Transcription Enhancer Factor 1 Binds Multiple Muscle MEF2 and A/T-Rich Elements during Fast-to-Slow Skeletal Muscle Fiber Type Transitions (original) (raw)
Abstract
In adult mouse skeletal muscle, β-myosin heavy chain (βMyHC) gene expression is primarily restricted to slow type I fibers; however, its expression can be induced in fast type II fibers in response to a sustained increase in load-bearing work (mechanical overload [MOV]). Our previous βMyHC transgenic and protein-DNA interaction studies have identified an A/T-rich element (βA/T-rich −269/−258) that is required for slow muscle expression and which potentiates MOV responsiveness of a 293-bp βMyHC promoter (β293wt). Despite the GATA/MEF2-like homology of this element, we found binding of two unknown proteins that were antigenically distinct from GATA and MEF2 isoforms. By using the βA/T-rich element as bait in a yeast one-hybrid screen of an MOV-plantaris cDNA library, we identified nominal transcription enhancer factor 1 (NTEF-1) as the specific βA/T-rich binding factor. Electrophoretic mobility shift assay analysis confirmed that NTEF-1 represents the enriched binding activity obtained only when the βA/T-rich element is reacted with MOV-plantaris nuclear extract. Moreover, we show that TEF proteins bind MEF2 elements located in the control region of a select set of muscle genes. In transient-coexpression assays using mouse C2C12 myotubes, TEF proteins transcriptionally activated a 293-bp βMyHC promoter devoid of any muscle CAT (MCAT) sites, as well as a minimal thymidine kinase promoter-luciferase reporter gene driven by three tandem copies of the desmin MEF2 or palindromic Mt elements or four tandem βA/T-rich elements. These novel findings suggest that in addition to exerting a regulatory effect by binding MCAT elements, TEF proteins likely contribute to regulation of skeletal, cardiac, and smooth muscle gene networks by binding select A/T-rich and MEF2 elements under basal and hypertrophic conditions.
The differentiation and maturation of skeletal muscle fibers into distinct phenotypes involve transcriptional activation of numerous unlinked muscle-specific genes encoding cytoskeletal, calcium-handling, metabolic, and contractile proteins that when assembled create an elaborate machine capable of accommodating a large array of functional demands (6, 42). Of the striated muscle contractile proteins, myosin heavy chain (MyHC) represents a major component of the myofibril sarcomeres (basic contractile unit) and is encoded by a multigene family that is tightly regulated throughout development. In the adult mouse hindlimb musculature, four MyHC isoforms (fast type IIb, IIx/d, IIa, and slow type I [or β]) are differentially expressed, and this expression pattern has been shown to contribute to the histochemical classification of four primary fiber types termed fast type IIb, IIx/d, and IIa and slow type I. Each of these fiber types displays unique functional properties with respect to size, metabolism, fatigability, and intrinsic contractile properties; the latter property is determined in part by their MyHC content. A well-established body of evidence gathered from studies utilizing animal and in vitro models has provided ample evidence that the amount and type of MyHC comprising a muscle's contractile apparatus is of functional significance and that physiological consequences occur as a result of alterations in MyHC composition, whether induced by physiological stimuli, disease, or mutation (natural or by gene targeting) (1, 2, 6, 41, 42). While considerable knowledge exists concerning MyHC function and the ability of adult muscle to alter its MyHC expression pattern in response to various stimuli, the in vivo identification of cis elements and trans factors that direct fiber-specific and perturbation-induced changes in MyHC expression remains incomplete.
To gain insight into how adult skeletal muscle adapts its MyHC phenotype to accommodate a sustained increase in workload (mechanical overload [MOV]), we have studied the regulated expression of the βMyHC gene. In adult mouse skeletal muscle, the βMyHC gene is primarily expressed in slow type I fibers; however, it can be induced by MOV in the fast type II plantaris muscle, a muscle that normally does not express this gene at any appreciable level (46, 47, 50, 53, 54). Our studies have documented this adaptation by measured increases in endogenous βMyHC mRNA, protein, and βMyHC transgene expression (46, 47, 50, 53, 54). A detailed transgenic analysis of both the mouse and human βMyHC promoters has delineated a minimal 293-bp human βMyHC promoter (termed β293wt) that we have shown mimics the expression pattern of the endogenous βMyHC gene during early development (fetal heart and hindlimb), in adult type I fibers, and in response to MOV (54). This analysis also revealed an 89-bp region (−293 to −205) that is required for slow muscle expression and MOV responsiveness. This region contains highly conserved distal muscle CAT (dMCAT; −290 to −284), A/T-rich (βA/T-rich −269/−258; also called GATA), C-rich (−244/−233), and proximal MCAT (pMCAT; −210/−203) elements (54). Independent mutation of the dMCAT (β293Mm; 6 lines) and the downstream nuclear factor of activated T-cell elements (NFAT, −179/−171; β293Nm, 11 lines) in the context of transgene β293wt reduced slow muscle expression levels but did not eliminate MOV responsiveness (50). In contrast, mutation of the βA/T-rich element (β293A/Tm; 21 independent lines) rendered this transgene silent under all conditions tested, including MOV, indicating that this element is required for constitutive slow muscle expression and possibly MOV responsiveness (51). Further support that the βA/T-rich element contributes to MOV induction of the transgene β293wt was obtained from our protein-DNA interaction studies, which showed enriched binding of two distinct nuclear proteins (44 and 48 kDa) only when using MOV-plantaris (MOV-P) nuclear extract (52). Despite the overlapping GATA/MEF2-like homology of the βA/T-rich element, the 44- and 48-kDa binding proteins were antigenically distinct from the GATA and MEF2 isoforms.
The absence of GATA and MEF2 proteins from the MOV-P-enriched human βA/T-rich binding complex was intriguing, since accumulating evidence has led to a proposed model implicating a primary role for GATA and MEF2 proteins in mediating skeletal muscle hypertrophy and the transcriptional activation of the slow fiber genes. This model postulates that perturbations such as low-frequency motor nerve stimulation, MOV, or exercise induce a sustained elevation in intracellular calcium levels, which in turn stimulates both the calcineurin- and calmodulin-dependent protein kinase signaling pathways and the subsequent transcriptional activation of slow fiber genes mediated by various members of the MEF2 and NFAT transcription factor families (12, 14, 32, 36, 43). In addition, other proteins have been proposed to play a role in mediating skeletal muscle hypertrophy and/or the slow muscle phenotype. For example, GATA2 expression is induced in skeletal muscle by MOV and in response to insulin-like growth factor 1 (IGF-1) and, thus, has been referred to as a marker of the hypertrophic response; however, its exact role has not been elucidated (34). In this regard, a recent immunohistochemical analysis of skeletal muscle from exercised mice and transgenic mice expressing a muscle-specific IGF-1 transgene revealed that GATA2 is not localized to the nucleus, where it would be expected to modulate the transcription of target genes (39). Another proposed mediator of muscle phenotype is the E-box-binding myogenic basic helix-loop-helix proteins, which have been linked to muscle fiber-type-specific expression by virtue of evidence gathered from transgenic analysis (58) and the correlative observation that myogenin is expressed at higher levels in adult slow type I fibers while MyoD is more abundant in adult fast type II fibers (21). In addition, recent evidence suggests a possible role for general transcription factor 3 (GTF3), also called MusTRD1, in regulating slow muscle gene expression based on its binding to a _bicoid_-like element located within the slow upstream regulatory element (SURE), a region required for slow muscle expression of the slow troponin I (sTnI) gene (9, 37). Notably, in our one-hybrid screening studies which used several distinct A/T-rich elements as bait, including the sTnI SURE _bicoid_-like element, we also confirmed that GTF3 is a specific binder of the _bicoid_-like site. However, subsequent electrophoretic mobility shift assays (EMSAs) revealed that GTF3 did not bind the βA/T-rich element (data not shown) and, thus, the specific βA/T-rich cognate binding factor(s) remains unknown.
To identify the functionally relevant βA/T-rich cognate binding factor, we performed a yeast one-hybrid screen of both adult skeletal muscle and MOV-P cDNA libraries, using the βA/T-rich element (5′-GGAGATATTTTT-3′) as bait. Herein we report the isolation of cDNAs encoding either nominal TEF-1 (NTEF-1) or related TEF-1b (RTEF-1b). This finding was not anticipated, since previous work had shown that all TEF proteins bind MCAT elements (5′-CATTCCT-3′) located within the promoter region of numerous muscle and nonmuscle genes. The vertebrate TEF genes encode a family of transcription factors that are characterized by the presence of an evolutionarily conserved TEA/ATTS DNA binding domain (TEAD) that is also found in plant (ABAA), fly (Scalloped), and yeast (TEC1) (22, 26) transcription factors. In vertebrates, the TEF proteins are encoded by a multigene family, and within this family additional diversity is brought about via alternative splicing. Northern blotting and in situ hybridization analyses of the various TEF-1 transcripts have revealed an overlapping expression pattern of NTEF-1/TEF-1/TEAD-1, RTEF-1/TEF-3/TEAD-4 and divergent TEF-1 (DTEF-1/TEF-5/TEAD-3) mRNA transcripts in skeletal and cardiac muscle, which raises the likely possibility that these factors serve redundant as well as distinct functions throughout development and in response to a variety of physiological signals.
The possibility that TEF-1 isoforms might play a more significant role than previously predicted in the regulation of striated muscle phenotype and MOV-induced fiber type shifts is intriguing and prompted us to study all adult TEF isoproteins (NTEF-1, RTEF1a, RTEF1b, and DTEF). Herein, we verify the specificity of NTEF-1 and RTEF-1b binding to the βA/T-rich element by competition and antibody supershift EMSAs. Scanning mutagenesis EMSA revealed that NTEF-1 and RTEF-1b binding predominantly involves strong interactions in the region of the βA/T-rich element that bears strong homology to consensus MEF2 binding sites. Importantly, we show that TEF-1 isoproteins can bind MEF2 elements located within the control region of other slow muscle and nonmuscle genes. In addition, binding of NTEF-1 at the βA/T-rich, cardiac/slow troponin C (cTnC), desmin MEF2, and the desmin palindromic myotube (pal-Mt) elements was enriched only when using nuclear extract isolated from MOV-P muscle. We confirmed the functional significance of NTEF-1 binding to the βMyHC A/T-rich element by using transient-coexpression assays in mouse C2C12 myotubes, where expression increased with βMyHC reporter constructs and constructs carrying tandem repeats of the βA/T-rich, desmin MEF2, and Pal-Mt elements. These data strongly implicate an important role for TEF transcription factors in directing slow-muscle-specific gene expression and in potentiating βMyHC induction in response to MOV. These unexpected findings will likely have global relevance to the regulation of muscle (skeletal, cardiac, and smooth) gene networks under basal and hypertrophic conditions, as well as gene expression in other tissues that express TEF proteins.
MATERIALS AND METHODS
Construction of yeast one-hybrid plasmids and yeast reporter lines.
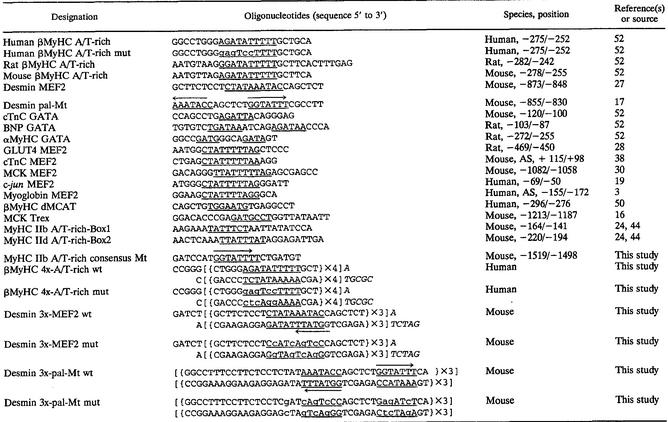
Complementary oligonucleotides representing four wild-type or mutant tandem repeats of the βMyHC A/T-rich element (βA/T-rich wild type, 5′-CTGGGAGATATTTTTGCT-3′; mutant, 5′-CTGGGgagTccTTTTGCT-3′ [mutated bases are lowercase]) were synthesized and annealed. The annealed oligonucleotides were inserted upstream of the HIS3 reporter gene promoter in pHISi-1 (Clontech) between the _Xba_I and _Mlu_I sites and upstream of the lacZ reporter gene promoter in pLacZi (Clontech) between the _Eco_RI and _Sal_I sites. Correct insert orientation and sequence were confirmed by restriction digest and automated sequencing of both strands (Applied Biosystems sequences, model 377). To obtain dual reporter yeast strains, wild-type and mutant (4x-βA/T-rich pHISi-1 and 4x-βA/T-rich pLacZi) plasmids were linearized by digestion with _Xho_I and _Nco_I, respectively, and sequentially integrated into the genome of yeast strain YM4271 following the manufacturer's instructions (Clontech).
One-hybrid screening.
The wild-type or mutant dual reporter strains were mated to Saccharomyces cerevisiae strain Y187 that was pretransformed with a GAL4-AD-human skeletal muscle cDNA fusion library in the yeast expression vector pACT2 (Clontech). The wild-type and mutant dual reporter plasmids were also used to screen a custom GAL4-AD-adult rat MOV-P skeletal muscle cDNA fusion library in the yeast expression vector pGAD10 (Clontech) using procedures described by the manufacturer. The MOV-P cDNA library was constructed from mRNA samples that were isolated and pooled from adult rat MOV-P muscles taken 14 and 49 days post-MOV. These two time points were chosen because they represent points at which βMyHC mRNAs begin to increase from very low levels (14 days) in the adult rat fast-twitch control plantaris muscle and the time point at which steady-state βMyHC mRNA levels are reached (49 days) (reference 53 and unpublished observation). Furthermore, since the adult rat plantaris muscle is comprised primarily of type II fibers (95%), we reasoned that it was possible that MOV-induced βMyHC expression could be initiated by one set of transcription factors (early) and maintained at steady-state levels by a different set of transcription factors. In theory, this paradigm would enable us to catch both sets of factors. Mated zygotes (control skeletal muscle library) or transformants (MOV library) were plated using stringent conditions on SD-His-Ura-Leu selective medium containing 30 mM 3-amino-1,2,3-triazole to suppress background growth. Positive clones were identified by robust growth on medium lacking essential amino acids (above) coupled with a positive result from a filter-lift β-galactosidase assay and no growth when using mutant reporter strains and plasmids. Plasmids from positive clones were extracted from yeast, transformed into DH5α bacterial cells (Invitrogen), and subjected to plasmid prep isolation (Qiagen) for sequencing. The resulting sequences of cDNA inserts were matched with known protein sequences from the PIR and SPTREMBL protein sequence databases.
Construction of plasmids used in transient-expression assays.
Desmin MEF2-dependent, pal-Mt-dependent, or βA/T-rich-dependent reporter constructs were generated by linking the Renilla luciferase gene carried in pRL-TK (Promega) to three wild-type or mutant copies of the desmin MEF2 or pal-Mt (Table 1) element or four wild-type or mutant copies of the βMyHC A/T-rich element (4x-βA/T-rich) (Table 1), respectively. Briefly, complementary wild-type or mutant oligonucleotides encoding either three tandem copies of the desmin MEF2 or desmin pal-Mt or four tandem copies of the βA/T-rich elements were synthesized, mixed at equimolar concentrations, and annealed in annealing buffer (10 mM Tris, 50 mM NaCl,1 mM EDTA). The annealed oligonucleotides were cloned into the _Bgl_II site at the 5′ end of the Renilla luciferase in pRL-TK, and the orientation and number of inserts in the resulting plasmids were verified via automated sequencing of both strands (Applied Biosystems sequencer model 377).
