Integrin Signaling through Arg Activates p190RhoGAP by Promoting Its Binding to p120RasGAP and Recruitment to the Membrane (original) (raw)
Abstract
The Rho family GTPases RhoA (Rho), Rac1, and Cdc42 are essential effectors of integrin-mediated cell attachment and spreading. Rho activity, which promotes formation of focal adhesions and actin stress fibers, is inhibited upon initial cell attachment to allow sampling of the new adhesive environment. The Abl-related gene (Arg) tyrosine kinase mediates adhesion-dependent inhibition of Rho through phosphorylation and activation of the Rho inhibitor p190RhoGAP-A (p190). p190 phosphorylation promotes its binding to p120RasGAP (p120). Here, we elucidate the mechanism by which p120 binding regulates p190 activation after adhesion. We show that p190 requires its p120-binding domain to undergo Arg-dependent activation in vivo. However, p120 binding does not activate p190RhoGAP activity in vitro. Instead, activation of p190 requires recruitment to the cell periphery. Integrin-mediated adhesion promotes relocalization of p190 and p120 to the cell periphery in wild-type fibroblasts, but not in _arg_−/− fibroblasts. A dominant-negative p120 fragment blocks p190:p120 complex formation, prevents activation of p190 by adhesion, and disrupts the adhesion-dependent recruitment of p190 to the cell periphery. Our results demonstrate that integrin signaling through Arg activates p190 by promoting its association with p120, resulting in recruitment of p190 to the cell periphery where it inhibits Rho.
INTRODUCTION
The careful regulation of cell migration and morphogenesis is essential for proper organismal development and homeostasis. Differentiating and migrating cells recognize specific environmental cues and respond by rearranging the structure of their cytoskeleton. Some of the most important morphogenetic and migratory cues are provided by adhesive substrates in the extracellular matrix (ECM), the semisolid protein meshwork that surrounds the cells. Cells attach and spread on ECM by using heterodimeric integrin receptors that bind to specific ECM proteins (e.g., fibronectin, laminin, collagen, and vitronectin) (van der Flier and Sonnenberg, 2001; Hynes, 2002). Clustering of integrin receptors upon attachment to the ECM promotes the recruitment and activation of cytoskeletal regulatory proteins, leading to changes in cell shape and movement (Jockusch et al., 1995; Schwartz et al., 1995; Zamir and Geiger, 2001a, b).
Rho family GTPases are important downstream effectors of integrin-mediated cell attachment and spreading (Ridley, 2000; Arthur et al., 2002; Frame and Brunton, 2002; Burridge and Wennerberg, 2004). Rho family GTPases act as molecular switches that cycle between an active GTP-bound form and an inactive GDP-bound form (Van Aelst and D'Souza-Schorey, 1997; Kaibuchi et al., 1999). In its active form, Rho stimulates both actin polymerization (Watanabe et al., 1997) and actomyosin contractility (Kimura et al., 1996). Integrin-mediated adhesion leads to an initial decrease in Rho activity (Ren et al., 1999) followed by a longer-lasting Rho activation (Gimond et al., 1999; Ren et al., 1999). This initial reduction of Rho-mediated contractility may give adherent cells freedom to sample their adhesive environment after initial contact. Subsequent Rho activation promotes actin stress fiber and focal adhesion formation (Ridley and Hall, 1992; Barry et al., 1997), allowing cells to anchor and crawl on the adhesive surface (Gimond et al., 1999).
Abl family nonreceptor tyrosine kinases, which include the mammalian Abl and Arg proteins, regulate cell spreading and migration on adhesive surfaces (Kain and Klemke, 2001; Woodring et al., 2002; Miller et al., 2004; Woodring et al., 2004; Moresco et al., 2005). We have previously identified p190RhoGAP-A (p190) as a major Arg substrate in the developing brain (Hernandez et al., 2004). Integrin-mediated adhesion leads to increased tyrosine phosphorylation of p190 in cells, and this event requires Arg kinase activity. Arg phosphorylates p190 on tyrosine 1105 (Y1105), which stimulates its ability to inhibit Rho in vivo.
Phosphorylation of Y1105 in the RasGAP-binding region of p190 promotes formation of a complex between p190 and the 120-kDa GTPase-activating protein for Ras (p120RasGAP or p120). Although phosphorylation of Y1105 is essential for complex formation, phosphorylation of Y1087 in p190 helps stabilize the interaction (Hu and Settleman, 1997), which is mediated by the binding of two Src homology (SH)2 domains in p120 to the phosphotyrosines 1087 and 1105 in p190 (Hu and Settleman, 1997; Roof et al., 1998; Hernandez et al., 2004). Increased p190 phosphorylation and the formation of the p190:p120 complexes correlate with increased disassembly of actin stress fibers in Src-overexpressing cells (Chang et al., 1995), suggesting that p120 binding regulates p190 activation. Despite this correlative evidence, the mechanism by which p190 becomes activated in response to adhesion is unclear.
We demonstrate here that Arg is required for the adhesion-dependent inhibition of Rho and describe the mechanism by which integrin-mediated adhesion activates p190RhoGAP activity. We show that the p120-binding region of p190 is required for Arg to stimulate its RhoGAP activity in vivo. However, p120 binding is not sufficient to activate p190 in vitro. Instead, p190 activation requires its recruitment to the cell periphery. We demonstrate that adhesion promotes p190 and p120 colocalization at the cell periphery in wild-type, but not arg −/− cells. A dominant-negative p120 fragment blocks formation of the p190:p120 complex and prevents adhesion-dependent p190 activation by blocking p190 recruitment to the cell periphery. Our results demonstrate that integrin signaling through Arg activates p190 by promoting its association with p120 and localization to the cell periphery so that it can inhibit Rho.
MATERIALS AND METHODS
Plasmids
Full-length Arg, p190, p120, and Rho constructs were generated as described previously (Hernandez et al., 2004). Hemagglutinin (HA)-tagged p190 fragments were generated from a full-length p190 template by using polymerase chain reaction (PCR). Red fluorescent protein (RFP) vector was generated by cutting monomeric RFP from the prSET vector (a generous gift of Roger Tsien, Department of Pharmacology, University of California, San Diego, CA) and ligating it into the pEYFP-N1 vector (Clontech, Mountain View, CA). The 2-3-2 fragment was generated from a full-length p120 template by using PCR and ligated into the RFP-N1 vector. p120-GFP was generated by ligating p120 and humanized Renilla green fluorescent protein (GFP) (Stratagene, La Jolla, CA) into the pK1 vector. The Paxillin, Cortactin, and Src expression constructs were generous gifts of Donna Webb (Department of Biological Sciences, Vanderbilt University, Nashville, TN), Tom Parsons (Department of Microbiology, University of Virginia, Charlottesville, VA), and John Cooper (Department of Cell Biology and Physiology, Washington University in St. Louis, St. Louis, MO), respectively.
Fibroblasts
Wild type and arg −/− fibroblasts were cultured as described previously (Miller et al., 2004). arg −/− fibroblasts were infected with pK1 retroviruses expressing yellow fluorescent protein (YFP) or Arg-YFP as described previously (Miller et al., 2004), and they were selected with puromycin (Sigma-Aldrich, St. Louis, MO) at concentrations up to 0.85 μg/ml.
Human Embryonic Kidney (HEK)293 Cell Transfection
HEK293 cells were plated on 10-cm plates with DMEM containing 10% fetal bovine serum and transfected with a minimum of 20 μg of plasmid DNAs per plate by using Lipofectamine 2000 (Invitrogen). More plasmid DNA was transfected for the HA-tagged 760-C p190 fragment construct to balance protein expression levels. When necessary, expression was controlled by using appropriate empty vectors. After 48 h, transfection efficiency was monitored by cotransfection of a GFP expression construct (Stratagene), and cells were used for Rho activity, or p190 or HA immunoprecipitation assays.
