Divergence of the genes on human chromosome 21 between human and other hominoids and variation of substitution rates among transcription units (original) (raw)
Abstract
The study of genomic divergence between humans and primates may provide insight into the origins of human beings and the genetic basis of unique human traits and diseases. Chromosome 21 is the smallest chromosome in the human genome, and some of its regions have been implicated in mental retardation and other diseases. In this study, we sequenced the coding and regulatory regions of 127 known genes on human chromosome 21 in DNA samples from human and chimpanzees and a part of the corresponding genes from orangutan, gorilla, and macaque. Overall, 3,003 nucleotide differences between human and chimpanzee were identified over ≈400 kb. The differences in coding, promoter, and exon–intron junction regions were 0.51 ± 0.02%, 0.88 ± 0.03%, and 0.85 ± 0.02%, respectively, much lower than the previously reported 1.23% in genomic regions, which suggests the presence of purifying selection. Significant variation in substitution rate among genes was observed by comparing the divergence between human and chimpanzee. Furthermore, by implementing a bioinformatics-based approach, we showed that the identification of genetic variants specific to the human lineage might lead to an understanding of the mechanisms that are attributable to the phenotypes that unique to humans, by changing the structure and/or dosage of the proteins expressed. A phylogenetic analysis unambiguously confirms the conclusion that chimpanzees were our closest relatives to the exclusion of other primates and the relative divergence of the Homo_–_Pan and that of (Homo_–_Pan)–Gorilla are 4.93 million years and 7.26 million years, respectively.
It was commonly recognized that human's closest relatives are the African great apes, i.e., chimpanzee and gorillas, even before the era of modern molecular biology (1). In the last two decades, DNA sequence and cytogenetic analyses have shown that the genomes of human and African great apes are strikingly similar (2,3) despite their great apparent phenotypic difference. A comparison of their genomes may shed light on understanding the genetic basis for distinct human traits, certain human diseases, and human reproductive biology (2).
Although the Human Genome Project has provided a wealth of genetic information about the human genome (4), the genomic study is still in its nascent stage for great apes, let alone other primates, such as Pan troglodytes (the chimpanzee) and Pan paniscus (the bonobo), which share nearly 99% of human genomic sequences (5). Some therefore argued that knowing the complete genome of at least one of these species will help to identify the genes that contribute to humaness (6,7). The sequencing of human chromosome 21, which is the smallest chromosome in the human genome and which has important regions related to mental retardation, is largely finished (8). In this study, we sequenced the coding and regulatory regions of 127 known genes on human chromosome 21 from DNA samples of human beings and a pool of 20 chimpanzee samples, which allows a direct comparison of the genomes of these two species. As outgroups, partial sequences of those genes were also determined for a gorilla, an orangutan, and a macaque. In this report, we try to address several different though related questions regarding the history and mechanism of human evolution by exploring the sequences of the homologous fragments of hominoid species, including (i) presence of various form of natural selection, (ii) variation of substitution rates, (iii) phylogeny of hominoid species, and (iv) the mutations that are specific to human lineage and their functional consequences.
Materials and Methods
Study Subjects. DNA samples from human (Homo sapiens), chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), and macaque (Macaca mulatta) were used in the study. The human samples were obtained from 31 Chinese individuals representing the major ethnic groups with proper informed consent. Genomic DNA of chimpanzee and macaque were prepared from peripheral blood of 20 chimpanzees and one macaque, respectively, which was approved by the Ethics Committee of Chinese National Human Genome Center (Shanghai). DNA samples of one gorilla and one orangutan were obtained from NIGMS Human Genetic Cell Repository (NG05251 and NA04272). The DNA concentration of each sample was brought to 40 ng/μl before being used.
DNA Amplification. The PCR and sequencing primers were designed by using Primer 3.0 (http://203.11.132.151/cgi-bin/primer/primer3_www.cgi) according to the human genome sequences available athttp://hgp.gsc.riken.go.jp./data_tools/data_chr21.html. The PCR products were ≈400–600 bp in size. PCR was performed in a 25-μl reaction volume for 2 min at 95°C and then 14 cycles of amplification with at 94°C for 20 s, 63°C decreased by 0.5°C increments every cycle for 20 s, and 72°C for 45 s.
Sequence Variation Identification. PCR products were sequenced in both directions with the PCR primers and sometimes with additional internal primers after purification with resin (Promega). The sequencing products were run on an automated DNA sequencer ABI 377 (Applied Biosystems) following the manufacturer's instructions. The computer program polyphred(9) was used to identify candidate variations, each of which was individually inspected manually.