TABLE 1.
Oligonucleotides used in this study
The β293wt promoter-reporter construct was generated by fusing the 5′ end of the bacterial chloramphenicol acetyltransferase (CAT) gene within pSVOCAT to the upstream promoter region (293 bp of human βMyHC promoter and 120 bp of 5′-untranslated region [includes exon 1]) of the human βMyHC gene. The construction of the plasmid with mutations at both the distal and proximal MCAT elements (β293 d/pMmut) was made by introducing a mutated pMCAT element into a plasmid made previously (β293Mm [50]), which already carried a mutated dMCAT element. The βA/T-rich element was mutated within plasmid β293wt to generate a reporter carrying a mutation only at the βA/T-rich element (β293A/Tmut). The pMCAT site and the βA/T-rich sites were mutated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Complementary oligonucleotide primers harboring mutations within the pMCAT and βA/T-rich elements were designed to meet the length and melting temperature requirements specified by the manufacturer and had the following sequences (mutated bases are lowercase and underlined): pMCAT element, 5′-CTCAGACCCTGCACAGTCgAcGttATAACAATGACGACCACTTCC-3′; βA/T-rich element, 5′-GTGAGGCCTGGCCTTGGGgagtccTTTTGCTGCACTTTGAGCC-3′. Base pair substitutions were incorporated at nucleotides that in our previous diethyl pyrocarbonate interference footprinting had revealed to be crucial protein-DNA contact sites (52). Unintentional transcription factor recognition sites were not created by these mutations, as assessed by cross-referencing the mutated primers against the Eukaryotic Transcription Factor database (TFD) available on the Wisconsin Package, Genetics Computer Group (Madison, Wis.). The PCR-mediated incorporation of mutant sequence was performed using 5 ng of double-stranded β293Mm (pMCAT) or β293wt (βA/T-rich) template using conditions recommended by the manufacturer. The PCR products were transformed into Epicurian Coli XL1-Blue supercompetent cells (Stratagene), plasmid DNA was isolated (Qiagen EndoFree anion-exchange columns), and successful incorporation of the mutation was verified via automated sequencing of both strands (Applied Biosystems model 377 sequencer).
Cell culture, transfections, and reporter gene assays.
Mouse skeletal muscle C2C12 myoblasts (American Type Culture Collection) were seeded at 5 × 104 cells/35-mm dish in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal bovine serum, 1% (vol/vol) sodium pyruvate, 100 IU of penicillin/ml, and 100 μg of streptomycin (GIBCO Invitrogen Corp.)/ml, in a humidified 10% carbon dioxide atmosphere. At 60 to 80% confluence, C2C12 myoblasts were transfected with various plasmid DNAs using FuGENE 6 transfection reagent according to the manufacturer's instructions (Roche Diagnostics Corporation). Each 35-mm dish was transfected with a total of 2.5 μg of plasmid DNA, which consisted of 2 μg of reporter plasmid, 100 ng of pXJ40 (TEF-1 expression plasmid as indicated), 200 ng of pRSV-βgal plasmid as an internal control, and pPAC0 plasmid as nonspecific DNA to equalize total transfected DNA. All transfections were repeated using three different preparations of plasmid DNA. C2C12 myoblasts were changed from growth medium to differentiation medium (5% heat-inactivated horse serum) 24 h posttransfection. Three days posttransfection, cells were harvested, extracts were prepared, and luciferase and β-galactosidase (β-Gal) activities in the cell extracts were assayed according to the manufacturer's instructions (Promega Corp.), or CAT assays were performed as described below. Protein concentrations of cell extracts were determined by the method of Bradford (7), and β-Gal (10 μg), luciferase (15 μg), and CAT (5 μg) activities were normalized for protein. Normalized luciferase or CAT activities (Renilla luciferase/β-Gal or CAT/β-Gal ratios) from all samples were compared. Luciferase assays were done using a TD20/20 (Turner Designs) luminometer following automatic injection of 100 μl of Renilla luciferase substrate mixture. The linear range of relative light units (RLU) using this instrument and assay system was determined to be between 0.001 and 10,000 RLU by developing a standard curve using recombinant luciferase. All raw RLU values were within this linear range. CAT assays were performed using the C2C12 extract prepared as described for β-Gal assays, with the addition of a second freeze-thaw followed by a 10-min 65°C incubation to inactivate endogenous phosphatases. CAT assays were done as described previously (31, 47). Incubation of C2C12 extract with 20 mM acetyl coenzyme A and [14C]chloramphenicol (0.05 mCi/ml; Perkin-Elmer) was 5 μg of total protein per 17 h at 37°C, and the percent conversion of [14C]chloramphenicol to the acetylated form was quantified using a PhosphorImager (Storm 860) with ImageQuant version 5.1 software. All data were normalized for β-Gal and are presented as CAT specific activity (picomoles per microgram of protein per minute).
Isolation of total RNA and Northern blot analysis.
Isolation of total cellular RNA and Northern blotting were performed as described by our investigators previously (47). Thirty micrograms of total RNA was loaded in each lane. A mouse-specific NTEF-1 cDNA probe was used to detect NTEF-1 transcripts in control plantaris (CP) versus MOV-P adult mouse muscle RNA. βMyHC transcripts were detected by using an oligonucleotide probe corresponding to the mouse βMyHC 3′-untranslated region, while loading equivalency was normalized by hybridization with a human 18S cDNA as described by Tsika et al. (47).
Preparation of nuclear protein extract from adult skeletal muscle.
Nuclear extracts were isolated from adult rat CP, MOV-P, and control soleus (CS) muscle as previously described (31, 52). Protein concentration was determined according to the method of Bradford (7).
Western blot analysis.
The total protein content in three different batches of CP and MOV-P nuclear extracts and cytosolic extracts from adult rat muscle tissue, C2C12 extract (Active Motif), and in vitro-synthesized (TnT) NTEF-1 and MEF2a protein was measured by using the method of Bradford (7). All protein extracts and positive and negative controls were fractionated on a 4 to 12% polyacrylamide gel electrophoresis (PAGE) Tris-base gel (Invitrogen Corporation) and transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories) by electroblotting at 30 V for 1 h. Amounts of total protein used were as follows: 30 μg of CP and MOV-P nuclear extract, 40 μg of CP and MOV-P cytosolic extract, 30 μg of C2C12 extract, and 0.5 μl of in vitro-synthesized NTEF-1 or MEF2a. A431 and BC3H1 cell lysates (40 μl) were used as positive controls for NTEF-1 and MEF2, respectively, and 4 μl of lysate not programmed with plasmid DNA was used as a negative control for in vitro-synthesized proteins (unprogrammed lysate [UL]). The polyvinylidene difluoride membrane was blocked overnight at 4°C with 5% nonfat dry milk in Tris-buffered saline buffer with 0.1% Tween 20 (TBST). Equal loading of cytosolic extract was visualized by using mouse monoclonal rasGAP antibody (1:200; Santa Cruz). The blots were incubated with the following primary antibodies for 1.5 h at room temperature: anti-TEF-1 (same as NTEF-1) monoclonal mouse immunoglobulin G (IgG; 1:250; BD Biosciences), anti-myoglobin polyclonal goat IgG (1:400; Santa Cruz Biotechnology, Inc.), anti-troponin I-SS polyclonal goat IgG (1:400; Santa Cruz Biotechnology, Inc.), or general anti-MEF2 rabbit polyclonal IgG (1:500; sc-313; Santa Cruz). The blots were then washed (three times for 5 min) with TBST buffer and further incubated with secondary antibodies as follows for 1 h at room temperature: a 1:2,000 dilution of goat anti-rabbit IgG, goat anti-mouse IgG conjugated with horseradish peroxidase (HRP) (Cell Signal Technology), or donkey anti-goat IgG-HRP (Santa Cruz Biotechnology, Inc.). After washing, HRP activity was detected using an enhanced chemiluminescence detection SuperSignal substrate (Pierce) and subjected to autoradiography. Each primary antibody was tested using both nuclear and cytosolic blotting representing three different extracts.
EMSAs.
All oligonucleotide probes used in this study are listed in Table 1. Electrophoretic mobility shift assays (EMSAs) were carried out as previously described (31). Binding reactions were performed using either 4 μg of CP or MOV-P nuclear extract and 20,000 cpm of labeled probe for 20 min at room temperature in a 25-μl total volume. Where indicated, either in vitro-transcribed and -translated (TnT) NTEF-1, RTEF-1a, RTEF-1b, DTEF-1, or MEF-2A were used in place of muscle nuclear extract (see figure legends for specific quantities). Binding reactions were resolved on a 5% nondenaturing polyacrylamide gel at 220 V for 2.5 h at 4°C. Supershift EMSAs were performed by first preincubating skeletal muscle nuclear extract or in vitro-synthesized proteins with 2 μl of the corresponding antibody for 30 min at room temperature followed by the addition of the 32P-labeled DNA probe. The antibodies used in the EMSA analysis were NTEF-1 (described above) and MEF2 antibodies as follows: MEF2a (glutathione _S_-transferase [GST]-MEF2a, human amino acids [aa] 129 to 263 [52, 60]) and MEF2b (GST-MEF2b [human aa 88 to 365] [18, 52]) from Y.-T. Yu; and MEF2c (human, amino terminus of MEF2C; sc-13917), MEF2 (polyclonal, general; sc-313), and MEF2 (monoclonal, general; sc-17785) from Santa Cruz Biotechnology, Inc. Following electrophoresis, the gels were dried and DNA-protein complexes were visualized by autoradiography at −80°C.
In vitro TnT.
In vitro-coupled TnT was performed using 1 μg of NTEF-1, RTEF-1a, RTEF-1b, DTEF-1, ETEF-1, or MEF-2a expression plasmids in the T7 TnT rabbit reticulocyte lysate kit according to the manufacturer's instructions (Promega). The expression plasmids corresponded to either NTEF-1 (pXJ40-TEF-1A; open reading frame [ORF] of human TEF-1), RTEF-1 (pXJ41-TEF-3; ORF of human TEF-3), DTEF-1 (pXJ40-TEF-5; ORF of human TEF-5), human RTEF-1b (pCITE-RTEF1b), or human MEF-2a (pcDNAI MEF-2a) (50, 52). Parallel TnT reactions were performed in the presence of [35S]methionine (Perkin-Elmer). The integrity and expected molecular weights of the protein products were verified by resolving the radiolabeled reaction products on a sodium dodecyl sulfate-12% PAGE (SDS-PAGE) gel. Parallel reactions of lysate not programmed with plasmid DNA served as negative controls (UL).
Statistical analysis.
Statistical analyses were performed by using the SPSS Graduate Pack 10.0 program (SPSS, Chicago, Ill.). A Levene's test for equality of variances was performed, followed by a two-tailed independent-sample t test to assess differences between group means. Where the Levene's test was rejected (significance of ≤0.05), the separate variance t test for means was used, where equal variances were not assumed. The lowest significance level accepted was P < 0.05. All data are reported as the mean ± standard error.
RESULTS
TEF-1 binds the βA/T-rich element in a yeast one-hybrid system.
Previously, our investigators have shown that a transgene comprised of 293 bp of human βMyHC 5′-flanking DNA (β293wt) was minimally sufficient to confer MOV-induced expression in both the fast-twitch plantaris and slow-twitch soleus muscles (54). Within this transgene we identified an A/T-rich element (βA/T-rich −269/−258) that when reacted with MOV-P nuclear extract formed an enriched binding complex with two unknown proteins (52). Mutation of the element led to the complete loss of expression, including MOV responsiveness, in all 21 transgenic lines examined, indicating an important role for the two proteins interacting with this element (51). To identify the βA/T-rich binding proteins, the βA/T-rich element was used as bait in a yeast one-hybrid system to screen (i) an adult human quadriceps muscle cDNA library, and (ii) an adult rat MOV-P muscle cDNA library. A yeast one-hybrid screen of the adult human skeletal muscle cDNA library was performed using four tandem copies of the wild-type βA/T-rich element as bait, and to control for nonspecific protein-DNA interaction four tandem copies of mutant βA/T-rich element were used. Following stringent selection criteria, positive clones were selected and additional assays were performed to eliminate false positives: (i) β-Gal and (ii) mating using a pHISi-1/pLacZi (dual reporter) strain containing four tandem copies of the βA/T-rich element, versus two negative controls (empty dual reporter strain and mutant βA/T-rich dual reporter strain). Screening of approximately 1.7 × 107 cDNA clones identified 33 that were positive for β-Gal activity and negative for growth using the mutant and empty dual reporter strain. Escherichia coli cells were transformed with the 33 plasmids extracted from positive yeast, and subsequent nucleotide sequence analyses of cDNA inserts showed that all 33 clones encoded TEF proteins: 21 NTEF-1 and 12 RTEF-1b. A yeast one-hybrid screen of the adult rat MOV-P cDNA library was also performed using four copies of either the wild-type or mutant βA/T-rich bait strains. Selection conditions and subsequent false-positive elimination assays were the same as described above. A total of approximately 2 × 106 cDNA transformants yielded five clones that were positive for β-Gal activity. E. coli cells were transformed with the five plasmids extracted from positive yeast, and subsequent sequence analysis revealed that these clones encoded NTEF-1.
Sequence analysis of clones containing the largest insert revealed a full-length cDNA representing a previously unidentified human ortholog for RTEF-1b (accession number AY101179). This cDNA insert was 1.7 kb with a 1,173-bp ORF. This cDNA encoded a protein with 391 aa residues corresponding to a predicted molecular mass of 44,039 kDa. Isolation and subsequent cloning of this cDNA into an in vitro TnT vector (pCITE4b; Promega Corp.) produced two proteins close in molecular mass following a TnT reaction (see Fig. 2, inset). The production of two proteins was presumably due to the presence of both the Kozak site provided by the TnT vector and the genuine initiation site within the cDNA.
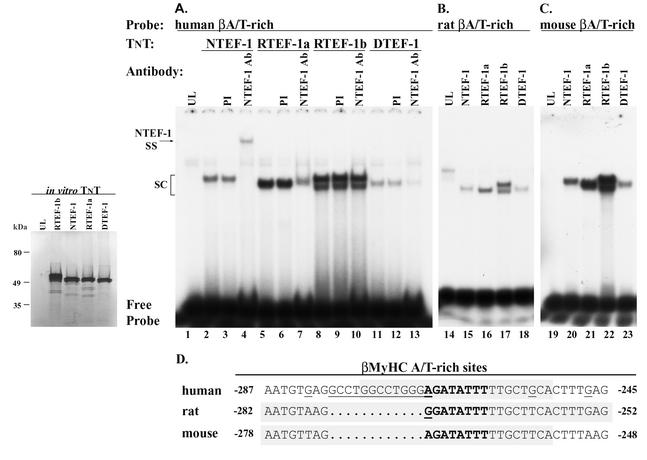
FIG. 2.
