A Common Haplotype of the Glucokinase Gene Alters Fasting Glucose and Birth Weight: Association in Six Studies and Population-Genetics Analyses (original) (raw)
Abstract
Fasting glucose is associated with future risk of type 2 diabetes and ischemic heart disease and is tightly regulated despite considerable variation in quantity, type, and timing of food intake. In pregnancy, maternal fasting glucose concentration is an important determinant of offspring birth weight. The key determinant of fasting glucose is the enzyme glucokinase (GCK). Rare mutations of GCK cause fasting hyperglycemia and alter birth weight. The extent to which common variation of GCK explains normal variation of fasting glucose and birth weight is not known. We aimed to comprehensively define the role of variation of GCK in determination of fasting glucose and birth weight, using a tagging SNP (tSNP) approach and studying 19,806 subjects from six population-based studies. Using 22 tSNPs, we showed that the variant rs1799884 is associated with fasting glucose at all ages in the normal population and exceeded genomewide levels of significance (_P_=10-9). rs3757840 was also highly significantly associated with fasting glucose (_P_=8×10-7), but haplotype analysis revealed that this is explained by linkage disequilibrium (_r_2=0.2) with rs1799884. A maternal A allele at rs1799884 was associated with a 32-g (95% confidence interval 11–53 g) increase in offspring birth weight (_P_=.002). Genetic variation influencing birth weight may have conferred a selective advantage in human populations. We performed extensive population-genetics analyses to look for evidence of recent positive natural selection on patterns of GCK variation. However, we found no strong signature of positive selection. In conclusion, a comprehensive analysis of common variation of the glucokinase gene shows that this is the first gene to be reproducibly associated with fasting glucose and fetal growth.
Blood glucose concentration is tightly regulated in humans, despite considerable variation in quantity, type, and timing of food intake. The appropriate release of insulin and other hormones prevents the damaging effects caused by hyper- or hypoglycemia. In the absence of food, blood glucose levels return to a tightly defined level of 3.5–5.5 mmol/liter in young adults.
Variation of fasting glucose levels is clinically important. Even in the normal range, the fasting glucose level is associated with future risk of type 2 diabetes and ischemic heart disease.1–3 During pregnancy, maternal fasting glucose concentration is an important determinant of offspring birth weight.4,5 Maternal hyperglycemia stimulates the release of fetal insulin, an important fetal growth factor. Factors such as age and obesity explain some of the variation in fasting glucose levels within a population, but, when these are accounted for, considerable variation between individuals still remains. Fasting glucose level is strongly heritable, and much of the interindividual differences in fasting glucose concentration can be explained by genetic variation.6–8
One of the principal regulators of fasting glucose concentrations is the enzyme glucokinase (GCK [MIM 138079]).9 GCK is predominantly expressed in the pancreas and liver and catalyzes the first rate-limiting reaction in glycolysis.9 Its lack of inhibition by its product and its sensitivity to changes in glucose concentrations within the physiological range make it ideally suited for its role as the pancreatic β-cell glucose sensor.9
The key role of GCK in glucose homeostasis is demonstrated by the impact of rare gene mutations that cause a subtype of diabetes known as “maturity-onset diabetes of the young” (MODY [MIM 606391]).10–12 GCK MODY is characterized by mild, stable hyperglycemia during fasting.13 GCK MODY mutations also affect birth weight. Children born to mothers with a GCK mutation are ∼600 g heavier at birth14; conversely, children who have a GCK MODY mutation but whose mothers do not are ∼500 g lighter at birth.14 If both the mother and the child have a GCK mutation, the effects cancel each other out and the baby is of normal birth weight.14
In the past, low–birth-weight babies had a high level of neonatal mortality, which was more marked before modern medical support was available. In contrast, high birth weight can pose a danger to the mother and to the child during birth. Although it is likely that natural selection has had a strong influence on birth-weight variation, several different models of selection are plausible. For example, it may be that variation influencing birth weight evolves under purifying selection, which eliminates alleles that increase or decrease birth weight beyond the optimal value for a given environment. Alternatively, if the population-average birth weight is not close to the optimal value, new variants that improve birth weight will be advantageous. Hence, variants that affect birth weight are interesting candidates for population-genetics studies aimed at understanding the evolution of birth weight.
The extent to which common variation of GCK explains normal variation of fasting glucose and birth weight is not known. We recently described an association between a variant of the GCK promoter, _GCK_-30 (rs1799884 [dbSNP]), and fasting glucose in young adults and pregnant women.15 We also demonstrated an association of the minor allele with increased offspring birth weight.15 It is not known whether further variation in the GCK gene alters fasting glucose or birth weight or the extent to which natural selection influenced variation of this gene. In the present study, we aimed to comprehensively define the role of variation of GCK on fasting glucose and birth weight, using a tagging SNP (tSNP) approach and studying 19,806 subjects from six different studies.
Material and Methods
Association Study
Determining the Linkage-Disequilibrium (LD) Structure of the GCK Gene Region
To determine the LD structure of GCK (45 kb) and the surrounding region (5 kb upstream and 19 kb downstream of GCK isoform 1), we first sequenced the exons of 43 unrelated U.K. white subjects. No common (>5% minor-allele frequency) coding variants were identified. We then used a directed resequencing approach, centering our PCR amplicons on SNPs reported in the November 2002 version of the Santa Cruz genome Web site. We initially focused on common SNPs in regions of conservation among humans, mice, and rats (using mVista), and we defined a cut-off of >100 bp that showed >75% matches. Next, to increase the average SNP density to 1 SNP per 2 kb, we chose additional amplicons randomly. This study was initiated before the availability of HapMap data.
Choice of tSNPs
We used the Tagger program to choose tSNPs.16 We forced in rs1799884, because of the previous association evidence, and rs34976798, which was discovered in our population-genetics survey (see table 1). The latter was chosen because it tags a high-frequency–derived haplotype in the non-African samples. A pairwise tSNP approach was equally efficient as a multimarker approach, so only single-marker tSNPs were used.
Table 1. .