Data Analysis. Sequence alignment was performed withbioedit software (10) and confirmed by visual inspection. The Kimura's two-parameter distance (11) was used to estimate the genomic divergence of each gene, whereas the synonymous substitution was used to estimate the age of divergence for coding sequence. The ratio of the rates of nucleotide substitutions that change amino acids (nonsynonymous substitutions, Ka) to those that do not (synonymous substitutions, Ks) was calculated by k-estimator 5.1 (12). The within-population polymorphisms were not included in divergence estimation. Phylogenetic trees were constructed by mega(13) using the neighbor-joining method (14) with 500 replications for bootstrapping.
Results and Discussion
The Sequence Divergence Between Human and Chimpanzee Is Significantly Lower in the Coding Regions Than Those Observed in Unclassified Genomic Regions. The sequence divergence between the species in the PCR primer binding sites might lead to the difficulties in amplifying some gene regions in chimpanzee and the presence of even more difficult regions in other primates, in which the divergence is higher. Nevertheless, for chimpanzee, 70% of the sequences were amplified and sequenced in 34 genes (26.8%), whereas 30–70% of the sequences were obtained in 72 genes (56.7%). The remainder of the 21 genes (16.5%) can be amplified in <30% of the sequences in chimpanzee. Overall, 3,003 nucleotide differences between human and chimpanzee were identified over a total of 399,264 bp from the 127 genes, which yielded an average 0.7% difference of genome sequence between the two species. There were 666 variants in the coding regions, among which 227 (34.1%) were nonsynonymous, 370 (55.5%) were synonymous, and 69 (10.4%) cannot be classified exactly in those genes where the ORFs are equivocal (seeTable 1). All of the data of sequence divergence between human and chimpanzee and sequences of chimpanzee and other hominoids can be accessed atwww.biosino.org/chgc-SNP.
Table 1. Divergence between human and chimpanzee.
| Location | Variations between human and chimpanzee (%) | Sequenced base pair (%) | Divergence, % |
|---|---|---|---|
| Promoter | 766 (25.5) | 86,834 (21.7) | 0.88 ± 0.03 |
| 5′UTR | 69 (2.3) | 6,906 (1.7) | 1.00 ± 0.10 |
| Coding region | 666 (22.2) | 130,149 (32.6) | 0.51 ± 0.02 |
| Synonymous | 370 (55.5) | 40,065.7 | 0.92 ± 0.05 |
| Nonsynonymous | 227 (34.1) | 87,824.3 | 0.26 ± 0.02 |
| Other | 69 (10.4) | 2,259 | 3.05 ± 0.36 |
| Coding boundary | 1,377 (45.9) | 161,945 (40.6) | 0.85 ± 0.02 |
| 3′UTR | 106 (3.5) | 11,427 (2.9) | 0.93 ± 0.09 |
| End | 19 (0.6) | 2,003 (0.5) | 0.95 ± 0.20 |
| Overall | 3,003 (100.0) | 399,264 (100.0) | 0.75 ± 0.01 |
Early DNA hybridization data showed that the divergence between human and chimpanzee was 1.6% (15). Chen and Li (16) compared autosomal intergenic nonrepetitive DNA segments and observed that the sequence divergence was 1.24%. In a study by Fujiyama et al.(17), it was found that human and chimpanzee had only 1.23% difference in randomly chosen genomic regions. Ebersberger et al.(18) estimated that the genome-wide average of the sequence divergence between the two species is 1.24% based on ≈19 Mb from 8,859 random fragments. In this study, the sequence divergence in coding sequences between human and chimpanzee was as low as 0.75 ± 0.01% overall, 0.51 ± 0.02% at the coding regions, 0.88 ± 0.03% at the promoter regions, and 0.85 ± 0.02% at the intronic regions near the exon/intron boundaries (seeTable 1). When the concatenated sequence of all of the genes is used, the number of substitutions per synonymous site (Ks) between the two species is 0.84 ± 0.04%, and the number of substitutions per nonsynonymous site is 0.25 ± 0.02%. The estimated divergence between human and chimpanzee of this study are not only much smaller than those from the aforementioned studies (P = 0 assuming binomial distribution), but also smaller than those reported in other genomic regions including those in 22q11.2 (1.35%), chromosome 12 contig 1 and 2 (1.19% and 1.20%), Xq13.3 (0.92%), Xc36 (1.32%), and SMCY (1.68%), though the difference of synonymous substitutions of the two studies are not statistically significant (P = 0.125, t test) (16). Interestingly, Ebersberger et al.(18) made an observation that the substitution rate is highest in chromosome 21 (1.5%) by comparing ≈19 Mb in human and chimpanzee. This is consistent with a similar observation made by Lercher et al.(19), who compared the Ks values between human and rodents. In light of these observation, the higher divergence in chromosome 21 genes would have been expected, which contradicts the observation made in this study.