EMSA analysis verifies specific binding of in vitro-synthesized TEF proteins to the human, rat, and mouse βA/T-rich elements. The inset reveals the correct size of [35S]methionine-labeled in vitro-synthesized human NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1. A rabbit reticulocyte lysate system was programmed with 1 μg of circular NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1 in the presence of [35S]methionine. The transcription-translation product was resolved by SDS-PAGE and exposed to film. Molecular mass markers (in kilodaltons) are shown to the left. The lane marked UL represents a parallel reaction not programmed with TEF-1 expression plasmid. (A) EMSA of 32P-labeled human βA/T-rich element reacted with in vitro-synthesized human NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1 protein (lanes 2, 5, 8, and 11). Antibody supershift EMSA was performed by preincubation of in vitro-synthesized human NTEF-1, RTEF-1a, RTEF-1b, or DTEF-1 protein with 2 μl of NTEF-1 antibody for 30 min at room temperature prior to addition of labeled probe (lanes 4, 7, 10, and 13). Addition of NTEF-1 antibody to binding reactions containing in vitro-synthesized NTEF-1, RTEF-1a, or RTEF-1b resulted in either a supershift (SS) or immunodepletion of the specific DNA-protein complex (SC), whereas reactions containing RTEF-1b were not altered (lane 10). Control reactions were performed with PI serum (lanes 3, 6, 9, and 12). (B) EMSA of 32P-labeled rat βA/T-rich element reacted with in vitro-synthesized human NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1 protein (lanes 15 to 18). (C) EMSA of 32P-labeled mouse βA/T-rich element reacted with in vitro-synthesized human NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1 protein (lanes 20 to 23). (D) Nucleotide sequence comparison showing conservation of the βA/T-rich element (gray stipple shows probes used in the EMSA) within the βMyHC proximal promoter of the human, rat, and mouse. Note that each of the in vitro-synthesized TEF proteins bound the βA/T-rich elements regardless of minor differences in core and flanking nucleotides.
Northern and Western analyses revealed increased levels of NTEF-1 in the MOV-P muscle.
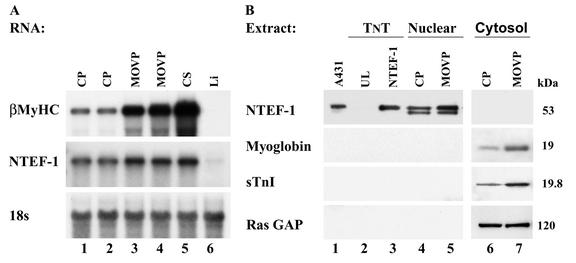
Because our yeast one-hybrid screen of an MOV-P cDNA library identified NTEF-1 as a factor that binds the βA/T-rich element, it was conceivable that NTEF-1 represented one of the two unknown proteins that formed an enriched binding complex at the βA/T-rich element only when using MOV-P nuclear extract (52). Therefore, to determine whether NTEF-1 is regulated by MOV, we performed both a Northern and Western analysis (Fig. 1). Northern blot analysis was performed using total RNA isolated from CS, liver (Li), and sham-operated CP and MOV-P muscles (Fig. 1A). As expected, high levels of βMyHC expression were detected in the slow-twitch CS muscle compared to the fast-twitch CP muscle (Fig. 1A, lane 5 versus lanes 1 and 2), and these transcripts were absent in nonmuscle (Li) tissue (Fig. 1A, lane 6). Consistent with a fast-to-slow fiber type shift, 7 weeks of MOV resulted in a large upregulation in βMyHC expression in MOV-P (Fig. 1A, lanes 1 and 2 versus lanes 3 and 4). Interestingly, qualitatively higher NTEF-1 mRNA levels were detected in the MOV-P than in the CP and resembled the levels detected in the CS (Fig. 1A). Hybridization to an 18S rRNA cDNA probe confirmed that all lanes were loaded evenly.
FIG. 1.
Northern blot analysis of NTEF-1 and βMyHC expression in CP and MOV-P muscle. (A) Total RNA isolated from CP, MOV-P, and CS (20 μg) muscle pooled from five mice was fractionated on a 1.5% agarose denaturing gel. Intensity of the hybridization signal was quantitated using a PhosphorImager and normalized to 18S values to account for loading differences between lanes. (B) NTEF-1 protein expression in CP and MOV-P muscle. Western blot analysis results are shown of rat CP and MOV-P nuclear extracts (30 μg; lanes 4 and 5) and cytoplasmic extract (40 μg; lanes 6 and 7) using mouse monoclonal NTEF-1, mouse monoclonal rasGAP antibody, polyclonal myoglobin, or polyclonal troponin I-SS. A431 extract (lane 1) and TnT NTEF-1 (lane 3) served as positive controls for the presence of NTEF-1 protein. UL (lane 2) was used as a control for nonspecific cross-reaction of the NTEF-1 antibody. NTEF-1 protein was increased in the MOV-P nuclear extract when compared to CP nuclear extract. Both myoglobin and sTnI protein were increased in MOV-P cytoplasmic extract, which is consistent with a fast-to-slow fiber type conversion following sustained periods of MOV. Efficient cellular fractionation is apparent, since rasGAP did not appear in nuclear extract while NTEF-1 was not detected in cytoplasmic extract.
To determine if the increase in NTEF-1 mRNA is also reflected as increased NTEF-1 protein levels in response to MOV, we performed Western blot analysis using CP and MOV-P nuclear extract (Fig. 1B). Using NTEF-1 antibody, a 53-kDa band was detected in A431 extract (Fig. 1B, lane 1) as well as in CP and MOV-P nuclear extracts (Fig. 1B, lanes 4 and 5) that corresponded to in vitro-synthesized NTEF-1 (Fig. 1B, TnT lane 3). The 53-kDa band representing NTEF-1 is qualitatively greater in the MOV-P nuclear extract than in the CP nuclear extract, suggesting that NTEF-1 is, in part, regulated at the level of transcription by MOV. In contrast, no band was detected in UL or cytoplasmic extracts (Fig. 1B, lanes 2, 6, and 7). Two markers were used, myoglobin and slow troponin I (sTnI), to verify that these extracts contained protein representative of a fast-to-slow fiber type conversion. Both were found to be qualitatively elevated in MOV-P cytoplasmic extract compared to CP extracts (Fig. 1B, lanes 6 and 7). Cellular fractionation was controlled by analysis of cytoplasmic rasGAP, which did not appear in CP or MOV-P nuclear extract and appeared qualitatively similar in both CP and MOV-P lanes, indicating that the lanes representing cytoplasmic extract were loaded evenly (Fig. 1B, lanes 6 and 7). Further, nuclear NTEF-1 was not detected in CP or MOV-P cytoplasmic extracts, indicating that our cytoplasmic extracts were not contaminated with nuclear proteins.
EMSA analyses verified the binding of TEF proteins to the human, rat, and mouse βA/T-rich elements.
TEF proteins are known to bind specifically to MCAT elements located within the promoter region of numerous muscle genes (22, 26). Therefore, our finding that NTEF-1 bound to the human βA/T-rich element was completely unexpected. A complete EMSA analysis was performed to confirm the specific binding of TEF proteins to the βA/T-rich element. These experiments were initiated by using in vitro-synthesized NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1 (Fig. 2, inset) and the βA/T-rich elements located within the human, rat, and mouse βMyHC proximal promoters (Fig. 2 and Table 1). As can be seen in Fig. 1B and the inset of Fig. 2, endogenous and in vitro-synthesized TEF-1 isoforms were approximately 53 kDa in size, which deviates slightly from our UV cross-linking study results that showed two βA/T-rich binding factors of 48 and 44 kDa (52). This discrepancy is likely due to irregularities in electrophoretic mobility following UV cross-linking, variations in sample salt concentration, or different levels of TEF protein modifications. A detectable binding complex did not form when the 32P-labeled human βA/T-rich element was reacted with 2 μl of UL (Fig. 2A, lane 1). Specific binding complexes formed when the 32P-labeled human βA/T-rich element was reacted with 2 μl of in vitro-synthesized NTEF-1, RTEF-1a, RTEF-1b, or DTEF-1 (Fig. 2A, lanes 2, 5, 8, and 11, respectively). Binding complexes were not altered by the addition of preimmune (PI) serum to the binding reaction mixtures (Fig. 2A, lanes 3, 6, 9, and 12, respectively), whereas addition of monoclonal TEF-1 (same as NTEF-1) antibody to binding reactions resulted in a supershift of NTEF-1 (Fig. 2A, lane 4) and a partial decrease in binding of RTEF-1a (Fig. 2A, lane 7) and DTEF-1 (Fig. 2A, lane 13), but not RTEF-1b (Fig. 2A, lane 10). Since the antibody used in this study was developed against aa 86 to 199 of NTEF-1, it is not surprising that this antibody also recognized RTEF-1a and DTEF-1, given the amino acid identity within this region between NTEF-1, RTEF-1a, and DTEF-1. In contrast, RTEF-1b contains a deletion of 43 aa (aa 119 to 161) within this region, which likely accounts for the observation that the anti-TEF-1 used in this study does not recognize RTEF-1b. As seen with the human βA/T-rich element, the rat and mouse 32P-labeled βA/T-rich elements also bound all in vitro-synthesized TEF proteins (NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1) (Fig. 2B and C). These results show that each distinct TEF protein binds specifically, but differentially, to the human, rat, and mouse βA/T-rich elements.
Competition scanning mutagenesis EMSA analysis determined the TEF binding site.
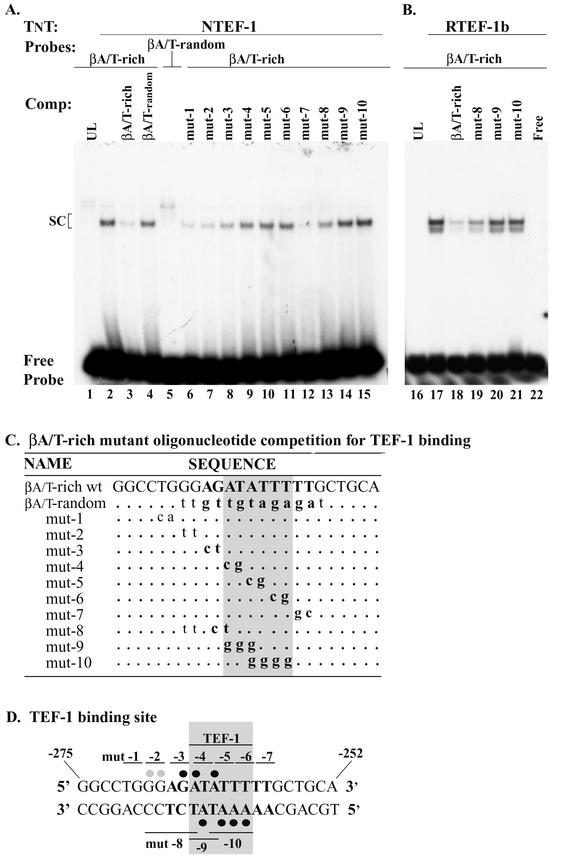
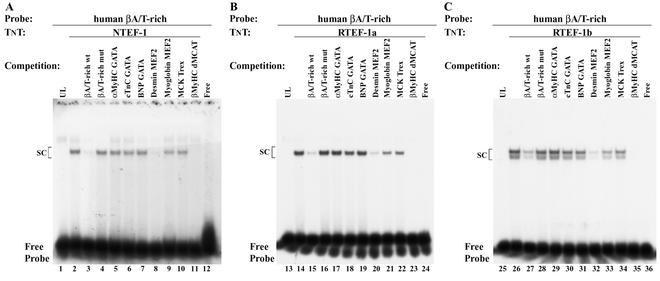
We performed both competition and scanning mutagenesis EMSA analysis to determine if the binding of NTEF-1 to the βA/T-rich site involved the same nucleotides as those previously identified by our investigators when using MOV-P nuclear extracts (Fig. 3D) (52). In competition EMSA analysis, we found that a binding complex did not form when the 32P-labeled human βA/T-rich element was reacted with UL, whereas a specific binding complex formed when this probe was reacted with in vitro-synthesized NTEF-1 (Fig. 3A, lane 1 versus lane 2). The addition of 100-fold molar excess cold wild-type βA/T-rich element to the binding reaction mixture abolished complex formation (Fig. 3A, lane 2 versus lane 3), whereas complex formation was not abolished by the addition of 100-fold molar excess cold βA/T-rich random element (composed of the same nucleotides as the wild-type βA/T-rich probe but in randomized order) (Fig. 3A, lane 4). As expected, when NTEF-1 was reacted with the 32P-labeled βA/T-rich random element, complex formation was not observed (Fig. 3A, lane 5).
FIG.3.
Competition EMSA determination of specific nucleotides involved in DNA-protein interactions at the βMyHC βA/T-rich element. (A) 32P-labeled βMyHC βA/T-rich oligonucleotide was incubated with 2 μl of in vitro-synthesized NTEF-1 (lanes 1 to 15). The specificity of NTEF-1 binding to the 32P-labeled human βA/T-rich oligonucleotide (lane 2) was demonstrated by addition of 100-fold molar excess cold competitor, either wild-type βA/T-rich (lane 3) or mutant βA/T-rich random (lane 4). The βA/T-rich random oligonucleotide is comprised of the same nucleotides as the wild-type βA/T-rich probe but in random order. As can be seen in lane 5, in vitro-synthesized NTEF-1 protein could not bind this oligonucleotide. Lanes 6 to 15, where indicated, show results when 100-fold molar excesses of various cold βA/T-rich mutant oligonucleotides (mut 1 to 10 in panel C) were added to the reaction prior to the addition of the probe. SC, specific complex. (B) 32P-labeled βMyHC βA/T-rich oligonucleotide was incubated with 2 μl of in vitro-synthesized RTEF-1b (lanes 17 to 21). The specificity of RTEF-1b binding to the 32P-labeled human βA/T-rich oligonucleotide (lane 17) was demonstrated by addition of 100-fold molar excess cold competitor, either wild-type βA/T-rich (lane 18) or mutant βA/T-rich random (lanes 19 to 21). Competition with mut-8, mut-9, and mut-10 revealed that strong interactions between the βA/T-rich probe and RTEF-1b occur in the MEF2-like homologue of the βA/T-rich probe. (C) Summary of competition EMSA with βA/T-rich mutant oligonucleotides. The βA/T-rich sequence is in boldface type. Wild-type βA/T-rich represents βA/T-rich sequence devoid of mutations. Mutated bases are in lowercase and bold type, and base pairs that are unchanged from the wild-type sequence are represented as dots. Nucleotides involved in binding are in gray stipple. (D) The βA/T-rich oligonucleotide is shown with βA/T-rich core sequence in boldface type and the binding site in gray stipple. The 2-bp numbering above the oligonucleotide sequence corresponds to that particular mutant oligonucleotide shown in panel C. Dimethyl sulfate and diethyl pyrocarbonate interference footprint of the βA/T-rich site is shown here to illustrate the DNA-protein binding profile when using MOV-P nuclear extract (45). Solid black circles represent strong binding, and gray circles denote weak binding. Sequence numbering begins at the 5′ end of the sense strand.