Rho Activity Assays
Rhotekin-agarose (Upstate Biotechnology, Charlottesville, VA) pull-down assays were performed on HEK293 cells as reported previously (Hernandez et al., 2004), by using Mg2+ lysis buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 1% NP-40, 10 mM MgCl2, 10% glycerol, protease, and phosphatase inhibitors). Active HA-RhoA was detected using an anti-HA monoclonal antibody (mAb) (clone 12CA5; Abcam, Cambridge, MA) and quantified using a densitometer and ImageQuant software (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Active Rho levels were normalized to total Rho in each sample, and the ratio of activated to total Rho was normalized to the control ratio for each condition tested.
The enzyme-linked immunosorbent assay (ELISA)-based G-LISA kit (Cytoskeleton, Denver, CO) was used to determine endogenously active RhoA levels in fibroblasts according to the manufacturer's instructions. In brief, wild-type, arg −/−, or arg −/− + Arg-YFP cells were trypsinized and held in suspension for 1 h on 1% agarose-coated plates. Cells were then plated on 10-cm dishes coated with 10 μg/ml fibronectin (Sigma-Aldrich) and blocked with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) (Invitrogen, Carlsbad, CA). After 0, 10, 20, 30, or 60 min, cells were lysed in 250 μl of G-LISA lysis buffer (supplemented with protease inhibitors), scraped into tubes, and snap-frozen in liquid nitrogen. Cell lysate was subsequently thawed, clarified for 2 min at 9000 × g, and protein concentration was normalized between the various time points. Equal total protein amounts were added to a 96-well dish coated with the Rho binding domain of Rho effector proteins (which bind active GTP-bound Rho) in duplicate and incubated at 4°C for 30 min with vigorous shaking. Active Rho levels were determined by subsequent incubations with anti-Rho antibody and secondary horseradish peroxidase-conjugated antibody for 45 min each with vigorous shaking at room temperature. After adding developing solution, active Rho was determined by measuring absorbance at 490 nm using an ELISA plate reader after subtraction from a sample containing only lysis buffer. Equal loading of total RhoA protein at each time point was determined via immunoblot by using anti-RhoA antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Relative Rho activity was determined by dividing absorbance at each time point by the absorbance of the zero time point. Experiments for each cell type were repeated at least three times.
Arg was detected by immunoblot as described previously (Koleske et al., 1998). p190 was detected using either an anti-p190 mAb (clone 30; BD Biosciences Transduction Laboratories, Lexington, KY) or anti-HA (Abcam) as indicated. p120 was detected using anti-p120 mAb (clone B4F8; Upstate Biotechnology).
Immunoprecipitations
HEK293 cells were lysed in modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 7.2, 150 mM NaCl, 0.25% deoxycholate, 1% NP-40, protease, and phosphatase inhibitors). Extracts were precleared with 50 μl of protein G Plus/A agarose beads (Calbiochem, San Diego, CA) for 30 min at 4°C. The protein concentration of each fraction was determined using the BCA detergent-compatible protein assay kit (Pierce Chemical, Rockford, IL). Two micrograms of anti-p190 mAb, anti-HA polyclonal antibody, anti-FLAG mAb (clone 2EL-1B11; Chemicon International, Temecula, CA), anti-Paxillin mAb (clone 349; BD Biosciences Transduction Laboratories), anti-Src mAb (H-12; Santa Cruz Biotechnology), or anti-Cortactin mAb (Abcam) was incubated with 500 μg of extract for 2 h at 4°C with gentle mixing. Then, 50 μl of protein G Plus/A agarose beads was added, and the immune complexes were incubated for 1 h at 4°C. Immunoprecipitates were washed three times with 0.5 ml of modified RIPA buffer, mixed with sample buffer, and separated by SDS-PAGE for immunoblot analysis. Coimmunoprecipitating proteins and loading controls were detected by stripping and reprobing the blot with anti-p120 mAb, anti-RFP polyclonal antibody (Chemicon International), and other specified antigens. Where specified, bands were quantified using a densitometer and ImageQuant software (GE Healthcare).
Recombinant Protein Production/Purification
Full-length histidine-tagged (amino terminal) rat p190RhoGAP-A (p190) and full-length untagged human p120RasGAP (p120) were cloned into the pFastBac1 donor plasmid (Invitrogen), which were subsequently used to generate bacmid DNA containing the His-p190 and p120 cDNA. Bacmid DNA was then transfected into Sf9 cells for generation of recombinant baculovirus encoding the genes. For protein expression, Hi-5 insect cells were infected with recombinant baculovirus(es) at a 0.5 multiplicity of infection and harvested after 48 h. Cells were lysed in 50 mM HEPES pH 7.25, 5 mM β-mercaptoethanol, 200 mM KCl, 0.01% NP-40, 5% glycerol, and protease inhibitors by using a French Press. Lysates were centrifuged at 10,000 × g for 10 min to pellet cellular debris. The supernatant was then centrifuged at 100,000 × g for 1 h, and this second supernatant was incubated with nickel-nitrilotriacetic acid (Ni-NTA) resin (QIAGEN, Valencia, CA) for 2 h. Protein was eluted with 20 mM HEPES, pH 7.25, 5 mM β-mercaptoethanol, 200 mM KCl, 0.01% NP-40, 5% glycerol, and 100 mM imidazole. Proteins were further purified to 80–90% purity by fast-performance liquid chromatography (FPLC) through an anion exchange Uno Q column (Bio-Rad, Hercules, CA) by using a 1–500 mM KCl linear gradient in FPLC buffer (20 mM HEPES, 0.01% NP-40, 5% glycerol, 0.5 mM EDTA, and 0.5 mM dithiothreitol [DTT]). Copurification of p190:p120 complex was achieved by simultaneously coinfecting Hi5 insect cells with baculovirus encoding His-p190 and untagged p120 and Arg. Eluate from Ni-NTA resin columns were then further purified by size exclusion chromatography (Superdex 200 HR-1030; GE Healthcare) as described previously (Tanis et al., 2003).
p190 Phosphorylation
The tyrosine kinase inhibitor PD180970 (Kraker et al., 2000) was included at 15 μM in the p190-expressing insect cells to prevent its tyrosine phosphorylation during expression. p190 was phosphorylated in vitro as described previously (Hernandez et al., 2004). Based on experiments run in parallel with [γ-32P]ATP, >60% of the p190 was phosphorylated in these studies.
GAP Assays
The radioactive filter-binding assay measuring the retention of [γ-32P]GTP-bound glutathione _S_-transferase (GST)-RhoA was carried out as reported previously (Zhang and Zheng, 1998). Briefly, recombinant GST-RhoA was preloaded with [γ-32P]GTP (10 μCi; 6000 Ci/mmol; PerkinElmer Life and Analytical Sciences, Boston, MA) in 160 μl of a buffer containing 50 mM HEPES, pH 7.5, 50 mM NaCl, 1 mg/ml BSA, 0.1 mM DTT, 0.1 mM EGTA, and 5 mM EDTA for 10 min at 37°C. MgCl2 was then added to a final concentration of 10 mM and incubated for 10 min on ice. The [γ-32P]-GTP-loaded GST-RhoA was diluted 25-fold into a buffer containing 25 mM HEPES, pH 7.5, 50 mM NaCl, 1 mM MgCl2, 0.1 mM GTP, 0.1 mM DTT, and 1 mg/ml BSA. Fifty microliters of 10 nM p190 or p190:p120 complex (1:1 M ratio at 10 nM each) was mixed with 950 μl of the loaded and diluted GST-RhoA and incubated at 30°C. At different time points, 100-μl aliquots of the reaction were terminated by filtering the reaction mixture through nitrocellulose filters, followed by washing with 10 ml of ice-cold buffer containing 50 mM HEPES, pH 7.5, and 10 mM MgCl2. The radioactivity retained on the filters was measured by scintillation counting.