The lower divergence at the intron regions was also observed at 32 loci (1.03 ± 0.07%) compared with intergenic regions (1.24%) by Chen and Li (16). But this estimation is still higher than what was observed in this study (0.85 ± 0.02%). The difference between these two estimations is significant (P = 0 assuming binomial distribution). This difference could be due to the fact that intronic regions near the exon/intron boundaries were studied, which tend to be more conservative considering their roles in splicing.
Therefore, we conclude that the higher level of similarity observed in the transcript units in this study is attributable to the presence of purifying natural selection exerted on the most important functional portions of the genes, including promoters, coding regions, and intronic regions near the exon–intron boundary. The coding regions are the most conserved part of the genome, followed by the exon–intron boundaries and promoter regions.
Variation of Substitution Rates Among Genes Was Observed. Despite high similarity of functional fragments, the overall sequence divergence, defined by the proportion of nucleotide difference of the two species between the species (p), varies significantly across the 106 genes in which at least 30% of the gene was successfully amplified and sequenced in both human and chimpanzee. The sequence divergence ranges from 0% to 2.5%. The largest divergences were observed in 10 genes (P > 1.0%), including PRED14 and KCNE1, where P > 2.0%, whereas the remaining 96 genes all showed divergence <1%. The variation of divergence at amino acid level was even more prominent with PRED14 and KCNE1 being the highest (see Table 2, data from all 127 genes are shown in Table 4, which is published as supporting information on the PNAS web site,www.pnas.org).
Table 2. Characteristics of 30 known genes.
| Divergence at part of genes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene name | GenBank accession no. | Length sequenced in human, bp | Percentage sequenced in chimpanzee | Overall divergence between human and chimp, % | Promoter, % | Coding, % | Exon-intron junction, % | Nonsynonymous change, no. | Synonymous change, no. | Ka | Ks | Ka/Ks | P* | Divergence at amino acid level, % |
| ADARB1 | AL163301 | 4,826 | 59.10 | 0.60 | 1.07 | 0.08 | 1.11 | 1 | 0 | 0.0006 | 0.0000 | NA | 1.0000 | 0.23 |
| AIRE | AP001754 | 6,393 | 68.37 | 1.17 | 1.72 | 1.01 | 0.63 | 5 | 4 | 0.0047 | 0.0084 | 0.5601 | 0.7078 | 1.68 |
| ATP5A | AP001694 | 2,079 | 100.00 | 1.06 | 0.42 | 1.53 | 1.69 | 0 | 0 | 0.0000 | 0.0000 | NA | 1.0000 | 0.00 |
| B3GALT5 | AL163280 | 3,886 | 63.29 | 1.63 | 1.96 | 1.41 | 2.78 | 6 | 7 | 0.0092 | 0.0225 | 0.4085 | 0.9110 | 1.95 |
| C210RF258 | AP001745-C | 5,402 | 49.44 | 0.82 | NS | 1.22 | 0.59 | 3 | 0 | 0.0036 | 0.0000 | NA | 1.0000 | 0.91 |
| CBR1 | AP001724 | 2,822 | 50.30 | 2.05 | 2.06 | 1.26 | 2.22 | 0 | 5 | 0.0000 | 0.0207 | 0.0000 | 0.9979 | 0.00 |
| CCT8 | AL163249 | 6,213 | 47.48 | 0.64 | 0.16 | 0.42 | 1.43 | 3 | 0 | 0.0062 | 0.0000 | NA | 1.0000 | 1.27 |
| COL6A2 | AP001759 | 2,628 | 73.48 | 0.21 | 0.00 | 0.35 | 0.00 | 4 | 0 | 0.0049 | 0.0000 | NA | 1.0000 | 1.06 |
| CRYAA | AP001748 | 3,235 | 36.75 | 1.43 | 1.75 | 0.48 | 0.83 | 1 | 0 | 0.0066 | 0.0000 | NA | 1.0000 | 1.43 |