We next performed a scanning mutagenesis EMSA analysis to identify nucleotides within the βA/T-rich element that interact with NTEF-1 (Fig. 3). Nucleotide substitutions were introduced 2 bp at a time, starting within the immediate 5′-flanking region and extending throughout the core βA/T-rich element and its immediate 3′-flanking region. These oligos were then added in 100-fold molar excess to binding reaction mixtures containing in vitro-synthesized NTEF-1 and the wild-type 32P-labeled human βA/T-rich element. Binding complexes were effectively competed away by the addition of human βA/T-rich mut-1, mut-2, mut-3, and mut-7 (Fig. 3A, lanes 6 to 8 and 12), but not by mut-4, mut-5, and mut-6 (Fig. 3A, lanes 9 to 11). The resulting data suggest that NTEF-1 binding to the human βA/T-rich element predominantly involves strong interactions in the region of the βA/T-rich site that bears strong homology to the consensus MEF2 (8 of 10 bp; antisense strand) binding site (Fig. 3A, lanes 6 to 12).
Because the βA/T-rich element is a composite site with strong homology to consensus GATA (5 of 6 bp; sense strand) at its 5′ end, we generated three additional human βA/T-rich mutant elements that were designed to eliminate binding at either the GATA-like site and its 5′-flanking region (mut-8), the core human βA/T-element (mut-9, which overlaps both the GATA and MEF2 sites), or the MEF2-like site alone (mut-9) (Fig. 3C and D). The addition of 100-fold molar excess cold human βA/T-rich mut-8 to binding reaction mixtures resulted in partial competition (Fig. 3A, lane 2 versus 13), whereas mut-9 and mut-10 did not compete for NTEF-1 binding (Fig. 3A, lane 2 versus lanes 14 and 15). In agreement with our results obtained with competition scanning mutagenesis, neither mut-9 nor mut-10 acted as competitors, illustrating that sequences spanning these two oligonucleotides contain important contact sites for NTEF-1.
The latter experiment was repeated using in vitro-synthesized RTEF-1b, since it was also identified as a cognate human βA/T-rich binding factor in yeast one-hybrid screens. A binding complex did not form when the wild-type human βA/T-rich element was reacted with UL, whereas a specific binding complex formed when in vitro-synthesized RTEF-1b was added to binding reaction mixtures (Fig. 3B, lane 16 versus 17). The addition of 100-fold molar excess cold wild-type human βA/T-rich element completely abolished complex formation (Fig. 3B, lane 17 versus 18). Partial competition of complex formation was observed by the addition of 100-fold molar excess cold human βA/T-rich mut-8 to the binding reaction mixture (Fig. 3B, lane 17 versus 19), while mut-9 and mut-10 did not compete (Fig. 3B, lane 17 versus lanes 20 and 21). These data support the notion that both NTEF-1 and RTEF-1b interact most strongly with the MEF2-like homology of the human βA/T-rich element (Fig. 3D).
Nominal TEF-1 binds the βA/T-rich element when using control and MOV-P nuclear extract.
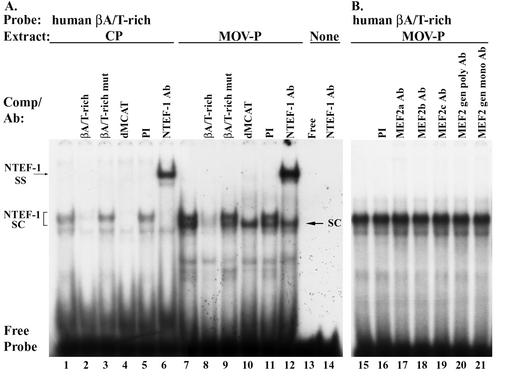
Having confirmed that in vitro-synthesized TEF protein can bind the human βA/T-rich element by EMSA analysis, we wished to determine if TEF protein was a component of the enriched binding activity found only in MOV-P nuclear extract (52). EMSA analysis of binding reactions containing the 32P-labeled human βA/T-rich element and 4 μg of CP nuclear extract revealed the formation of a binding complex that was specific (specific complex [SC]) and which appeared as an enriched doublet when 4 μg of MOV-P nuclear extract was used (Fig. 4A, lane 1 versus 7). The addition of 100-fold molar excess cold wild-type βA/T-rich element to binding reaction mixtures containing CP (Fig. 4A, lane 1 versus 2) or MOV-P (Fig. 4A, lane 7 versus 8) nuclear extract completely abolished complex formation, whereas addition of 100-fold molar excess cold mutant βA/T-rich element (Fig. 4A, lanes 3 and 9) did not compete for complex formation, indicating that this complex was specific. Interestingly, when a 100-fold molar excess of the human βMyHC dMCAT element, to which all TEF proteins avidly bind (50), was added to the binding reaction containing CP nuclear extract, complex formation was completely abolished (Fig. 4A, lane 4), whereas when MOV-P nuclear extract was used only the upper band of the enriched doublet was competed away (Fig. 4A, lane 10). Binding complexes were not altered by the addition of PI serum to binding reactions (Fig. 4A, lanes 5 and 11). Addition of monoclonal NTEF-1 antibody to binding reactions containing CP or MOV-P nuclear extract resulted in a supershift of the upper band of the specific complex (Fig. 4A, lanes 6 and 12), whereas the lower band of the enriched doublet was not supershifted when using MOV-P nuclear extracts. Lane 13 shows free probe, while lane 14 shows that when NTEF-1 antibody was reacted with free probe a complex did not form. These data provide conclusive evidence that NTEF-1 accounts for the upper band of the enriched doublet that forms at the βA/T-rich element when using CP and MOV-P nuclear extracts.
FIG. 4.
Competition and antibody EMSA analysis of CP and MOV-P skeletal muscle nuclear extract binding at the human βA/T-rich element. (A) The 32P-labeled βA/T-rich element was incubated with 4 μg of either CP (lanes 1 to 6) or MOV-P (lanes 7 to 12) nuclear extract. Unlabeled competitor oligonucleotides harboring wild-type βA/T-rich (lanes 2 and 8), mutant βA/T-rich (lanes 3 and 9), and dMCAT (lanes 4 and 10) binding elements were added at 100-fold molar excess. Because all TEF proteins avidly bind the dMCAT element, it was used as a positive control for TEF protein binding. Antibody supershift EMSAs were performed by preincubation of CP or MOV-P nuclear extract with 2 μl of either PI serum (lanes 5 and 11) or monoclonal NTEF-1 antibody (lanes 6 and 12) for 30 min at room temperature. Binding complex formation at the βA/T-rich element is enriched when using MOV-P nuclear extract (lanes 7 to 12) versus CP nuclear extracts (lanes 1 to 6). The specific binding complex was determined to be NTEF-1 based on inhibition of complex formation by addition of 100-fold molar excess cold dMCAT element (NTEF-1 SC; lanes 4 and 10) and the formation of a supershifted complex by addition of NTEF-1 antibody (NTEF-1 SS; lanes 6 and 12). An additional enriched DNA-protein complex formed only when using MOV-P nuclear extract (SC; arrow). Free probe represents excess unreacted radiolabeled oligonucleotide. Lane 14, 32P-labeled βA/T-rich element with NTEF-1. (B) MEF2 antibody supershift EMSA was performed by preincubation of 7.5 μg of MOV-P nuclear extract with 2 μl of PI serum (lane 16) or MEF2a (lane 17), MEF2b (lane 18), MEF2c (lane 19), MEF2 general polyclonal (lane 20), or MEF2 general monoclonal antibody (lane 21) for 30 min at room temperature.
Since TEF proteins have recently been shown to physically interact with MEF2 (29), we performed supershift EMSA analyses to determine if MEF2 protein represents a component of the enriched doublet that forms at the human βA/T-rich element when using MOV-P nuclear extract. When the 32P-labeled human βA/T-rich element was reacted with MOV-P nuclear extract, an enriched complex was formed that was not altered by the addition of PI serum or by the addition of MEF2a, MEF2b, MEF2c, MEF2d, or general MEF2 antibody (Fig. 4B, lanes 16 to 21). In this analysis, the enriched doublet observed in Fig. 4A was obscured, since the amount of MOV-P nuclear protein used was nearly doubled (7.5 μg versus 4 μg), and the autoradiograph was overexposed in an effort to detect any supershifted complex. Furthermore, the different antibodies used were selected to assure the detection of any MEF2 isoprotein present. Specificity of MEF2a and MEF2b antibodies has been demonstrated previously (18, 52, 60), and the MEF2c, general MEF2 polyclonal, and general MEF2 monoclonal antibodies are broadly reactive with all MEF2 isoforms. Thus, these data confirm our previous results by showing that MEF2 proteins are not a component of the enriched doublet that forms at the human βA/T-rich complex when using MOV-P nuclear extract. Efforts to identify the factor(s) comprising the higher-mobility complex of the enriched doublet are in progress.
NTEF-1 transactivates βMyHC reporter genes in C2C12 myotubes.
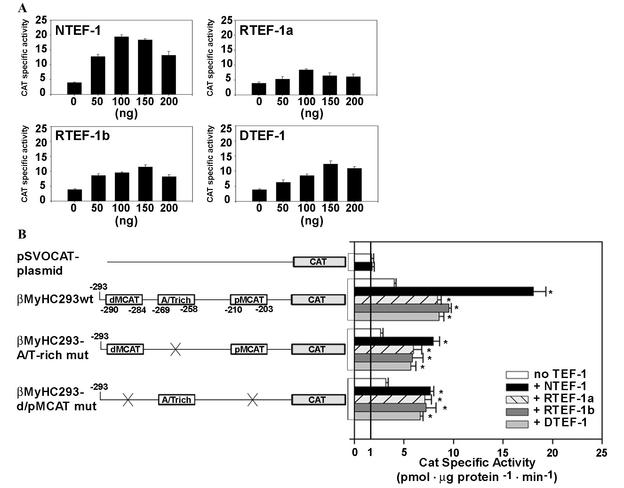
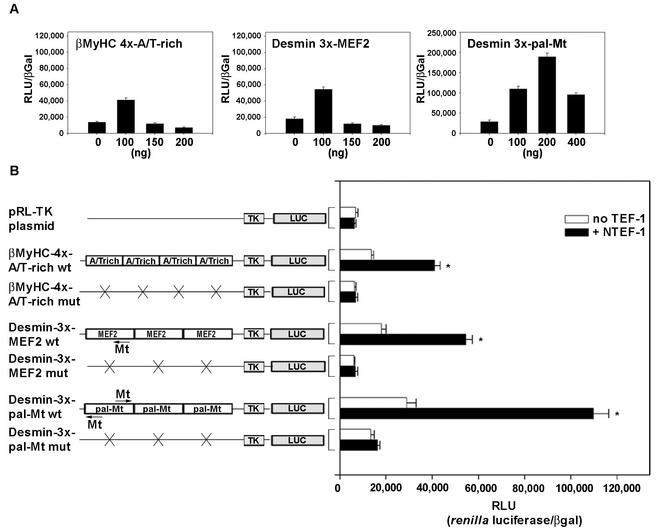
To determine the functional significance of NTEF-1 binding to the human βA/T-rich element, we conducted transient-expression assays in which TEF-1 expression vectors (pXJ40-TEF-1; cytomegalovirus [CMV]-driven expression vector) were cotransfected with both wild-type and mutant versions of a 293-bp βMyHC CAT reporter gene into C2C12 myoblasts that were subjected to differentiation media to form myotubes (Fig. 5). A dose-response curve using NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1 revealed that 100 ng of TEF expression vector was the optimal amount to use in cotransfection experiments without the effects of squelching influencing human βMyHC reporter gene expression levels (Fig. 5A).
FIG. 5.
Response of wild-type 293-bp human βMyHC promoter-reporter gene to increasing amounts of TEF protein. (A) C2C12 myoblasts were transfected with the 293-bp βMyHC reporter gene (2 μg) and increasing amounts of TEF expression vector (CMV-driven pXJ40-NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1) and subjected to differentiation media to form myotubes. Increasing amounts of TEF-1 protein expression led to squelching. Fold increases in CAT specific activity (picomole per microgram of protein per minute) were expressed as the mean ± standard error (n = 3). (B) TEF transactivates βMyHC reporter genes devoid of dMCAT and pMCAT elements in C2C12 myotubes. The addition of 100 ng of TEF isoforms (CMV-driven pXJ40-NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1) significantly transactivated the 293-bp βMyHC promoter (βMyHC293 wt) compared to levels seen using vector alone (pSVOCAT) or to basal levels (-TEF) observed when using reporter constructs without cotransfection with TEF isoforms. Constructs harboring mutations of either the βA/T-rich element (βMyHC293-A/T-rich mut) or dMCAT and pMCAT elements (βMyHC293-d/pMCAT mut) resulted in significant increases above basal levels, indicating that 293 bp of βMyHC with the wild-type βA/T-rich element is capable of transactivating the reporter gene. All data were normalized using β-Gal activity to accommodate variations in transfection efficiency. Data are reported as CAT specific activity (in picomoles per microgram of protein per minute) and are expressed as the mean ± standard error (n = 20 for basal [-TEF] transfections; n = 11 for cotransfections using NTEF-1; n = 7 for cotransfections using RTEF-1a, RTEF-1b, and DTEF-1 and for transfections using vector alone). *, P < 0.001.
The basal expression level (CAT specific activity [in picomoles per microgram of protein per minute]) of wild-type β293wt in C2C12 myotubes was significantly higher than that of the pSVOCAT plasmid (2.4-fold). Mutation of the βA/T-rich element (β293A/T-rich mut) decreased basal expression by 33.3% compared to the expression levels of wild-type β293wt. Since βMyHC transactivation can be mediated by TEF protein binding at MCAT elements, we introduced mutations into the distal and proximal MCAT elements (β293d/pMCAT mut), which resulted in a 20% lower level of expression compared to wild-type β293wt expression (Fig. 5B).
CAT specific activity of the pSVOCAT plasmid was low and exhibited only a nonsignificant increase when cotransfected with 100 ng of NTEF-1 (Fig. 5B). Expression levels of wild-type β293wt increased 4.9-fold above basal levels of cotransfection with NTEF-1 and increased from 2.1- to 2.4-fold when cotransfected with RTEF-1a, RTEF-1b, or DTEF-1 (Fig. 5B). When β293A/T-rich mut was cotransfected with NTEF-1, its expression levels increased by 3.1-fold, which was likely due to NTEF-1 binding to the dMCAT and pMCAT elements. Similarly, when β293A/T-rich mut was cotransfected with either RTEF-1a, RTEF-1b, or DTEF-1, expression levels increased from 2.1- to 2.2-fold (Fig. 5B). Furthermore, cotransfection of the β293d/pMCAT mut with the TEF proteins revealed a 2.1- to 2.3-fold increase in expression, indicating the importance of TEF proteins binding to the βA/T-rich element (Fig. 5B). These experiments done in mouse C2C12 myotubes indicate the functional importance of the βA/T-rich element in the context of its natural promoter, since its mutation decreased basal expression by 33.3% and the binding of TEF proteins at the βA/T-rich element conferred a 2.1- to 2.4-fold increase in expression (Fig. 5B). Similar results were obtained using rat βMyHC constructs (unpublished observation).
MEF2 elements compete for TEF protein binding.