Initial tSNP Association Results for Fasting Glucose in the EFS[Note]
| Mean (SD) Glucose Level [No. of Subjects] for Genotype | |||||
|---|---|---|---|---|---|
| SNP | Alleles(1/2) | 11 | 12 | 22 | ANOVA P |
| rs3217943 | C/T | 4.48 (.47) [84] | 4.53 (.44) [602] | 4.53 (.43) [783] | .594 |
| rs2537184 | G/A | 4.53 (.44) [372] | 4.53 (.44) [744] | 4.51 (.44) [377] | .985 |
| rs882020 | T/C | 4.68 (.36) [16] | 4.53 (.46) [324] | 4.53 (.43) [1,117] | .371 |
| rs2268575 | C/T | 4.67 (.44) [37] | 4.52 (.45) [438] | 4.52 (.43) [976] | .120 |
| rs2971679 | T/C | 4.53 (.47) [42] | 4.53 (.45) [403] | 4.53 (.44) [1,039] | .995 |
| rs2971677 | A/C | 4.51 (.45) [39] | 4.52 (.45) [398] | 4.53 (.43) [1,020] | .870 |
| rs2971676 | A/G | 4.46 (.36) [17] | 4.52 (.45) [232] | 4.53 (.44) [1,240] | .765 |
| rs4724290 | T/C | 4.49 (.43) [108] | 4.53 (.44) [590] | 4.53 (.44) [735] | .714 |
| rs2041547 | C/T | 4.55 (.43) [390] | 4.53 (.45) [744] | 4.49 (.41) [355] | .212 |
| rs2971671 | C/T | 4.60 (.46) [68] | 4.56 (.44) [477] | 4.51 (.43) [887] | .054 |
| rs2284777 | C/T | 4.52 (.34) [19] | 4.52 (.43) [336] | 4.53 (.43) [1,144] | .890 |
| rs758988 | C/A | 4.52 (.44) [1,028] | 4.54 (.45) [421] | 4.51 (.37) [44] | .727 |
| rs2284770 | A/T | 4.50 (.45) [460] | 4.54 (.44) [673] | 4.52 (.41) [320] | .345 |
| rs2284769 | G/C | 4.67 (.29) [19] | 4.51 (.45) [291] | 4.53 (.44) [1,190] | .234 |
| rs1799884 | T/C | 4.53 (.51) [55] | 4.58 (.44) [449] | 4.50 (.43) [988] | .010 |
| rs1476891 | A/G | 4.53 (.44) [683] | 4.52 (.45) [614] | 4.55 (.45) [158] | .775 |
| rs3757840 | G/T | 4.48 (.42) [350] | 4.53 (.43) [714] | 4.57 (.48) [407] | .016 |
| rs34976798a | G/T | 4.68 (.28) [6] | 4.49 (.44) [173] | 4.53 (.44) [1,270] | .347 |
| rs2908287 | G/A | 4.52 (.43) [842] | 4.52 (.44) [546] | 4.58 (.46) [100] | .408 |
| rs2331003 | A/G | 4.53 (.42) [940\ | 4.53 (.44) [490] | 4.56 (.42) [67] | .847 |
| rs1003573 | C/T | 4.53 (.43) [549] | 4.52 (.43) [656] | 4.54 (.46) [233] | .919 |
| rs917791 | G/A | 4.55 (.46) [193] | 4.51 (.43) [698] | 4.54 (.40) [551] | .311 |
Fasting glucose initial study: 1,552 subjects from the Exeter Family Study (EFS)
We genotyped 1,552 subjects (median age 33 years) from the population-based EFS, to test for nominal associations (P<.05) of tSNPs. The EFS has been described in detail elsewhere.17,18 The clinical characteristics of these and the replication studies’ subjects are presented in table 2. Subjects from all studies with fasting glucose over the normal range (>6 mmol/liter) were excluded. Briefly, the EFS is a study of newborn babies and their parents from a geographically defined region of Exeter, United Kingdom.17,18 Fasting glucose was measured using dry-slide technology on Vitros 950 analyzers (Ortho Clinical) before December 2001 and with manufacturer’s standard reagents on Modular analyzers (Roche Diagnostics) thereafter.
Table 2. .
Characteristics of Subjects Examined in the Fasting Glucose Study[Note]
| Finding for | ||||||
|---|---|---|---|---|---|---|
| Characteristica | EFS | PEB Adults | BCG | InChianti | BWHHS | ALSPAC Children |
| No. (% male) | 1,552 (50) | 356 (52) | 626 (53) | 1,022 (43) | 2,652 (0) | 839 (54) |
| Median age, in years (IQR) | 32.0 (7.0) | 34.1 (7.0) | 25.0 (1.0) | 71.0 (12.0) | 68.0 (9.0) | 8.0 (.0) |
| Median BMI, in kg/m2 (IQR) | 24.8 (5.6) | 25.9 (5.4) | 24.3 (7.3) | 27.0 (5.5) | 27.6 (5.0) | 16.5 (4.0) |
| Median fasting glucose, in mmol/liter (IQR) | 4.5 (.6) | 4.7 (.6) | 4.6 (.6) | 4.8 (.7) | 5.6 (.8) | 4.9 (.7) |
| Median fasting blood insulin, in pmol/liter (IQR) | 55.3 (38.5) | 56.7 (36.8) | 39.6 (25.8) | 66.4 (48.3) | Not applicable | 37.7 (44.6) |
Fasting glucose replication studies
We used five population-based studies for replication of nominally significant tSNPs. We studied 356 parents (median age 35 years) from the Plymouth EarlyBird study (PEB).19 The PEB is a nonintervention prospective study of school-aged children and their parents. Fasting glucose was measured on a Cobas Integra 700 analyzer (Roche Diagnostics).
We studied 626 subjects (median age 25 years) from the Barry Caerphilly Growth study (BCG).20 These subjects are from the follow-up of consecutively born infants, from the towns of Barry and Caerphilly in South Wales, born between 1972 and 1974. Between 1997 and 1999, all the original children who had completed the 5-year follow-up were traced, and they completed a questionnaire and attended a screening clinic, where growth and oral glucose tolerance test measurements were taken.
We studied 1,022 subjects (median age 71 years) from the Invecchiare in Chianti study (InChianti), a prospective population-based cohort of elderly Italians from two towns (Greve in Chianti and Bagno a Ripoli) from the Chianti region of Tuscany. The study includes randomly selected participants aged 65–102 years and 30 men and women from each age decade between 20 and 70 years. The data collection started in September 1998 and was completed in March 2000. Fasting glucose concentration was determined by an enzymatic colorimetric assay, with use of a modified glucose-oxidase-peroxidase method and a Roche-Hitachi 917 analyzer (Roche Diagnostics).
We studied 2,652 subjects (median age 68 years) from the British Women’s Health and Heart Study (BWHHS). The BWHHS is a cohort of women aged 60–79 years, randomly selected from 23 British towns.21 Baseline data were collected between April 1999 and March 2001.
We studied 839 children (median age 8 years) from the Avon Longitudinal Study of Parents and Children (ALSPAC) (the only subjects from the ALSPAC study for whom fasting glucose measurements were available). Glucose was measured by the glucose-oxidase method on a YSI 2300 stat plus analyzer. Further details of the study are described in the “Birth-weight study” section. Blood samples were taken after a minimum 6-h fast. Plasma glucose was measured by a glucose oxidase Trinder method,22 with use of a Hitachi Modular analyzer.