| DSCR2 | AL163279-C | 2,886 | 42.69 | 0.89 | 0.63 | 1.32 | 3.36 | 2 | 0 | 0.0014 | 0.0000 | NA | 1.0000 | 0.96 |
| HSF2BP | AP001752-C | 4,793 | 37.03 | 0.73 | 0.78 | 0.96 | 0.51 | 1 | 0 | 0.0015 | 0.0000 | NA | 1.0000 | 2.88 |
| IFNAR1 | AP001716 | 5,421 | 37.63 | 0.88 | 1.24 | 0.69 | 0.41 | 3 | 1 | 0.0024 | 0.0019 | 1.2602 | 0.2459 | 1.54 |
| IFNGR2 | AP001717 | 3,856 | 41.57 | 0.69 | NS | 0.87 | 0.33 | 2 | 2 | 0.0062 | 0.0144 | 0.4322 | 0.6486 | 1.30 |
| KCNE1 | AP001720-C | 2,506 | 67.32 | 1.36 | 1.03 | 2.51 | NS | 6 | 4 | 0.0221 | 0.0303 | 0.7278 | 0.5774 | 4.51 |
| KIAA0539 | AP001713-C | 7,481 | 63.20 | 1.08 | 0.38 | 1.20 | 1.40 | 4 | 12 | 0.0033 | 0.0323 | 0.1031 | 0.3164 | 0.90 |
| MX1 | AL163285 | 8,143 | 66.66 | 0.94 | 0.84 | 0.93 | 1.05 | 6 | 7 | 0.0065 | 0.0146 | 0.4443 | 0.8844 | 1.28 |
| MX2 | AL163285 | 5,949 | 81.74 | 1.32 | 1.76 | 0.74 | 1.43 | 8 | 5 | 0.0071 | 0.0080 | 0.8797 | 0.4859 | 1.37 |
| PCNT | AP001760 | 22,136 | 76.29 | 0.73 | 0.00 | 0.92 | 0.62 | 35 | 37 | 0.0073 | 0.0137 | 0.5280 | 0.9953 | 1.33 |
| PDXK | AP001752 | 5,302 | 47.98 | 1.26 | 1.83 | 0.30 | 0.64 | 1 | 0 | 0.0036 | 0.0000 | NA | 1.0000 | 0.89 |
| PRED14 | AP001679 | 1,456 | 56.80 | 1.45 | 0.24 | 2.52 | NS | 4 | 0 | 0.0343 | 0.0000 | NA | 1.0000 | 7.55 |
| PRED22 | AP001693-C | 5,031 | 74.18 | 0.75 | 0.37 | 0.39 | 1.05 | 2 | 0 | 0.0050 | 0.0000 | NA | 1.0000 | 1.18 |
| PRSS7 | AL163218-C | 2,414 | 74.23 | 0.73 | 0.56 | 0.58 | 1.47 | 2 | 0 | 0.0009 | 0.0000 | NA | 1.0000 | 1.74 |
| SAMSN-1 | AL163206-C | 4,916 | 67.05 | 1.06 | 2.38 | 0.47 | 0.51 | 3 | 1 | 0.0050 | 0.0065 | 0.7732 | 0.0625 | 1.05 |
| SH3BGR | AL163280 | 3,339 | 63.10 | 0.76 | 1.07 | 0.38 | 0.69 | 2 | 0 | 0.0029 | 0.0000 | NA | 1.0000 | 1.13 |
| TFF3 | AP001746-C | 2,946 | 22.57 | 1.50 | NS | 0.60 | 0.98 | 1 | 0 | 0.0056 | 0.0000 | NA | 1.0000 | 1.80 |
| TMPRSS2 | AL163286-C | 6,879 | 59.25 | 0.91 | 0.71 | 0.24 | 1.44 | 2 | 0 | 0.0036 | 0.0000 | NA | 1.0000 | 0.73 |
| TRPC7 | APP001754 | 15,120 | 74.79 | 1.03 | 0.83 | 1.10 | 0.96 | 17 | 19 | 0.0054 | 0.0139 | 0.3912 | 0.9958 | 1.56 |
| TTC3 | AP001728 | 18,213 | 84.85 | 0.67 | 0.70 | 0.36 | 0.88 | 14 | 6 | 0.0040 | 0.0029 | 1.3919 | 0.1820 | 0.76 |
| USP16 | AL163249 | 7,752 | 77.52 | 0.53 | 0.22 | 0.89 | 0.49 | 8 | 6 | 0.0046 | 0.0073 | 0.6245 | 0.7304 | 1.52 |
| WDR4 | AB039887 | 6,097 | 57.68 | 1.02 | 1.07 | 0.36 | 0.73 | 2 | 0 | 0.0039 | 0.0000 | NA | 1.0000 | 1.07 |
The overall sequence divergence, such as p, is affected by numerous factors such as variation of mutation rates among regions and variation of magnitude of natural selection among genes. However, the variation of synonymous substitution rates among genes reflects the variation of the intergenic substitution rate. Lercher et al.(19) found that, by comparing human and rodent genomes, divergence varies drastically across the genome, whereas the local similarity of the divergence may extend to whole mouse chromosomes. In this study, among the genes with 30% or more sequences obtained in both human and chimpanzee samples, the variation of synonymous substitution rate is substantial, ranging from 0% to 3.23% (0.73 ± 0.05%), with the substitution rate of KIAA0539 being the highest.
