene transcription mediated by stress-activated MAPKs could exist among eukaryotic cells.
Results
RNA Pol II holoenzyme is recruited to osmoresponsive genes in response to stress
Hot1 induces a subset of stress-responsive genes under the control of the Hog1 MAPK in response to osmostress (Rep et al., 1999). Recently, we have shown that the Hot1 activator targets the Hog1 MAPK to osmoresponsive promoters (e.g. STL1) upon stress (Alepuz et al., 2001). However, the underlying mechanism by which Hot1 induces transcription and its regulation by the MAPK is not clearly established.
To address this question, we utilized chromatin immunoprecipitation (ChIP) to follow the binding of the transcription machinery to the STL1 promoter before and after osmostress. The STL1 gene is a prototypical Hot1-regulated gene. It is highly expressed in response to osmostress and is completely dependent on the presence of Hot1 and Hog1 MAPK (Posas et al., 2000; Rep et al., 2000). Chromatin from a yeast strain expressing functional epitope-tagged components of the RNA Pol II holoenzyme from their natural locus was immunoprecipitated with antibodies against the HA epitope and analyzed by PCR. We first probed when the Srb–mediator would occupy the STL1 promoter. As shown in Figure 1, Srb10, Srb11 and Rgr1 are found at the STL1 promoter only after osmotic stress. A similar picture was obtained with the core RNA Pol II (Rpb1) and its associated general transcription factors TFIIB (Sua7) and TFIIH (Kin28) (Figure 1). Virtually no signal was detectable in ChIP assays from normal growing cells, whereas stress treatment quickly elicited a strong signal. Thus, the RNA Pol II complex is recruited to the STL1 promoter only in response to stress.
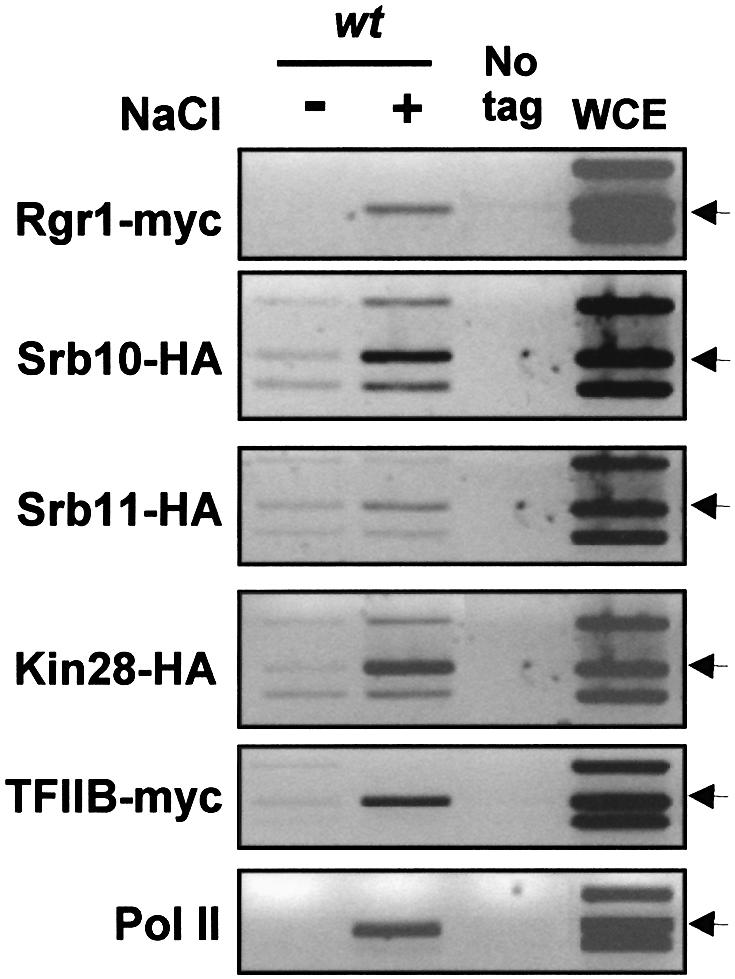
Fig. 1. Hog1 mediates recruitment of the transcription machinery to stress-responsive promoters in response to stress. Osmostress induces the recruitment of mediator to osmostress-regulated genes as detected by ChIP analysis. Strains containing genomic tags of Rgr1-myc (K9671), Srb10-HA (PAY172), Srb11-HA (PAY257), Kin28-HA (PAY168), TFIIB-myc (K8407) and Rpb1-myc (P156) were grown, and samples for ChIP analyses were taken before (–) and 10 min after (+) the addition of NaCl to a final concentration of 0.4 M. Immuno precipitations were performed using mouse anti-myc or anti-HA monoclonal antibodies. PCR was realized with primers spanning the TATA box of _STL1 (_arrows) and two pairs of control oligonucleotides spanning the GAL1 and FUS1 gene regions (upper and lower bands, respectively). Control lanes show DNA amplified from extracts without tagged protein (K699, no tag) or prior immunoprecipitation (whole-cell extract, WCE). The same WCE and no tag is presented for experiments carried out in parallel.
Recruitment of the transcription machinery to Hot1-dependent genes depends on active Hog1 and the presence of Hot1
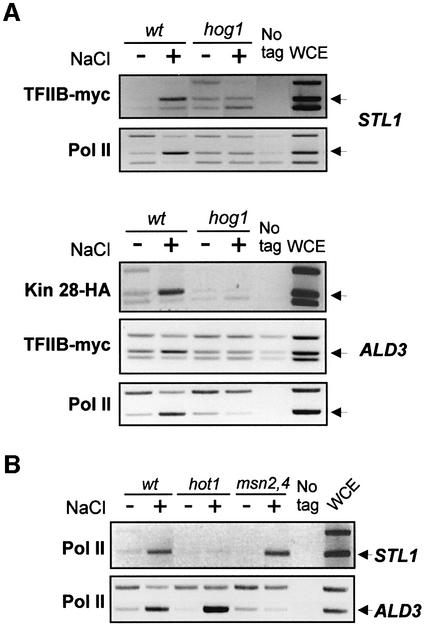
To dissect the role of Hot1 and Hog1 MAPK in the recruitment of the RNA Pol II holoenzyme to osmoresponsive promoters, we analyzed the recruitment of the complex in _hog1_Δ- and _hot1_Δ-deficient cells as described above. As shown in Figure 2A, Hog1 is essential for binding of TFIIB and RNA Pol II to the STL1 promoter.
Fig. 2. Recruitment of RNA Pol II holoenzyme to promoters depends on specific activators and Hog1 MAPK activity. (A) Hog1 is necessary for TFIIH, TFIIB and Pol II osmotic-stress-dependent association with STL1 and ALD3 promoters. TFIIB-myc strains PAY220 (wt) and PAY217 (_hog1_Δ), and Kin28-HA strains PAY168 (wt) and PAY173 (_hog1_Δ) were grown and samples for ChIP analyses were taken as in Figure 1. Pol II binding was detected by using a mouse monoclonal antibody against Rpb1 (8WG16, Covance). Immunoprecipitated samples were processed for ChIPs as described in Materials and methods. Binding to STL1 and/or ALD3 promoters was determined by PCR. (B) Association of Pol II with STL1 and ALD3 promoters requires the presence of specific activators. Cross-linked cell extracts from non-stressed (–) or osmotically stressed (+) wild-type strain (K699) or strains containing a hot1 mutation (UG43) or msn2 msn4 mutations (YM24) were immunoprecipitated using 8WG16 antibody against Pol II. Binding of Pol II to STL1 or ALD3 promoters was assayed as before.
The ALD3 gene is strongly responsive to osmostress and depends on Hog1. However, ALD3 expression is not mediated by Hot1 but by the Msn2 and Msn4 transcription factors (Rep et al., 2000). Association of Kin28, TFIIB and Pol II to the ALD3 promoter also correlated with stress induction and Hog1 signaling (Figure 2A).