Birth-weight study
We examined ALSPAC subjects for the association of maternal and offspring GCK polymorphisms with offspring birth weight. ALSPAC is a prospective study of pregnancies recruited from all pregnant women in three Bristol-based District Health Authorities with expected delivery dates between April 1991 and December 1992. We have reported elsewhere the association of one GCK polymorphism with offspring birth weight in 1,159 of the ALSPAC mother-child pairs.15 In the present study, we extended this earlier analysis by including >6,500 mother-child pairs. Birth weights were taken from hospital records, and gestational age was estimated from the last menstrual period and ultrasound data. For the present study, subjects born after gestation of <36 wk, nonwhites, and twins were excluded from the analysis.
Association-study statistics
All standard statistical analyses were performed using Stata SE version 9 (StataCorp). We used analysis of variance (ANOVA) and linear regression to test for association of fasting glucose and birth weight with genotype. For LD statistics and tSNP identification, we used Tagger.16 For the haplotype analyses, we used qtphase from the unphased suite of programs.23 Fixed-effects meta-analysis statistics and plots were generated using Stats Direct version 2.4.2 (Sale), with a weighted-mean-difference approach. For the association of genotype with fasting glucose, we included only normal, glucose-tolerant subjects in our analyses and therefore excluded anyone with a fasting glucose >6 mmol/liter. For the studies of elderly subjects, this meant exclusion of a high proportion of subjects (13.9% from InChianti and 36.5% from the BWHHS). We therefore also performed the analyses excluding only World Health Organization–defined diabetic subjects (fasting glucose >7 mmol/liter). For the birth-weight studies, we analyzed (1) the effect of maternal genotype on birth weight both with and without correction for fetal genotype and (2) the effect of fetal genotype on birth weight both with and without correction for maternal genotype.
Power
We used Quanto for power calculations.24 For the initial study of all tSNPs, with the assumption of an additive model and an allele frequency of 20%, we had ∼80% power to detect a per-allele effect on fasting glucose of >0.055 mmol/liter, at P<.05. We did not correct for the testing of multiple SNPs; instead, we included all nominally significant SNPs (_P_<.05) in the replication studies. For birth weight, with the assumption of an additive model and an allele frequency of 20%, the ALSPAC cohort provided ∼80% power to detect per-allele differences of >26 g for both maternal and fetal genotype analyses at the P<.05 level of significance.
Genotyping method
All genotyping was performed by Kbioscience. Details of the company and the KASPar genotyping method can be found at the Kbioscience Web site.
Genotyping quality control
In the initial study of the EFS subjects, we analyzed 23 tSNPs, and all were in Hardy-Weinberg equilibrium (HWE) (_P_>.01) except rs2537185 (_P_=4×10-9). The overall success rate for the 23 tSNPs in the initial study was 94.6%; the minimum success rate was 92.1% for rs2971671. The overall genotype error rate in the initial part of the study was 0.6% (18 errors from 3,243 duplicate genotypes). Six of these duplicate errors were for SNP rs2537185 (6/141=4.3% error rate). Given this error rate and the departure from HWE, rs2537185 was excluded from further analysis. For the genotyping of rs1799884 and rs3757840, all cohorts were in HWE (all _P_>.10), and the LD statistics between the two SNPs were similar across all studies (_r_2 range 0.18–0.22; D′ range 0.98–1.00).
Population-Genetics Study
DNA samples
We used a separate set of samples to test for evolutionary neutrality in the GCK region. We surveyed sequence variation in samples from three major ethnic groups: 16 Hausa from Yaounde (Cameroon), 16 Han Chinese from Taiwan, and 16 individuals from central Italy; we used a European sample separate from that used in the study SNP survey, because these same population samples, together with the Hausa and Han Chinese samples, were surveyed elsewhere at 50 unlinked noncoding (therefore, putatively neutrally evolving) regions25,26 and at other candidate diabetes genes.27 This is important because it helps distinguish between differences in patterns of sequence variation due to demography alone and those due to selection and demography. SNPs associated with fasting glucose or with possible evidence of selection were genotyped in the Human Genome Diversity Project (HGDP) panel of 1,052 individuals (of whom 927 are unrelated) from 52 populations. Sequence data were also collected from one common chimpanzee, one gorilla, and one orangutan, to determine the putative ancestral allele at each polymorphic site.
PCR amplification and DNA sequencing
Primers for PCR amplification and sequencing were designed using PRIMER.28 The survey included a segment of 17,355 bp centered on rs1799884 in the GCK promoter and spanning the entire gene. PCR primers were designed to amplify a 2,000–2,400-bp fragment, with at least 200 bp overlap between amplicons. Sequencing was performed using internal sequencing primers with the ABI Big Dye Terminator v. 3.1 Cycle Sequencing kit and with ABI 3700 or 3730 automated sequencers. The Phred-Phrap-Consed package was used to assemble and analyze all sequences and score genotypes.29
Genotyping of HGDP
On the basis of the fasting glucose association results and the survey of population-genetics variation, three SNPs were genotyped in the HGDP panel: rs3757840, rs1799884, and rs13239289. SNPs rs1799884 and rs13239289 were genotyped using the 5′ nuclease assay (TaqMan [Applied Biosystems]). SNP rs3757840 was genotyped by RFLP analysis with _Bgl_II. Primer sequences and PCR conditions for both the 5′ nuclease assays and the RFLP assay are available from the authors.
Data analysis
Summary statistics of population variation were calculated using the online applications SLIDER and MAXDIP.25,30 The HKA test was performed using an online application. Haplotypes were inferred, using PHASE2,31 in each population sample separately. Coalescent simulations for the haplotype test were performed with the assumption of values of 4_Nr_ estimated from the data for each population sample.32 For all tests, gene conversion was assumed to occur twice as often as crossover, and the mean conversion tract length was 500 bp.
Results
LD Structure of the Glucokinase Region
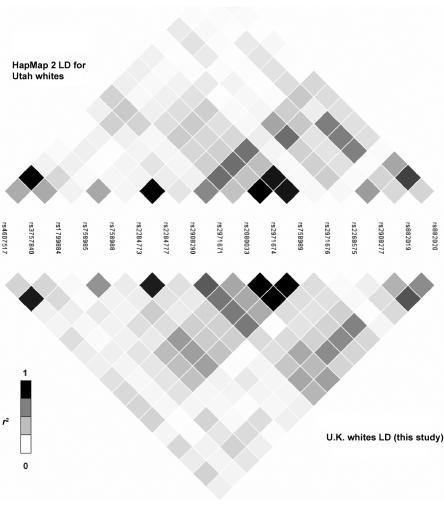
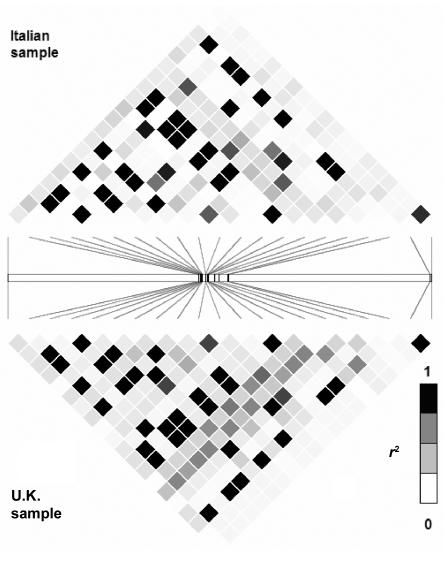
The LD structure of the 116-kb region of GCK that is based on 84 SNPs genotyped for 43 unrelated U.K. white subjects is shown in figure 1. We compared the LD structure determined by our resequencing of the GCK region with data from HapMap 2 and from the Italian sample of the population-genetics survey. Seventeen SNPs were in both the HapMap 2 and our data set; figure 2 compares the data sets. Twenty SNPs were identified by both the tSNP association study and the population-genetics survey of the GCK region; figure 3 compares the data sets. Patterns of LD were very similar across all three sets of samples.
