DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development (original) (raw)
Abstract
We here identify a protein (AlfA; actin like filament) that defines a new family of actins that are only distantly related to MreB and ParM. AlfA is required for segregation of Bacillus subtilis plasmid pBET131 (a mini pLS32-derivative) during growth and sporulation. A 3-kb DNA fragment encoding alfA and a downstream gene (alfB) is necessary and sufficient for plasmid stability. AlfA-GFP assembles dynamic cytoskeletal filaments that rapidly turn over (_t_1/2<∼45 s) in fluorescence recovery after photobleaching experiments. A point mutation (alfA D168A) that completely inhibits AlfA subunit exchange in vivo is strongly defective for plasmid segregation, demonstrating that dynamic polymerization of AlfA is necessary for function. During sporulation, plasmid segregation occurs before septation and independently of the DNA translocase SpoIIIE and the chromosomal Par proteins Soj and Spo0J. The absence of the RacA chromosome anchoring protein reduces the efficiency of plasmid segregation (by about two-fold), suggesting that it might contribute to anchoring the plasmid at the pole during sporulation. Our results suggest that the dynamic polymerization of AlfA mediates plasmid separation during both growth and sporulation.
Keywords: actin, bacterial cytoskeleton, DNA segregation, sporulation
Introduction
Dynamic protein polymers and ATP-dependent motor proteins act together to drive chromosome segregation in eukaryotic cells. In bacteria, two families of actin-like proteins that assemble cytoskeletal filaments involved in cell growth or DNA segregation have been characterized (Carballido-Lopez and Errington, 2003b; Gerdes et al, 2004). Proteins of the MreB family are essential in many bacteria where they are required for maintaining cell shape and protein localization (reviewed in Errington, 2003a; Carballido-Lopez and Errington, 2003b; Moller-Jensen and Lowe, 2005; Thanbichler et al, 2005). ParM of plasmid R1 (Gerdes et al, 2004) defines a second bacterial actin family. ParM works together with the DNA-binding protein ParR and a DNA site parC, to relocate plasmids from the center of the cell to the cell poles (Jensen and Gerdes, 1997; Gerdes et al, 2004). Although ParM, MreB and yeast actin only share ∼11% amino acid identity, their three-dimensional structures are very similar (van den Ent et al, 2001, 2002). In the presence of ATP, ParM assembles into dynamic filaments that closely resemble the two-stranded, helical filaments of actin (Moller-Jensen et al, 2002; van den Ent et al, 2002). Polymerization of ParM displays dynamic instability and occurs between two plasmid molecules, providing the driving force for moving them apart (Moller-Jensen et al, 2003; Garner et al, 2004).
Unlike DNA segregation during vegetative growth, where chromosomes and plasmids separate before cell division, chromosome segregation during Bacillus subtilis sporulation occurs both before and after septation and is orchestrated by a complex development program. The first stage of this process begins before septation when a region of DNA near the origin of replication migrates to the cell pole and the chromosomes condense into an elongated structure (axial filament) (Ryter, 1965; Setlow et al, 1991; Webb et al, 1997; Errington et al, 2001; Figure 1A). Each chromosome in the axial filament condenses into two visually distinct domains separated by a gap large enough to accommodate the polar septum (Setlow et al, 1991; Pogliano et al, 2002; Figure 1B). The smaller domain located at the extreme cell pole contains approximately ⅓ of the chromosome near the oriC region (Wu and Errington, 1994, 1998; Webb et al, 1997). These events depend upon the Soj/Spo0J partitioning proteins and the developmentally regulated chromosome remodeling protein RacA (Ireton et al, 1994; Sharpe and Errington, 1996; Ben-Yehuda et al, 2003; Wu and Errington, 2003). Relocalization of the cell division machinery also occurs during this stage of development when the medial ring of the cell division protein FtsZ relocalizes via a spiral intermediate into two rings, one near each cell pole (Figure 1B, blue rings; Ben-Yehuda and Losick, 2002). Polar septation bisects one of the chromosomes, trapping the small domain in the forespore cell and the larger domain in the mother cell. Sequential activation of the polar division sites and cell-specific gene expression ensures that only one forespore is formed (reviewed in Stragier and Losick, 1996; Rudner and Losick, 2001; Errington, 2003b).
Figure 1.
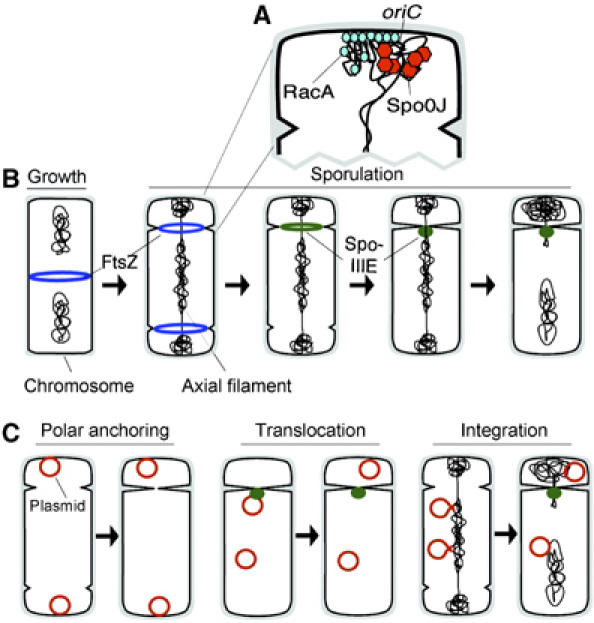
A model depicting DNA segregation and cell division during sporulation. (A) Enlargement of a cell pole, early during sporulation, depicting how the Soj/Spo0J proteins act to condense DNA surrounding the replication origin and how this region is further compacted and tethered to the pole by RacA (Ben-Yehuda et al, 2003, 2005; Wu and Errington, 2003). (B) A vegetative B. subtilis cell containing two chromosomes and a medial FtsZ ring (1st cell, blue ring). At the onset of sporulation, chromosomes rearrange into an axial filament where each chromosome condenses into two visually distinct domains separated by a gap (second cell, black lines). FtsZ relocalizes as two rings that are positioned near the poles between the gaps in the axial filament (second cell, blue rings). As division occurs at one of the FtsZ complexes, a ring of SpoIIIE localizes to the leading edge of the invaginating septum (third cell, green ring). Septation then delivers the DNA translocase SpoIIIE to the chromosome (fourth cell, green dot), where it completes chromosome partitioning (fifth cell). (C) Three models depicting how low copy number plasmids could be actively partitioned into the forespore (see text for details).