The βA/T-rich element is a composite GATA/MEF2-like element. Our previous study revealed that oligonucleotides containing consensus GATA sites could not compete for complex formation when the human βA/T-rich element was reacted with CP or MOV-P nuclear extract, whereas oligonucleotides containing MEF2 sites partially competed (52). To ascertain if GATA or MEF2 sites could interfere with TEF protein binding to the human βA/T-rich element, we performed competition EMSAs using oligonucleotides containing either GATA or MEF2 sites (Fig. 6). A binding complex did not form when the 32P-labeled human βA/T-rich element was reacted with UL (Fig. 6, lanes 1, 13, and 25), whereas specific complexes did form when the 32P-labeled human βA/T-rich element was reacted with in vitro-synthesized NTEF-1 (Fig. 6A, lane 1 versus 2), RTEF-1a (Fig. 6B, lane 13 versus 14), or RTEF-1b (Fig. 6C, lane 25 versus 26). The addition of 100-fold molar excess cold wild-type βA/T-rich element to the binding reaction mixture abolished complex formation (Fig. 6, lanes 2 versus 3, 14 versus 15, and 26 versus 27), while the addition of 100-fold molar excess cold βA/T-mut element did not compete (Fig. 6, lanes 2 versus 4, 14 versus 16, and 26 versus 28). Addition of 100-fold excess cold αMyHC GATA, cTnC GATA, B-type natriuretic peptide (BNP) GATA, or Trex (shown not to bind NTEF-1 [16, 50]) to binding reaction mixtures did not abolish complex formation (Fig. 6A, lanes 2 versus 5 to 7 and lane 10; B, lanes 14 versus 17 to 19 and lane 22; C, lanes 26 versus 29 to 31 and lane 34). The addition of either 100-fold molar excess cold desmin MEF2 or βMyHC dMCAT, to which all TEF proteins avidly bind (50), to binding reaction mixtures completely abolished complex formation (Fig. 6A, lanes 2 versus 8 or 11; B, lanes 14 versus 20 or 23; C, lanes 26 versus 32 or 35), while addition of the myoglobin MEF2 element partially competed for complex formation (Fig. 6A, lane 2 versus 9; B, lane 14 versus 21; C, lane 26 versus 33). The failure of the αMyHC, cTnC, and BNP GATA elements to compete for TEF protein-βA/T-rich complex formation supports the notion that TEF proteins do not bind consensus GATA sites; however, effective competition by the desmin and myoglobin MEF2 elements suggests the possibility that TEF proteins can bind these MEF2 elements (Fig. 6).
FIG. 6.
EMSA analysis of in vitro-transcribed and translated NTEF-1, RTEF-1a, and RTEF-1b proteins. 32P-labeled βA/T-rich element was reacted with 2 μl of in vitro-synthesized NTEF-1 (A, lane 2), RTEF-1a (B, lane 14), and RTEF-1b (C, lane 26). For competition assays, the following nonradioactive oligonucleotides were added to the binding reactions at 100-fold molar excess prior to the addition of the 32P-labeled βA/T-rich probe: βA/T-rich wild type (lanes 3, 15, and 27); βA/T-rich mut (lanes 4, 16, and 28); α-MyHC GATA (lanes 5, 17, and 29); cTnC GATA (lanes 6, 18, and 30); BNP GATA (lanes 7, 19, and 31); desmin MEF2 (lanes 8, 20, and 32); myoglobin MEF2 (lanes 9, 21, and 33); MCK Trex (lanes 10, 22, and 34); βMyHC dMCAT (lanes 11, 23, and 35); free probe (lanes 12, 24, and 36). Notably, the addition of the desmin MEF2 element to binding reactions containing NTEF-1 (lane 8), RTEF-1a (lane 20), and RTEF-1b (lane 32) resulted in an unexpected inhibition of complex formation.
EMSA analysis reveals that TEF proteins bind MEF2 elements located within muscle and nonmuscle genes.
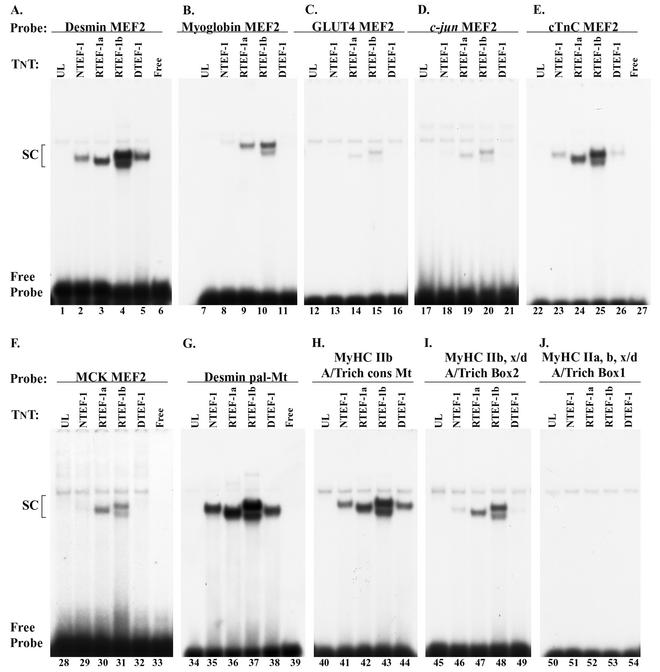
Because the desmin MEF2 element acted as a potent competitor of TEF protein binding to the human βA/T-rich element, we performed an EMSA analysis using in vitro-synthesized TEF protein to determine if TEF proteins could bind the desmin MEF2 element, as well as a variety of other MEF2 elements (Fig. 7). As can be seen clearly in Fig. 7, when the 32P-labeled desmin MEF2, cTnC MEF2, desmin pal-Mt element, fast myosin heavy chain IIb (MyHC IIb) Mt element consensus (GGTATTT), and MyHC IIb and IIx/d A/T-rich box 2 were reacted with in vitro-synthesized TEF proteins a complex formed (Fig. 7A, E, G, H, and I). On the other hand, only in vitro-synthesized RTEF1a and -1b bound the 32P-labeled myoglobin, GLUT4, c_-jun_, and MCK MEF2 elements (Fig. 7B, C, D, and F), and none of the in vitro-synthesized TEF proteins bound the 32P-labeled A/T-rich box 1 site, which is present in all three fast MyHC promoter regions (Fig. 7J). Taken together, these data show that TEF proteins differentially bind to a select set of MEF2 and A/T-rich elements and raise intriguing questions concerning the full range of physiological functions served by the TEF proteins.
FIG. 7.
EMSA analysis of TEF protein binding to a variety of MEF2 and A/T-rich elements. Binding reaction mixtures contained 2 μl of in vitro-synthesized NTEF-1, RTEF-1a, RTEF-1b, or DTEF-1 protein and the following 32P-labeled MEF2 oligonucleotides: desmin MEF2 (A), myoglobin MEF2 (B), GLUT4 MEF2 (C), c-jun MEF2 (D), cTnC MEF2 (E), MCK MEF2 (F), desmin pal-Mt (G), MyHC IIb A/T-rich cons Mt (H), MyHC IIb and IIx/d A/T-rich box 2 (I), and MyHC IIb, x/d, a, A/T-rich box 1 (J). With the exception of the MyHC IIb, x/d, a, A/T-rich box 1, TEF protein bound differentially to all of the elements tested. The sequence position (numbering) for each oligonucleotide is shown in Table 1. The desmin pal-Mt denotes an element located within the desmin enhancer that overlaps the desmin MEF2 site, is present as a palindrome, and is required for high-level expression of desmin reporter genes in myotubes (Mt) (17).
Enriched binding of nominal TEF-1 to several MEF2 elements when using MOV-P nuclear extract.
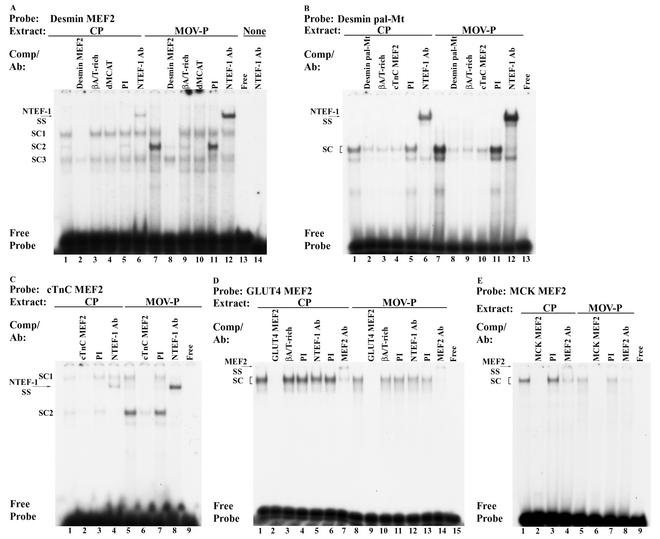
Because our EMSA analysis revealed binding of in vitro-synthesized TEF isoproteins to the cTnC and desmin MEF2 elements as well as the desmin pal-Mt element (Fig. 7A, E, and G), it was important to determine whether these elements were bound by NTEF-1 when reacted with CP or MOV-P nuclear extract. EMSA analysis of binding reactions containing the 32P-labeled desmin MEF2 element and 4 μg of CP or MOV-P nuclear extract revealed the formation of three binding complexes (SC1, SC2, and SC3) (Fig. 8A, lanes 1 and 7). Addition of 100-fold molar excess cold wild-type desmin MEF2 element abolished formation of complex SC1 and SC2 when using CP and MOV-P nuclear extract, indicating that only these two complexes were specific to the desmin element (Fig. 8A, lanes 2 and 8). In contrast, addition of 100-fold molar excess cold human βA/T-rich or dMCAT elements competed for complex SC2 only, indicating that this complex was comprised of TEF protein (Fig. 8A, lanes 3 and 4 and lanes 9 and 10). Importantly, complex SC2 was greatly enriched only when using MOV-P nuclear extract (Fig. 8A, lane 1 versus 7). Binding complexes were not altered by addition of PI serum (Fig. 8A, lanes 5 and 11), whereas addition of monoclonal NTEF-1 antibody to binding reaction mixtures containing CP or MOV-P nuclear extract resulted in a supershift of complex SC2 (Fig. 8A, lanes 6 and 12). Lane 13 shows free probe, while lane 14 shows that NTEF-1 antibody reacted with free probe did not form a complex.
FIG. 8.
EMSA analysis of CP and MOV-P skeletal muscle nuclear extract binding at the human βA/T-rich element. The 32P-labeled oligonucleotides used were desmin MEF2 (A), desmin pal-Mt (B), cTnC MEF2 (C), GLUT4 MEF2 (D), and MEK MEF2 (E) elements incubated with 4 μg of either CP (lanes 1 to 6) or MOV-P (lanes 7 to 12) nuclear extract. Unlabeled competitor oligonucleotides harboring wild-type desmin MEF2, βA/T-rich, dMCAT, desmin pal-Mt, cTnC MEF2, GLUT4 MEF2, and MCK MEF2 binding elements were added at 100-fold molar excess. All TEF proteins avidly bind the dMCAT element, which was used as a positive control for TEF protein binding. Antibody supershift EMSAs were performed by preincubation of CP or MOV-P nuclear extract with 2 μl of PI serum, NTEF-1, or general monoclonal MEF2 antibody (MEF2 Ab) for 30 min at room temperature. Note the enriched binding of NTEF-1 at the desmin MEF2, desmin pal-Mt, and cTnC MEF2 elements only when using MOV-P nuclear extract (A to C), whereas MEF2 comprises the binding complex formed at the GLUT4 and MCK MEF2 elements, and this binding decreases when using MOV-P nuclear extract (D and E).
EMSA analysis of binding reactions containing the 32P-labeled desmin pal-Mt element and 4 μg of CP or MOV-P nuclear extract revealed formation of one binding complex (SC) (Fig. 8B, lanes 1 and 7). Addition of 100-fold molar excess cold wild-type desmin pal-Mt element abolished complex formation when using CP and MOV-P nuclear extract, indicating the specificity of this complex to the desmin pal-Mt element (Fig. 8B, lanes 2 and 8). Interestingly, addition of 100-fold molar excess cold βA/T-rich or cTnC MEF2 elements abolished complex formation, indicating that this complex was comprised of TEF protein (Fig. 8B, lanes 3 and 4 and lanes 9 and 10). Importantly, complex SC was greatly enriched only when using MOV-P nuclear extract (Fig. 8B, lane 1 versus 7). Binding complexes were not altered by addition of PI serum (Fig. 8B, lanes 5 and 11). Addition of monoclonal NTEF-1 antibody to binding reactions containing CP or MOV-P nuclear extract resulted in a supershift of this complex (Fig. 8B, lanes 6 and 12). Lane 13 shows free probe.
When 32P-labeled cTnC MEF2 element was reacted with 4 μg of CP or MOV-P nuclear extract, two specific complexes (SC1 and SC2) formed (Fig. 8C, lanes 1 and 5). Addition of 100-fold molar excess cold wild-type cTnC MEF2 element abolished formation of complex SC1 and SC2 when using CP and MOV-P nuclear extract, indicating that these complexes were specific (Fig. 8C, lanes 2 and 6). Notably, complex SC2 was greatly enriched only when using MOV-P nuclear extract (Fig. 8C, lane 1 versus 5). Binding complexes were not altered by addition of PI serum (Fig. 8C, lanes 3 and 7), while addition of NTEF-1 antibody resulted in a supershift of complex SC2 (Fig. 8C, lanes 4 and 8). These data provide conclusive evidence that NTEF-1 specifically binds the desmin MEF2 and pal-Mt elements as well as the cTnC MEF2 element and that this binding is notably enriched only when using MOV-P nuclear extract.
MCK and GLUT4 MEF-2 elements bind MEF2 protein when reacted with adult skeletal muscle nuclear extract.
Previous work has shown that the MCK and GLUT4 MEF2 elements bind MEF2 protein (28, 30, 33, 45), while our present EMSA analysis has shown that these two MEF2 elements can bind in vitro-synthesized RTEF-1a and RTEF-1b, albeit weakly (Fig. 7C and F). Because these two elements are capable of binding both MEF2 and TEF proteins, we performed an EMSA analysis to determine whether these elements bound NTEF-1 when using CP or MOV-P nuclear extract (Fig. 8D and E). EMSA analysis of binding reactions containing the 32P-labeled GLUT4 MEF2 element and 4 μg of CP or MOV-P nuclear extract revealed formation of a single binding complex (SC1) that decreased when using MOV-P nuclear extract (Fig. 8D, lane 1 versus 8). Addition of 100-fold molar excess cold wild-type GLUT4 element abolished complex formation when using CP and MOV-P nuclear extracts (Fig. 8D, lanes 2 and 9). In contrast, the addition of 100-fold excess cold human βA/T-rich element did not compete for complex formation, indicating the specificity of complex formation for the GLUT4 element and that this complex is likely not comprised of TEF-1 protein (Fig. 8D, lanes 3 and 10). Binding complexes were not altered by addition of PI serum (Fig. 8D, lanes 4, 6, 11, and 13) or by addition of monoclonal NTEF-1 antibody (Fig. 8D, lanes 5 and 12) to binding reactions containing CP or MOV-P nuclear extract, whereas the addition of general monoclonal MEF2 antisera immunodepleted complex formation (Fig. 8D, lanes 7 and 14). Lane 15 shows free probe.
Similarly, EMSA analysis of binding reactions containing the 32P-labeled MCK MEF2 element and 4 μg of CP or MOV-P nuclear extract revealed formation of a single binding complex that decreased when using MOV-P nuclear extract (Fig. 8E, lane 1 versus 5). Addition of 100-fold molar excess cold wild-type MCK element abolished complex formation when using CP and MOV-P nuclear extract (Fig. 8E, lanes 2 and 6). Binding complexes were not altered by addition of PI serum (Fig. 8E, lanes 3 and 7), while the addition of general monoclonal MEF2 antibody (Fig. 8E, lanes 4 and 8) to binding reactions containing CP or MOV-P nuclear extract immunodepleted complex formation. Lane 9 shows free probe. When considered with our results shown in Fig. 4, these data reveal that both NTEF-1 and MEF2 are present within adult CP and MOV-P nuclear extracts and that binding site preference clearly exists.