Immunofluorescence Microscopy
Coverslips were coated with either 0.1% poly-l-lysine (PLL) (Sigma-Aldrich) or 10 μg/ml fibronectin (Sigma-Aldrich) by shaking overnight at 4°C and blocked with a 1% BSA solution (Invitrogen) for 1 h at 37°C. Wild-type, arg −/−, or arg −/− + Arg-YFP fibroblasts were either left untransfected or transfected with RFP, 2-3-2-RFP, p120-GFP, and/or p190FF-YFP by using Lipofectamine 2000 (Invitrogen), and then allowed to attach to coverslips for 10, 20, 30, or 45 min at 37°C. Cells were washed, fixed, and extracted as described previously (Miller et al., 2004). After extraction, cells were blocked overnight at 4°C in a blocking buffer (2% BSA, 0.1% Triton X-100, and 0.1% sodium azide in PBS). Cells were stained with either anti-p190 or anti-p120 antibody, Alexa 594-labeled secondary antibody (Invitrogen), and in some cases p190-stained cells were reblocked overnight with blocking buffer, and stained with anti-p120 antibody, Alexa 488-labeled secondary antibody (Invitrogen), and Alexa 350-phalloidin (Invitrogen). Cells were imaged on a Nikon TE2000-S microscope by using 40× Nikon PlanApo 1.0 or 100× Nikon PlanFuor 1.3 oil objectives. Images were processed using Adobe Photoshop (Adobe Systems, Mountain View, CA) and NIH ImageJ (http://rsb.info.nih.gov/ij). To ensure specific staining for the protein of interest, wild-type cells plated on fibronectin were fixed as described above, blocked, stained with anti-p190 antibody, Alexa 594-labeled secondary antibody, then reblocked, and stained only with Alexa 488-labeled secondary antibody, which revealed only faint fluorescence in the green channel in the thickest part of the cell body. The same control was performed with anti-p120 antibody. p190 and p120 were considered localized at the cell periphery when staining intensity was significantly higher at the periphery than within the cell body. Localization was quantified, where indicated, in 50 or more cells for each condition.
Arg Is Required for Adhesion-dependent Inhibition of Rho
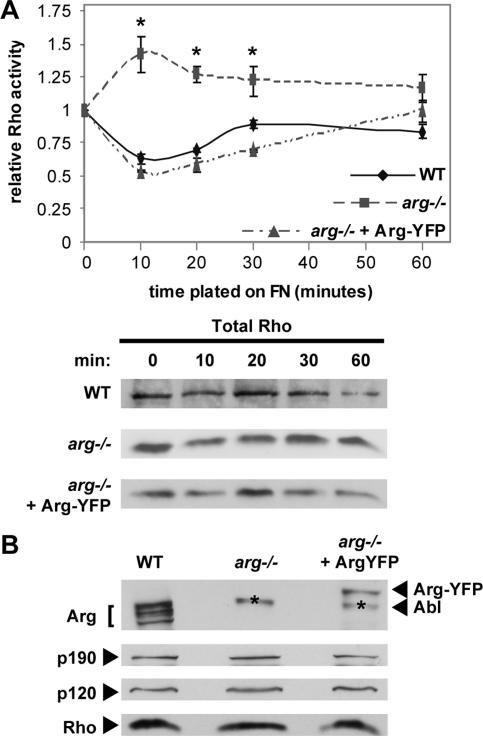
We previously demonstrated that Arg is required for the adhesion-dependent phosphorylation of p190RhoGAP in fibroblasts plated on fibronectin (FN) (Hernandez et al., 2004). We used an ELISA-based assay to test whether Arg could mediate adhesion-dependent changes in Rho activity. Adhesion of wild-type fibroblasts to FN leads to a 40% reduction in active Rho levels by 10 min, which are restored to normal levels by 60 min, as reported previously by others (Ren et al., 1999; Arthur et al., 2000) (Figure 1A). This adhesion-dependent Rho inhibition is not observed in arg −/− fibroblasts. In contrast, Rho activity levels increased by 40% in arg −/− fibroblasts at 10 min after adhesion, returning to baseline by 60 min (Figure 1A). Retrovirally mediated reexpression of an Arg-yellow fluorescent protein fusion (arg −/− + Arg-YFP cells) restored adhesion-dependent Rho inhibition to the arg −/− cells, whereas expression of YFP alone had no effect (our unpublished data). Control experiments revealed that wild-type, arg −/−, and arg −/− + Arg-YFP cells express p190RhoGAP, p120RasGAP, and Rho at similar levels (Figure 1B) and that total Rho levels are unchanged by adhesion in all cell types tested (Figure 1A). These results indicate that Arg mediates the adhesion-dependent inhibition of Rho activity during cell spreading.
Figure 1.
Arg is required for adhesion-dependent Rho inhibition. (A) Top, relative Rho activity plotted as a function of time. Active Rho levels were assessed in wild-type (WT, diamonds), arg −/− (squares), and arg −/− fibroblasts expressing an Arg-yellow fluorescent protein (arg −/− + Arg-YFP) (triangles) held in suspension (0 min) or plated on FN for 10, 20, 30, or 60 min by using an ELISA assay. Relative Rho activity was determined by dividing the absorbance reading at each time point by the absorbance of the 0 time point for each cell type. Mean ± SE, n ≥ 3. Analysis of variance between all cell types: 10-min time point, p = 0.0003; 20-min time point, p = 0.0001; 30-min time point, p = 0.0026; and 60-min time point, p = 0.0486. Post hoc Student-Newman-Keuls test for each time point (*p < 0.05). Bottom, total Rho was determined for each cell type at each time point to ensure equal loading for the ELISA assay. One hundred micrograms of total protein extract was immunoblotted for RhoA. (B) One hundred micrograms of protein extract from WT, arg −/−, and arg −/− + Arg-YFP were immunoblotted for Arg, p190, p120, and Rho. Our current lot of anti-Arg antisera cross-reacts slightly with endogenous Abl, and these cross-reacting bands in arg −/− and arg −/− + Arg-YFP lysate are indicated with an asterisk (*).
The p120-binding Region of p190 Is Required for Its Activation by Arg
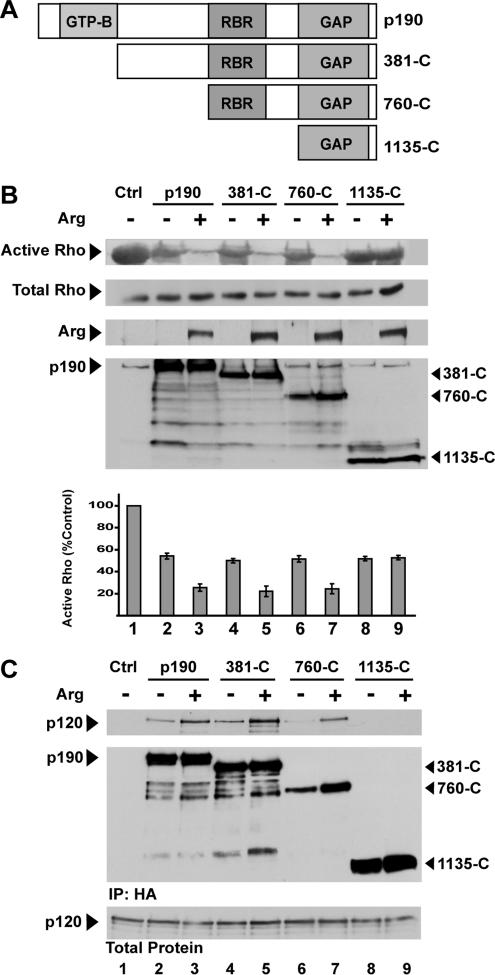
Our previous findings have shown that expression of p190 in HEK293 cells leads to a reduction in active Rho levels, as measured by Rhotekin pull-down assays (Ren and Schwartz, 2000; Hernandez et al., 2004). p190 activity is further stimulated by coexpression of Arg, leading to quantitatively larger decreases in active Rho levels. We previously showed that a p190 point mutant defective for p120 binding is not activated by Arg (Hernandez et al., 2004). Using a Rhotekin pull-down assay, we tested several p190 fragments (Figure 2A) to determine which domains in p190 are required for stimulation by Arg. The p190 C-terminal half (760-C) containing both the p120_R_asGAP-_b_inding _r_egion (RBR) and the GAP domain (Figure 2A) has basal RhoGAP activity (Figure 2B, compare lanes 1 and 6) that is further stimulated by Arg (Figure 2B, compare lanes 6 and 7). In contrast, the isolated GAP domain of p190 has basal RhoGAP activity (Figure 2B, compare lanes 1 and 8), but it is not stimulated by Arg in this assay (Figure 2B, compare lanes 8 and 9). These experiments suggest that the RBR is required for Arg to activate p190.