The second stage of partitioning begins after the septum constricts around the DNA that spans the two chromosome domains and depends upon SpoIIIE, a member of the conserved FtsK/SpoIIIE motor proteins that translocate DNA across membranes (Errington et al, 2001; Sherratt, 2003). SpoIIIE localizes to the leading edge of the invaginating septum (Figure 1B, green ring) and forms a complex that pumps the larger chromosome domain into the forespore (Figure 1B, green spot; Wu and Errington, 1994; Bath et al, 2000; Liu et al, 2006). Consequently, in translocase-defective spoIIIE mutants, complete DNA segregation into the forespore is never achieved (Wu and Errington, 1994; Wu et al, 1995; Errington et al, 2001).
Here we aim to characterize the mechanisms by which plasmid DNA is faithfully inherited by spores of B. subtilis. Early in the spore formation pathway, the smaller forespore occupies approximately ⅛th of the parental cell volume (Stragier and Losick, 1996; Errington, 2003b). Therefore, low copy plasmids must be actively partitioned into forespores in order to be stably inherited during sporulation. Plasmids might have evolved to utilize the cellular machinery used to partition chromosomes during sporulation. For example, plasmids could interact with RacA or Soj/Spo0J and become positioned at the cell pole before septation (Figure 1C, polar anchoring). Such a polar anchoring mechanism was recently proposed for B. subtilis phage phi29, whose segregation was shown to depend upon Soj/Spo0J (Meijer et al, 2005). Alternatively, plasmids could translocate into the forespore after septation by direct interaction with SpoIIIE (Figure 1C, translocation). Plasmids could also transiently become integrated into the chromosome and achieve forespore positioning by ‘piggybacking' along with the chromosome (Figure 1C, integration).
We have used the plasmid pLS32 to study segregation of stable, low copy number plasmids in B. subtilis. pBET131 is a mini-pLS32 derivative containing a 7.2-kb region of pLS32 DNA (Tanaka and Ogura, 1998; Tanaka et al, 2005) and is present at a copy number of two per chromosome equivalent. Its replication origin has been characterized, making this plasmid a useful candidate for addressing the mechanisms of plasmid segregation (Tanaka and Ogura, 1998; Tanaka et al, 2005). Our results show that pBET131 segregation depends upon a novel bacterial actin that assembles into dynamic cytoskeletal filaments.
Results
Plasmid pBET131 is actively partitioned during growth and sporulation
We began by measuring the stability of the low copy number plasmid pBET131 in B. subtilis during both growth and sporulation. Assuming that there are four plasmid copies per cell just before cell division (based on previous copy number measurements (Tanaka and Ogura, 1998)), random segregation during growth would yield 6% plasmid free cells per generation. However, pBET131 is stably inherited in the absence of selection during exponential growth with an observed loss rate of only 0.5% per generation (Table I). In contrast, a mini-plasmid (pJSA5) containing only the origin of replication and the replication gene repN from pLS32 within a 1.5-kb region of DNA (_ori_1.5 kb) was nearly 4.5 times less stable, displaying a loss rate of 2.3% per generation. Since these two plasmids have a similar copy number (Table I), pBET131 is most likely actively partitioned during vegetative growth.
Table 1.
Plasmid stability during growth
| Strain | Plasmid | Relevant genotype | Relative copy no.a | CmR at _t_0 (%) | CmR at _t_15 (%) | Plasmid loss rate (%)b |
|---|---|---|---|---|---|---|
| NHB92 | pBET131 | alfA+ | 1.0 | 88 | 80 | 0.5 |
| NHB91 | pJSA5 | Origin only | 1.4 | 45 | 10 | 2.3 |
| NHB93 | pNCH113 | alfA D168A | 1.2 | 75 | 22 | 3.5 |
| EBS691 | pEB255 | 3.0 kb alf region | 1.9 | 84 | 77 | 0.5 |
| aCopy number of plasmids relative to pBET131 in strains growing in LB at 30°C. See Supplementary data for method of copy number determination. | ||||||
| b% CmR colonies at _t_0—% CmR colonies at 15 generations of growth in the absence of selection (_t_15)/number of generations. |
To quantify segregation of pBET131 into spores, cells were first grown and sporulated in the presence of chloramphenicol to select for the plasmid. Spores were germinated on LB plates lacking chloramphenicol and the resulting colonies were then tested for plasmid-encoded chloramphenicol resistance (CmR). The percentage of CmR spores was then adjusted to reflect the percentage of CmR cells at the onset of sporulation (_t_0) (Table II). pBET131 was inherited by a significant fraction of spores (58%), while the mini-plasmid pJSA5 containing only the origin of replication was inherited by only 18%. This three fold difference in the rates of inheritance suggested that pBET131 is also actively segregated during sporulation. As a control, when CmR was placed on the chromosome (KP646), 100% of germinated spores produced CmR colonies (data not shown).