NTEF-1 preferentially binds the desmin pal-Mt element, while MEF2 prefers the MCK MEF2 element.
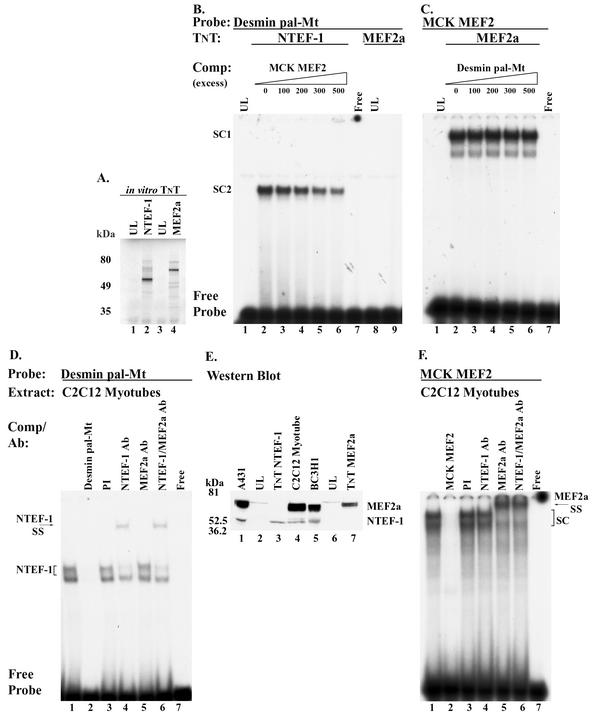
Our present work shows that TEF protein binds both the desmin MEF2 element, which contains an Mt sequence (GGTATTT), and the pal-Mt element when using in vitro-synthesized TEF protein and CP and MOV-P nuclear extracts (Fig. 7A and G; Fig. 8A and B). In contrast, even though in vitro-synthesized RTEF-1 proteins can bind the MCK MEF2 element, only MEF2 protein binds this element when reacted with CP or MOV-P nuclear extract (Fig. 7F; Fig. 8E). It is noteworthy that the desmin pal-Mt element is contained within each of the three tandem sequences used to generate MEF2-sensor mice, mice frequently used to assess the transcriptional activating function of MEF2 proteins in determining basal muscle fiber phenotype and muscle fiber type plasticity induced by various modes of muscle activity, including MOV (15, 35, 55-57). Therefore, it was important to determine if in vitro-synthesized MEF2 protein was capable of binding the pal-Mt element. EMSA analysis of binding reactions containing the 32P-labeled desmin pal-Mt element and in vitro-synthesized NTEF-1 (Fig. 9A inset, lane 2) revealed the formation of a specific binding complex that was partially competed away only when cold MCK MEF2 element was added to the binding reaction mixtures in great excess (300- to 500-fold excess) (Fig. 9B, lanes 1 to 6). Interestingly, when the 32P-labeled desmin pal-Mt element was reacted with in vitro-synthesized MEF2a (Fig. 9A inset, lane 4), a complex was not formed (Fig. 9B, lane 8 versus 9). In contrast, when the 32P-labeled MCK MEF2 element was reacted with in vitro-synthesized MEF2a, a robust low-mobility complex was formed that was not competed for by the addition of a 500-fold molar excess cold desmin pal-Mt element (Fig. 9C, lanes 1 to 6).
FIG. 9.
EMSA analysis of TEF binding to desmin pal-Mt and MCK MEF2 elements. (A) The inset shows the correct size of [35S]methionine-labeled in vitro-synthesized NTEF-1 and MEF2a. Rabbit reticulocyte lysate system was programmed with 1 μg of circular NTEF-1 or MEF2a in the presence of [35S]methionine. The transcription-translation product was resolved by SDS-PAGE and exposed to film. Molecular mass markers (in kilodaltons) are shown to the left. Lanes 1 and 3, marked UL, represent parallel reactions not programmed with NTEF-1 or MEF2a expression plasmid. (B) EMSA of 32P-labeled desmin pal-Mt element reacted with in vitro-synthesized human NTEF-1 protein (lanes 2 to 6) or MEF2a protein (lane 9). Unlabeled competitor oligonucleotide harboring a wild-type MCK MEF2 element (lanes 2 to 6) was added at a 0- to 500-fold molar excess. (C) EMSA of 32P-labeled MCK MEF2 element reacted with in vitro-synthesized MEF2a protein (lanes 2 to 6). Unlabeled competitor oligonucleotide harboring a wild-type desmin pal-Mt element (lanes 2 to 6) was added at a 0- to 500-fold molar excess. (D) EMSA of 32P-labeled desmin pal-Mt element reacted with 4 μg of C2C12 myotube nuclear extract (lanes 1 to 7). Antibody supershift EMSA was performed by preincubation of 4 μg of C2C12 myotube nuclear extract with 2 μl of PI serum (lane 3), NTEF-1 (lane 4), MEF2a (lane 5), or NTEF-1 and MEF2a (lane 6) antibody for 30 min at room temperature. (E) Western blot analysis using A431 extract (lane 1), TnT NTEF-1 product (lane 3), 30 μg of C2C12 myotube nuclear extract (lane 4), BC3H1 extract (lane 5), and TnT MEF2a product (lane 7). A431 and TnT NTEF-1 served as a positive control for the presence of NTEF-1 protein in C2C12 myotube nuclear extract, while BC3H1 extract and TnT MEF2a were controls for MEF2 protein. UL (lanes 2 and 6) was used as a control for nonspecific cross-reaction of the NTEF-1 or MEF2a antibody. NTEF-1 protein was increased in the MOV-P nuclear extract compared to the CP nuclear extract. (F) EMSA of 32P-labeled MCK MEF2 element reacted with 4 μg of C2C12 myotube nuclear extract (lanes 1 to 7). Antibody supershift EMSA was performed by preincubation of 4 μg of C2C12 myotube nuclear extract with 2 μl of PI serum (lane 3), NTEF-1 (lane 4), MEF2a (lane 5), or NTEF-1 and MEF2a (lane 6) antibody for 30 min at room temperature. Free probe represents excess unreacted radiolabeled oligonucleotide.
In a recent study, the desmin pal-Mt element was shown to form a specific binding complex that was not abolished by 50-molar excess cold consensus MCK MEF2 element when reacted with myogenic cell (C2,7) nuclear extract (17). Interestingly, our present work shows that NTEF-1 can bind the desmin pal-Mt element, which is only partially abolished by the addition of 500-fold molar excess cold consensus MCK MEF2 element (Fig. 9B, lanes 1 to 6). Thus, to determine whether NTEF-1 represents the previously reported unknown desmin pal-Mt binding factor (MtBF), we performed an EMSA analysis using nuclear extract isolated from C2C12 myotubes. When the 32P-labeled desmin pal-Mt element was reacted with 4 μg of C2C12 nuclear extract, two specific binding complexes formed that were abolished by addition of 100-fold molar excess cold Mt element to the binding reaction mixture (Fig. 9D, lane 1 versus 2). Binding complexes were not altered by addition of PI serum, while the addition of monoclonal NTEF-1 antibody to binding reactions resulted in a supershift of only the top band (Fig. 9D, lane 4). The addition of MEF2 antibody did not supershift or immunodeplete either complex, indicating that MEF2 protein was not a component of either binding complex (Fig. 9D, lane 5). The addition of both anti-NTEF-1 and anti-MEF2 antibody to binding reactions resulted in a supershift of the top band, similar to that seen when only NTEF-1 was added, indicating that NTEF-1 is the sole protein comprising this binding complex (Fig. 9D, lanes 4 and 6). Lane 7 shows free probe.
It was possible that we did not detect MEF2 binding to the desmin pal-Mt element, because MEF2 protein was not in our C2C12 nuclear extract. To determine if our isolated C2C12 nuclear extract contained MEF2 and NTEF-1 protein, we performed a Western blot analysis using C2C12 nuclear extract and anti-NTEF-1 and anti-MEF2 antibodies (Fig. 9E). Using NTEF-1 antibody, a 53-kDa band was detected in A431 and BC3H1 extracts (Fig. 9E, lanes 1 and 5) as well as in C2C12 nuclear extract (Fig. 9E, lane 4) that corresponded to in vitro-synthesized NTEF-1 (Fig. 9E, TnT lane 3). Likewise, the use of MEF2a antibody detected a band of approximately 65 kDa that corresponded to in vitro-synthesized MEF2A (Fig. 9E, TnT lane 7) and was present in all the cell types of nuclear extracts examined (Fig. 9E, lanes 1, 4, and 5). Therefore, when these data are considered with our EMSA analysis shown in Fig. 8B, they indicate that the desmin pal-Mt element does not bind in vitro-synthesized MEF2a protein or MEF2 proteins within CP, MOV-P, and C2C12 nuclear extracts.
To determine if our isolated C2C12 nuclear extract contained MEF2 that was capable of binding a genuine MEF2 element, we performed an EMSA analysis using the MCK MEF2 element as probe and C2C12 myotube nuclear extract (Fig. 9F). When the 32P-labeled MCK MEF2 element was reacted with 4 μg of C2C12 nuclear extract, a specific binding complex formed that was abolished by addition of 100-fold molar excess cold MCK MEF2 element to the binding reaction mixture (Fig. 9F, lane 1 versus 2). Binding complexes were not altered by addition of PI serum or NTEF-1 antibody (Fig. 9F, lanes 3 and 4), while the addition of MEF2 antibody to binding reactions resulted in a supershift of the specific binding complex (Fig. 9F, lane 5). The addition of both NTEF-1 and MEF2 antibodies to binding reactions resulted in a supershift of the specific complex, similar to that seen when only MEF2a antibody was added, indicating that MEF2a is the sole protein comprising this binding complex (Fig. 9D, lanes 5 and 6). Lane 7 shows free probe.
βMyHC βA/T-rich element, desmin MEF2, and pal-Mt sites are capable of driving the expression of a heterologous promoter in C2C12 myotubes.
Because our yeast one-hybrid screen and EMSA analyses identified TEF protein as the enriched βA/T-rich binding activity in MOV-P nuclear extracts, it was important to determine if the βA/T-rich element alone could confer TEF responsiveness onto a heterologous promoter. The luciferase activity of the pRL-TK (thymidine kinase) plasmid was low and exhibited only a nonsignificant increase when cotransfected with 100 ng of NTEF-1 (Fig. 10B). The basal expression level of four tandem wild-type βA/T-rich elements was twofold higher than that of pRL-TK, and its expression levels increased an additional threefold when cotransfected with 100 ng of NTEF-1 (Fig. 10). Mutation of βA/T-rich elements decreased basal expression levels similar to those of pRL-TK plasmid, with or without the addition of NTEF-1 (Fig. 10B).
FIG. 10.
Response of wild-type concatenated βMyHC A/T-rich and desmin MEF2 and pal-Mt elements to increasing amounts of TEF protein. (A) C2C12 myoblasts were transfected with βMyHC 4x-AT-rich wt, desmin 3x-MEF2 wt, or desmin 3x-pal-Mt wt reporter constructs (2 μg) and increasing amounts of TEF expression vector (CMV-driven pXJ40-NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1), and subjected to differentiation media to form myotubes. Increasing amounts of TEF-1 protein expression led to squelching. Luciferase-normalized RLU (RLU/β-Gal ratio) were expressed as the mean ± standard error (n = 6). (B) TEF transactivates βMyHC A/T-rich and desmin MEF2 and pal-Mt elements in C2C12 myotubes. The addition of 100 ng of TEF isoforms (CMV-driven pXJ40-NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1) significantly transactivated wild-type A/T-rich, MEF2, and pal-Mt elements compared to levels seen using vector alone (pRL-TK) or basal levels (-TEF) observed when using reporter constructs without cotransfection with TEF isoforms. Mutation of the A/T-rich and MEF2 elements in reporter constructs significantly reduced expression to levels observed when using pRL-TK vector alone, and mutation of the pal-Mt elements significantly reduced expression 85% in cotransfection experiments. All data were normalized using β-Gal activity to accommodate variations in transfection efficiency. Data are reported as luciferase-normalized RLU (RLU/β-Gal ratio) and are expressed as the mean ± standard error (n = 11 for A/T-rich and MEF2 plasmid; n = 7 for pal-Mt constructs). *, P < 0.001.
Because MEF2 has been shown by others (27) to bind the desmin MEF2 element and herein we have shown that TEF proteins bind this element, it was necessary to determine if TEF proteins could transactivate the TK promoter carrying tandem copies of wild-type desmin MEF2 elements. The basal expression level of three concatenated wild-type desmin MEF2 elements was 2.6-fold higher than that of pRL-TK, and its expression levels increased 3-fold further when cotransfected with 100 ng of NTEF-1 (Fig. 10B). Mutation of desmin MEF2 elements decreased basal expression levels to levels similar to those of the pRL-TK plasmid, with or without the addition of NTEF-1. Likewise, the basal expression level of three concatenated wild-type desmin pal-Mt elements was 4.2-fold higher than that of pRL-TK, and its expression levels increased 3.8-fold further when cotransfected with 100 ng of NTEF-1 (Fig. 10B). Mutation of desmin pal-Mt elements decreased basal expression levels to levels similar to those of pRL-TK plasmid, with or without the addition of NTEF-1. These data provide clear evidence that binding of NTEF-1 to the desmin MEF2 and pal-Mt elements or βMyHC βA/T-rich element is of functional significance, and they also demonstrate that the βA/T-rich element is capable of driving the expression of a heterologous promoter in C2C12 myotubes.
DISCUSSION
This study reports the novel finding that NTEF-1 represents the enriched binding activity that forms at the βA/T-rich element when reacted with MOV-P nuclear extract (52), and it verifies the functional importance of this binding in transient-expression studies by using βMyHC reporter genes and C2C12 myotubes. Our data reveal that nuclear and in vitro-synthesized TEF proteins bind specifically to a variety of muscle gene MEF2 elements that include the desmin MEF2 and pal-Mt elements, regulatory sites that comprise the MEF2-dependent transgene within the widely studied desmin MEF2-sensor transgenic mice, suggesting that the expression of this transgene likely reflects the transcriptional activity of both TEF and MEF2 proteins. Furthermore, these novel data provide evidence suggesting that TEF may represent the previously unidentified myoglobin A/T-binding factor 35 (ATF35) and desmin MtBF (17, 18). Importantly, these data also provide new supporting evidence suggesting that TEF proteins may play a more comprehensive role than anticipated in the regulation of striated muscle gene expression and in response to various modes of altered muscle activity, including MOV, exercise, and zero gravity.
The βMyHC βA/T-rich element is a target binding site for TEF proteins.