Figure 1. .
LD structure of glucokinase. _r_2 values between the 84 SNPs across a 116-kb region are presented. Arrows indicate tSNPs used in association study.
Figure 2. .
Comparison of LD structure determined in this study with that determined from HapMap 2 data. The LD structures are based on the _r_2 values between the 17 SNPs that are present in both data sets.
Figure 3. .
Comparison of LD structure determined for 16 Italians from the Natural Selection study with that determined from U.K. samples used for the tSNP-association study. The LD structure is based on the _r_2 and values between 20 SNPs that are present in both data sets.
tSNP Performance
Of the 23 genotyped tSNPs, 22 passed our stringent quality-control criteria. These 22 SNPs captured all 84 SNPs, with a mean _r_2 of 0.837. Of the captured SNPs, 73% SNPs were captured with an _r_2>0.8, 89% were captured with an _r_2>0.5, and 98% were captured with an _r_2>0.2.
Initial tSNP Fasting Glucose Association Analysis
The results for the initial tSNP association analysis are shown in table 1. Two SNPs, rs3757840 and rs1799884, were nominally significant.
Fasting Glucose Replication Studies
We next attempted to replicate the associations in an additional 5,495 subjects from five further studies. The results for both SNPs in the individual cohorts are shown in table 3 and figure 4. Overall, there was strong replication of the SNPs with fasting glucose in these cohorts (overall combined P values: for rs1799884, _P_=1×10-9; for rs3757840, _P_=8×10-7). The result was very similar when samples with impaired fasting glucose (i.e., a cut-off of 7 mmol/liter rather than 6 mmol/liter) were included.
Table 3. .
Fasting Glucose Results for rs1799884 and rs3757840 for All Cohorts in the Present Study[Note]
| Mean (SD) Glucose Level [No. of Subjects] for | ||||||||
|---|---|---|---|---|---|---|---|---|
| rs1799884 | rs3757840 | |||||||
| Study | GG | GA | AA | Dominant P | CC | CA | AA | Linear Trend P |
| ALSPAC Children | 4.90 (.32) [506] | 4.98 (.32) [207] | 4.95 (.35) [21] | .008 | 4.87 (.32) [174] | 4.94 (.32) [368] | 4.95 (.32) [180] | .02 |
| BCG | 4.57 (.38) [426] | 4.64 (.37) [180] | 4.48 (.45) [18] | .13 | 4.57 (.41) [127] | 4.59 (.36) [324] | 4.64 (.40) [166] | .10 |
| EFS | 4.50 (.43) [988] | 4.58 (.44) [449] | 4.53 (.51) [55] | .01 | 4.48 (.42) [350] | 4.53 (.43) [714] | 4.57 (.48) [407] | .02 |
| PEB | 4.65 (.69) [224] | 4.75 (.41) [94] | 4.66 (.48) [14] | .08 | 4.61 (.44) [81] | 4.69 (.45) [172] | 4.79 (.48) [76] | .01 |
| BWHHS | 5.49 (.39) [1,671] | 5.54 (.37) [678] | 5.60 (.28) [69] | .001 | 5.48 (.41) [594] | 5.51 (.38) [1,238] | 5.53 (.35) [578] | .03 |
| InChianti | 4.77 (.49) [657] | 4.82 (.51) [321] | 4.95 (.47) [44] | .03 | 4.76 (.51) [178] | 4.79 (.49) [456] | 4.80 (.50) [293] | .40 |
Figure 4. .
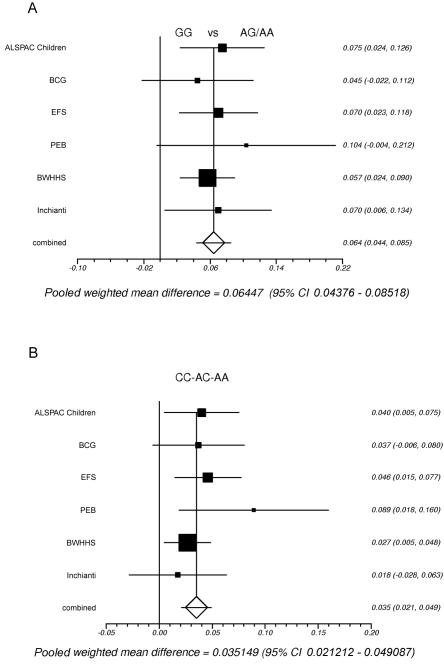
A, Meta-analysis of the fasting glucose association results of rs1799884 AA and AG versus GG for all studies. Studies are ordered by ascending age of the cohort. Heterogeneity _P_=.94; combined _P_=1×10-9. B, Meta-analysis of the fasting glucose association results of rs3757840. The effect size per allele is presented. Heterogeneity _P_=.56; combined _P_=8×10-7.
Are There Two Independent Effects?
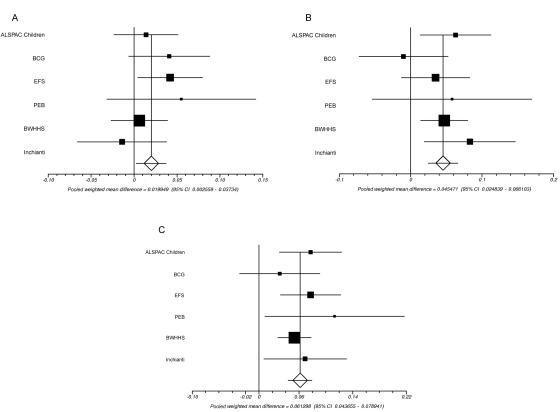
The SNPs rs1799884 and rs3757840 are 2,148 bp apart and are in LD (_D_′=1; _r_2=0.2). To determine whether these SNPs have independent effects on fasting glucose, we performed haplotype analyses using the unphased program.23 The haplotype results are presented in table 4 and figure 5. Alleles from haplotypes formed by these two SNPs are given as rs1799884, then rs3757840. The pairwise comparison of the GA and the GC haplotypes (a test of rs3757840) was only marginally significant (fig. 5_A_) (combined effect _size_=0.02 mmol/liter; combined _P_=.03). In contrast, the AA versus GA comparison (a test of rs1799884) was highly significant (fig. 5_B_) (combined effect _size_=0.05 mmol/liter; _P_=1×10-5), and the GC versus AA comparison (a test of both SNPs) was also significant (fig. 5_C_) (combined effect _size_=0.06 mmol/liter; _P_=1×10-11). This strongly suggests that the rs1799884 SNP statistically explains the association of the rs3757840 variant with fasting glucose. Regression analyses, which analyzed the effect of one SNP on fasting glucose while adjusting for the other, produced similar results (data not shown).