We evaluated the homogeneity of overall divergence, Ka and Ks of the 106 genes with 30% or more sequence in both species by a permutation approach. This was achieved by first generating two homologous sequences of the size that is equal to the summation of all fragments studied and they differ by the amount of average divergence observed in this study. Each nucleotide was then subsequently assigned randomly to one of the 106 genes with different but known fragment sizes. A χ2 statistic, the summation of the ratios of the squared difference between the divergence of the simulated data and the expected divergence for each gene and the expected divergence, was then calculated. This process was repeated 1,000,000 times, and we obtained the distribution of the χ2 statistic assuming homogeneity of the substitution rate across the genes (the null hypothesis). The χ2 value for the observed data were then estimated by replacing the simulated divergences with their observed counterparts. The probability of the observation and those with more skewed values can be therefore obtained under the null hypothesis. Following this procedure, we found that variations of substitution rates (overall divergence, Ks, and Ka) across the chromosomes are statistically significant, with the probability of 0.00001, 0.00078, and 0.00060, respectively, assuming no variation. This observation rejects the hypothesis of homogeneous substitution rates among genes.
The correlation between the sequence divergence and synonymous substitution rate is poor (ρ = 0.344). Therefore, the variation of Ka cannot be solely explained by the variation in Ks (i.e., the variation of neutral substitution rate alone), indicating the presence of intergenic variation of natural selection.
**Lack of Statistical Power in Detecting Positive Darwinian Selection.**The ratio of the number of nonsynonymous substitution per site (Ka) and the number of synonymous substitution per site (Ks) between human and chimpanzee was estimated for each gene and listed inTable 2, where S is the number of synonymous substitution and A is the number of nonsynonymous substitution. Under the assumption of selection neutrality, the value of Ka/Ks is expected to be 1, whereas a gene that underwent positive Darwinian selection would lead to a larger Ka/Ks ratio and a gene that experienced purifying selection would show a lower ratio (20). In this set of genes, 17 had Ka/Ks values >1 (see Table 2, data from all 127 genes are shown in Table 4) including IFNAR1 (Ka/Ks = 1.26), TTC3 (1.39), ADARB1 (A = 1/S = 0), CRYAA (1/0), PDXK (1/0), TMPRSS2 (1/0), CCT8 (2/0), WDR4 (3/0), COL6A2 (4/0), DSCR2 (2/0), HSF2BP (1/0), PRED14 (4/0), PRSS7 (2/0), C21ORF258 (3/0), SH3BGR (2/0), PRED22 (2/0), and TFF3 (1/0). The probability of the events that are equal to and more biased than the observation can be estimated under the hypothesis of Ka = Ks by using the binomial distribution, but none of genes is statistically significant to reject the hypothesis (seeTable 2). Among these genes, four genes showed lowest probabilities. They are TTC3 (P = 0.333), CCT8 (P = 0.316), COL6A2 (P = 0.280), and PRED14 (_P_= 0.312). Although the observation of Ka/Ks >1 is often used as evidence to support the presence of positive Darwinian selection (21), this approach suffers from the presence of substantial purifying selection in the gene and lack of statistical power as exemplified in this study. However, this observation does not preclude the presence of the positive Darwinian selection that might have exerted an affect on some of those genes.