Using the yeast one-hybrid system to screen both control and MOV-P adult skeletal muscle cDNA libraries, we identified NTEF-1 and RTEF-1b as genuine βMyHC βA/T-rich binding proteins. This finding was unexpected, because the βA/T-rich element (5′-GGAGATATTTTT-3′) is a composite GATA/MEF2-like element and the typical target sites for TEF protein binding are MCAT elements (5′-CATTCCT-3′). Although the nucleotide composition between these two regulatory elements is quite divergent, differences among genuine TEF protein binding sites are not new, as exemplified by the simian virus 40 enhancer GTIIC and _Sph_I/II elements (22, 26). Likewise, it has been shown that multiple different factors can bind the same A/T-rich element, such as the βA/T-rich studied herein, and that this binding may differ in accordance with developmental stage and/or cell type. For example, both MEF2 and an unknown factor termed ATF-35 were found to bind the A/T-rich element within the myoglobin gene, while HF-1b bound the rat slow myosin light chain 2 A/T-rich element (18, 61). Also, accumulating evidence has established that the nucleotides flanking cis elements, such as the MCAT, E-box, and MEF2, can exert a profound effect on transcription factor binding specificity and affinity (5, 20, 25). This latter point was clearly illustrated when transgenic mice carrying transgenes comprised of tandem arrays of CArG box elements from two different genes displayed different expression patterns which could partially be attributed to differences in flanking nucleotides (10).
Although the binding at the βMyHC βA/T-rich element that we identified in this study is unique, we have provided multiple lines of evidence that TEF proteins bind specifically to this element. First, our direct and supershift EMSA results provide conclusive evidence that TEF specifically interacts with the human βA/T-rich element by showing binding of in vitro-synthesized NTEF-1, RTEF-1a, RTEF-1b, and DTEF-1 proteins and that this observation is not unique to the human βA/T-rich element, since each of the TEF isoproteins also bound both the rat and mouse βA/T-rich elements (Fig. 2). Second, our competition scanning mutagenesis EMSA analyses provide evidence that both in vitro-synthesized NTEF-1 and RTEF-1b interact strongly with the nucleotides comprising the MEF2-like homology of the human βA/T-rich element, with weaker interactions occurring across the GATA-like homology (Fig. 3). Importantly, this result confirmed our previous footprint analysis using MOV-P nuclear extract (52). Third, in competition EMSAs, oligonucleotides containing either the αMyHC, cTnC, or BNP GATA elements did not compete for binding of in vitro-synthesized NTEF-1, RTEF-1a, or RTEF-1b to the human βA/T-rich element, as would be expected based on our previous findings that GATA protein, whether in vitro-synthesized or within CP or MOV-P nuclear extracts, did not bind this element (Fig. 6) (52). Most important in this EMSA evidence that TEF proteins are binding the βA/T-rich element were the observations that the high-affinity TEF binding human βMyHC dMCAT element (50) completely abolished complex formation, whereas the MCK Trex element (16), an MCAT-like element which does not bind TEF, could not compete for binding (Fig. 6). Complete elimination of complex formation when using the dMCAT was also observed when using CP and MOV-P nuclear extract (Fig. 4A).
Our antibody EMSA also revealed that NTEF-1 comprised the specific enriched binding complex that formed when using MOV-P nuclear extract, indicating that NTEF-1 binding to the βA/T-rich element likely plays a role in βMyHC induction by MOV. Consistent with this notion, NTEF-1 expression was increased in response to MOV, as assessed by Northern and Western analyses (Fig. 1). Furthermore, others have implicated NTEF-1 in conferring inducible expression (muscle loading and hormonal stimuli) to several muscle-specific genes, including the βMyHC gene (26). Another notable observation important to our ongoing studies was the formation of an additional higher-mobility specific binding complex that formed only when using MOV-P, but not CP, nuclear extract (Fig. 4A). Furthermore, this complex was neither competed for by the dMCAT element nor supershifted by NTEF-1 antibody, indicating that this complex was not comprised of a TEF protein (Fig. 4A). The identity of the protein(s) that forms this complex remains to be determined but likely corresponds to the unknown 44-kDa protein previously reported by our investigators (52). Although this protein binds the A/T-rich element independently, it is conceivable that it functions collaboratively with NTEF-1 to modulate muscle-specific induction of βMyHC in response to MOV. An example of collaborative interactions with the TEF proteins has been shown for the muscle-specific transcription of the chicken cTnT gene, which required the concurrent binding of NTEF-1 to the MCAT core sequence and poly(ADP-ribose) polymerase to the MCAT 5′-adjacent sequence (8). Likewise, we have shown the cobinding of Max, poly(ADP-ribose) polymerase, and NTEF-1 at the dMCAT element as a requirement of high-level expression of a βMyHC transgene, and this binding complex is enriched under MOV conditions (50). Other TEF interaction proteins that have been reported are TONDU, a mammalian homologue of the Drosophila melanogaster vestigial gene and Yes-associated protein (49). In fact, the requirement for interacting protein modulation of TEF transcriptional activity has been repeatedly reported (4, 22, 26, 49). Clear illustrations of this exist in experiments using cells that do not normally contain TEF protein. In these cells, cotransfections with TEF did not result in activation of reporter genes, whereas in cells that do normally express TEF protein cotransfection with increasing amounts of TEF resulted in squelching (down-regulation of reporter gene expression), indicating that TEF cofactors are not only required but also are only available in limited amounts (22, 26).
Finally, our cell culture experiments done in C2C12 myotubes demonstrate the functional importance of the βA/T-rich element in directing TEF-dependent expression of the βMyHC (Fig. 5; Fig. 10). Although mutation of the βA/T-rich element in the context of the 293-bp βMyHC promoter/CAT reporter gene did not completely eliminate expression as observed in our transgenic mouse studies (51), it did significantly lower basal expression levels in C2C12 myotubes (Fig. 5A). This difference can likely be accounted for based on tighter regulatory control mechanisms imposed on the 293-bp βMyHC transgene when in the context of native chromatin. In our transient-expression assays, TEF-dependent regulation of the βMyHC promoter was easily identified based on two prominent features directly attributable to TEF regulation, including (i) increased expression above basal levels of a 293-bp βMyHC/CAT reporter construct devoid of MCAT elements in response to overexpression of NTEF-1 (Fig. 5B) and (ii) squelching of reporter gene activity concurrent with increasing amounts of cotransfected TEF protein (Fig. 5A). This was further substantiated in our experiments using constructs harboring tandem βA/T-rich elements fused to a minimal TK promoter devoid of TEF binding sites, which revealed significant TEF-dependent activation that was completely eliminated by mutation of βA/T-rich elements (Fig. 10B). Furthermore, this TEF-dependent expression decreased as the amount of TEF protein increased (Fig. 10A). These data provide substantial evidence in support of TEF proteins specifically binding to the βMyHC proximal promoter βA/T-rich element and that this interaction likely contributes to βMyHC induction in response to MOV.
The binding of TEF proteins to MEF2 elements is unexpected.
Previous studies have demonstrated that the desmin MEF2 element binds both nuclear (C2C12) and in vitro-translated MEF2 protein (27) yet, despite this, when the desmin MEF2 element was used as a competitor in our EMSAs we found that it completely abolished complex formation between the βA/T-rich element and in vitro-synthesized TEF protein (Fig. 6). These seemingly contradictory results were surprising, since our previous work showed that in vitro-synthesized MEF2c as well as MEF2 protein within CP or MOV-P nuclear extracts were not components of the enriched binding complex that formed at the βA/T-rich element (52), which was again confirmed by our present antibody EMSA result (Fig. 4B). Nevertheless, this finding was intriguing since it suggested that the desmin MEF2 element could bind TEF protein. Notably, the desmin MEF2 element is overlapped by the Mt element (GGTATTT), which is required for high-level expression in myotubes (17). The factor that binds this element (MtBF) was not identified; however, their EMSA analysis using C2,7 myotube nuclear extract showed a mobility pattern that closely resembled our EMSA pattern when using the βA/T-rich element and in vitro-synthesized TEF proteins, as well as CP and MOV-P nuclear extracts (17) (Fig. 2A and 4). As anticipated, we found that TEF protein bound avidly to the desmin MEF2 and pal-Mt elements, suggesting that the unknown MtBF is TEF (Fig. 7A and G). The binding of TEF protein to the desmin MEF2 and Mt elements was shown to be of functional significance in our transient-coexpression assays, wherein constructs harboring tandem desmin MEF2 or pal-Mt elements fused to a minimal TK promoter displayed significant increases in expression in response to overexpressed NTEF-1 protein, and this expression was completely eliminated by mutation of these elements (Fig. 10B). Further, TEF-dependent expression decreased as the amount of TEF protein increased (Fig. 10A). Of particular interest is that the Mt element is present as a palindrome (pal-Mt) in the desmin MEF2 transgene harbored within the MEF2-sensor mouse, a transgenic model that has been used to monitor the transcriptional activation function of MEF2 proteins under conditions that induce skeletal muscle hypertrophy and fiber type shifts (15, 35, 55-57).
Given the high degree of nucleotide sequence homology shared among MEF2 elements and the fact that numerous MEF2 elements contain overlapping Mt sites (Table 1), it was logical to suspect that TEF proteins could interact with other muscle gene MEF2 elements. This deduction proved true, as can be seen in our EMSA analyses which demonstrate binding of in vitro-synthesized TEF proteins at MEF2 elements located within the control region of fast (GLUT4, MCK, MyHC IIb, and MyHC IIx/d) and slow (βMyHC, myoglobin, and cTnC) muscle genes, as well as the nonmuscle c-jun gene (Fig. 7A to I). Of interest, the myoglobin MEF2 element used herein has been reported to bind MEF2 as well as an unknown factor called ATF35, which our data would suggest may be TEF. We base this on several considerations: (i) an initial estimated mass of 49 kDa based on UV cross-linking studies corresponds to the approximate mass of TEF proteins (13, 18, 22, 26, 50); (ii) when using Sol8 or C2C12 myotube versus myoblast nuclear extract, ATF35 binding was more robust, a characteristic also reported for the desmin pal-Mt element, which we showed herein binds TEF protein; and (iii) in competition EMSAs, high levels of unlabeled MCK MEF2 element are required to compete for ATF35 binding (18), which is similar to our results using the desmin pal-Mt element (Fig. 9B).
This broad-spectrum EMSA analysis of both fast and slow muscle gene MEF2 elements revealed that each TEF protein bound differentially to the same MEF2 element as well as between the different MEF2 elements, suggesting that the TEF proteins likely serve independent and overlapping functions (Fig. 7). The differential binding of TEF protein between MEF2 elements was also seen when using nuclear extracts isolated from adult skeletal muscle. In addition to showing enriched NTEF-1 binding at the βMyHC βA/T-rich element, a gene representative of slow-fiber expression, we also observed enriched NTEF-1 binding at the desmin pal-Mt element as well as the cTnC and desmin MEF2 elements only when using MOV-P nuclear extract (Fig. 4; Fig. 8A to C). In contrast, even though both RTEF-1a and RTEF-1b can bind the MCK and GLUT4 MEF2 elements, genes representative of fast glycolytic fibers, when these two elements were reacted with CP and MOV-P nuclear extracts only MEF2 protein bound, and this binding decreased in response to MOV (Fig. 8D and E). Our result suggests that a likely determinant of TEF versus MEF2 protein interaction with these two elements is binding affinity, as demonstrated in our EMSAs, wherein the binding of in vitro-synthesized TEF protein to the desmin pal-Mt element was only partially disrupted by competition with excess MCK MEF2 element and the desmin pal-Mt element could not compete for in vitro-synthesized MEF2a binding to the MCK MEF2 element (Fig. 9A to C). Furthermore, although our Western analysis showed that C2C12 myotube nuclear extract contains both TEF and MEF2 proteins, we showed by EMSA analysis that only NTEF-1 binds the desmin pal-Mt element (Fig. 9D), whereas only MEF2 protein binds the MCK MEF2 element (Fig. 9E).
Are the desmin MEF2-sensor transgenic mice detecting the transcriptional activity of MEF2 and/or TEF protein?
Myofiber hypertrophy and the induction of slow muscle genes is a common adaptation of skeletal muscle to a chronic increase in MOV (workload). Recent studies have focused attention on the role of calcineurin and calcium- and calmodulin-dependent protein kinase (CaMK) signaling pathways in conferring slow-fiber-specific gene expression and their involvement in the adaptive response to various modes of muscle activity. The present model of slow-fiber-specific gene expression predicts that calcium-mobilizing stimuli evoked by chronic motor nerve activity, exercise, or MOV result in the activation of the calcineurin and CaMK signaling pathways, which impinge on the downstream modulators MEF2 and NFAT, which in turn participate in the activation of the slow fiber gene program (12, 14, 32, 36, 43). Transcriptional activation by MEF2 has been shown to be more efficient following its dephosphorylation by activated calcineurin, an outcome in response to various modes of muscle activity, including MOV as used herein (12, 14, 32, 36, 43). Additional evidence that MEF2 participates in slow muscle gene expression is provided by experiments utilizing the desmin-sensor mice, wherein a desmin MEF2-dependent reporter gene is up-regulated in response to overexpression of constitutively activated calcineurin or by increased muscle activity (57). In addition, Wu et al. (57) have reported that slow muscle expression of the sTnI reporter gene in transgenic mice requires the SURE region NFAT and MEF2 elements. On the other hand, others have provided evidence that slow muscle expression of the sTnI gene requires the binding of GTF3/MusTRD1 to an additional sequence within the SURE region (9, 37). It is likely that accurate slow muscle expression of the sTnI gene involves the integrated input from all of these elements. Nevertheless, it is interesting that the MEF2 element within SURE is overlapped by a consensus Mt site, an element that we repeatedly show binds in vitro-synthesized and nuclear TEF proteins (Fig. 7A, G, and H; Fig. 8A and B; Fig. and 9D). Thus, our data showing avid TEF binding to the desmin MEF2 and pal-Mt elements raise the likely possibility that the desmin MEF2-sensor mice are detecting enhanced TEF protein activity in addition to MEF2. In this regard, our antibody EMSA results rule out the formation of a TEF-1/MEF2 heterodimer at both the βA/T-rich element (Fig. 4B) and the desmin pal-Mt element (Fig. 9D). It is important that while the desmin MEF2 reporter has been shown to be expressed in transgenic mouse day 9.5 somites, both MEF2 and TEF proteins are expressed in the developing mouse somites at that time (35, 59). The importance to NTEF-1 expression during embryonic development is further illustrated by the finding that disruption of the NTEF-1 gene results in cardiac defects resulting in embryonic lethality (11). In addition, TEF expression has been detected at the two-cell stage and in embryonic and adult stem cells, suggesting a role for these proteins during early developmental periods, and perhaps in the repair of damaged tissues, including striated muscle (23, 40).
It is intriguing to speculate that in adult skeletal muscle MEF2 and TEF proteins may compete for MEF2 site occupancy, depending on the activity state of a given skeletal muscle. For example, MOV may induce TEF protein binding at the desmin MEF2/Mt element, whereas voluntary wheel running or motor nerve pacing may induce MEF2 activity. Clearly, additional work will be required to clarify whether TEF or MEF2 occupies these MEF2 binding sites in fast versus slow muscle and under various conditions of muscle activity. Nevertheless, we have provided sufficient lines of evidence that confirm NTEF-1 binding to the βMyHC proximal promoter βA/T-rich element, its functional significance, and that this binding is enriched under MOV conditions. Furthermore, our present and past work has shown the differential binding of TEF proteins to a variety of MEF2, A/T-rich, and MCAT elements under control, non-weight-bearing, and MOV conditions (48, 50), suggesting that TEF proteins may serve a broad role in striated and smooth muscle gene regulation under basal and hypertrophic conditions. On the basis of these findings, it becomes clear that the delineation of whether TEF proteins serve specific and/or redundant functions is important for a better understanding of normal muscle physiology and its adaptive responses.