Recruitment of Hog1 to stress-responsive promoters depends on the presence of specific activators. Hot1 is required for binding of Hog1 to STL1, and Msn2/Msn4 are required for Hog1 binding to ALD3 (Alepuz et al., 2001). As revealed by ChIP analysis, binding of Pol II (Rpb1) to STL1 was dependent on the presence of Hot1 and independent of Msn2 and Msn4, whereas binding of Rpb1 to ALD3 was totally dependent on the presence of Msn2 and Msn4 (Figure 2B). The stable recruitment of RNA Pol II holoenzyme seems to correlate closely with the promoter anchorage of Hog1 by specific factors. Therefore, our data suggest that binding of RNA Pol II to osmoresponsive promoters must be a function of both an active Hog1 MAPK and the presence of specific activators.
Phosphorylation of Hot1 activator by the MAPK is not required for gene expression
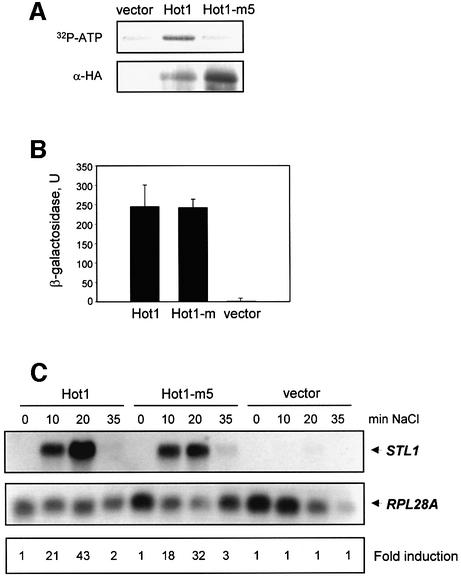
The activity of the MAPK was required for Hot1-mediated binding of the RNA Pol II complex to the STL1 promoter, and gene expression upon stress. A possible mechanism of Hot1 regulation is through direct phosphorylation by the MAPK. To test this possibility, we expressed and purified from yeast an HA-tagged wild-type Hot1 and a mutant allele of Hot1 (Hot1-m5) that contains mutations in all putative phosphorylation sites for the MAPK (i.e. Ser30, Ser70, Ser153, Ser360 and Ser410 to Ala). After immunoprecipitation, HA-tagged Hot1 and Hot1-m5 were subjected to an in vitro phosphorylation assay together with active Hog1 (see Materials and methods). As shown in Figure 3A, wild-type Hot1 was phosphorylated by Hog1 whereas the mutated allele Hot1-m5 was not.
Fig. 3. Hot1 phosphorylation by Hog1 is not required for STL1 activation. (A) Hot1-m5 mutant is not phosphorylated in vitro by Hog1. HA-tagged Hot1 or Hot1-m5 proteins were purified from yeast and incubated with active Hog1 and radioactive ATP (see Materials and methods). Phosphorylated proteins were resolved by SDS–PAGE and transferred to membrane. In vitro phosphorylated proteins were detected by autoradiography (upper panel). HA-tagged Hot1 proteins were detected by immunoblot using anti-HA monoclonal antibodies (lower panel). (B) Hot1 mutant and Hot1 wild type interact with Hog1. Two-hybrid analysis was realized in L40 strain transformed with a LexA-Hog1 plasmid and empty pGAD424 (vector), or containing a wild-type Hot1 (pUG603; Hot1) or an unphosphorylatable Hot1 mutant (pPA89; Hot1-m). β-galactosidase was measured as described in Materials and methods. (C) Hot1-m5 induces STL1 gene expression upon osmostress. Cell cultures of a hot1 mutant strain (PAY181) transformed with the pRS316 plasmid containing Hot1 (pPA97), Hot1-m5 (pPA106) or empty vector were incubated with 0.4 M NaCl at the indicated times. Total RNA was assayed by northern blot analysis for transcript levels of STL1 and RPL28 as a loading control. Quantification data come from the same original blot for each strain and relate to the values at zero time (see Materials and methods).
We then tested whether elimination of the phosphorylation sites affected the binding of Hot1 to the MAPK. Previously, it was shown by two-hybrid analysis that a Gal4 fusion protein containing most of Hot1 was able to interact with Hog1 (Rep et al., 1999). We created a mutant allele of that Gal4 fusion protein with mutations on the phosphorylation sites for the MAPK (Hot1-m). As shown in Figure 3B, two-hybrid analyses indicated that Hot1 was able to interact with Hog1, and binding was not affected by mutation of the Hog1 phosphorylation sites in Hot1. Similar results were obtained with a hog1-K/N mutant (data not shown).
To analyze the role of Hog1 phosphorylation in Hot1 transcriptional activity, we transformed a _hot1_Δ strain with an empty vector or centromeric plasmids carrying wild-type HOT1 or the mutant allele HOT1-m5 expressed under the native HOT1 promoter. Yeast cells were exposed to osmostress, and expression of STL1 was followed by northern blotting. As shown in Figure 3C, expression of STL1 was not induced in a _hot1_Δ strain carrying the control vector but was strongly induced upon stress in a strain carrying wild-type HOT1. Unexpectedly, both the level of expression and the kinetics of expression of STL1 upon stress in the HOT1-m5 strain were similar to those in the wild-type strain. Thus, these results clearly indicate that phosphorylation of Hot1 by the MAPK is not essential for regulated gene expression upon stress.
Artificial binding of Hot1 to promoters is not sufficient for RNA Pol II recruitment and activation
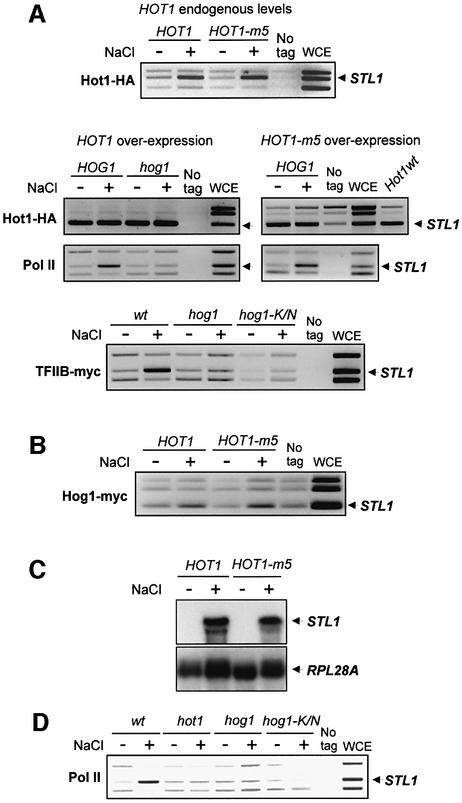
There are indications for interdependent promoter binding of Hog1 and Hot1 activator (Alepuz et al., 2001). This situation made it difficult to separate the contribution by the DNA binding factor from that of the kinase in the recruitment of the RNA Pol II. Recently, we observed that an increase in HOT1 dosage will lead to Hog1-independent promoter binding of this factor yet without resulting in any significant elevation of transcription (Figure 4A and C). Moreover, even with Hot1 constitutively present at the STL1 promoter, binding of the mediator, TFIIB and the core RNA Pol II was absolutely dependent on stress and the presence of catalytically competent MAPK (Figure 4A; data not shown).