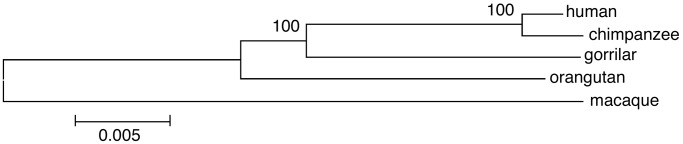
Phylogeny of Hominoid and Estimation of Human/Pan Divergence. Other primate species were also included in this study by amplifying and sequencing the corresponding regions of a gorilla, an orangutan, and a macaque. A neighbor-joining tree was reconstructed by using the 44,076 bp of successfully amplified and sequenced regions from all of the five species. The tree supported the Homo_–_Pan clade with a 100% bootstrap value (Fig. 1), further confirming the conclusion that chimpanzees are our closest relatives (1,17,22,23). The divergence between human and other hominoid species can be obtained based on the number of synonymous substitutions estimated from the 28.5-kbp coding sequences that were obtained for all four species including human, chimpanzee, gorilla, and orangutan. The divergence based on the number of synonymous substitutions are 0.72 ± 0.12%, 1.11 ± 0.14%, and 2.00 ± 0.19% for_Homo_–Pan, Homo_–_Gorilla, and_Homo_–orangutan, respectively. The relative divergence of the_Homo_–Pan clade and that of (Homo_–_Pan)–Gorilla clade are 4.93 million years and 7.26 million years, respectively, which were calibrated by a 14-million-year divergence between human and orangutan. These estimates are in general consistent with those in earlier studies (17,24). The time span between the two speciation events, i.e., (Homo_–_Pan)–Gorilla and_Homo_–Pan, was ≈2.33 million years, which is slightly lower than that based on mtDNA (24), but it is consistent with those based on both synonymous distance of coding regions and 53 intergenic regions by Chen and Li (16).
Fig. 1.
Phylogeny of the five species. Genetic distances were estimated by a Kimura two-parameter matrix, and polygenetic trees were constructed by the neighbor-joining method.
Human-Specific Substitutions. Substitutions that are specific to the human lineage could play an important role in contributing to the origin of human species and therefore are of special interest. In this study, there are 310 such substitutions that are specific to the human lineage, human-specific single-nucleotide polymorphisms (HS-SNPs), using the parsimony principle from 44,076 bp of nucleotides sequenced completely in DNA samples both from human, chimpanzee and gorilla, or orangutan, where the sequences of the latter two species were used as outgroups. The other 322 substitutions are specific to chimpanzee, whereas the specificity of the remaining 146 substitutions cannot be determined. The substitutions that are specific to the human lineage may be attribute to the phenotype of humanness by altering (i) structure and/or (ii) dosage of the proteins expressed. Therefore, we systematically examined the HS-SNPs in these two categories by using bioinformatics tools. First, all of the segments that contain HS-SNPs in the promoter regions of the genes were inspected by matching the original and mutated sequences against putative transcription binding sites (matinspector V2.2 based on transfac 4.0,http://transfac.gbf.de/TRANSFAC/). The HS-SNPs that show possible altered binding of the transcriptional factors were flagged and carefully examined. Second, all of the HS-SNPs in the coding regions that show amino acid replacement were also flagged, and their roles in determining second and tertiary structure of protein were further interrogated by employing two well documented algorithms (www.chgc.sh.cn/PP/andhttp://www-igbmc.u-strasbg.fr/Computer/ClustalW/clustalw-article.html). The predicted structures of the protein from different species were then contrasted and any significant differences were flagged. Our approaches are by no means intended to be exhaustive and reliable in studying the functional consequence of HS-SNPs, and the quality of the observations we made is largely limited by the efficiency, accuracy, and reliability of the bioinformatics tools and the our knowledge of the genes. However, such analyses do provide the first, though rudimentary, approaches to explore the functional consequences of the HS-SNPs. In the following discussion, we present several human specific substitutions that may have implications in human evolution.
There are five sequence variations between human and the other three hominoids over a 1-kb sequence upstream the first exon of DYRK1A (as shown onTable 3), of which three variations may change the putative transcription binding sites. This gene is the human homolog of the Drosophila minibrain gene (25), which is located in the critical region of the Down's syndrome (DS), overexpressed in DS fetal brain. It also causes mental retardation and motor anomalies in transgenic animals (26–28).