Table 2.
Plasmid stability during sporulation
| Strain | Plasmid | Genotype | CmR at _t_0 (%)a | CmR spores adjusted (%)b,c |
|---|---|---|---|---|
| EBS109 | pBET131 (alfA+) | wt | 93 | 58 |
| NHB11 | pJSA5 (origin only) | wt | 79 | 18 |
| NHB10 | pNCH106 (AlfA-GFP) | wt | 94 | 50 |
| NHB22 | pNCH113(alfA D168A) | wt | 64 | 34 |
| EBS443 | pBET131 (alfA+) | racA | 93 | 32 |
| EBS646 | pJSA5 (origin only) | racA | 44 | 16 |
| EBS650 | pNCH113(alfA D168A) | racA | 52 | 17 |
| a107–268 colonies were counted. | ||||
| bAdjusted values calculated as (% CmR spores × 100)/% CmR colonies at _t_0. | ||||
| c191–392 colonies were counted. |
Development of a GFP expression assay for plasmid segregation into the forespore
We attempted to use GFP-LacI tagging to localize pBET131 during sporulation, but found that the presence of the GFP-LacI tag destabilized the plasmid. We therefore developed a different GFP-based assay to accurately monitor plasmid segregation during sporulation. This assay relies upon the well-characterized transcription factor σF, which becomes active only in the forespore, where it transcribes genes required for development such as the spoIIQ gene (Stragier and Losick, 1996). When GFP is expressed from the spoIIQ promoter (P_FS_-gfp), GFP fluorescence is detected only in the forespore (Webb et al, 1995). We placed the P_FS_-gfp reporter on plasmid pBET131 (to create pGAP100) and on the smaller unstable plasmid pJSA5 (to create pJSA6), so that GFP expression would indicate the presence of the plasmids in the forespore (Figure 2A). Wild-type B. subtilis cells containing pGAP100 or pJSA6 were sporulated in the presence of the membrane stain FM 4-64, and samples were taken at various times after the start of sporulation and visualized by fluorescence microscopy (Figure 2B and C). As expected, before polar septation, the cells did not express GFP (not shown). After 2 h of sporulation (t 2), 39% of pGAP100 sporangia expressed GFP in the forespore, and this number increased to 50% by t 4. At the same time points, only 5% (t 2) and 7% (t 4) of pJSA6 sporangia expressed GFP, supporting our conclusion that pBET131 is significantly more stable during growth and sporulation than plasmids containing only the origin of replication. In comparison, 82% (t 2) and 94% (t 4) of sporangia from the control strain with P_FS_-gfp on the chromosome expressed GFP (Figure 2D). These results are in agreement with those obtained from testing the antibiotic resistance of germinated spores (Table II) and indicate that our fluorescence assay efficiently reports plasmid segregation.
Figure 2.
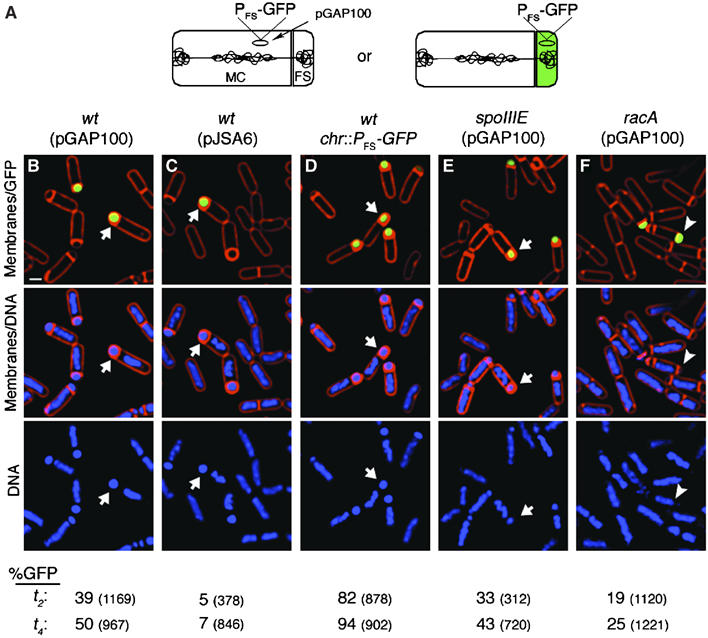
Cell biological assays for plasmid segregation. (A) A GFP reporter for plasmid segregation that can distinguish between sporangia with the pGAP100 plasmid in the mother-cell (MC) (left) or in the forespore (FS) (right). Only plasmids that reside in the forespore will express GFP from the forespore-specific promoter (_P_FS). (B–F) Segregation of DNA during sporulation using a GFP-based reporter assay. Cells were sporulated in the presence of the membrane stain FM 4-64 (red), harvested at _t_2 (shown) and _t_4 (images not shown) and stained with the DNA stain DAPI (blue). The percentage of cells expressing GFP (green) at _t_2 and t_4 is reported at the bottom. (B) GFP expressed from the forespore specific reporter (P FS -GFP) on pBET131 derivative pGAP100 in a wild type-strain (arrow; strain EBS111). Scale bar=1 μm. (C) GFP expressed from P FS -GFP on pJSA6, a derivative containing only the origin of replication of pBET131 (EBS114). (D) GFP expressed from P FS -GFP integrated near the oriC region of the chromosome in a wild-type strain (KP646). (E) P FS -GFP expression from pGAP100 in spoIIIE73–11 (EBS375) or in (F) Δ_racA strain (EBS102). In the racA strain, forespores lacking chromosomal DNA can still trap pGAP100 (arrowhead). (Bottom) Percentage of forespores with GFP at _t_2 or _t_4. Total sporangia scored are indicated in parentheses and the strains corresponding to the data sets remain in the same column as indicated for the images.