Figure 2.
Stimulation of p190 by Arg requires the p120-binding domain. (A) Domain structure of p190RhoGAP (p190). Domains indicated are as follows: GTP-B, GTP-binding domain; RBR, p120RasGAP-binding region; GAP, GTPase-activating protein domain. Truncation mutants lacking the GTP-binding domain (381-C); both the GTP-binding domain and the middle sequence (760-C); and a truncation mutant expressing only the GAP domain (1135-C) were generated. (B) Rhotekin pull-down assays were performed from lysates of HEK293 cells transfected with expression vectors for HA-Rho alone (Ctrl) or together with p190 +/− Arg, or the various p190 truncation mutants +/− Arg as indicated. Active Rho pull-down (top) and total cell lysate immunoblots (middle) were probed as indicated. After quantifying bands by densitometry, active Rho levels in each sample were normalized to total Rho in the sample and this was normalized to the control ratio. Mean ± SE, n ≥ 3 (bottom). (C) HA-tagged p190 was immunoprecipitated with anti-HA antibody from lysates of HEK293 cells that had been untransfected (Ctrl) or transfected with expression vectors for full-length p190 or the various truncation mutants as indicated. Immunoprecipitates on the same blot were probed for p120 (top) and p190 (middle) as indicated. Total protein lysates (25 μg of protein) were probed for p120 (bottom).
We also tested whether the responsiveness of p190 and p190 fragments to Arg stimulation correlates with their ability to bind p120 (Figure 2C). Expression of Arg with full-length p190 increased the amount of p120 coimmunoprecipitating with p190 (Figure 2C, compare lanes 2 and 3). Arg similarly promotes an increased association of p120 with the p190-381-C and p190-760-C fragments, both of which contain the RBR in addition to the RhoGAP domain (Figure 1C, compare lanes 4 and 5 and lanes 6 and 7). Not surprisingly, Arg does not stimulate p120 binding to the p190-1135-C fragment containing just the isolated RhoGAP domain (Figure 2C, compare lanes 8 and 9). Together, these experiments suggest that Arg-dependent stimulation of p190RhoGAP activity requires binding to p120.
p120 Binding Does Not Stimulate p190RhoGAP Activity In Vitro
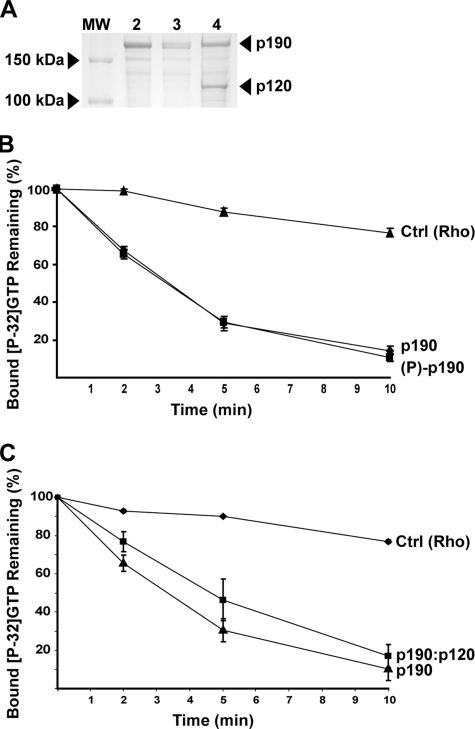
To test whether phosphorylation enhances p190RhoGAP activity, we prepared unphosphorylated p190RhoGAP by expressing it in insect cells in the presence of the tyrosine kinase inhibitor PD180970 (Kraker et al., 2000), and phosphorylating a portion of this p190RhoGAP in vitro by using purified recombinant Arg, as described previously (Figure 3A) (Hernandez et al., 2004). Our previous studies indicate that >60% of the p190RhoGAP is phosphorylated under these conditions (Hernandez et al., 2004). The RhoGAP activity of purified recombinant p190 is not altered by Arg phosphorylation (Figure 3B). We next examined whether the binding of p120 to p190 can stimulate p190 RhoGAP activity. After infecting insect cells with baculoviruses expressing His-tagged p190, p120, and Arg, we purified a complex containing p190 and p120 at a 1:1 stoichiometry (Figure 3A, lane 4). The complex did not contain detectable amounts of Arg, indicating that Arg does not stably associate with the p190:p120 complex under these conditions. The p190:p120 complex has specific RhoGAP activity that is nearly identical to purified unphosphorylated p190 (Figure 3C). Similar results were obtained when Src was used to promote complex formation during expression of p190 and p120 in insect cells (Settleman, unpublished data). These observations suggest that p120 binding cannot activate p190 RhoGAP activity in vitro.
Figure 3.
Binding of p120 does not activate p190RhoGAP in vitro. (A) Coomassie stain of purified p190 (lane 2), in vitro phosphorylated p190 (lane 3), and purified p190:p120 complex (lane 4). Standard protein molecular masses are shown for 150 and 100 kDa (lane 1). (B) GTP hydrolysis by RhoA was monitored over time in the absence of p190 (triangles), or in the presence of p190 (diamonds) or in vitro phosphorylated p190 (squares). (C) GTP hydrolysis by RhoA was measured over time in the absence of p190 (diamonds), or in the presence of p190 (triangles) or p190:p120 complex (1:1 M ratio; squares). Mean ± SE, n ≥ 3.
A Dominant-Negative p120 Fragment Blocks Arg-dependent Activation of p190
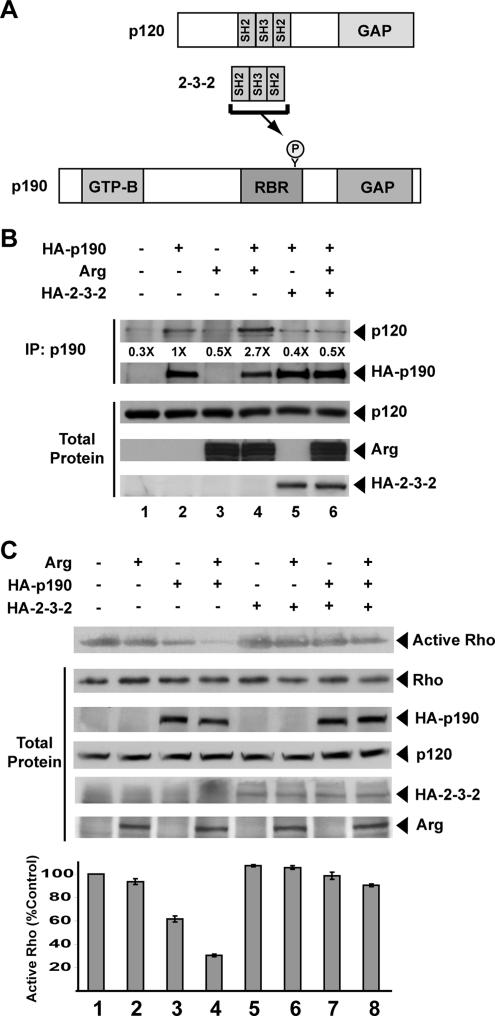
The p120 N-terminal half contains a tandem arrangement of SH2, SH3, and SH2 domains (Figure 4A). Phosphorylation of p190 on Y1105 in the p120-binding region creates a binding site for one of the two SH2 domains in p120RasGAP (Hu and Settleman, 1997; Roof et al., 1998). We tested whether an SH2-SH3-SH2 (2-3-2)-domain–containing p120 fragment (2-3-2 fragment) (Figure 4A) could act in a dominant-negative manner to block p120 binding to p190.