Table 4. .
Haplotype Results for the Association of Fasting Glucose against rs1799884 and rs3757840 for All Cohorts[Note]
| Mean (SD) Glucose Level [No. of Subjects] for Haplotype | Difference in Glucose (95% CI) [_P_] for Pairwise Haplotype Comparisona | ||||||
|---|---|---|---|---|---|---|---|
| Study | GC | GA | AA | Global P | GA and GC | AA and GA | AA and GC |
| ALSPAC Children | 4.91 (.01) [707] | 4.92 (.01) [455] | 4.99 (.02) [241] | .009 | .014 (−.02, .05) [.46] | .063 (.01, .11) [.013] | .077 (.03, .12) [.001] |
| BCG | 4.58 (.02) [571] | 4.62 (.02) [439] | 4.61 (.03) [208] | .21 | .041 (−.01, .09) [.08] | −.01 (−.07, .05) [.70] | .03 (−.03, .09) [.31] |
| EFS | 4.50 (.01) [1,293] | 4.55 (.01) [886] | 4.58 (.02) [511] | .002 | .04 (.00, .08) [.03] | .08 (.03, .12) [.0008] | .04 (−.01, .08) [.15] |
| PEB | 4.67 (.03) [277] | 4.72 (.04) [178] | 4.78 (.05) [103] | .09 | .055 (−.03, .14) [.21] | .06 (−.05, .02) [.31] | .11 (.06, .16) [.03] |
| BWHHS | 5.50 (.01) [2,301] | 5.51 (.01) [1,487] | 5.55 (.01) [786] | .008 | .01 (−.02, .04) [.64] | .05 (.01, .08) [.005] | .05 (.02, .08) [.0007] |
| InChianti | 4.79 (.02) [817] | 4.78 (.02) [666] | 4.86 (.03) [384] | .03 | −.01 (−.07, .04) [.66] | .08 (.02, .15) [.0109] | .07 (.01, .13) [.03] |
Figure 5. .
Meta-analysis of the pairwise differences between haplotypes of rs1799884 and rs3757840 across fasting glucose. A, GA versus GC. B, AA versus GA. C, AA versus GC. Heterogeneity _P_=.40, .41, and .65, respectively.
Birth-Weight Analysis for rs1799884
We next examined the effect of rs1799884 on birth weight, using 6,366 mothers and 7,232 offspring from the ALSPAC study. Results are shown in table 5. We have replicated our previous results, which shows that the presence of a maternal A allele at rs1799884 is associated with an increase of 27 g in offspring birth weight (_P_=.02). Correction for fetal genotype did not affect the magnitude of the association (26 g); we still found no evidence of a fetal genotype effect, either before (_P_=.72) or after (_P_=.56) correction for maternal genotype. When we combined this result with our data published elsewhere,15 the maternal genotype effect size was 32 g (95% CI 11–53 g) greater birth weight for A allele carriers at rs1799884 (_P_=.002).
Table 5. .
ALSPAC Study Birth-Weight Association Results for the rs1799884 Genotype[Note]
| Mean Birth Weight (SE) [No. of Subjects] for rs1799884 | ||||
|---|---|---|---|---|
| Genotype | GG | GA | AA | Pa |
| Maternal | 3,476 (6.7) [4,269] | 3,501 (10.1) [1,905] | 3,522 (31.8) [192] | .02 |
| Offspring | 3,500 (6.3) [4,896] | 3,507 (9.6) [2,090] | 3,518 (30.9) [246] | .72 |
Population-Genetics Analyses
The resequencing survey of 48 unrelated individuals from three major ethnic groups, centered on the fasting glucose–associated SNP rs1799884, yielded 112 polymorphisms: 105 biallelic SNPs and 7 insertion/deletions.
Summary statistics of sequence variation are presented in table 6. Polymorphism levels are summarized in terms of the number of polymorphic sites (S) and nucleotide diversity per bp (π). Information about LD levels is summarized by ρH01, which is a composite likelihood estimator of the population crossover–rate parameter (4_Nr_ , where N is the effective population size and r is the crossover rate between adjacent nucleotides).25,30 Tajima’s D summarizes information about the spectrum of allele frequencies in each sample33; a value near zero is expected under an equilibrium-neutral model, whereas negative and positive values indicate, respectively, an excess of rare and intermediate frequency variants.
Table 6. .
Summary Statistics of Observed Variation in the Resequencing Survey
| Population | Sa | πb(%) | Tajima’s Dc | ρH01d(%) |
|---|---|---|---|---|
| Hausa | 47 | .060 | −.491 | .0397 |
| Italians | 81 | .122 | .159 | .0212 |
| Chinese | 75 | .116 | .268 | .0270 |
Contrary to the pattern observed in most studies of human variation,26,34–39 GCK polymorphism levels in the Hausa sample from sub-Saharan Africa are lower than those observed in both non-African samples. We compared these results with those of the 50 noncoding regions surveyed in the same samples.26 Five of these regions were more variable in the Italian than in the Hausa sample, but none of them showed a difference as large as that observed at GCK. When the Hausa and the Chinese samples were compared, only one noncoding region showed a difference as large as that observed at GCK for the same samples. Hence, polymorphism levels at GCK in the Hausa sample appear to be unusually low relative to those observed in the non-African samples, as might be expected if positive natural selection drove a variant to fixation in the Hausa but not in the other samples. To test whether polymorphism levels in the Hausa are low relative to the expectations of a standard neutral model, we used the HKA test40 to compare levels of polymorphism and sequence divergence between human and chimpanzee at GCK with those observed at 50 noncoding regions surveyed in the same samples. This test detected a nonsignificant deficit of polymorphism levels in the Hausa (_P_=.148) and a nonsignificant excess of polymorphism levels in the Italians and Chinese (_P_=.068 and _P_=.105, respectively). Likewise, the allele-frequency spectrum is not unusual, compared with the 50 noncoding regions in the same samples and compared with the expectations of the standard neutral model.