SpoIIIE does not translocate pBET131 into the forespore
SpoIIIE is a member of a highly conserved family of DNA transport proteins that could potentially play a role in translocating plasmid DNA into the forespore. To test this, we assayed segregation of pGAP100 into forespores in spoIIIE73–11 (Sharp and Pogliano, 1999) and spoIIIE36 (not shown) (Wu and Errington, 1994) mutant cells. Both alleles are defective in chromosome translocation, yet the proteins localize to the septum and maintain strict compartmentalization of gene expression. In addition, the large domain of the axial filament remains in the mother cell, and forespore-specific reporter genes present in this domain are not expressed (Wu and Errington, 1998). As spoIIIE mutants do not produce spores, we relied upon the GFP reporter assay and fluorescence microscopy to monitor pGAP100 segregation into forespores (Figure 2A). After 4 h of sporulation, the percentage of cells expressing GFP from the plasmid (43%; Figure 2E) was similar to the wild type (50%; Figure 2B). This suggests that SpoIIIE does not translocate pBET131 into forespores.
Testing if plasmids enter the forespore after polar septation: a plasmid with a molecular memory of mother cell gene expression
The results obtained using translocation defective spoIIIE mutants indicated that SpoIIIE does not transport pBET131 across the septum. However, these results do not rule out the possibility that this plasmid enters the forespore after septation by an alternate translocase, perhaps by homologs of SpoIIIE encoded on the chromosome of B. subtilis, or by an unrecognized class of proteins. We therefore developed a system to precisely distinguish between pre- and post-septational plasmid segregation that provides a direct answer regardless of whether the process requires SpoIIIE or an unidentified protein. This ‘molecular memory' assay is comprised of a reporter of forespore-specific gene expression that can only be expressed if it has previously been exposed to mother cell-specific gene expression (Figure 3). The reporter consists of a kanamycin resistance cassette (kan) flanked by loxP sites inserted between a forespore-specific promoter (_P_FS) and the GFP gene, to generate _P_FS-_loxP_-_kan_-_loxP_-gfp. The loxP sites are substrates for the Cre recombinase (Figure 3A; Austin et al, 1981; Sauer, 1993; Van Duyne, 2001), which can be expressed from a mother cell-specific promoter (_P_MC). Thus, only when the reporter is present in the mother cell after septation will the kanamycin cassette be excised and the reading frame of GFP be restored. Subsequent translocation of recombinant plasmids into the forespore will result in forespore-specific production of GFP.
Figure 3.
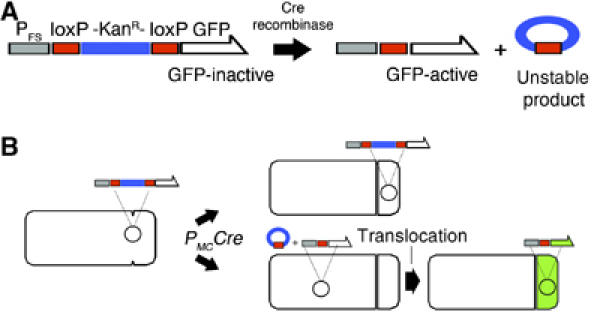
Assay for plasmid segregation postseptation. (A) ‘Molecular memory' cassette. Cre recombinase activates the forespore specific GFP reporter by splicing out the loxP flanked kan resistance gene thereby restoring the reading frame of GFP. The circular recombinant product cannot replicate and will be lost from the cell. (B) Cartoon depicting two possible scenarios of DNA segregation during sporulation. Expression of the Cre recombinase in the mother cell will activate the reporter gene only if it resides in the same cell. When the reporter is trapped in the forespore and Cre is expressed in the mother cell (top cell), GFP is not expressed. When the reporter is present in the mother cell where Cre is expressed, excision of the kan cassette and subsequent translocation of the activated reporter to the forespore will then result in GFP expression.
We first tested this system by placing the P_FS_-loxP-kan-loxP-gfp cassette on the chromosome near oriC, a region that is always in the forespore after septation. We then expressed Cre in the mother cell, the forespore, or not at all during sporulation (Table III). The efficiency of cassette excision was determined by testing spores for kanamycin resistance (lost after Cre-mediated excision) and by using fluorescence microscopy to visualize forespore-specific GFP expression (gained after excision). As expected, in the absence of Cre, the kan cassette is not excised, GFP is not expressed, and 100% of the spores are kanamycin resistant (KanR) (Table III, EBS38). When Cre is expressed in the forespore, 95% of the forespores express GFP and only 4% remain KanR, demonstrating that in most forespores, Cre is active and able to excise the cassette (JP969). In contrast, when Cre is expressed in the mother cell, 3% of the forespores express GFP, and 96% remain KanR (EBS40), indicating that in most cells, mother cell expressed Cre is unable to excise the kan cassette in the forespore. Residual excision (3%) in the forespore when Cre is expressed in the mother cell is probably due to leakage of a few molecules from the mother cell into the forespore (in theory, only four molecules of Cre are needed to create a functional enzyme, making this assay extraordinarily sensitive). This low level of inappropriate excision (3%) has little impact on the experiments described here.
Table 3.