Fig. 4. Binding of Hot1 to promoters is not sufficient for RNA Pol II recruitment and activation. (A) Promoter-bound Hot1 is not sufficient for initiating Pol II holoenzyme recruitment. Yeast strains were transformed with a centromeric plasmid expressing Hot1-m5 or Hot1-m5- HA at endogenous levels (upper panel), or with multicopy plasmid overexpressing Hot1-HA (left middle panels) or Hot1-m5-HA (right middle panels). ChIPs were performed to determine binding of Hot1, Pol II and TFIIB to STL1 promoter (arrow). The binding of overexpressed wild-type Hot1 (Hot1wt) was analyzed in parallel to overexpressed Hot1-m5 binding to compare the affinity of both proteins with the STL1 promoter (right middle panels). Rpb1-myc (Pol II) binding was analyzed in strains P156 (HOG1) and PAY217 (hog1) (middle panels); Hot1-HA or Hot1-m5-HA binding was analyzed in PAY181(HOG1) and strain PAY218 (_hog1_Δ). TFIIB-myc binding was analyzed in strains K8407 (wt) and PAY218 transformed with an empty vector or with a plasmid containing a kinase dead Hog1 version (hog1-K/N) (lower panel). (B) Hog1 binds to STL1 promoter upon stress in cells overexpressing Hot1. Binding of Hog1-myc was analyzed by ChIP in the PAY181 (_hot1_Δ) strain cotransformed with plasmids overexpressing Hot1-HA or Hot1-m5-HA. K699 strain was used as a control (no tag). (C) STL1 expression in cells with constitutively bound Hot1 and Hot1-m5. Wild-type cells with multicopy plasmids overexpressing Hot1 and Hot-m5 were grown in minimal medium and treated with 0.4 M NaCl for 20 min. Total RNA was probed with fragments of STL1 and RPL28A (as a loading control). (D) Hog1 kinase activity is necessary for the initial step of recruitment of the transcription machinery. Rpb1-myc (Pol II) association with the STL1 promoter (arrow) was measured in strains P156 (wt), PAY226 (hot1) and PAY228 transformed with a control vector (hog1) or a plasmid containing the kinase dead Hog1 (hog1-K/N). ChIPs were performed to determine binding to STL1 promoter.
We have shown that the Hot1-m5 allele was unphosphorylatable by Hog1. We then investigated whether binding of Hog1 or RNA Pol II was affected by mutation of the phosphorylation sites in Hot1 in the overexpression system. Binding of Hot1-m5 was similar to that of the wild type when expressed at endogenous levels (Figure 4A, upper panel). Upon overexpression, as for the wild type, binding of Hot1-m5 was constitutive on the STL1 promoter and, more importantly, recruitment of RNA Pol II and binding of Hog1 to the promoter also depended on stress (Figure 4A and B). These results were consistent with the induction of STL1 expression by wild-type Hot1 and mutant Hot1-m5 observed upon stress (Figure 4C). Thus, in a system where binding of Hot1 is unaffected by Hog1, recruitment of RNA Pol II is still dependent on Hog1 activity and independent of the phosphorylation state of the Hot1 activator.
To analyze the role of Hog1 kinase activity in the recruitment of RNA Pol II to STL1, we transformed a _hog1_Δ strain with an empty vector or a vector expressing the catalytically inactive Hog1 enzyme (hog1-K/N). Recruitment of Pol II was completely dependent on Hog1 kinase activity (Figure 4D).
Hog1 interacts with the RNA Pol II holoenzyme
The Hog1 MAPK regulated Hot1-mediated transcription by a mechanism other than direct phosphorylation of the activator. To gain a better understanding of Hog1-mediated responses we attempted two different approaches: a biochemical identification of direct interactors for Hog1, and a genetic screening to unravel elements required for Hog1-mediated gene expression (described below). It has been reported that interaction of MAPKs with substrates is mediated via conserved docking domains (Sharrocks et al., 2000). Therefore, we fused the C-terminal region of Hog1 (residues 265–435), which contains a hypothetical docking site (DS), to glutathione _S_-transferase (GST). Cell extracts were prepared from cells carrying GST or GST–Hog1DS, and GST pull-down experiments were carried out with gluthatione–Sepharose beads. Coprecipitating proteins were resolved by SDS–PAGE and silver stained, and prominent bands were analyzed by high-accuracy peptide mass mapping using MALDI analysis (described in Materials and methods). Two of the most abundant proteins present in the coprecipitation with Hog1DS corresponded to the α and β subunits of the RNA polymerase (Rpb1 and Rpb2).
Binding of Hog1 to Rpb1 was confirmed by direct coprecipitation experiments. Yeast cells containing a chromosomally myc-tagged Rpb1 were transformed with plasmids that expressed GST-tagged full-length Hog1, Hog1DS (residues 265–435) or Hog1ΔDS (residues 1–301, corresponding to the kinase domain). As shown in Figure 5A, Rpb1 coprecipitated with full-length Hog1 and with the C-terminal region (Hog1DS) but not with Hog1ΔDS. Whereas binding of Rpb1 to the full-length Hog1 was stress dependent, binding to Hog1DS was constitutive. Thus, binding of Rpb1 to Hog1 is mediated by the non-catalytic region that comprises the MAPK docking site. To assess whether binding of Hog1 with the core of the RNA Pol II was direct, we performed an in vitro binding assay using purified Hog1 from Escherichia coli with Rpb1-myc from yeast (described in Materials and methods). As shown in Figure 5C, Hog1 was able to interact with purified Rpb1.
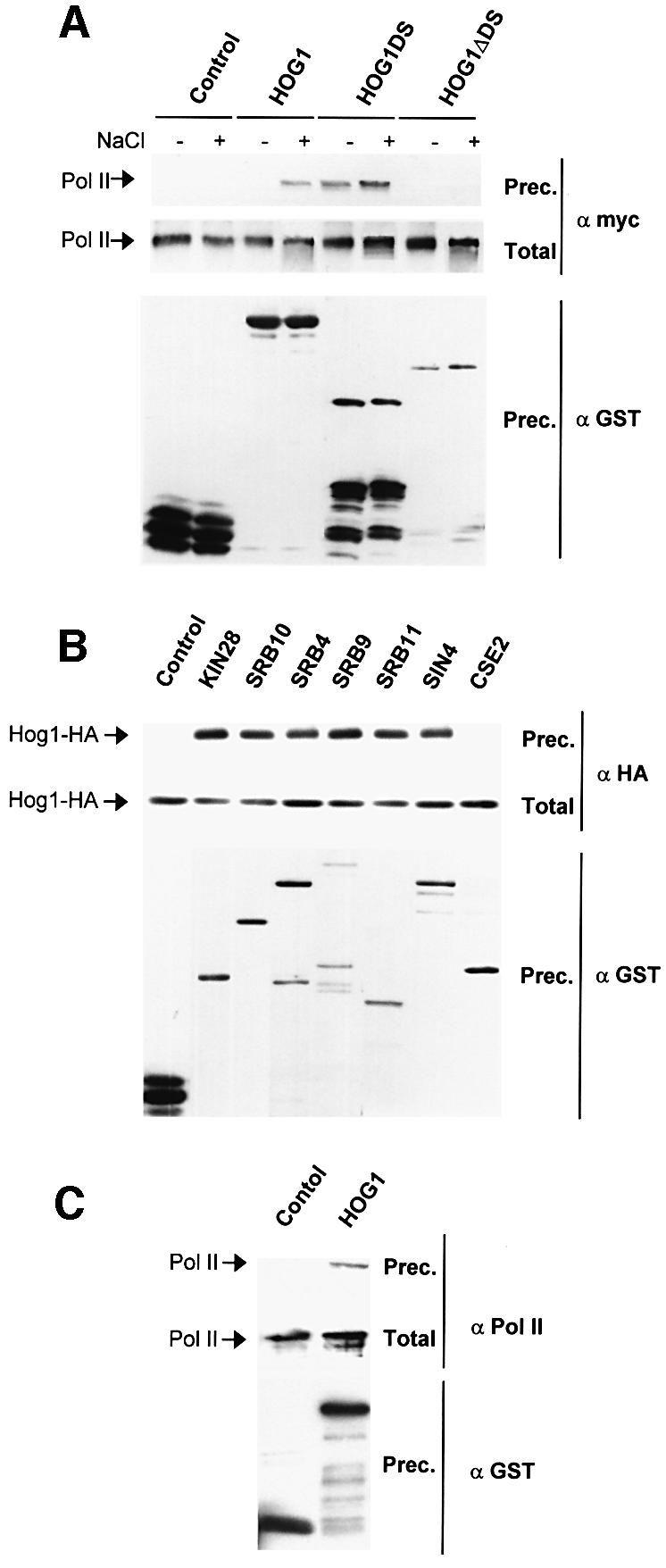
Fig. 5. In vivo and in vitro binding of Hog1 to RNA Pol II holoenzyme. (A) Hog1 physically interacts with the largest subunit of the Pol II in vivo. A myc-tagged Rpb1 strain expressed GST, GST–Hog1 or GST–Hog1DS under the PTEF1 promoter, or GST–Hog1ΔDS under the PGAL1 promoter. Cells were grown in the presence of glucose or galactose and samples were taken before (–) or 10 min after (+) treatment with NaCl. GST proteins were pulled down by glutathione– Sepharose 4B and the presence of Rpb1-myc (Pol II) was probed by immunoblotting using anti-myc (upper panel). Total extract represents <20% of total input protein (middle panel). The amount of precipitated GST proteins was detected using anti-GST (lower panel). (B) Hog1 interacts with general components of the transcription machinery. Wild-type strain TM141 was transformed with a plasmid expressing HA-Hog1 under the PGAL1 promoter and a plasmid expressing GST or a GST-containing protein. Cells were grown in the presence of galactose and samples were taken 10 min after the addition of 0.4 M NaCl. GST proteins were purified as above and HA-Hog1 was detected by western blotting using HA antibodies. Total extracts (middle panel) and GST proteins (lower panel) are shown. (C) Hog1 physically interacts with the largest subunit of the Pol II in vitro. The GST–Hog1 was purified from E.coli and incubated with semipure RNA Pol II holoenzyme. The presence of Rpb1 was probed by immunobloting using 8WG16 antibody against Pol II (upper panel). Total extracts (middle panel) and GST proteins (lower panel) are shown.