Figure 4.
p190 activation in vivo requires p120 binding. (A) p120 binds to p190 in a phosphotyrosine-dependent manner. The model depicts binding of the p120-2-3-2 fragment to p190, which inhibits the binding of p120 to p190. (B) p190 was immunoprecipitated from lysates of HEK293 cells that had been transfected with expression vectors for HA-p190, Arg, and/or the HA-tagged 2-3-2 fragment as indicated. Immunoprecipitates on the same blot were probed for p120 and HA-p190 (top two panels) as indicated. Total protein lysates (100 μg of protein) were probed for p120, Arg, or HA-2-3-2 fragment (bottom three panels). (C) Rhotekin pull-down assays were performed from lysates of HEK293 cells transfected with expression vectors for HA-Rho alone (lane 1), or together with HA-p190, Arg, and/or the 2-3-2 fragment as indicated (lanes 2–8). Active Rho pull-down (top) and total cell lysate immunoblots (total protein panels) were probed and quantitated as described in Figure 2. Mean ± SE, n ≥ 3 (bottom).
As we have shown previously, coexpression of Arg with HA-tagged-p190 (HA-p190) leads to an approximately threefold increased binding of p120 to p190 (Figure 4B, top, compare lanes 2 and 4). Expression of the 2-3-2 fragment blocks this Arg-dependent association of p120 with p190 (Figure 4B, top, compare lanes 4 and 6). Control experiments show that although a monomeric red fluorescent protein fusion to the 2-3-2 fragment (2-3-2-RFP) binds to p190, it does not bind the phosphotyrosine-containing proteins Paxillin, Src, or Cortactin (Supplemental Figure 1). These findings suggest that 2-3-2-RFP interacts selectively with p190 to block its interaction with p120.
We next tested whether the 2-3-2 fragment could influence Arg-dependent p190 activation in vivo. As shown above (Figure 2), expression of HA-p190 in HEK293 cells leads to a reduction of active Rho levels to ∼60% of control untransfected HEK293 cells (Figure 4C, compare lanes 1 and 3) as measured by a Rhotekin pull-down assay. Cotransfection of Arg with HA-p190 leads to a further reduction in active Rho levels to ∼30% of control untransfected HEK293 (Figure 4C, compare lanes 1 and 4). Interestingly, the 2-3-2 fragment blocks p190 activity in cells expressing HA-p190 alone or HA-p190 and Arg, resulting in Rho activity that is comparable with control untransfected HEK293 cells (Figure 4C, compare lane 1 with lanes 7 and 8). Together, these experiments indicate that a dominant-negative p120 fragment can block activation of p190RhoGAP by Arg.
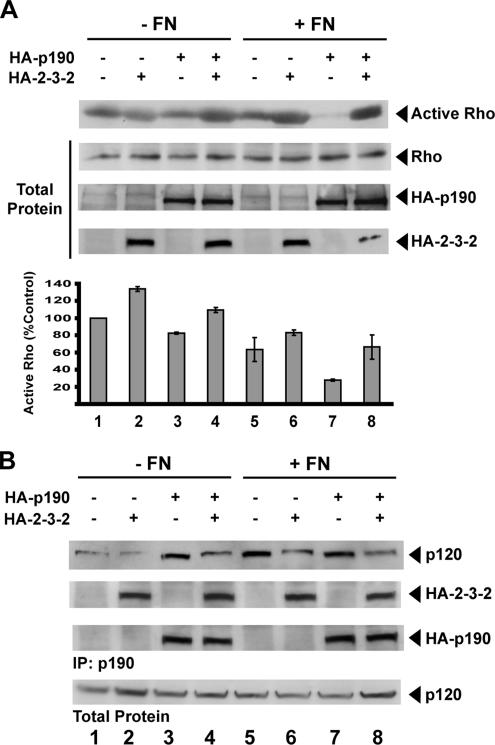
Disruption of p190:p120 Complex Formation Blocks Adhesion-dependent Rho Inhibition
p190 mediates the adhesion-dependent inhibition of Rho in fibroblasts as they attach and spread on FN (Arthur et al., 2000; Hernandez et al., 2004). We asked whether interfering with binding of p120 to p190 by using the 2-3-2 fragment could disrupt this adhesion-dependent p190 activation.
As expected, plating of HEK293 cells on FN reduces the amount of active Rho to 65% of control cells not plated on FN (Figure 5A, compare lanes 1 and 5). Expression of HA-p190 in cells plated on FN leads to a further decrease of active Rho levels to 28% of control cells not plated on FN (Figure 5A, compare lanes 1 and 7). Coexpression of the 2-3-2 fragment with HA-p190 blocks much of the p190-dependent decrease in Rho activity observed after FN adhesion. This results in a reduction of active Rho to only 66% of control levels, a level similar to that observed in untransfected cells plated on FN (Figure 5A, compare lanes 5 and 8). Expression of the 2-3-2 fragment leads to slight increases in Rho activity under all experimental conditions. Experiments performed in parallel confirmed that expression of the 2-3-2 fragment reduces the amount of p120 associating with p190 (Figure 5B).
Figure 5.
Dominant-negative p120 blocks adhesion-dependent p190 activation. (A) Rhotekin pull-down assays were performed from lysates of nonstimulated (−FN; lanes 1–4) or FN-stimulated (+FN; lanes 5–8) HEK293 cells. Cells were transfected with expression vectors for HA-Rho alone (lanes 1 and 5) or together with HA-p190 and/or the 2-3-2 fragment as indicated (lanes 2–4 and 6–8). Active Rho was quantified as in Figure 2. Mean ± SE, n ≥ 3 (bottom). (B) p190 was immunoprecipitated from lysates of nonstimulated (−FN; lanes 1–4) or FN-stimulated (+FN; lanes 5–8) HEK293 cells. Cells were transfected with expression vectors for GFP alone (control; lanes 1 and 5) or HA-p190 and/or 2-3-2 fragment as indicated (lanes 2–4 and 6–8). Pull-downs (top three panels) were probed as indicated. Total protein lysates (100 μg of protein) were probed for p120RasGAP (bottom).
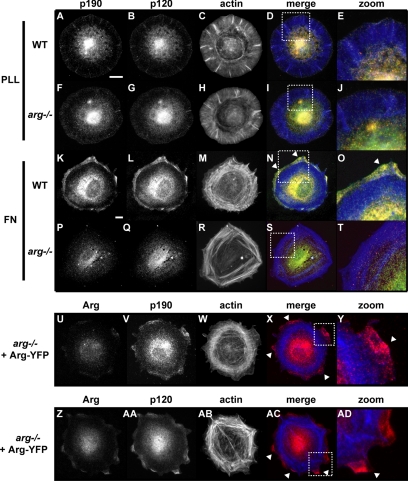
Arg Is Required for Adhesion-dependent p190 and p120 Colocalization at the Cell Periphery
Although p120 binding is required for p190 activation in cells, purified p190 and the p190:p120 complex have similar RhoGAP activities in vitro (Figure 3C). These observations suggest that although p190 activation is not observed in vitro, p190:p120 complex formation may be necessary for p190 activation in vivo.