Efficiency of excision by Cre when P_FS_-loxP-kan-loxP-gfp is located on the chromosome near the origin or terminus
| Chromosomal location of P_FS_-loxP-kan-loxP-gfp | ||||||
|---|---|---|---|---|---|---|
| Origin (amyE) | Terminus (cotC) | |||||
| Cre expression | Strain | KanRa spores (%) | GFPb _t_4 (%) | Strain | KanR spores (%) | GFPb _t_4 (%) |
| None | EBS38 | 100 | 0 | EBS397 | 100 | 0 |
| Forespore | JP969 | 4 | 95 | EBS400 | 3 | 91 |
| Mother cell | EBS40 | 96 | 3 | EBS402 | 16 | 62 |
| aKanR indicates the failure to excise the cassette. A total of 150–250 colonies were counted for each strain. | ||||||
| bGFP expression depends on excision of the loxP-kan-loxP cassette. GFP _t_4 is the percentage of forespores expressing GFP 4 h after the start of sporulation. A total of 410–896 cells were counted for each strain. |
As an additional test of the efficiency of this system, we determined if Cre expression in the mother cell occurs with sufficient kinetics to excise a gene that resides initially in the mother cell and is then translocated into the forespore. We placed the P_FS_-loxP-kan-loxP-gfp cassette near the chromosomal terminus at the cotC locus (Table III). Following polar septation, this region is trapped in the mother cell, and is subsequently translocated into the forespore by SpoIIIE. As expected, the reporter was not activated in the absence of Cre (0% forespores with GFP by _t_4, EBS397) and after DNA translocation it became available as a substrate for Cre expressed in the forespore (91% forespores with GFP by _t_4, EBS 400). When Cre was expressed in the mother cell, GFP in the forespore was readily observed by _t_4 (62%), and the percentage of KanR spores produced by this strain was 16% (Table III, EBS402), demonstrating that this assay can efficiently (84%) detect postseptational DNA translocation.
Next, we applied this assay to the plasmid pBET131, to determine if it enters the forespore before or after septation. The P_FS_-gfp cassette of pGAP100 was replaced with the P_FS_-loxP-kan-loxP-gfp to create pGAP101. We then tested excision in strains expressing Cre in the forespore, the mother cell, or not at all. After sporulation, we monitored the percentage of spores that gave rise to CmR colonies (encoded on the plasmid backbone, serving as a reporter of plasmid inheritance) and KanR colonies (to monitor excision), and compared these to the percentage of forespores expressing GFP. Based on the above control experiments, we expected the following results: (1) When Cre is not present in the strain, GFP would never be expressed and all of the spores inheriting the plasmid would be KanR. (2) When Cre is expressed in the forespore, all of the plasmids that enter the forespore should be subjected to the Cre recombinase, so approximately 95% of the plasmid-containing forespores will be KanS and produce GFP. (3) When Cre is expressed in the mother cell, the result will depend on whether the plasmid is inherited before or after septation. If the plasmids enter after septation, then ∼62% of the forespores that inherit plasmids (∼50%) should express GFP and ∼84% of plasmid-containing spores should be KanS, given the efficiency of Cre recombination in our control experiments (EBS402; Table III). However, if the plasmids enter before septation, then few plasmid containing forespores should express GFP or become KanS, except for the ∼3% level of inappropriate excision that we observed in our control experiments.
The results are summarized in Table IV. When Cre was absent from the strain (JP959), 53% of the spores inherited the plasmid and none expressed GFP. When Cre was expressed in the forespore (JP963), 46% of the spores inherited the plasmid and 44% expressed GFP, in good agreement with our expectations. In striking contrast, when Cre was expressed in the mother cell (JP962), a similar number of forespores (49%) inherited the plasmid, but only 1.2% expressed GFP by _t_4. Moreover, nearly all of the spores containing a plasmid (49%) were also KanR (49%), indicating very little excision of the kan cassette. These results suggest that pBET131 enters the forespore before septation, so that the reporter construct is never exposed to mother cell specific gene expression.
Table 4.
Efficiency of excision when P_FS_-loxP-kan-loxP-gfp is located on the plasmid and Cre is expressed in forespore or mother cell
| Cre expression | Strain | CmRa spores (%) | KanRa spores (%) | CmRb _t_0 (%) | GFPc _t_2 (%) | GFPc _t_4 (%) |
|---|---|---|---|---|---|---|
| None | JP959 | 53 | 53 | 88 | 0 | 0 |
| forespore | JP963 | 46 | 4 | 88 | 40 | 44 |
| Mother cell | JP962 | 49 | 49 | 92 | 0.8 | 1.2 |
| aA total of 225–333 colonies counted. | ||||||
| bA total of 106–120 colonies counted. | ||||||
| cA total of 321–828 sporangia counted; GFP expression was examined after 2 h (_t_2) or 4 h (_t_4) after the start of sporulation. |
Effect of RacA and Soj/Spo0J on plasmid segregation
During sporulation RacA plays an important role in both condensing chromosomes and in anchoring them to the cell pole (Ben-Yehuda et al, 2003; Wu and Errington, 2003). We therefore tested if RacA also plays a significant role in plasmid segregation. Using GFP expression to measure segregation of pGAP100 into forespores, we found that by _t_4, only 25% of forespores expressed GFP from the plasmid (Figure 2F), suggesting that like the effect of RacA on chromosome segregation, plasmid segregation is also affected about two-fold. A similar result was obtained by assaying CmR spores (Table II; compare EBS443 versus EBS109). This reduction was modest but very reproducible and statistically significant (P<0.001).
We also measured plasmid segregation in a soj/spo0J deletion mutant, using both GFP reporters and antibiotic resistance assays. In contrast to the phage phi29, whose inheritance is dependent upon Spo0J and Soj (Meijer et al, 2005), deleting these genes did not effect plasmid segregation into forespores. In strain EBS125, pGAP100 was inherited by 30% of forespores by _t_2 (_n_=502) and by 43% by _t_4 (_n_=496), very similar to wild type.