We next tested whether Hog1 interactions extended to other components of the transcription initiation complex. One of the general factors closely associated with the activation of the core polymerase is TFIIH. Wild-type cells expressing an HA-Hog1 protein were transformed with a plasmid expressing GST-tagged Kin28, the catalytically important subunit of TFIIH. A GST pull-down from osmotically challenged yeast cell extracts showed that Hog1 is also closely associated with this general factor (Figure 5B), and supported the notion that Hog1 interactions might take place with larger entities and not with the individual components. We undertook similar GST pull-down assays with parts of the Srb–mediator that identify different subcomplexes of the holoenzyme (Woychik and Hampsey, 2002). The results (Figure 5B) provided strong evidence that Hog1 can indeed interact with certain forms of the RNA polymerase holoenzyme. One exception was provided by Cse2/Med9, a component of one of the proposed Rgr1 mediator modules, documenting a certain level of selectivity in the observed interactions between the MAPK and the RNA Pol II holoenzyme.
Binding of the Hog1 MAPK to the RNA Pol II holoenzyme is unlikely to be mediated by specific activators, because this interaction was not affected in a strain deficient in _msn1_Δ, _msn2_Δ, _msn4_Δ and _hot1_Δ (Figure 6A). Furthermore, binding of Hot1 to components of the RNA Pol II holoenzyme was assayed in parallel to Hog1. As shown in Figure 6B, Hog1 was able to interact with Kin28 and Sin4, whereas Hot1 was unable to interact with these components. Thus, our data suggest that the recruitment of RNA Pol II holoenzyme to Hot1-dependent promoters is carried out by Hog1 and not by Hot1, further indicating that Hog1 acts as an adaptor for Hot1 in the assembly of the RNA Pol II complexes at the STL1 promoter.
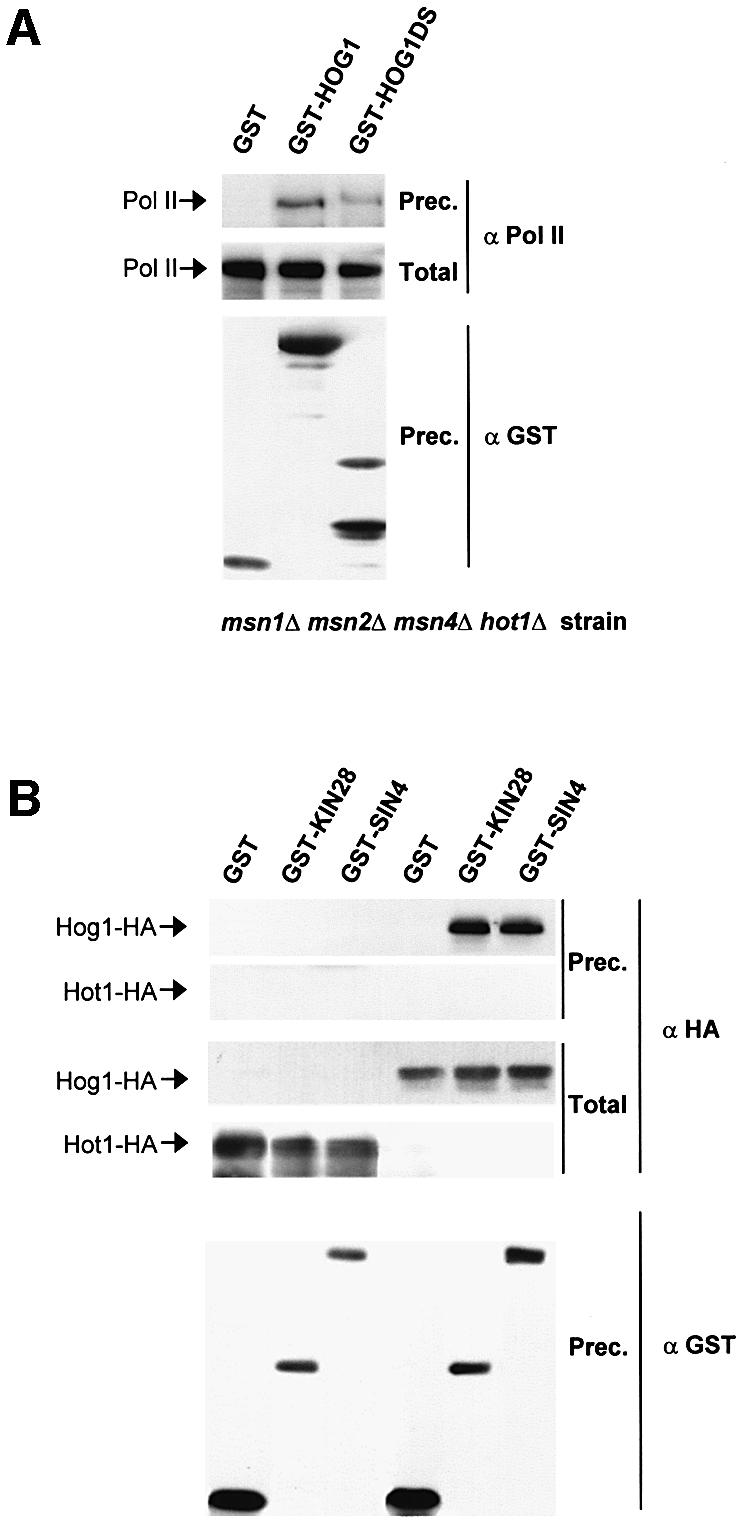
Fig. 6. In vivo binding of Hog1 to RNA Pol II is not mediated by specific activators. (A) Hog1 interacts with RNA Pol II in a mutant strain deficient in the transcription factors Hot1, Msn1, Msn2 and Msn4. The YMR120 strain (hot1 msn1 msn2 msn4) was transformed with a plasmid expressing GST, GST–Hog1 or GST–Hog1DS under the PTEF1 promoter. Cells were grown to mid-log phase and treated with 0.4 M NaCl for 10 min. Coprecipitation experiments were carried out as in Figure 5A. The presence of Rpb1 was probed by immunoblotting by using 8WG16 antibody against Pol II (upper panel). Total extract represents <20% of total input protein (middle panel). The amount of precipitated GST proteins was detected using anti-GST (lower panel). (B) Hot1 does not interact with general components of the transcription machinery. Wild-type strain TM141 was transformed with a plasmid expressing HA-Hot1 (left lanes) or HA-Hog1 (right lanes) and a plasmid expressing GST or a GST-containing protein. Cells were grown and treated with NaCl as before. GST proteins were purified as above and HA-Hot1 or HA-Hog1 was detected by western blotting using antibodies against HA (upper panels). Total extract represents <20% of total input protein (middle panels). The amount of precipitated GST proteins was detected using anti-GST (lower panel).