We examined whether Arg-mediated complex formation affects p190 localization in cells. After plating wild-type or _arg_−/− fibroblasts on PLL- or FN-coated coverslips for 45 min, cells were fixed and stained for p190, p120, and F-actin. Cells plated on PLL display a weak colocalization of p190 and p120 that overlaps with radial F-actin–rich structures (Figure 6, A–E). Adhesion of wild-type cells to FN leads to a significant recruitment of p190 and p120 to the cell periphery (Figure 6, K and L), and overlay of these images shows extensive colocalization of p190 and p120 (Figure 6, N and O). These p190:p120 clusters form just outside the extensive network of cortical F-actin rings. Colocalization of p190 and p120 is barely detectable in _arg_−/− fibroblasts plated on PLL (Figure 6, I and J). Neither p190 nor p120 relocalize to the cell periphery in _arg_−/− fibroblasts plated on FN (Figure 6, P and Q), but Arg-YFP reexpression in _arg_−/− fibroblasts restores adhesion-dependent relocalization of p190 and p120 to the cell periphery (Figure 6V, AA). Although Arg localizes to the cell periphery in cells that have attached for 2 h to FN (Miller et al., 2004), we did not observe significant Arg-YFP colocalization with p190 or p120 during the first 30 min of cell attachment and spreading (Supplemental Figure 2). Together, these data indicate that Arg is required for integrin-mediated recruitment of p190:p120 clusters to the cell periphery.
Figure 6.
Arg is required for p190:p120 colocalization at the cell periphery. WT (A–E, K–O), _arg_−/− (F–J, P–T), or _arg_−/− + Arg-YFP cells (U–AD) were plated on coverslips coated with either 0.1% PLL (A–J) or 10 μg/ml FN (K–AD), and stained for p190 (A, F, K, P, and V), p120 (B, G, L, Q, and AA), and F-actin (C, H, M, R, W, and AB). Merged images (D, I, N, and S) and enlargements (E, J, O, and T) of each channel in top grid show p190 in red, p120 in green, and F-actin in blue. Merged images and enlargements in bottom grids show either p190 (X–Y) or p120 (AC and AD) in red and F-actin (X and Y, AC and AD) in blue. Enlarged regions are indicated by white dotted boxes in merge panels. White arrowheads highlight regions of p190:p120 colocalization (N and O), or individual p190 (X and Y) or p120 localization (AC and AD). Bar in A (applies to A–D, F–I) and K (applies to K–N, P–S, U–X, Z–AC), 10 μm.
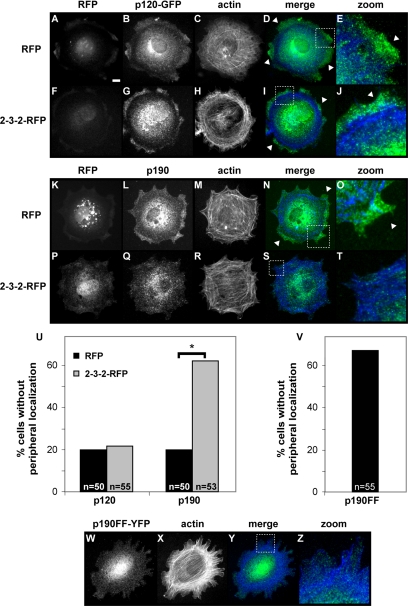
Disruption of p190 Binding to p120 Blocks Adhesion-dependent Recruitment of p190 to the Cell Periphery
Our finding that p190 and p120 colocalize to the cell periphery suggests that p120 binding may recruit p190 to the periphery. We tested whether the dominant-negative 2-3-2 fragment of p120 fused to monomeric red fluorescent protein (2-3-2-RFP) could block adhesion-dependent localization of p190 to the cell periphery. Wild-type cells were transfected with a 2-3-2-RFP expression vector, plated on FN, and immunostained for p190. Because anti-p120 antibodies cross-react with the 2-3-2 fragment, we also cotransfected cells with RFP or 2-3-2-RFP and p120-GFP expression vectors to monitor p120 localization.
Expression of 2-3-2-RFP does not affect adhesion-dependent localization of p120 to the cell periphery (Figure 7, G, I, and J). Interestingly, adhesion-dependent localization of p190 to the cell periphery is disrupted in 62% of the 2-3-2-RFP–expressing cells (n = 53) (Figure 7U) and p190 localizes diffusely throughout the cell body in these cells (Figure 7, Q, S, and T). Diffuse p190 distribution is only observed in 20% (n = 50) of RFP-transfected cells (Figure 7, L, N, and O). Thus, the 2-3-2-RFP fragment can disrupt adhesion-dependent p190 recruitment to the cell periphery. These studies indicate that Arg-dependent phosphorylation of p190 promotes its binding to p120 and translocation to the cell periphery. In agreement with these findings, a p190 mutant containing phenylalanine substitutions in its two tyrosine phosphorylation sites required for p120 binding (Y1087F and Y1105F) does not exhibit adhesion-dependent relocalization to the cell periphery (Figure 7, V–Z).
Figure 7.
The 2-3-2 fragment inhibits p190 recruitment to the cell periphery. Wild-type cells were transfected with either RFP (A and K) or 2-3-2-RFP (F and P), plated on FN-coated coverslips, and in some cases immunostained for p190 (L and Q). p120 localization in RFP or 2-3-2-RFP transfected cells was assessed by cotransfection with p120-GFP (B and G). p190FF-YFP was transfected into wild-type cells to asses its localization (W). F-actin was stained in all cell types (C, H, M, R, and X). Merged images and enlargements show either p120 (D, E, I, and J), p190 (N and O, S and T), or p190FF (Y and Z) in green and F-actin in blue. Enlargement regions are indicated by white dotted boxes in merge panels. White arrowheads highlight regions of p120 (D and E, I and J) or p190 localization (N and O). p120, p190, and p190FF localization at the cell periphery was quantified. (U and V) Black bars indicate RFP-transfected cells for p190 and p120 localization, and gray bars indicate 2-3-2-RFP transfected cells, with number of cells (n ≥ 50) indicated in each instance. Student's t test between p190 localization in RFP versus 2-3-2-RFP–expressing cells, *p < 0.0001. Bar in A, 10 μm.
DISCUSSION
We demonstrate here that Arg is required for adhesion-dependent inhibition of Rho and elucidate the mechanism by which Arg mediates Rho inhibition. We have previously identified p190 as an Arg substrate. Using p190 truncation mutants, we establish that the RBR of p190 is required for Arg to stimulate RhoGAP activity. Arg-dependent phosphorylation of Y1105 in the RBR of p190 promotes p120 binding (Hu and Settleman, 1997; Roof et al., 1998; Hernandez et al., 2004). Expression of a dominant-negative 2-3-2 fragment of p120 blocks binding of p120 to p190 and prevents p190 activation by adhesion. We also show that Arg is required for p190 and p120 recruitment to the cell periphery after adhesion to FN, and p190 localization is disrupted by the 2-3-2 fragment. Together, our results indicate that p120 binding regulates p190 RhoGAP activity in vivo by promoting the recruitment of a p190:p120 complex to the cell periphery where it can inhibit Rho.
Phosphorylation of p190 on Y1105 promotes its association with p120 (Hu and Settleman, 1997; Roof et al., 1998; Hernandez et al., 2004). We have previously reported that adhesion to FN stimulates p190 phosphorylation in wild-type, but not _arg_−/− cells, and this phosphorylation is restored to these cells by Arg reexpression (Hernandez et al., 2004). Burridge and colleagues have shown that integrin stimulation with RGD-containing peptides stimulates p190 in wild-type, but not _src_−/− _yes_−/− _fyn_−/− (SYF) cells (Arthur et al., 2000). Arg phosphorylates p190 directly on Y1105 (Hernandez et al., 2004), a site that is also phosphorylated in p190 immunopurified from Src-overexpressing cells (Roof et al., 1998). Together, these studies suggest that both Arg and Src family kinases are required for integrin-dependent phosphorylation and activation of p190.