Identification of an actin-like protein (AlfA) required for pBET131 segregation
pBET131 was substantially more stable during both growth (Table I) and sporulation (Table II) than smaller plasmids (such as pJSA5) that have a similar copy number. We noticed that pBET131 encodes an open reading frame very distantly related to MreB (17% identical), actin (16%) and ParM (15%) (Figure 4, AlfA). Phylogenetic analysis indicates that it represents a new family of bacterial actins (Figure 4A). AlfA contains residues conserved in actins across kingdoms, including an aspartic acid in the phosphate 2 domain, which is involved in nucleotide hydrolysis (Bork et al, 1992; Holmes et al, 1993; van den Ent et al, 2001; Figure 4B).
Figure 4.
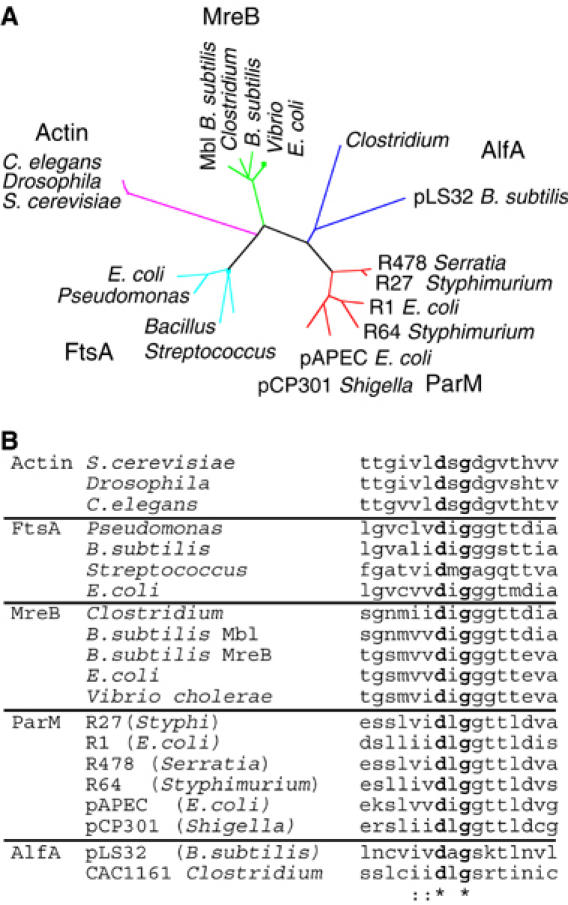
Identification of an actin related protein encoded by plasmid pLS32. (A) Phylogenetic analysis showing the relationship between MreB, ParM, FtsA, actin and AlfA. The tree was calculated using the neighbor-joining method. Sequences were aligned using TCoffee and ClustalW and the tree constructed using Phylip version 3.6 (Felsenstein, 1989). Sequence abbreviations and accession numbers are as follows. Actin family members (purple): Drosophila melanogaster (AAM50595), Saccharomyces cerevisiae (NP_116614), Caenorhabditis elegans (CAA34718). MreB family members (green): B. subtilis (MreB AAA22397 and Mbl AAA67878), Clostridium tetani (NP_781022), Vibrio cholerae (AAF93588), Escherichia coli (AAA58054). FtsA family members (cyan): B. subtilis (AAA22456), Pseudomonas aeruginosa (AAG07796), Streptococcus pneumoniae (AAL00315), Escherichia coli (NP_414636). ParM family members (red): plasmid R478 Serratia marcescens (NP_941097), plasmid R27 Salmonella typhi StbA (AAD54035), plasmid R1 E. coli (P11904), plasmid R64 Salmonella typhimurium (BAB91612), plasmid pAPEC-02-R E. coli (AAT37581), plasmid pCP301 Shigella flexneri (AAL72301). AlfA family members (purple): plasmid pLS32 B. subtilis (BAA24871), CAC1161 Clostridium acetobutylicum (AAK79133). (B) Alignment of the phosphate 2 motif for selected protein sequences belonging to actin, FtsA, ParM, MreB and AlfA. The aspartic acid is conserved in actins across kingdoms. Sequences and accession numbers are the same as in (A).
To test the role of AlfA in plasmid segregation, we changed the conserved aspartic acid (D168) to alanine (alfA D168A). The analogous mutation in ParM completely eliminates its ability to function in vivo (Jensen and Gerdes, 1997; Moller-Jensen et al, 2002). During exponential growth, pNCH113 (pBET131 alfA D168A) was significantly (seven-fold) less stable than the wild-type plasmid, displaying a loss rate of 3.5% per generation (Table I). This plasmid was as unstable as smaller plasmids (pJSA3 or pJSA5) containing only the origin of replication (_ori_1.5 kb), indicating that AlfA is required for plasmid segregation during exponential growth. A 3079 bp DNA fragment containing the alfA gene along with surrounding DNA was subcloned into pJSA3 to create pEB255. When grown for 15 generations without antibiotic selection, this plasmid was as stable as pBET131, demonstrating that the alfA region is sufficient for plasmid segregation (Table I).