Binding of Hog1 to a promoter stimulates transcription in response to stress in a mediator-dependent manner
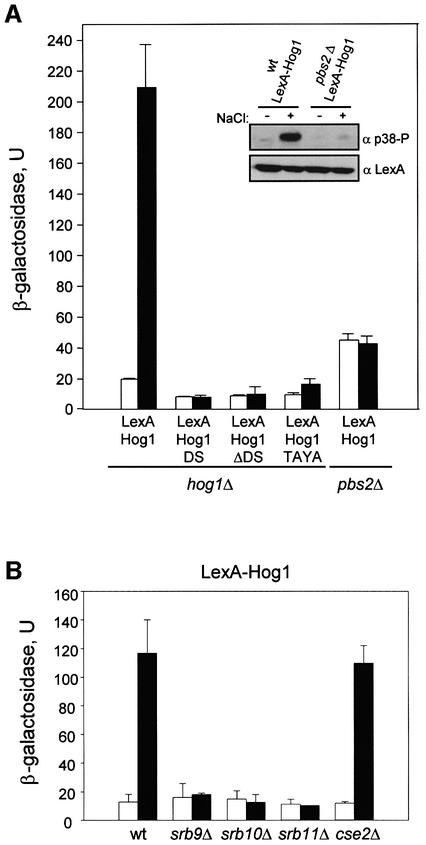
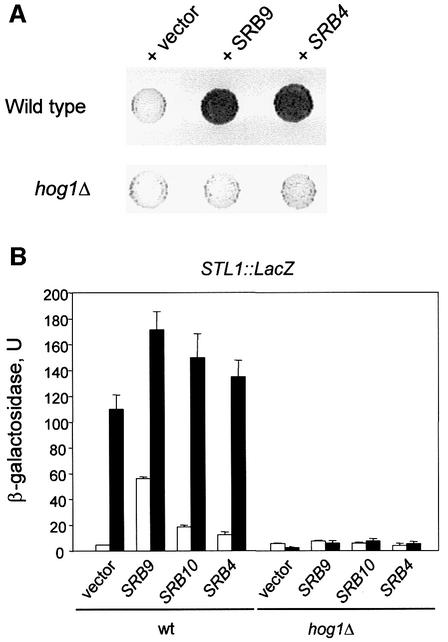
To study whether selective recruitment of the RNA Pol II holoenzyme by Hog1 was sufficient to induce transcriptional activation, we tethered Hog1 to the LexA-LacZ reporter system present in the L40 strain (Ptashne and Gann, 1997). When bound to a promoter as a LexA fusion protein, Hog1 was able to induce transcription from the reporter system to >10-fold under osmostress conditions (Figure 7A). The difference in transcription between normal and stressed cells seemed to depend on Hog1 phosphorylation, because it was lost in cells deficient in the MAPKK responsible for Hog1 phosphorylation and activation, i.e. _pbs2_Δ cells (see inset). It is also worth noting that the complete active Hog1 protein was required for transcriptional activity, because neither the Hog1DS nor the Hog1ΔDS proteins fused to a LexA were able to activate transcription. Also, a catalytically inactive mutant that could not be phosphorylated by Pbs2 (Hog1TAYA, which contains mutations Thr174 and Tyr176 to Ala) was unable to induce gene expression upon stress (Figure 7A). Activation of transcription by the LexA-GAL4 activator was higher than that observed with Hog1, but unaffected by osmostress (data not shown). We then tested whether integrity of Srb–mediator was required for Hog1-mediated gene induction. We deleted some genes encoding non-essential components of the Srb–mediator complex such as SRB9, SRB10, SRB11 or CSE2 in an L40 strain. Hog1-dependent gene induction upon stress was almost abolished in the tested strains, except for the cse2 mutant (Figure 7B). It is worth noting that GST–Cse2 also did not interact with Hog1 in the pull-down experiments. Thus, transcription not only requires the Hog1 catalytic activity, but also the presence of the C-terminal docking domain that could capture the RNA polymerase. Taken together, our data indicate that the Hog1DS protein, which does not have kinase activity, coprecipitates the RNA Pol II holoenzyme but cannot activate transcription. On the other hand, Hog1ΔDS is catalytically active but cannot bind RNA Pol II and does not induce transcription.
Fig. 7. Artificially recruited Hog1 stimulates transcription in response to stress in a mediator-dependent manner. (A) Transcription activation from LexA-Hog1 in response to stress. L40 (_LexA_-lacZ reporter strain) containing _hog1_Δ or _pbs2_Δ mutations were transformed with constructs expressing LexA-Hog1, LexA-Hog1DS, LexA-Hog1ΔDS or LexA- Hog1TAYA. β-galactosidase activity was assayed in cells grown to mid-log phase before (open bars) or after (filled bars) a brief osmotic stress (0.4 M NaCl for 30 min). β-galactosidase values are given in nmol/min/mg. To test the degree of LexA-Hog1 phosphorylation, total cell extracts were resolved in SDS–PAGE and immunoblotted with α-LexA antibody (Santa Cruz Biotechnology Inc) to detect LexA-Hog1 (lower panel) and with antiphospho-p38 MAPK antibody (New England BioLabs) to detect phosphorylated Hog1 (upper panel). Data shown are averages of five transformants. (B) Mutations in Srb– mediator genes reduce transcriptional activation by LexA-Hog1. Wild-type L40 or L40 strains containing mutations in srb9, srb10, srb11 or cse2 were transformed with the pBTM116 plasmid expressing LexA-Hog1. β-galactosidase activity was assayed before (open bars) or after (filled bars) a brief osmotic stress as in (A).
Expression of STL1 is induced by overexpression of Srb–mediator components in a HOG1-dependent manner
To complement our biochemical approach (described above), genetic screening was performed to identify elements required for Hog1-mediated STL1 gene expression. We created a reporter strain containing an integrated STL1::LacZ construct (see Materials and methods) and searched for genes whose overexpression resulted in constitutive STL1 expression. From this screen we identified plasmids overexpressing SRB9, SRB4 and SRB10 that increased the basal level of STL1 expression in the absence of stress and even raised the normal levels of STL1 in response to stress. The effects caused by the overexpression of SRB genes disappeared in _hog1_Δ cells (Figure 8A and B). Similarly, an increased level of SRB9 and SRB10 was able to induce expression of the LexA-LacZ reporter system by the LexA-Hog1 construct (data not shown). Thus, the results from the STL1 promoter fully reflect the results obtained with the artificial LexA promoter system, and both datasets implicate the same Srb–mediator proteins in allowing gene expression upon Hog1 recruitment.
Fig. 8. STL1 transcription is upregulated by stress and overexpression of Srb–mediator in a _HOG1_-dependent manner. (A) Srb–mediator modulates _HOG1_-mediated STL1 gene expression. Yeast cells containing an integrated STL1::LacZ reporter construct were transformed with a multicopy genomic library and positive clones were selected by their ability to induce STL1. Representative filter β-galactosidase assay demonstrating induction of STL1::LacZ by several positive clones from the screening and their dependence of HOG1 is shown. (B) A wild-type (TM141) and a _hog1_Δ mutant strain (TM233) carrying an integrated STL1::lacZ reporter construct were transformed with the multicopy plasmid pRS425 either empty (vector) or carrying SRB9, SRB10 or SRB4 genes. Cells were grown and β-galactosidase activity was assayed before (open bars) or after (filled bars) a brief osmotic stress (0.4 M NaCl for 30 min).