One possible explanation for these findings is that Src family kinases and Arg differentially phosphorylate p190 depending on the mode of integrin stimulation (i.e., soluble peptides versus adhesion to FN-coated surfaces). Alternatively, Src family kinases may serve as links between integrin receptors and Arg kinase activity. Src family kinases can phosphorylate and activate Abl and Arg in vitro and have been shown to mediate signaling between the platelet-derived growth factor receptor and Abl family kinases (Plattner et al., 1999; Dorey et al., 2001; Tanis et al., 2003). Finally, it is possible that Src family kinases or Arg play a structural, but noncatalytic role in p190 activation. These models can be tested by measuring p190 activation in SYF or Arg-deficient cells reconstituted with Src or Arg mutants.
Arg stimulates p190 activity through phosphorylation of Y1105 in the p120-binding region (Hernandez et al., 2004). Because this phosphorylation event promotes binding of p120 to p190, we examined whether the binding of p120 stimulates p190 activity. Our finding that the RhoGAP activities of p190 and the p190:p120 complex are similar argues that complex formation does not activate p190 in vitro.
Active Rho localizes to the plasma membrane where it regulates actomyosin contractility and cytoskeletal rearrangements (Ridley, 2000; Pertz et al., 2006). Previous studies have shown that p190 relocalizes to the cell periphery in response to adhesion or growth factor stimulation (Chang et al., 1995; Nakahara et al., 1998; Brouns et al., 2000; Tsubouchi et al., 2002). This recruitment of p190 correlates with a reduction of F-actin stress fibers, which likely reflects decreased Rho activity (Chang et al., 1995; Nakahara et al., 1998). Our finding that adhesion to FN stimulates p190:p120 colocalization at the cell periphery, and that p190 localization is disrupted by the 2-3-2 fragment, suggests that p120 activates p190 by promoting its recruitment to the membrane to inhibit Rho.
p120 has both pleckstrin homology (PH) and Ca2+-dependent phospholipid binding (CaLB) domains that may facilitate interactions with the plasma membrane (Gawler et al., 1995a, b; Drugan et al., 2000; Koehler and Moran, 2001). We do not observe significant localization of p120 to the cell periphery in the absence of integrin-mediated adhesion, suggesting that integrin-mediated adhesion may be required to generate phosphatidylinositol species that act as binding targets for the PH and CaLB domains in p120. Our finding that the 2-3-2 fragment blocks p190 localization to the periphery without affecting p120 localization strongly suggests that p120 serves as a peripheral binding target for p190. Interestingly, the 2-3-2 fragment did not block p190 relocalization in every cell. It is also possible that p190 interacts via additional surfaces with other targets at the cell periphery that facilitate its recruitment. One likely additional target for p190 is RhoE, which is associated with the membrane and can bind and activate p190 (Wennerberg et al., 2003). It is also likely that p190 binding stabilizes p120 localization at the cell periphery, because p120 levels are reduced at the periphery in _arg_−/− cells plated on FN (Figure 6Q).
Active Rho promotes cell contractility through the activation of actomyosin contractility (Ridley, 2001). Our finding that Arg is required for the formation of p190:p120 complexes at the cell periphery suggests that Arg activity may locally regulate Rho activity during cell adhesion and spreading. It is important to note that the experiments presented here were performed during initial cell attachment and spreading on FN-coated surfaces. The initial global reduction in Rho activity in wild-type fibroblasts observed by us and others (Ren et al., 1999; Arthur et al., 2000) likely allows cells to spread and sample their new adhesive environment. Cells initially spread in all directions, and the p190 relocalization we observe along the entire cell periphery is consistent with a uniform circumferential Rho inhibition during this spreading phase. Recently, Hahn and colleagues observed that active Rho primarily localizes to protrusion sites in cells exhibiting directed cell migration (Pertz et al., 2006). The localization of active Rho to these protrusive sites underscores the need for a dynamic cycling of Rho activity for efficient directed cell motility. A local inhibitor of Rho, such as p190, is likely critical for localized Rho inactivation during Rho activity cycling. We have previously shown that Arg promotes lamellipodial protrusion in fibroblasts after they adhere and spread on FN (Miller et al., 2004). We propose that Arg-dependent formation of p190:p120 clusters may relax Rho-mediated contractility locally to allow the initiation or persistence of these cellular protrusions. In support of this, Arthur and Burridge have previously shown that p190 overexpression promotes cell edge protrusion as cells migrate into wounds induced in a fibroblast monolayer, whereas cells expressing a dominant negative p190 exhibit deficient membrane protrusion and cell spreading (Arthur and Burridge, 2001). Future imaging studies should reveal how the formation of p190:p120 clusters affect Rho activity locally and how this affects cell edge protrusion.
Supplementary Material
[Supplemental Material]
ACKNOWLEDGMENTS
We thank Walter Boron for use of the ELISA plate reader, Mark Parker and Xianyun Ye for technical assistance, Scott Boyle and Justin Peacock for helpful discussions, and Koleske laboratory members for critical comments on the manuscript. This work was supported by a predoctoral fellowship from the Anna Fuller Fund to W.D.B., a predoctoral National Research Service Award (NS42365) to S.E.H., a grant from the National Institute of General Medical Sciences (CA62142) to J. S., and grants from the National Institute of Neurological Disorders and Stroke (NS39475), National Alliance for Research on Schizophrenia and Depression, and the Leukemia and Lymphoma Society of America to A.J.K.
Abbreviations used:
2-3-2 fragment
SH2-SH3-SH2-domain–containing p120RasGAP fragment
Arg
Abl-related gene
FN
fibronectin
HEK
human embryonic kidney
p120
120-kDa Ras GTPase-activating protein
p190
190-kDa Rho GTPase-activating protein
PLL
poly-l-lysine
RBR
RasGAP-binding region of p190RhoGAP.