Visual inspection of the inferred haplotypes for each population sample revealed a haplotype class defined by at least seven SNPs (rs13306391, rs34976798, 65255 [nucleotide position from start of AC006454, submitted to dbSNP], rs13239289, rs34807880, rs34380942, and rs34780996) at high frequency in the non-African samples (27 of 32 inferred haplotypes) and fixed in the Hausa sample. To determine whether positive natural selection acted on this haplotype class, we used the haplotype test,32 which determines whether there is a deficit of variation within a haplotype class, given its frequency and relative to neutrality expectations. Briefly, given the number of variable sites in a population sample, coalescent simulations under neutrality are performed to estimate the probability that a haplotype subset of a given size has as few or fewer polymorphic sites than in the observed haplotype distribution. This test did not detect a significant departure in either the Italian or the Chinese data (_P_>.2).
We also used the results of a recent analysis of the phase I data of the International HapMap Project to assess the evidence of positive natural selection (Haplotter).41,42 The analysis is based on a test statistic, called the “integrated haplotype score” (IHS), which measures how unusual the haplotypes around a given SNP are, relative to the genomewide HapMap data; the larger the absolute value of IHS, the more unusual the haplotypes. The GCK gene does not show an unusual clustering of SNPs with large IHS values (_P_=.99 in all HapMap population samples). We also searched for three SNPs of interest: rs1799884 and rs3757840, which are associated with variation in birth weight and fasting glucose, and rs13239289, which defines the high-frequency haplotype in the Italians and Chinese. Because rs3757840 and rs13239289 are not in the HapMap phase I data, we used surrogate SNPs rs7810158 and rs3757838 (_r_2=0.74 and _r_2=1, respectively). None of the SNPs is associated with an unusual haplotype structure on a genomewide scale.
It was shown elsewhere that birth weight is inversely correlated with heat stress, which raises the possibility that genetic variation in birth weight may be shaped by selective pressures related to climate variables.43 To assess whether such spatially varying selective pressures acted on variation in the GCK promoter, we genotyped three SNPs in the HGDP panel. Two of the SNPs (rs1799884 and rs3757840) were chosen because they show a significant association with variation in birth weight and fasting glucose. The third SNP (rs13239289) was chosen among those that define the haplotype fixed in the Hausa sample and at near-fixation frequency in the non-African samples. We tested the null hypothesis of no correlation between SNP allele frequency in each HGDP population and a set of climate-related variables for the same populations by using the Spearman rank correlation test. However, a correlation with some of these variables might be expected simply as a result of the history of human migrations and may not necessarily reflect the action of spatially varying selection. To determine whether the observed correlations are unusual compared with empirical genomewide patterns, we compared the rank correlation score for each SNP with the distribution of rank correlation scores for a set of microsatellite loci genotyped in the HGDP panel for each climate variable, as described in the work of Thompson et al.44; the proportion of microsatellites with more-extreme rank correlation scores can be considered an empirical P value for the correlation observed at the SNPs. As shown in table 7, rs13239289 has a strong correlation with temperature variables, which is unusual in comparison with the microsatellite distribution. However, this SNP—as determined by genotyping rs34976798, which defines the same haplotype and has an _r_2 of 1.0 with rs13239289 in the 48 samples—is not associated with fasting glucose (_P_=.35) in the tSNP analysis (table 1.)
Table 7. .
Correlation between Allele Frequency and Climate Variables in the HGDP Panel
| Findings for SNP | ||||||
|---|---|---|---|---|---|---|
| rs13239289 | rs3757840 | rs1799884 | ||||
| ClimateVariableaand Season | r | P | r | P | r | P |
| Minimum: | ||||||
| Summer | −.277 | .2176 | −.076 | .6648 | −.081 | .7916 |
| Winter | −.542 | .0209 | −.161 | .4034 | −.103 | .7580 |
| Maximum: | ||||||
| Summer | −.223 | .4184 | −.239 | .1761 | −.146 | .5966 |
| Winter | −.549 | .0126 | −.223 | .3068 | −.029 | .9445 |
| Surface: | ||||||
| Summer | −.234 | .2915 | −.200 | .2159 | −.140 | .5664 |
| Winter | −.564 | .0126 | −.211 | .3182 | −.096 | .7748 |
Discussion
We performed a tSNP association analysis of the glucokinase gene to identify common genetic variants associated with fasting glucose and birth weight. Using six studies, we consistently replicated the association of a haplotype, defined by rs1799884_—previously called “_GCK(-30)”—with fasting glucose and birth weight. Previous smaller studies of GCK by ourselves and others15,45,46 that analyzed only rs1799884 have found a similar increase in fasting glucose associated with the A allele. Our results provide evidence that this variant alters fasting glucose at genomewide levels of significance. Not all individual studies provided significant evidence of association, and some provided stronger evidence for rs3757840, a SNP partially correlated with rs1799884, with an _r_2 of 0.22. Our results highlight the importance of using multiple replication studies to establish the true effect of common variants on common phenotypes. Our results also indicate that association signals may be detected with SNPs with _r_2 values as low as 0.22.
Our tSNP analysis found no evidence that GCK variation, other than the variation in the haplotype that includes rs1799884, affects GCK activity. The tSNP with the highest _r_2 with _rs1799884_—_rs3757840_—was associated with fasting glucose, but the association is explained statistically by LD with rs1799884 (_r_2=0.22). We also note that the tSNP rs2971671, with the second highest _r_2 with rs1799884 (_r_2=0.19), was close to reaching nominal significance in our initial study (_P_=.054). The identification of the signal of rs1799884 with use of a SNP with a low _r_2 highlights the potential of using a tSNP approach for association studies in identification of common variants for complex traits. This is important, since the utility in identification of common disease variants of the HapMap project and whole-genome association studies has been questioned.47
We have replicated our previous finding that the maternal rs1799884 genotype influences offspring birth weight. The association of maternal rs1799884 with offspring birth weight (27 g) is, as predicted, from the correlation between maternal glucose concentration and offspring birth weight—on the basis of regression of maternal pregnant glucose and birth weight in the EFS mothers, a 0.08 mmol/liter increase in fasting glucose would be expected to increase birth weight by 21 g. We found no evidence that fetal genotype is associated with offspring birth weight (A allele effect _size_=4 g; _P_=.72).
The population-genetics analyses of GCK was centered on the rs1799884 SNP associated with fasting glucose and revealed a few interesting patterns. An extended haplotype defined by the derived allele at 17 SNPs occurs at high frequency (84%) in the two non-African samples and is fixed in the Hausa sample. Accordingly, the Hausa sample exhibits lower polymorphism levels compared with the non-African samples. This could be consistent with a complete selective sweep in the Hausa sample and a nearly complete sweep in the non-African samples. However, a statistical test aimed at determining whether the deficit of polymorphism in the Hausa is inconsistent with evolutionary neutrality failed to reject the null hypothesis (_P_=.148). Likewise, the haplotype test did not yield significant results in the non-African samples. Although these results may be due to limited power of the tests, we conclude that there is no evidence in the resequencing data to indicate recent positive natural selection. Interestingly, however, one of the SNPs defining the haplotype fixed in the Hausa and at high frequency in the non-African samples shows a strong correlation with climate variables. Given the reported correlation between birth weight and indices of heat stress,45 it is tantalizing to speculate that GCK variation linked to this haplotype may have been shaped by spatially varying selective pressures related to climate.