Mammalian p38α coprecipitates with RNA Pol II from epithelial cells
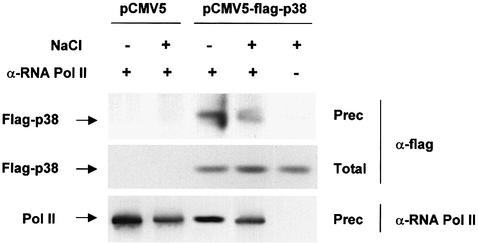
The mammalian p38 MAPK is the functional homolog of the yeast Hog1 MAPK. We have shown that Hot1-mediated transcription requires binding of the Hog1 MAPK to specific promoters, and that the MAPK recruits the RNA Pol II holoenzyme. We then tested whether p38 was also able to interact with the mammalian RNA Pol II holoenzyme. HeLa cells were transiently transfected with either an empty vector or a vector carrying a Flag-tagged p38α (Flag-p38). Cells were subjected to a brief osmotic shock (0.3 M NaCl for 20 min), and RNA Pol II was immunoprecipitated from cell extracts using specific monoclonal antibodies against RNA Pol II (8GW16). The presence of p38 in the precipitates was probed by an anti-Flag antibody. As shown in Figure 9, p38 was able to coprecipitate with RNA Pol II irrespective of the environmental conditions. Thus, our results suggested that binding of p38 SAPKs, from both yeast and mammals, to the RNA Pol II might indicate a common mechanism of interaction with the transcriptional machinery.
Fig. 9. Co-immunoprecipitation of RNA Pol II and p38. HeLa cells were transfected with either pCMV5 or pCMV5-Flag-p38. Transfected cells were serum starved and, when indicated, stimulated with 0.3 M NaCl for 20 min. Lysates from the transfected cells were immunoprecipitated with anti-RNA Pol II antibody (8WG16), and the immunoprecipitates were subjected to immunoblot analysis for flag-p38 using the anti-Flag M2 monoclonal antibody.
Discussion
There are many examples where direct modification of a transcription factor by SAPKs has been proven to constitute an essential step for activation of transcription (Kyriakis and Avruch, 2001). However, our initial results (Alepuz et al., 2001) suggested that this may not be true for Hot1, one of the transcription factors involved in the high osmolarity response in yeast. In an artificial promoter system, it was shown that a lexA–Hot1 fusion protein needs SAPK activity for effective activation of a lexA operator system even if DNA binding is constitutive in this system and even if certain phosphorylation site mutants are used. These results suggested a novel mechanism for MAPK-dependent activation, but left several questions unanswered. First, it did not completely resolve the problem of how much Hot1 phosphorylation was affected by the site-specific mutations. Secondly, it did not indicate at what steps during assembly of the initiation complex kinase activity might be needed. Finally, it did not provide any evidence as to whether the kinase could physically interact with any of general transcription complexes. The results reported here now provide a much clearer picture on all three important points. In addition, we also found evidence that the proposed mechanisms might apply even to higher eukaryotes.
Related to the first problem, we show that the direct phosphorylation of Hot1 by the MAPK is indeed not critical for regulation and activation per se. We have now performed these experiments with full-length Hot1 and with physiologically relevant promoters. Mutations in all five putative Hog1 phosphorylation sites in Hot1 clearly abolish Hot1 phosphorylation by Hog1 in vitro. Never theless, the mutations do not affect interaction between the two proteins and do not affect binding of Hog1 to target promoters. More importantly, Hot1-mediated gene expression was only marginally affected upon stress. Thus, activation of gene expression by Hot1 must revolve around a mechanism other than phosphorylation of the activator by the MAPK.
Binding of RNA Pol II to Hot1-mediated promoters only occurs in response to osmostress and depends on the MAPK. Therefore, we propose that the activated MAPK is the entity that directly recruits the RNA Pol II holoenzyme to targeted promoters. Several lines of evidence support this proposition. A biochemical characterization of Hog1 interacting proteins by mass spectrometry yielded Rpb1 and Rpb2 proteins (α and β subunits of RNA Pol II) as direct interactors for the MAPK. Further studies demonstrated that only Hog1, and not Hot1, is able to precipitate the RNA Pol II complex. The functional relevance of the interaction of Hog1 with RNA Pol II was further exemplified by the fact that artificial recruitment of Hog1 to DNA is able to induce gene expression upon stress, and that overexpression of SRB proteins leads to gene expression in a _HOG1_-dependent manner. Moreover, our data suggest that, apart from recruitment of RNA Pol II, Hog1 kinase activity may also play a role in the activation of the holoenzyme. Data supporting this idea come from experiments where either complete Hog1 or subdomains of the kinase were artificially tethered to a heterologous promoter via a DNA-binding domain. The promoter association of an inactive kinase turned out to be insufficient to induce gene expression. At the least, phosphorylation of Hog1 and the presence of the C-terminal docking domains are necessary preconditions for full transcription. Promoter binding of Hog1 domains that are only capable of interacting with the RNA Pol II holoenzyme in vitro is insufficient for transcription.
Taken together, our data allow us to propose a mechanism for regulation of gene expression upon stress, in which Hot1 protein acts as an anchor for the MAPK. Hog1 is the key factor for inducing gene expression by directly recruiting the RNA Pol II holoenzyme. Recently, other independent studies of yeast have suggested that the successful recruitment of a preinitiation complex might be kinase mediated. It was shown that Snf1 kinase also associates with Srb–mediator elements of the RNA Pol II holoenzyme, and that recruitment of Snf1 to a heterologous promoter induces transcription depending on the presence of Srb–mediator proteins (Kuchin et al., 2000). Furthermore, recruitment of RNA Pol II, TFIIB and TFIIH to SBF-dependent promoters requires Cdk1 kinase activity (Cosma et al., 2001). However, this is the first report of direct recruitment of the RNA Pol II complex by a MAPK to targeted promoters.
Several indications suggest that this mechanism of gene regulation by Hog1 is not restricted to Hot1-dependent genes. It was reported that recruitment of Hog1 to Msn2/4 promoters was dependent on the presence of the transcription factor and of active MAPK (Alepuz et al., 2001). We have shown here that binding of RNA Pol II to an Mns2/4-dependent promoter is independent of Hot1, but is completely dependent on the presence of Msn2/4. Consistent with our model, binding of RNA Pol II also requires the presence of the Hog1 MAPK. Thus, different activators under the control of the Hog1 MAPK could be using a similar mechanism to induce gene expression upon stress.
Sko1 regulates genes that are induced upon osmostress by recruiting the Cyc8 (Ssn6)–Tup1 corepressor complex to target promoters (Proft and Serrano, 1999; Garcia-Gimeno and Struhl, 2000; Proft et al., 2001). Recently, it has been reported that Sko1 phosphorylation by Hog1 is critical for switching Sko1 from a repressor to a transcriptional activator (Proft et al., 2001; Proft and Struhl, 2002). Thus, two different steps seem to be required for Sko1 to induce gene expression. The first step consists in the release of repression, which involves phosphorylation of Sko1 by the MAPK and the recruitment of SAGA and SWI/SNF complexes via Tup1 to the promoter. In the second step, in which Sko1 acts as an activator, Hog1 activity is essential but direct phosphorylation of Sko1 is not relevant (Proft and Struhl, 2002). Thus, based on our model, we propose that, once relieved from the repressing state, Sko1 could be working like Hot1, recruiting Hog1 and RNA Pol II to induce gene expression.
The mammalian SAPK homolog of Hog1 is the p38 MAPK which can functionally replace Hog1 in yeast, suggesting a high degree of complementation (Galcheva-Gargova et al., 1994). It has not yet been demonstrated that p38 is recruited to target promoters by specific transcription factors; however, we have detected interaction of p38 with RNA Pol II enzyme on HeLa cells. Although further studies are required to determine whether p38 can induce gene expression by direct contact with RNA Pol II, this is the first evidence that suggests a novel common mechanism for regulation of gene expression by SAPKs among eukaryotes.