Acknowledgments
This work was supported by Public Health Service grants AR41464 and AR47197 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases.
We thank Y.-T. Yu for the MEF2a and MEF2b antibodies, J. Molkentin for the MEF2a expression vector, and I. Davidson for the NTEF, RTEF-1a, and DTEF expression vectors.
N. Karasseva and G. Tsika contributed equally and should be considered equal primary authors.
REFERENCES
- 1.Acakpo-Satchivi, L. J. R., W. Edelmann, C. Sartorius, B. D. Lu, P. A. Wahr, S. C. Watkins, J. M. Metzger, and L. A. Leinwand. 1997. Growth and muscle defects in mice lacking adult myosin heavy chain genes. J. Cell Biol. 139**:**1219-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barany, M. 1967. ATPase activity of myosin correlated with speed of muscle shortening. J. Gen. Physiol. 5**:**197-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassel-Duby, R., D. Hernandez, M. A. Gonzalez, J. K. Krueger, and R. S. Williams. 1992. A 40-kilodalton protein binds specifically to an upstream sequence element essential for muscle-specific transcription of the human myoglobin promoter. Mol. Cell. Biol. 12**:**5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belandia, B., and M. G. Parker. 2000. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J. Biol. Chem. 275**:**30801-30805. [DOI] [PubMed] [Google Scholar]
- 5.Black, B., and E. N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14**:**167-196. [DOI] [PubMed] [Google Scholar]
- 6.Booth, F. W., and K. M. Baldwin. 1996. Muscle plasticity: energy demand and supply processes, p. 1075-1123. In L. B. Rowell and J. T. Shepherd (ed.), Handbook of physiology. Exercise: regulation and integration of multiple systems. American Physiology Society, Bethesda, Md.
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72**:**248-254. [DOI] [PubMed] [Google Scholar]
- 8.Butler, A. J., and C. P. Ordahl. 1999. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol. Cell. Biol. 19**:**296-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvo, S., D. Vullhorst, P. Venepally, J. Cheng, I. Karavanova, and A. Buonanno. 2001. Molecular dissection of DNA sequences and factors involved in slow muscle-specific transcription. Mol. Cell. Biol. 21**:**8490-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, P. S., L. Li, J. McAnally, and E. N. Olson. 2001. Muscle specificity encoded by specific serum response factor-binding sites. J. Biol. Chem. 276**:**17206-17212. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Z., G. A. Friedrich, and P. Soriano. 1994. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 8**:**2293-2301. [DOI] [PubMed] [Google Scholar]
- 12.Chin, E. R., E. N. Olson, J. A. Richardson, Q. Yang, C. Humphries, J. M. Shelton, H. Wu, W. Zhu, R. Bassel-Duby, and R. S. Williams. 1998. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 12**:**2499-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chodosh, L. A. 1996. UV crosslinking of proteins to nucleic acids, p. 12.5.1-12.5.8. In F. M. Ausubel et al. (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 14.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109**:**S67-S79. [DOI] [PubMed] [Google Scholar]
- 15.Dunn, S. E., A. R. Simard, R. Bassel-Duby, R. S. Williams, and R. N. Michel. 2001. Nerve activity-dependent modulation of calcineurin signaling in adult fast and slow skeletal muscle fibers. J. Biol. Chem. 276**:**45243-45254. [DOI] [PubMed] [Google Scholar]
- 16.Fabre-Suver, C., and S. D. Hauschka. 1996. A novel site in the muscle creatine kinase enhancer is required for expression in skeletal but not cardiac muscle. J. Biol. Chem. 271**:**4646-4652. [DOI] [PubMed] [Google Scholar]
- 17.Gao, J., Z. Li, and D. Paulin. 1998. A novel site, Mt, in the human desmin enhancer is necessary for maximal expression in skeletal muscle. J. Biol. Chem. 273**:**6402-6409. [DOI] [PubMed] [Google Scholar]
- 18.Grayson, J., R. S. Williams, Y.-T. Yu, and R. Bassel-Duby. 1995. Synergistic interactions between heterologous upstream activation elements and specific TATA sequences in a muscle-specific promoter. Mol. Cell. Biol. 15**:**1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, T.-H., and R. Prywes. 1995. Regulatory role of MEF2D is serum induction of the c-jun promoter. Mol. Cell. Biol. 15**:**2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauschka, S. 1994. The embryonic origin of muscle, p. 3-73. In A. G. Engal and C. Franzini-Armstrong (ed.), Myology: basic and clinical. McGraw-Hill, New York, N.Y.
- 21.Hughes, S. M., J. M. Taylor, S. J. Tapscott, C. M. Gurley, W. J. Carter, and C. A. Peterson. 1993. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscles controlled by innervation and hormones. Development 118**:**1137-1147. [DOI] [PubMed] [Google Scholar]
- 22.Jacquemin, P., and I. Davidson. 1997. The role of the TEF transcription factors in cardiogenesis and other developmental processes. Trends Cardiol. Med. 7**:**192-197. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko, K. J., E. B. Cullinan, K. E. Latham, and M. L. DePamphillis. 1997. Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development 124**:**1963-1973. [DOI] [PubMed] [Google Scholar]
- 24.Lakich, M. M., T. T. Diagana, D. L. North, and R. G. Whalen. 1998. MEF-2 and Oct-1 bind to two homologous promoter sequence elements and participate in the expression of a skeletal muscle-specific gene. J. Biol. Chem. 273**:**15217-15266. [DOI] [PubMed] [Google Scholar]
- 25.Larkin, S. B., I. K. Farrance, and C. P. Ordahl. 1996. Flanking sequences modulate the cell specificity of M-CAT elements. Mol. Cell. Biol. 16**:**3742-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin, S. B., and C. P. Ordahl. 1999. Multiple layers of control in transcriptional regulation by MCAT elements and the TEF-1 protein family, p. 307-329. In R. P. Harvey and N. Rosenthal (ed.), Heart development. Academic Press, San Diego, Calif.
- 27.Li, H., and Y. Capetanaki. 1994. An E box in the desmin promoter cooperates with the E box and MEF-2 sites of a distal enhancer to direct muscle-specific transcription. EMBO J. 13**:**3580-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, M.-L., A. L. Olson, N. P. Edgington, W. S. Moye-Rowley, and J. E. Pessin. 1994. Myocyte enhancer factor 2 (MEF2) binding site is essential for C2C12 myotube-specific expression of the rat GLUT4/muscle-adipose facilitative glucose transporter gene. J. Biol. Chem. 269**:**28514-28521. [PubMed] [Google Scholar]
- 29.Maeda, T., M. P. Gupta, and A. F. R. Stewart. 2002. TEF-1 and MEF2 transcription factors interact to regulate muscle-specific promoters. Biochem. Biophys. Res. Commun. 294**:**791-797. [DOI] [PubMed] [Google Scholar]
- 30.Martin, J. F., J. J. Schwarz, and E. N. Olson. 1993. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc. Natl. Acad. Sci. USA 90**:**5282-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy, J. J., D. R. Vyas, G. L. Tsika, and R. W. Tsika. 1999. Segregated regulatory elements direct β-myosin heavy chain expression in response to altered muscle activity. J. Biol. Chem. 274**:**14270-14279. [DOI] [PubMed] [Google Scholar]
- 32.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27**:**40-47. [DOI] [PubMed] [Google Scholar]
- 33.Mora, S., and J. E. Pessin. 2000. The MEF2A isoform is required for striated muscle-specific expression of the insulin-responsive GLUT4 glucose transporter. J. Biol. Chem. 275**:**16323-16328. [DOI] [PubMed] [Google Scholar]
- 34.Musaro, A., K. J. A. McCullagh, F. J. Naya, E. N. Olson, and N. Rosenthal. 1999. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 400**:**581-585. [DOI] [PubMed] [Google Scholar]
- 35.Naya, F. J., C. Wu, J. A. Richardson, P. Overbeek, and E. N. Olson. 1999. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development 126**:**2045-2052. [DOI] [PubMed] [Google Scholar]
- 36.Olson, E. N., and R. S. Williams. 2000. Calcineurin signaling and muscle remodeling. Cell 101**:**689-692. [DOI] [PubMed] [Google Scholar]
- 37.O'Mahoney, J. V., K. L. Guven, J. Lin, J. E. Joya, C. S. Robinson, R. P. Wade, and E. C. Hardeman. 1998. Identification of a novel slow-muscle-fiber enhancer binding protein, MusTRD1. Mol. Cell. Biol. 18**:**6641-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parmacek, M. S., H. S. Ip, F. Jung, T. Shen, J. F. Martin, A. J. Vora, E. N. Olson, and J. M. Leiden. 1994. A novel myogenic regulatory circuit controls slow/cardiac troponin C gene transcription in skeletal muscle. Mol. Cell. Biol. 14**:**1870-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul, A. C., and N. Rosenthal. 2002. Different modes of hypertrophy in skeletal muscle fibers. J. Cell Biol. 156**:**751-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramalho-Santos, M., S. Yoon, Y. Matsuzaki, R. C. Mulligan, and D. A. Melton. 2002. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298**:**597-600. [DOI] [PubMed] [Google Scholar]
- 41.Sartorius, C. A., B. D. Lu, L. Acakpo-Satchivi, R. P. Jacobsen, W. C. Byrnes, and L. A. Leinwand. 1998. Myosin heavy chains IIa and IId are functionally distinct in the mouse. J. Cell Biol. 141**:**943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiaffino, S., and C. Reggiani. 1996. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 76**:**371-432. [DOI] [PubMed] [Google Scholar]
- 43.Schiaffino, S., and A. L. Serrano. 2002. Calcineurin signaling and neural control of skeletal muscle fiber type and size. Trends Pharmacol. Sci. 23**:**569-575. [DOI] [PubMed] [Google Scholar]
- 44.Takeda, S., D. L. North, T. Diagana, Y. Miyagoe, M. M. Lakich, and R. G. Whalen. 1995. Myogenic regulatory factors can activate TATA-containing promoter elements via an E-box independent mechanism. J. Biol. Chem. 270**:**15664-15670. [DOI] [PubMed] [Google Scholar]
- 45.Thai, M. V., S. Guruswamy, K. T. Cao, J. E. Pessin, and A. L. Olson. 1998. Myocyte enhancer factor 2 (MEF2)-binding site is required for GLUT4 gene expression in transgenic mice. J. Biol. Chem. 273**:**14285-14292. [DOI] [PubMed] [Google Scholar]
- 46.Tsika, G. L., J. L. Wiedenman, L. Gao, J. J. McCarthy, K. Sheriff-Carter, I. D. Rivera-Rivera, and R. W. Tsika. 1996. Induction of β-MHC transgene in overloaded skeletal muscle is not eliminated by mutation of conserved elements. Am. J. Physiol. Cell. Physiol. 271**:**C690-C699. [DOI] [PubMed] [Google Scholar]
- 47.Tsika, R. W., S. D. Hauschka, and L. Gao. 1995. M-creatine kinase gene expression in mechanically overloaded skeletal muscle of transgenic mice. Am. J. Physiol. Cell Physiol. 269**:**C665-C674. [DOI] [PubMed] [Google Scholar]
- 48.Tsika, R. W., J. McCarthy, N. Karasseva, Y. Ou, and G. L. Tsika. 2002. Divergence in species and regulatory role of β-myosin heavy chain proximal promoter muscle-CAT elements. Am. J. Physiol. Cell Physiol. 283**:**C1761-C1775. [DOI] [PubMed] [Google Scholar]
- 49.Vassilev, A., K. J. Kaneko, H. Shu, Y. Zhao, and M. L. DePamphilis. 2001. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15**:**1229-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vyas, D. R., J. J. McCarthy, G. L. Tsika, and R. W. Tsika. 2001. Multiprotein complex formation at the β-myosin heavy chain distal muscle CAT element correlates with slow muscle expression but not mechanical overload responsiveness. J. Biol. Chem. 276**:**1173-1184. [DOI] [PubMed] [Google Scholar]
- 51.Vyas, D. R., J. J. McCarthy, G. L. Tsika, and R. W. Tsika. 2000. Muscle-specific transcription of β-myosin heavy chain transgene requires an A/T-rich element. Basic Appl. Myol. 10**:**87-88. [Google Scholar]
- 52.Vyas, D. R., J. J. McCarthy, and R. W. Tsika. 1999. Nuclear protein binding at the β-myosin heavy chain A/T-rich element is enriched following increased skeletal muscle activity. J. Biol. Chem. 274**:**30832-30842. [DOI] [PubMed] [Google Scholar]
- 53.Wiedenman, J. L., I. Rivera-Rivera, D. Vyas, G. Tsika, L. Gao, K. Sheriff-Carter, X. Wang, L. Y. Kwan, and R. W. Tsika. 1996. β-MHC and SMLC1 transgene induction in overloaded skeletal muscle of transgenic mice. Am. J. Physiol. Cell Physiol. 270**:**C111-C121. [DOI] [PubMed] [Google Scholar]
- 54.Wiedenman, J. L., G. L. Tsika, L. Gao, J. J. McCarthy, I. D. Rivera-Rivera, D. Vyas, K. Sheriff-Carter, and R. W. Tsika. 1996. Muscle-specific and inducible expression of 293-base pair β-myosin heavy chain promoter in transgenic mice. Am. J. Physiol. Reg. Integ. Comp. Physiol. 271**:**R688-R695. [DOI] [PubMed] [Google Scholar]
- 55.Wu, H., F. J. Naya, T. A. McKinsey, B. Mercer, J. M. Shelton, E. R. Chin, A. R. Simard, R. N. Michel, R. Bassel-Duby, E. N. Olson, and R. S. Williams. 2000. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 19**:**1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, H., and E. N. Olson. 2002. Activation of the MEF2 transcription factor in skeletal muscles from myotonic mice. J. Clin. Investig. 109**:**1327-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, H., B. Rothermel, S. Kanatous, P. Rosenberg, F. J. Naya, J. M. Shelton, K. A. Hutcheson, J. M. DiMaio, E. N. Olson, R. Bassel-Duby, and R. S. Williams. 2001. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 20**:**6414-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan, Z., L. A. Serrano, S. Schiaffino, R. Bassel-Duby, and R. S. Williams. 2001. Regulatory elements governing transcription in specialized myofiber subtypes. J. Biol. Chem. 276**:**17361-17366. [DOI] [PubMed] [Google Scholar]
- 59.Yockey, C. E., G. Smith, S. Izumo, and N. Shimizu. 1996. cDNA cloning and characterization of murine transcriptional enhancer factor-1-related protein 1, a transcription factor that binds to the M-CAT motif. J. Biol. Chem. 271**:**3727-3736. [DOI] [PubMed] [Google Scholar]
- 60.Yu, Y.-T. 1996. Distinct domains of myocyte enhancer binding factor-2A determining nuclear localization and cell type-specific transcriptional activity. J. Biol. Chem. 271**:**24675-24683. [PubMed] [Google Scholar]
- 61.Zhu, H., V. T. Nguyen, A. B. Brown, A. Pourhosseini, A. V. Garcia, M. van Bilsen, and K. R. Chien. 1993. A novel, tissue-restricted zinc finger protein (HF-1b) binds to the cardiac regulatory element (HF-1b/MEF-2) in the rat myosin light-chain 2 gene. Mol. Cell. Biol. 13**:**4432-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]