We also asked whether the variants contributing to fasting glucose and birth-weight variation in our association studies were associated with a selective advantage. However, neither our resequencing data nor an analysis of the HapMap phase I data suggests that this is the case. Given the small phenotypic effect of rs1799884 on birth weight (i.e., ∼32 g), it is perhaps not surprising that no fitness effect appears to be associated with this variant.
Other important conclusions can be drawn from our study. The effect of rs1799884 on fasting glucose is constant in groups of subjects whose median age varied from 8 years to 71.5 years (heterogeneity _P_=.94) when a cutoff of 6 mmol/liter is taken (i.e., normal glucose-tolerant subjects only).
In conclusion, we have comprehensively assessed the role of the glucokinase gene in determining fasting glucose level and birth weight. Birth weight is a trait strongly influenced by evolutionary pressure, but a comprehensive analysis of GCK did not reveal any consistent signals of recent positive selection. We have reproducibly, and at genomewide levels of significance, identified a common haplotype of the glucokinase gene that affects fasting glucose and birth weight.
Acknowledgments
The population genetics work in this study was funded by National Institutes of Health grant R01 DK056670. M.N.W. is a Vandervell Foundation research fellow. BWHHS is funded by grants from the U.K. Department of Health and the British Heart Foundation. We thank P. Whincup, G. Wannamethee, and Rita Patel, who have contributed to the direction and management of data in this study. D.A.L. is funded by a U.K. Department of Health Career Scientist Award. V.J.C. is supported by National Research Service Award postdoctoral fellowship DK66974. We are extremely grateful to all the families who took part in this study, the midwives who helped in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The U.K. Medical Research Council, the Wellcome Trust, and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors, and M.E.P. and T.M.F. will serve as guarantors for the contents of this work.
Web Resources
The URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Haplotter, http://hg-wen.uchicago.edu/selection/haplotter.htm
- HKA, http://genapps.uchicago.edu/hka/index.html
- Kbioscience, http://www.kbioscience.co.uk/
- MAXDIP, http://genapps.uchicago.edu/maxdip/index.html
- mVista, http://genome.lbl.gov/vista/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for GCK and MODY)
- SLIDER, http://genapps.uchicago.edu/slider/index.html
References
- 1.Bjornholt J, Erikssen G, Aaser E, Sandvik L, Nitter-Hauge S, Jervell J, Erikssen J, Thaulow E (1999) Fasting blood glucose: an underestimated risk factor for cardiovascular death: results from a 22-year follow-up of healthy nondiabetic men. Diabetes Care 22:45–49 [DOI] [PubMed] [Google Scholar]
- 2.Bjornholt J, Erikssen G, Liestol K, Erikssen J, Thaulow E (2001) Prediction of type 2 diabetes in healthy middle-aged men with special emphasis on glucose homeostasis: results from 22.5 years’ follow-up. Diabet Med 18:261–267 10.1046/j.1464-5491.2001.00488.x [DOI] [PubMed] [Google Scholar]
- 3.Charles MA, Fontbonne A, Thibult N, Warnet JM, Rosselin GE, Eschwege E (1991) Risk factors for NIDDM in white population: Paris prospective study. Diabetes 40:796–799 [DOI] [PubMed] [Google Scholar]
- 4.Farmer G, Russell G, Hamilton-Nicol DR, Ogenbede HO, Ross IS, Pearson DW, Thom H, Kerridge DF, Sutherland HW (1988) The influence of maternal glucose metabolism on fetal growth, development and morbidity in 917 singleton pregnancies in nondiabetic women. Diabetologia 31:134–141 10.1007/BF00276845 [DOI] [PubMed] [Google Scholar]
- 5.Breschi MC, Seghieri G, Bartolomei G, Gironi A, Baldi S, Ferrannini E (1993) Relation of birthweight to maternal plasma glucose and insulin concentrations during normal pregnancy. Diabetologia 36:1315–1321 10.1007/BF00400812 [DOI] [PubMed] [Google Scholar]
- 6.Mills GW, Avery PJ, McCarthy MI, Hattersley AT, Levy JC, Hitman GA, Sampson M, Walker M (2004) Heritability estimates for beta cell function and features of the insulin resistance syndrome in UK families with an increased susceptibility to type 2 diabetes. Diabetologia 47:732–738 10.1007/s00125-004-1338-2 [DOI] [PubMed] [Google Scholar]
- 7.Freeman MS, Mansfield MW, Barrett JH, Grant PJ (2002) Heritability of features of the insulin resistance syndrome in a community-based study of healthy families. Diabet Med 19:994–999 10.1046/j.1464-5491.2002.00843.x [DOI] [PubMed] [Google Scholar]
- 8.Snieder H, Boomsma D, van Doornen LJ, Neale MC (1999) Bivariate genetic analysis of fasting insulin and glucose levels. Genet Epidemiol 16:426–426 [DOI] [PubMed] [Google Scholar]
- 9.Matschinsky FM (1990) Glucokinase as glucose sensor and metabolic signal generator in pancreatic β-cell and hepatocytes. Diabetes 30:647–752 [DOI] [PubMed] [Google Scholar]
- 10.Vionnet N, Stoffel M, Takeda J, Yasuda K, Bell GI, Zouali H, Lesage S, Velho G, Iris F, Passa PH, Froguel PH, Cohen D (1992) Nonsense mutation in the glucokinase gene causes early-onset non-insulin-dependent diabetes mellitus. Nature 356:721–722 10.1038/356721a0 [DOI] [PubMed] [Google Scholar]
- 11.Froguel P, Vaxillaire M, Sun F, Velho G, Zouali H, Butel MO, Lesage S, Vionnet N, Clement K, Fougerousse F, Tanizawa Y, Weissenbach J, Beckmann JS, Lathrop GM, Passa P, Permutt MA, Cohen D (1992) Close linkage of glucokinase locus on chromosome 7p to early-onset non-insulin-dependent diabetes mellitus. Nature 356:162–164 10.1038/356162a0 [DOI] [PubMed] [Google Scholar]
- 12.Hattersley AT, Turner RC, Permutt MA, Patel P, Tanizawa Y, Chiu KC, O’Rahilly S, Watkins PJ, Wainscoat JS (1992) Linkage of type 2 diabetes to the glucokinase gene. Lancet 339:1307–1310 10.1016/0140-6736(92)91958-B [DOI] [PubMed] [Google Scholar]
- 13.Stride A, Vaxillaire M, Tuomi T, Barbetti F, Njolstad PR, Hansen T, Costa A, Conget I, Pedersen O, Sovik O, Lorini R, Groop L, Froguel P, Hattersley AT (2002) The genetic abnormality in the beta cell determines the response to an oral glucose load. Diabetologia 45:427–435 10.1007/s00125-001-0770-9 [DOI] [PubMed] [Google Scholar]