When we measured the ability of the plasmid containing the alfA D168A point mutation (pNCH113) to enter spores (Table II), we found that it was about two-fold less stable during sporulation (34 versus 58% for pBET131). This small but reproducible decrease was statistically significant (P<0.001) and indicated that AlfA also plays a role in segregation during sporulation. Although pJSA5 and pNCH113 (alfA D168A) were equally unstable during exponential growth, pNCH113 (alfA D168A) was slightly (two-fold) but reproducibly (P<0.001) more stable during sporulation than pJSA5 (34% of spores inherited pNCH113 (strain NHB22) versus 18% for pJSA5 (strain NHB11; Table II). This difference suggested that other factors, in addition to AlfA, might play a role in pBET131 segregation during sporulation. Since we had also noticed a small (two-fold) but reproducible difference in pBET131 inheritance in a racA mutant, we tested the ability of pNCH113 (alfA D168A) to be inherited during sporulation in a racA mutant. In the absence of RacA, both plasmids (pJSA5 and pNCH113) were equally unstable (P<0.001) (16 versus 17%; Table II). These findings are consistent with a model in which both AlfA and RacA contribute to plasmid segregation during sporulation.
AlfA assembles dynamic filaments in vivo
To understand how AlfA functions, we constructed a plasmid containing a GFP fusion to the C-terminus of AlfA and recombined it with pBET131 to produce a plasmid (pNCH106) expressing both untagged AlfA and the AlfA-GFP fusion protein. This plasmid was as stable as pBET131 during both exponential growth (not shown) and during sporulation (Table II), indicating that the AlfA-GFP fusion protein does not interfere with segregation. Remarkably, AlfA-GFP formed filaments in nearly every cell (90%; _n_=278). These filaments usually spanned the entire cell length (Figure 5A–C).
Figure 5.
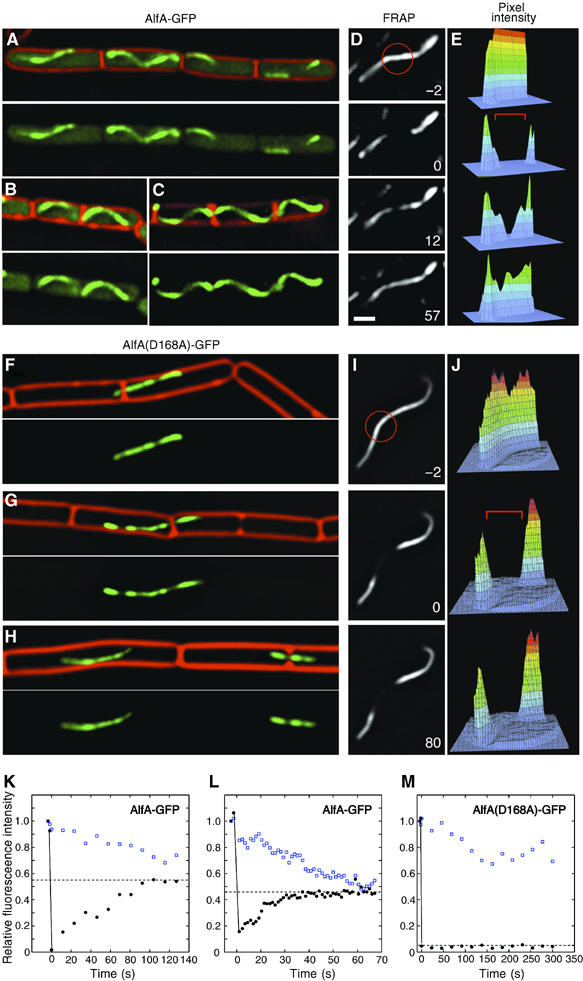
AlfA-GFP assembles dynamic filamentous structures. Strains were grown on an agarose pad containing 10% LB at 30°C. Three examples of wild type (A–C) NHB10 (pNCH106 AlfA-GFP) and mutant (F–H) NHB55 (pNCH135 AlfA(D168A)-GFP) are shown. Cell membranes were stained red with FM 4-64. All images are to the same scale. White bar equals 1 micron. (D, E, I, J) FRAP demonstrates that wild-type AlfA-GFP filaments (D, E) are dynamic but filaments assembled by AlfA(D168A)-GFP (I, J) are static. After collecting a prebleach image (−2 s), a small region (red circle) of the filament was exposed to light from an argon laser (488 nm) for 0.5 s and images collected at the indicate times (sec) (D, I). Three-dimensional plots (E, J) show pixel intensity data corresponding to each image. The bleached zone is indicated by a red bracket. (K–M) Quantitative FRAP analysis for wild type (K–L) or AlfA(D68A)-GFP (M). Relative fluorescence intensity of the bleached region (black circles) or an unbleached region (open blue squares) of the filament is plotted versus time. The dashed line indicates the extent of final recovery observed by the end of the experiment. The observed decrease in fluorescence for the unbleached region is due to both photobleaching and to incorporation of bleached subunits into the unbleached region of the polymer.
We investigated AlfA-GFP dynamics using fluorescence recovery after photobleaching (FRAP) in cells growing on an agarose pad at 30°C. A region of a filament was irreversibly bleached by high intensity laser light (Figure 5D and E) and recovery of fluorescence was monitored with time. Bleaching and recovery are shown quantitatively by three-dimensional graphs corresponding to each image (Figure 5D and E). When the middle of the filament in Figure 5D was bleached, fluorescence was partially recovered after 12 s and was fully recovered by 57 s (Figure 5D and E). In comparison, an unbleached region of the same filament (open squares) slowly decreased in intensity over time due to a combination of photobleaching from repeated image acquisition and from incorporation of bleached subunits from the cytoplasmic pool (Figure 5K). The half-life of recovery ranged from ∼45 s (Figure 5K) for the filament shown in Figure 5D to as short as ∼10 s (Figure 5L). This rapid recovery indicates that AlfA filaments are dynamic, continuously exchanging subunits with a cytoplasmic pool.