Table 3. Putative transcript factor binding sites in promoter region of_DYRK1A_.
| Position* | Variation human/Pan | Human binding site | Chimpanzee binding site |
|---|---|---|---|
| 297872 | G/A | V$GFI1_01 | V$GFI1_01 |
| aagaagaaAATCgtatctgtagct | aagaagaaAATCatatctgtagct | ||
| 297947 | T/C | V$DELTAEF1_01 tctgACCTtgt | Lost the binding site |
| 297976 | G/A | No binding site | V$AP1_Q4: agTGACtagcc |
| 298278 | G/T | No binding site | No binding site |
| 298428 | G/A | V$HNF3B_01: AtttaTGTTttcttt | V$HNF3B_01: atttaTATTttcttt |
| Lost the binding site | V$TATA_01: ataTAAAttcaatct |
Another example was observed within the coding sequence of IFNAR1_gene, a type I IFN receptor belonging to the class II cytokine receptor family (29,30). Multiple sequence alignment of IFNAR1 from human, chimpanzee, and other animals indicated that one proline residue of a highly conserved ISPP amino acid segment is missing from the human protein primary structure, whereas it is unchanged in all of the other animals from chicken to chimpanzee. Both the secondary and the tertiary structures of the human and chimpanzee_IFNAR1 are predicted when the algorithms mentioned earlier are used. It is striking to notice that the amino acid segment containing this conserved sequence is likely to be the only region that bears significant difference between the human and nonhuman hominoids. In the case of chimpanzee, the PPG region is part of a helix structure, whereas in the case of human, the PG region is a part of a random coil (data not shown). This structural variation certainly needs experimental confirmation, and its functional implication is yet to be explored. It was reported that great apes could be infected with human hepatitis viruses, but usually do not progress to cirrhosis or hepatocellular carcinomas as often seen in human (31). Therefore, we would like to speculate a possibility of formulating a useful index for predicting the long-term efficacy of IFN therapy against chronic hepatitis C virus infection (32) on the confirmation of the functional consequence of the difference.
Although the establishment of biological significance of the aforementioned variations has a long way to go, the fact that they are present only in human suggests that this kind of genomic change might be most important in the development of human traits. To this end, it is worth pointing out that a simple comparison between chimpanzee and human sequences may lead to the implication of the divergences responsible for humanness. Human traits and specific adaptations in the human lineage, such as those changes in brain architecture could be rooted in both recent and more distant evolution history (33).
Supplementary Material
[Supporting Table]
Acknowledgments
We thank the members and associates of the Chinese National Human Genome Center at Shanghai for their enthusiastic support and great contribution to this project. We give special thanks to the Shanghai Zoo and the Kunming Institute of Zoology for the chimpanzee and macaque samples that they kindly provided. We also thank Drs. Bing Su, Zihe Rao, Joshua Akey, and Jingchu Luo for their professional comments and instructions, and Drs. Yixue Li and Haiwei Fan from the Shanghai Center for Bioinformation Technology for the database establishment of this project. This work was supported by Chinese National Natural Science Foundation Grant 39993420, National Key Project for Basic Research from Chinese Ministry of Science and Technology Grant G1998051019, Chinese High-Tech Program (863) Grants 2001AA224021 and 2002BA711A10, and Shanghai Science and Technology Development Fund Grant 00DJ14003.
Abbreviations: SNP, single-nucleotide polymorphism; Ka, nonsynonymous mutations per nonsynonymous site; Ks, synonymous mutations per synonymous site.
References
- 1.Jones, S., Martin, R. D. & Pilbeam, D. R., eds. (1992) The Cambridge Encyclopedia of Human Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 2.Varki, A. (2000) Genome Res. 10 1065-1070. [DOI] [PubMed] [Google Scholar]
- 3.Gagneux, P. & Varki, A. (2001) Mol. Phylogenet. Evol. 18 2-13. [DOI] [PubMed] [Google Scholar]