- 14.Hattersley AT, Beards F, Ballantyne E, Appleton M, Harvey R, Ellard S (1998) Mutations in the glucokinase gene of the fetus result in reduced birth weight. Nat Genet 19:268–270 10.1038/953 [DOI] [PubMed] [Google Scholar]
- 15.Weedon MN, Frayling TM, Shields B, Knight B, Turner T, Metcalf BS, Voss L, Wilkin TJ, McCarthy A, Ben-Shlomo Y, Smith GD, Ring S, Jones R, Golding J, Byberg L, Mann V, Axelsson T, Syvanen AC, Leon D, Hattersley AT (2005) Genetic regulation of birth weight and fasting glucose by a common polymorphism in the islet cell promoter of the glucokinase gene. Diabetes 54:576–581 [DOI] [PubMed] [Google Scholar]
- 16.de Bakker PIW, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D (2005) Efficiency and power in genetic association studies. Nat Genet 37:1217–1223 10.1038/ng1669 [DOI] [PubMed] [Google Scholar]
- 17.Knight B, Shields BM, Turner M, Powell RJ, Yajnik CS, Hattersley AT (2005) Evidence of genetic regulation of fetal longitudinal growth. Early Hum Dev 81:823–831 10.1016/j.earlhumdev.2005.06.003 [DOI] [PubMed] [Google Scholar]
- 18.Knight B, Shields BM, Hattersley AT (2006) The Exeter Family Study of Childhood Health (EFSOCH): study protocol and methodology. Paediatr Perinat Epidemiol 20:172–179 10.1111/j.1365-3016.2006.00701.x [DOI] [PubMed] [Google Scholar]
- 19.Wilkin TJ, Voss LD, Metcalf BS, Mallam K, Jeffery AN, Alba S, Murphy MJ (2004) Metabolic risk in early childhood: the EarlyBird Study. Int J Obes Relat Metab Disord Suppl 28:S64-S69 10.1038/sj.ijo.0802807 [DOI] [PubMed] [Google Scholar]
- 20.Frayling TM, Hattersley AT, McCarthy A, Holly J, Mitchell SM, Gloyn AL, Owen K, Davies D, Davey-Smith G, Ben-Shlomo Y (2002) A putative functional polymorphism in the IGF-1 gene: association studies with type 2 diabetes, adult height, glucose tolerance, and fetal growth in UK populations. Diabetes 51:2313–2316 [DOI] [PubMed] [Google Scholar]
- 21.Lawlor DA, Bedford C, Taylor M, Ebrahim S (2003) Geographical variation in cardiovascular disease, risk factors, and their control in older women: British Women’s Heart and Health Study. J Epidemiol Community Health 57:134–140 10.1136/jech.57.2.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trinder P (1969) Determination of blood glucose using 4-amino phenazone as oxygen acceptor. J Clin Pathol 22:246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudbridge F (2003) Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 25:115–121 10.1002/gepi.10252 [DOI] [PubMed] [Google Scholar]
- 24.Gauderman WJ (2002) Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol 155:478–484 10.1093/aje/155.5.478 [DOI] [PubMed] [Google Scholar]
- 25.Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A (2001) Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet 69:831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voight BF, Adams AM, Frisse LA, Qian YD, Hudson RR, Di Rienzo A (2005) Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci USA 102:18508–18513 10.1073/pnas.0507325102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vander Molen J, Frisse LM, Fullerton SM, Qian Y, del Bosque-Plata L, Hudson RR, Di Rienzo A (2005) Population genetics of CAPN10 and GPR35: implications for the evolution of type 2 diabetes variants. Am J Hum Genet 76:548–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lincoln SE, Daly MJ, Lander ES (1991) PRIMER: a computer program for automatically selecting PCR primers, release 0.5, Cambridge, MA [Google Scholar]
- 29.Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 10.1093/nar/25.14.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson RR (2001) Two-locus sampling distributions and their application. Genetics 159:1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ (1994) Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics 136:1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, Kruglyak L (2004) Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol 2:E286 10.1371/journal.pbio.0020286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cann RL, Stoneking M, Wilson AC (1987) Mitochondrial DNA and human evolution. Nature 325:31–36 10.1038/325031a0 [DOI] [PubMed] [Google Scholar]
- 36.Nachman MW, Bauer VL, Crowell SL, Aquadro CF (1998) DNA variability and recombination rates at X-linked loci in humans. Genetics 150:1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, et al (2001) Haplotype variation and linkage disequilibrium in 313 human genes. Science 293:489–493 (erratum 293:1048) 10.1126/science.1059431 [DOI] [PubMed] [Google Scholar]
- 38.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507 10.1126/science.1840702 [DOI] [PubMed] [Google Scholar]
- 39.Nachman MW (2001) Single nucleotide polymorphisms and recombination rate in humans. Trends Genet 17:481–485 10.1016/S0168-9525(01)02409-X [DOI] [PubMed] [Google Scholar]
- 40.Hudson RR, Kreitman M, Aguade M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voight BF, Kudaravalli S, Wen X, Pritchard JK (2006) A map of recent positive selection in the human genome. PLoS Biol 4:e72 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wells JCK, Cole TJ (2002) Birth weight and environmental heat load: a between-population analysis. Am J Phys Anthropol 119:276–282 10.1002/ajpa.10137 [DOI] [PubMed] [Google Scholar]
- 44.Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A (2004) CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet 75:1059–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose CS, Ek J, Urhammer SA, Glumer C, Borch-Johnsen K, Jorgensen T, Pedersen O, Hansen T (2005) A -30G→A polymorphism of the β-cell-specific glucokinase promoter associates with hyperglycemia in the general population of whites. Diabetes 54:3026–3031 [DOI] [PubMed] [Google Scholar]
- 46.Marz W, Nauck M, Hoffmann MM, Nagel D, Boehm BO, Koenig W, Rothenbacher D, Winkelmann BR (2004) G(-30)A polymorphism in the pancreatic promoter of the glucokinase gene associated with angiographic coronary artery disease and type 2 diabetes mellitus. Circulation 109:2844–2849 10.1161/01.CIR.0000129306.44085.C4 [DOI] [PubMed] [Google Scholar]
- 47.Terwilliger JD, Hiekkalinna T (2006) An utter refutation of the “Fundamental Theorem of the HapMap.” Eur J Hum Genet 14:426–437 [DOI] [PubMed] [Google Scholar]