Materials and methods
Strains, media and genetic techniques
The following yeast strains were used for coprecipitation assays: P156 (MAT_α RPB1-myc18::TRP1 leu2-3_,112 ura3-1 his3-11 trp1-1 can100), YMR120 (MATa hot1::kanMX4 msn1::URA3 msn2-3::HIS3 msn4-1::TRP1 leu2-3, 112 ura3-1 his3-11 trp1-1 can100) and the wild-type strain TM141 (MATa ura3 leu2 trp1 his3). For analyzing LacZ expression, we used the strain L40 (MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::LexA-lacZ) and its derivatives YEN60 (pbs2::LEU2), YEN61 (srb9::HIS3), YEN58 (srb11::HIS3), YEN106 (srb10:: KAN-MX) and YEN104 (cse2::KAN-MX). Plasmid Yip358R containing the reporter STL1::lacZ was transformed into a wild-type TM141 and the derivative hog1 mutant TM233 (_MAT_α ura3 leu2 trp1 his3 lys2 hog1::TRP1) strains, creating YEN2 and YEN7, respectively. Genomic disruptions were made by long flanking homology PCR-based gene disruption. All strains for ChIP and northern blot assays were derivatives of K699 (leu2-3, 112 ura3-1 his3-11 trp1-1 can100) obtained through genetic crosses or transformation. Tagging of genomic open reading frames with HA or myc epitopes was carried out using a PCR-based strategy. Yeast strains PAY172 (MATa SRB10-HA6::HIS3), PAY257 (_MAT_α SRB11-HA6::TRP1), PAY168 (MATa KIN28-HA6::HIS3), PAY173 (MATa KIN28-HA6::HIS3 hog1::TRP1), PAY220 (MATa TFIIB-myc18::TRP1), PAY217 (MATa TFIIB-myc18::TRP1 hog1::kanMX4), PAY228 (MATa RPB1-myc18::TRP1 hog1::kanMX4) and PAY218 (MATa TFIIB-myc18::TRP1 hog1::kanMX4 ash1::URA3) were obtained in this work. Other strains used for ChIPs, also derivatives of K699, were K9671 (_MAT_α RGR1-myc18::TRP1 ash1::HIS3), K8407 (MATa TFIIB-myc18::TRP1 ash1::HIS3), P156 (_MAT_α RPB1-myc18:: TRP1), K4327 (MATa hog1::TRP1), UG43 (_MAT_α hot1::kanMX4) and YM24 (MATa msn2-3::HIS3 msn4-1::TRP1). For northern blotting, protein in vitro phosphorylation and ChIP experiments, we used the strain PAY181 (MATa hot1::KAN-MX). YPD medium contains 10 g/l yeast extract, 20 g/l peptone and 20 g/l dextrose. Selective medium contains 1.7 g/l yeast nitrogen base (Difco), 5 g/l (NH4)2SO4, 20 g/l dextrose and supplements (100 mg/l each amino acids, uracil or adenine as appropriate, except where indicated). X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solid medium contains selective medium buffered with MES [2-(_N_-morpholino) ethanoesulfonic acid] at pH 7 plus 0.1 mg/ml X-gal. All yeast growth was at 30°C.
Plasmids
The pRS426TEG1 (PTEF1-GST, URA3+, 2 µm) and pRS426GAG1 (PGAL1-GST, URA3+, 2 µm) vectors were a gift from M.Takekawa (unpublished work). Wild-type KIN28, SRB10, SRB4, SRB9, SRB11, SIN4 and CSE2 genes were cloned into pRS426TEG1. Full-length HOG1 or the docking site region of this kinase (Hog1DS: from amino acid 265 to 435) was fused to GST under the TEF1 promoter, while the construct lacking the docking site domain (Hog1ΔDS: from amino acid 1 to 301) was fused to GST under GAL1 promoter. For HA tagging, HOG1 was cloned into the yeast vector YCpIF16 (PGAL1-HA, TRP1, CEN). Full-length HOG1, _HOG1_DS, _HOG1_ΔDS or HOG1TAYA (encodes mutations to Thr174 and Tyr176 to Ala) were cloned into the pBTM116 plasmid to fuse them to the LexA binding domain. STL1::LacZ fusion was constructed by insertion of the –825/+4 nucleotides of the STL1 gene into the integrative plasmid YIp358R. SRB9, SRB4 and SRB10 were cloned with their own promoters in the multicopy plasmid pRS425 (Stratagene) by PCR. The pLexA-Hog1 and pUG603 plasmids used for two-hybrid analysis were as described (Rep et al., 1999). pPA89 was obtained from pUG603 by mutating all Hog1 putative phosphorylation sites in Hot1 to alanine (Hot1-m5). Plasmids pPA97 and pPA106 contained full-length Hot1 and Hot1-m5, respectively, with three HA epitope tags, and were cloned in the vector pRS316 (CEN, URA3). Plasmids pPA112, pPA113 and pPA155 contained the same HA-Hot1 or Hot1-m5 proteins as above, but in multicopy 2 µm yeast vectors. Mammalian expression plasmid pCMV5-Flag-p38 was as described previously (Takekawa et al., 1997).
ChIP assays
ChIP PCR assays were performed as described previously (Alepuz et al., 2001). In all ChIP experiments, yeast cultures were grown to early log phase (OD600 of 0.6–1.0) before the cells were exposed to osmotic stress.
Expression and purification of epitope-tagged proteins
GST fusion proteins encoding PBS2(EE) and HOG1 were constructed using pGEX-4T (Pharmacia), expressed in E.coli DH5, and purified using glutathione–Sepharose beads (Pharmacia) in buffer B as described (Bilsland-Marchesan et al., 2000). For the in vitro phosphorylation experiments, HA-tagged Hot1 (pPA112) and Hot1-m5 (pPA155) were expressed in PAY181 yeast strain, and Hot1 proteins were immunoprecipitated with anti-HA monoclonal antibody 12CA5 and protein A– Sepharose beads (Roche). The beads were washed extensively with washing buffer (50 mM Tris–HCl pH 7.5, 0.1% NP-40, 150 mM NaCl plus antiproteases and phosphatase inhibitors) and resuspended in kinase buffer [50 mM Tris–HCl pH 7.5, 2 mM dithiothreitol (DTT)].
In vitro phosphorylation experiments
One microgram of recombinant GST–HOG1 from E.coli was activated by phosphorylation using 0.5 µg of GST–PBS2(EE) in the presence of kinase buffer and ATP as described (Bilsland-Marchesan et al., 2000). After 15 min at 30°C, HA-tagged Hot1 or Hot1-m5 proteins, purified from 5 mg of total protein from Saccharomyces cerevisae, were added to the previous mixture together with [γ-32P]ATP (0.2 µCi/µl). The mixture was then incubated for 5 min at 30°C, and the reactions were terminated by the addition of 2× SDS loading buffer. Labeled proteins were resolved by SDS–PAGE and then transferred to membrane. HA-tagged proteins were probed by immunoblotting with an anti-HA monoclonal antibody, and labeled proteins were detected by autoradiography.
GST pull-down experiments
To characterize Hog1 interacting proteins, we purified proteins associated with the GST fusion Hog1DS. Purification was started from 3 l of cell culture, grown at OD660 = 1. GST pull-down experiments were carried out using glutathione–Sepharose 4B in buffer A (50 mM Tris–HCl pH 7.5, 15 mM EDTA, 15 mM EGTA, 0.1% Triton X-100, 150 mM NaCl, 2 mM DTT plus antiproteases and phosphatase inhibitors). Pulled-down proteins were resolved on 6% SDS–PAGE. After silver staining, proteins in gel were digested with trypsin and identified by MALDI-TOF mass spectrometry as described previously (Paradela et al., 2000). In vivo interactions were determined by GST pull-down experiments. When necessary, protein expression from the GAL1 promoter was induced for 4 h with 2% galactose, and cells were either subjected to stress (0.4 M NaCl, 5 min) or untreated. One milligram of yeast extract in buffer A was incubated with 50 µl of glutathione–Sepharose beads overnight at 4°C. The beads were washed extensively with buffer A, resuspended in loading buffer and resolved by SDS–PAGE. For in vitro binding assay, a GST fusion protein encoding Hog1 was expressed in E.coli and purified using glutathione–Sepharose beads. Approximately 1 µg of GST and GST– Hog1 bound to beads was incubated with 0.5 µg of semipure RNA Pol II holoenzyme (kindly provided by Dr C.Gustafsson, Karolinska Institute, Sweden) for 3 h and then washed extensively.