Footnotes
REFERENCES
- Arthur W. T., Burridge K. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol. Biol. Cell. 2001;12:2711–2720. doi: 10.1091/mbc.12.9.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur W. T., Noren N. K., Burridge K. Regulation of Rho family GTPases by cell-cell and cell-matrix adhesion. Biol. Res. 2002;35:239–246. doi: 10.4067/s0716-97602002000200016. [DOI] [PubMed] [Google Scholar]
- Arthur W. T., Petch L. A., Burridge K. Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr. Biol. 2000;10:719–722. doi: 10.1016/s0960-9822(00)00537-6. [DOI] [PubMed] [Google Scholar]
- Barry S. T., Flinn H. M., Humphries M. J., Critchley D. R., Ridley A. J. Requirement for Rho in integrin signalling. Cell Adhes. Commun. 1997;4:387–398. doi: 10.3109/15419069709004456. [DOI] [PubMed] [Google Scholar]
- Brouns M. R., Matheson S. F., Hu K. Q., Delalle I., Caviness V. S., Silver J., Bronson R. T., Settleman J. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 2000;127:4891–4903. doi: 10.1242/dev.127.22.4891. [DOI] [PubMed] [Google Scholar]
- Burridge K., Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- Chang J. H., Gill S., Settleman J., Parsons S. J. c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following epidermal growth factor stimulation. J. Cell Biol. 1995;130:355–368. doi: 10.1083/jcb.130.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorey K., Engen J. R., Kretzschmar J., Wilm M., Neubauer G., Schindler T., Superti-Furga G. Phosphorylation and structure-based functional studies reveal a positive and a negative role for the activation loop of the c-Abl tyrosine kinase. Oncogene. 2001;20:8075–8084. doi: 10.1038/sj.onc.1205017. [DOI] [PubMed] [Google Scholar]
- Drugan J. K., Rogers-Graham K., Gilmer T., Campbell S., Clark G. J. The Ras/p120 GTPase-activating protein (GAP) interaction is regulated by the p120 GAP pleckstrin homology domain. J. Biol. Chem. 2000;275:35021–35027. doi: 10.1074/jbc.M004386200. [DOI] [PubMed] [Google Scholar]
- Frame M. C., Brunton V. G. Advances in Rho-dependent actin regulation and oncogenic transformation. Curr. Opin. Genet. Dev. 2002;12:36–43. doi: 10.1016/s0959-437x(01)00261-1. [DOI] [PubMed] [Google Scholar]
- Gawler D. J., Zhang L. J., Moran M. F. Mutation-deletion analysis of a Ca(2+)-dependent phospholipid binding (CaLB) domain within p120 GAP, a GTPase-activating protein for p21 ras. Biochem. J. 1995a;307:487–491. doi: 10.1042/bj3070487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawler D. J., Zhang L. J., Reedijk M., Tung P. S., Moran M. F. CaLB: a 43 amino acid calcium-dependent membrane/phospholipid binding domain in p120 Ras GTPase-activating protein. Oncogene. 1995b;10:817–825. [PubMed] [Google Scholar]
- Gimond C., van Der Flierqq A., van Delft S., Brakebusch C., Kuikman I., Collard J. G., Fassler R., Sonnenberg A. Induction of cell scattering by expression of beta1 integrins in beta1-deficient epithelial cells requires activation of members of the rho family of GTPases and downregulation of cadherin and catenin function. J. Cell Biol. 1999;147:1325–1340. doi: 10.1083/jcb.147.6.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez S. E., Settleman J., Koleske A. J. Adhesion-dependent regulation of p190RhoGAP in the developing brain by the Abl-related gene tyrosine kinase. Curr. Biol. 2004;14:691–696. doi: 10.1016/j.cub.2004.03.062. [DOI] [PubMed] [Google Scholar]
- Hu K. Q., Settleman J. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 1997;16:473–483. doi: 10.1093/emboj/16.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Jockusch B. M., Bubeck P., Giehl K., Kroemker M., Moschner J., Rothkegel M., Rudiger M., Schluter K., Stanke G., Winkler J. The molecular architecture of focal adhesions. Annu. Rev. Cell Dev. Biol. 1995;11:379–416. doi: 10.1146/annurev.cb.11.110195.002115. [DOI] [PubMed] [Google Scholar]
- Kaibuchi K., Kuroda S., Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu. Rev. Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- Kain K. H., Klemke R. L. Inhibition of cell migration by Abl family tyrosine kinases through uncoupling of Crk-CAS complexes. J. Biol. Chem. 2001;276:16185–16192. doi: 10.1074/jbc.M100095200. [DOI] [PubMed] [Google Scholar]
- Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., Yamamori B., Feng J., Nakano T., Okawa K., Iwamatsu A., Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Koehler J. A., Moran M. F. Regulation of extracellular signal-regulated kinase activity by p120 RasGAP does not involve its pleckstrin homology or calcium-dependent lipid binding domains but does require these domains to regulate cell proliferation. Cell Growth Differ. 2001;12:551–561. [PubMed] [Google Scholar]
- Koleske A. J., Gifford A. M., Scott M. L., Nee M., Bronson R. T., Miczek K. A., Baltimore D. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 1998;21:1259–1272. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- Kraker A. J., Hartl B. G., Amar A. M., Barvian M. R., Showalter H. D., Moore C. W. Biochemical and cellular effects of c-Src kinase-selective pyrido[2,3-d]pyrimidine tyrosine kinase inhibitors. Biochem. Pharmacol. 2000;60:885–898. doi: 10.1016/s0006-2952(00)00405-6. [DOI] [PubMed] [Google Scholar]
- Miller A. L., Wang Y., Mooseker M. S., Koleske A. J. The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J. Cell Biol. 2004;165:407–419. doi: 10.1083/jcb.200308055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco E. M., Donaldson S., Williamson A., Koleske A. J. Integrin-dependent dendrite branch stabilization requires Abl family kinases. J. Neurosci. 2005;25:6105–6118. doi: 10.1523/JNEUROSCI.1432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Mueller S. C., Nomizu M., Yamada Y., Yeh Y., Chen W. T. Activation of beta1 integrin signaling stimulates tyrosine phosphorylation of p190RhoGAP and membrane-protrusive activities at invadopodia. J. Biol. Chem. 1998;273:9–12. doi: 10.1074/jbc.273.1.9. [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R. L., Hahn K. M. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- Plattner R., Kadlec L., DeMali K. A., Kazlauskas A., Pendergast A. M. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 1999;13:2400–2411. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. D., Kiosses W. B., Schwartz M. A. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. D., Schwartz M. A. Determination of GTP loading on Rho. Methods Enzymol. 2000;325:264–272. doi: 10.1016/s0076-6879(00)25448-7. [DOI] [PubMed] [Google Scholar]
- Ridley A. Rho GTPases. Integrating integrin signaling. J. Cell Biol. 2000;150:F107–F109. doi: 10.1083/jcb.150.4.f107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11:471–477. doi: 10.1016/s0962-8924(01)02153-5. [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Roof R. W., Haskell M. D., Dukes B. D., Sherman N., Kinter M., Parsons S. J. Phosphotyrosine (p-Tyr)-dependent and -independent mechanisms of p190 RhoGAP-p120 RasGAP interaction: Tyr 1105 of p190, a substrate for c-Src, is the sole p-Tyr mediator of complex formation. Mol. Cell. Biol. 1998;18:7052–7063. doi: 10.1128/mcb.18.12.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. A., Schaller M. D., Ginsberg M. H. Integrins: emerging paradigms of signal transduction. Annu. Rev. Cell Dev. Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- Tanis K. Q., Veach D., Duewel H. S., Bornmann W. G., Koleske A. J. Two distinct phosphorylation pathways have additive effects on Abl family kinase activation. Mol. Cell. Biol. 2003;23:3884–3896. doi: 10.1128/MCB.23.11.3884-3896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi A., Sakakura J., Yagi R., Mazaki Y., Schaefer E., Yano H., Sabe H. Localized suppression of RhoA activity by Tyr31/118-phosphorylated paxillin in cell adhesion and migration. J. Cell Biol. 2002;159:673–683. doi: 10.1083/jcb.200202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L., D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- van der Flier A., Sonnenberg A. Function and interactions of integrins. Cell Tissue Res. 2001;305:285–298. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Madaule P., Reid T., Ishizaki T., Watanabe G., Kakizuka A., Saito Y., Nakao K., Jockusch B. M., Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K., Forget M. A., Ellerbroek S. M., Arthur W. T., Burridge K., Settleman J., Der C. J., Hansen S. H. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr. Biol. 2003;13:1106–1115. doi: 10.1016/s0960-9822(03)00418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodring P. J., Litwack E. D., O'Leary D. D., Lucero G. R., Wang J. Y., Hunter T. Modulation of the F-actin cytoskeleton by c-Abl tyrosine kinase in cell spreading and neurite extension. J. Cell Biol. 2002;156:879–892. doi: 10.1083/jcb.200110014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodring P. J., et al. c-Abl phosphorylates Dok1 to promote filopodia during cell spreading. J. Cell Biol. 2004;165:493–503. doi: 10.1083/jcb.200312171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir E., Geiger B. Components of cell-matrix adhesions. J. Cell Sci. 2001a;114:3577–3579. doi: 10.1242/jcs.114.20.3577. [DOI] [PubMed] [Google Scholar]
- Zamir E., Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 2001b;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- Zhang B., Zheng Y. Regulation of RhoA GTP hydrolysis by the GTPase-activating proteins p190, p50RhoGAP, Bcr, and 3BP-1. Biochemistry. 1998;37:5249–5257. doi: 10.1021/bi9718447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material]