- 4.International Human Genome Sequencing Consortium (2001) Nature 409 860-921. [DOI] [PubMed] [Google Scholar]
- 5.Goodman, M., Porter, C. A., Czelusniak, J., Page, S. L., Schneider, H., Shoshani, J., Gunnell, G. & Groves, C. P. (1998) Mol. Phylogenet. Evol. 9 585-598. [DOI] [PubMed] [Google Scholar]
- 6.McConkey, E. H. & Goodman, M. (1997) Trends Genet. 13 350-351. [DOI] [PubMed] [Google Scholar]
- 7.McConkey, E. H., Fouts, R., Goodman, M., Nelson, D., Penny, D., Ruvolo, M., Sikela, J., Stewart, C. B., Varki, A. & Wise, S. (2000) Mol. Phylogenet. Evol. 15 1-4. [DOI] [PubMed] [Google Scholar]
- 8.Hattori, M., Fujiyama, A., Taylor, T. D., Watanabe, H., Yada, T., Park, H. S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D. K., et al.(2000) Nature 405 311-319. [DOI] [PubMed] [Google Scholar]
- 9.Nickerson, D. A., Tobe, V. O. & Taylor, S. L. R. (1997) Nucleic Acids Res. 25 2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, T. A. (1999) Nucleic Acids. Symp. Ser. 41 95-98. [Google Scholar]
- 11.Kimura, M. (1980) J. Mol. Evol. 16 111-120. [DOI] [PubMed] [Google Scholar]
- 12.Comeron, J. M. (1999) Bioinformatics 15 763-764. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, S., Tamura, K. & Nei, M. (1994) Comput. Appl. Biosci. 10 189-191. [DOI] [PubMed] [Google Scholar]
- 14.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4 406-425. [DOI] [PubMed] [Google Scholar]
- 15.Sibley, C. G. & Ahlquist, J. E. (1987) J. Mol. Evol. 26 99-121. [DOI] [PubMed] [Google Scholar]
- 16.Chen, F. C. & Li, W. H. (2001) Am. J. Hum. Genet. 68 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiyama, A., Watanabe, H., Toyoda, A., Taylor, T. D., Itoh, T., Tsai, S. F., Park, H. S., Yaspo, M. L., Lehrach, H., Chen, Z., et al.(2002) Science 295 131-134. [DOI] [PubMed] [Google Scholar]
- 18.Ebersberger, I., Metzler, D., Schwarz, C. & Pääbo S. (2002) Am. J. Hum. Genet. 70 1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lercher, M. J., Williams, E. J. B. & Hurst, L. D. (2001) Mol. Biol. Evol. 18 2032-2039. [DOI] [PubMed] [Google Scholar]
- 20.Kimura. (1983) The Neural Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 21.Hughes, A. L. & Nei, M. (1988) Nature 335 167-170. [DOI] [PubMed] [Google Scholar]
- 22.Ruvolo, M. (1997) Mol. Biol. Evol. 14 248-265. [DOI] [PubMed] [Google Scholar]
- 23.Satta, Y., Klein, J. & Takahata, N. (2000) Mol. Phylogenet. Evol. 14 259-275. [DOI] [PubMed] [Google Scholar]
- 24.Horai, S., Hayasaka, K., Kondo, R., Tsugane, K. & Takahata, N. (1995) Proc. Natl. Acad. Sci. USA 92 532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guimera, J., Casas, C., Pucharcos, C., Solans, A., Domenech, A., Planas, A. M., Ashley, J., Lovett, M., Estivill, X. & Pritchard, M. A. (1996) Hum. Mol. Genet. 5 1305-1310. [DOI] [PubMed] [Google Scholar]
- 26.Guimera, J., Casas, C., Estivill, X. & Pritchard, M. (1999) Genomics 57 407-418. [DOI] [PubMed] [Google Scholar]
- 27.Kentrup, H., Becker, W., Heukelbach, J., Wilmes, A., Schurmann, A., Huppertz, C., Kainulainen, H. & Joost, H. G. (1996) J. Biol. Chem. 271 3488-3495. [DOI] [PubMed] [Google Scholar]
- 28.Altafaj, X., Dierssen, M., Baamonde, C., Marti, E., Visa, J., Guimera, J., Oset, M., Gonzalez, J. R., Florez, J., Fillat, C. & Estivill, X. (2001) Hum. Mol. Genet. 10 1915-1923. [DOI] [PubMed] [Google Scholar]
- 29.Gibbs, V. C., Takahashi, M., Aguet, M. & Chuntharapai, A. (1996) J. Biol. Chem. 271 28710-28716. [DOI] [PubMed] [Google Scholar]
- 30.Oritani, K., Kincade, P. W., Zhang, C., Tomiyama, Y. & Matsuzawa, Y. (2001) Cytokine Growth Factor Rev. 12 337-348. [DOI] [PubMed] [Google Scholar]
- 31.Muchmore, E., Popper, H., Peterson, D. A., Miller, M. F. & Lieberman, H. M. (1988) J. Med. Primatol. 17 235-246. [PubMed] [Google Scholar]
- 32.Fukuda, R., Ishimura, N., Ishihara, S., Tokuda, A., Satoh, S., Sakai, S., Akagi, S., Watanabe, M. & Fukumoto, S. (1996) J. Gastroenterol. 31 806-811. [DOI] [PubMed] [Google Scholar]
- 33.Goodman, M. (1999) Am. J. Hum. Genet. 64 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supporting Table]