β-galactosidase assays
Exponentially growing cells (OD660 = 0.5–0.8), either subjected to osmotic stress (0.4 M NaCl for 30 min) or untreated, were permeabilized by ethanol–toluene treatment, and β-galactosidase was measured as described (Proft et al., 2001). Results are presented as mean values obtained from at least three independent transformants measured in duplicate.
Two-hybrid assay
The two-hybrid analysis was carried out essentially as described (Bilsland-Marchesan et al., 2000) using pGAL424 and pBTM116 as the activation domain (AD) plasmid and the LexA DNA-binding domain (DB) plasmid, respectively. The L40 reporter strain was cotransformed with LexA-Hog1 and AD-Hot1 (pUG603), AD-Hot1-m (pPA89) or the empty vector pGAD424. β-galactosidase activity was measured as described (Alepuz et al., 2001).
Northern blot analysis
Total RNA was isolated from YPD- or SCD-grown yeast cells that were either untreated or subjected to the indicated osmotic stress conditions. The probes used were PCR fragments containing the entire open reading frame of STL1 (1.7 kbp) and RPL28 (0.95 kbp). Signals were quantified using a Fujifilm BAS-5000 phosphoimager.
Cell culture, transient transfection and co-immunoprecipitation assay
HeLa cells were grown at 37°C in a 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Cells were transiently transfected with the appropriate expression plasmids using calcium phosphate coprecipitation. Transfected cells were cultured for 24 h in serum-free medium and, where indicated, treated with 0.3 M NaCl for 20 min.
The cells were lysed in lysis buffer (10 mM HEPES pH 7.9, 150 mM NaCl, 0.1 mM EDTA, 10% glycerol, 0.1% NP-40, 0.1 mM Na3VO4, 0.2 mM NaF, 10 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 10 µg/ml leupeptin and 10 µg/ml pepstatin). Then, 2 mg aliquots of cell extract were incubated with anti-RNA Pol II antibody overnight at 4°C, mixed with 90 µl of Gamma-Bind Plus Sepharose beads (Amersham Pharmacia Biotech.) and incubated for an additional 2 h at 4°C. The immunoprecipitates were washed seven times with lysis buffer and subjected to immunoblot analysis for flag-p38 using the anti-Flag M2 monoclonal antibody (Sigma).
Acknowledgments
Acknowledgements
We are grateful to P.Cosma, S.Paniza, K.Nasmyth, B.Piña, S.D.Hanes, M.Takekawa, G.Gil, J.Aramburu and H.Saito for plasmids and strains, C.Gustafsson for purified RNA Pol II extracts, M.Proft for helpful discussions and sharing results before publication, and A.Jovanovic and M.Carmona for technical assistance. P.M.A. was the recipient of an EMBO Long-Term Fellowship. M.Z. is the recipient of a Ramón Areces PhD fellowship. This work was supported by grants from the Ministerio de Ciencia y Tecnología PM99-0028, ‘Distinció de la Generalitat de Catalunya per a la Promoció de la Recerca Universitaria. Joves Investigadors’ DURSI (Generalitat de Catalunya), the EMBO YIP program (F.P.) and the Austrian Government (Austrofan) (G.A.), and by an ‘Acción Integrada Hispano-Austriaca’, HU2001-0010 (F.P. and G.A.).
References
- Alepuz P.M., Jovanovic,A., Reiser,V. and Ammerer,G. (2001) Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell, 7, 767–777. [DOI] [PubMed] [Google Scholar]
- Bilsland-Marchesan E., Ariño,J., Saito,H., Sunnerhagen,P. and Posas,F. (2000) RCK2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase HOG1. Mol. Cell. Biol., 20, 3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma M.P., Panizza,S. and Nasmyth,K. (2001) Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell, 7, 1213–1220. [DOI] [PubMed] [Google Scholar]
- de Nadal E., Alepuz,P.M. and Posas,F. (2002) Dealing with osmostress through MAP kinase activation. EMBO Rep., 3, 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E., Casadomé,L. and Posas,F.(2003) Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol. Cell. Biol., 23, 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galcheva-Gargova Z., Derijard,B., Wu,I.H. and Davis,R.J. (1994) An osmosensing signal transduction pathway in mammalian cells. Science, 265, 806–808. [DOI] [PubMed] [Google Scholar]
- Garcia-Gimeno M.A. and Struhl,K. (2000) Aca1 and Aca2, ATF/CREB activators in Saccharomyces cerevisiae, are important for carbon source utilization but not the response to stress. Mol. Cell. Biol., 20, 4340–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev., 66, 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin S., Treich,I. and Carlson,M. (2000) A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl Acad. Sci. USA, 97, 7916–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis J.M. and Avruch,J. (2001) Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev., 81, 807–869. [DOI] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang,C.L. and Olson,E.N. (2002) MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci., 27, 40–47. [DOI] [PubMed] [Google Scholar]
- Nehlin J.O., Carlberg,M. and Ronne,H. (1992) Yeast SKO1 gene encodes a bZIP protein that binds to the CRE motif and acts as a repressor of transcription. Nucleic Acids Res., 20, 5271–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradela A., Alvarez,I., Garcia-Peydro,M., Sesma,L., Ramos,M., Vazquez,J. and Lopez De Castro,J.A. (2000) Limited diversity of peptides related to an alloreactive T cell epitope in the HLA-B27-bound peptide repertoire results from restrictions at multiple steps along the processing-loading pathway. J. Immunol., 164, 329–337. [DOI] [PubMed] [Google Scholar]
- Posas F., Chambers,J.R., Heyman,J.A., Hoeffler,J.P., de Nadal,E. and Arino,J. (2000) The transcriptional response of yeast to saline stress. J. Biol. Chem., 275, 17249–17255. [DOI] [PubMed] [Google Scholar]
- Proft M. and Serrano,R. (1999) Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol. Cell. Biol., 19, 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M. and Struhl,K. (2002) Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell, 9, 1307–1317. [DOI] [PubMed] [Google Scholar]
- Proft M., Pascual-Ahuir,A., de Nadal,E., Arino,J., Serrano,R. and Posas,F. (2001) Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J., 20, 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. and Gann,A. (1997) Transcriptional activation by recruitment. Nature, 386, 569–577. [DOI] [PubMed] [Google Scholar]
- Rep M., Reiser,V., Gartner,U., Thevelein,J.M., Hohmann,S., Ammerer,G. and Ruis,H. (1999) Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol., 19, 5474–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Krantz,M., Thevelein,J.M. and Hohmann,S. (2000) The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem., 275, 8290–8300. [DOI] [PubMed] [Google Scholar]
- Robinson M.J. and Cobb,M.H. (1997) Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol., 9, 180–186. [DOI] [PubMed] [Google Scholar]
- Sharrocks A.D., Yang,S.H. and Galanis,A. (2000) Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci., 25, 448–453. [DOI] [PubMed] [Google Scholar]
- Takekawa M., Posas,F. and Saito,H. (1997) A human homolog of the yeast SSK2/SSK22 MAP kinase kinase kinases, MTK1, mediates stress-induced activation of the p38 and JNK pathways. EMBO J., 16, 4973–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A.C. and Struhl,K. (1992) ACR1, a yeast ATF/CREB repressor. Mol. Cell. Biol., 12, 5394–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N.A. and Hampsey,M. (2002) The RNA polymerase II machinery: structure illuminates function. Cell, 108, 453–463. [DOI] [PubMed] [Google Scholar]