Dynamic assembly of AlfA correlates with its ability to segregate plasmid DNA
A plasmid (pNCH135) that expresses the untagged wild type AlfA together with the AlfA(D168A)-GFP mutant protein was also highly unstable, indicating that the mutant protein is dominant to the wild type. When strains containing this plasmid were grown in LB chloramphenicol at 30°C, fewer than 40% of cells contained GFP due to rapid plasmid loss. Unlike the wild-type AlfA-GFP, the filaments assembled by AlfA(D168A)-GFP rarely extended from one end of the cell to the other, were generally straight and not helical like those of wild type, and were often trapped by invaginating septa (Figure 5F–H). In FRAP experiments, these polymers were not dynamic; instead, after photobleaching, fluorescence did not recover after either 80 s (Figure 5I and J) or after 5 min (Figure 5M), indicating that the mutant protein assembles into a relatively static polymer. These experiments also demonstrate that photobleaching GFP fused to AlfA is irreversible under these conditions, in agreement with control experiments in which the entire cell was bleached and fluorescence did not recover (not shown). Since plasmids containing the point mutation are defective for segregation, our quantitative FRAP experiments demonstrate a direct correlation between AlfA polymer dynamics and the ability to segregate plasmid DNA.
AlfB is required for plasmid DNA segregation
Immediately downstream of alfA is an open reading of 93 amino acids (alfB) (Figure 6A) that we suspected might also be involved in plasmid segregation. A variant of pBET131 in which the alfB coding region was replaced with a kanamycin resistance gene (pFG6001) was highly unstable in B. subtilis. After eight generations of growth without antibiotic selection, >99% of cells had lost the plasmid. Transformation of B. subtilis with equal quantities of pFG6001, pBET131, or pJSA3 yielded 10–20 fold fewer colonies from pFG6001 than from the other two plasmids (Figure 6B). The pFG6001 transformants give rise to plasmid-free cells at a very high rate and form small colonies when grown on LB plates containing chloramphenicol to select for the plasmid (Figure 6C). The alfB gene is therefore likely to be part of the AlfA segregation system, where it could play a role analogous to ParR of plasmid R1, a DNA binding protein that tethers plasmid DNA to the ParM filaments and controls expression of the parMR operon.
Figure 6.
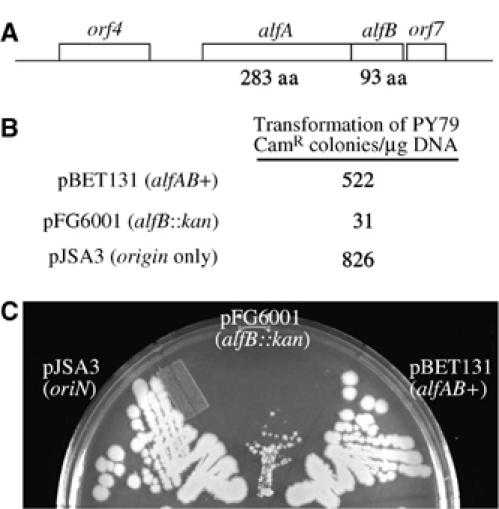
The alf region contains two genes required for plasmid segregation. (A) A schematic showing the 3 kb alfAB region sufficient for stabilizing a mini-pLS32 replicon. Mutations in either alfA or alfB disrupt segregation. The role of upstream (orf4) or downstream regions (orf7) is unknown. (B) Plasmids containing alfB∷kan mutations are unstable and only poorly transform into B. subtilis. PY79 was transformed with equal amounts (500 ng) of plasmid DNA (pBET131 alfAB+, pJSA3 (_ori_1.5 kb), pFG6001 alfB∷kan) and plated on LB cam plates. The number of transformants obtained is the average for two experiments and the differences between pBET131 and pFG6001 were reproduced at least five times. (C) Strains containing pFG6001 grow poorly on LB cam plates. PY79 containing pBET131 (wild type), pJSA3 (_ori_1.5 kb), or pFG6001 (alfB∷kan) were streaked onto LB cam plates and incubated overnight at 30°C.
Discussion
We here report the identification of a plasmid encoded protein (AlfA) that is distantly related to the bacterial actins MreB and ParM (Figure 4) and necessary for stability of plasmid pBET131. The distant phylogenetic relationship between AlfA and actin is supported by our finding that an AlfA-GFP fusion protein forms dynamic filaments that resemble the filaments formed by the bacterial actin ParM. In addition, the point mutation alfA D168A, which would be expected to disrupt the ATPase active site, destabilizes pBET131 and renders the normally dynamic AlfA-GFP filaments static. The rapid turnover of wild type AlfA-GFP in FRAP experiments is consistent with actin dynamics in eukaryotes (for example see Theriot and Mitchison, 1991). However, the mechanism of AlfA subunit exchange was not apparent from our experiments since the bleached spots appeared to recover uniformly with no obvious polarity. One possible explanation is that AlfA filaments consist of bundles of overlapping polymers as has been proposed for B. subtilis Mbl (Carballido-Lopez and Errington, 2003a). In this case, recovery would be expected to occur simultaneously over the length of the bleached zone and without obvious polarity.
A 3 kb DNA fragment containing the alfAB region was sufficient to stabilize a plasmid containing only the origin of replication from pLS32, suggesting that it includes a minimal plasmid segregation system. Deletion of alfB, a gene immediately downstream of and most likely translationally coupled to alfA, resulted in a plasmid that was extremely unstable; a cell transformed with this plasmid barely grew under selection for the plasmid and the plasmid could not be maintained at all in the absence of selection. We suspect that AlfB works together with AlfA as part of a DNA segregation system, possibly as a DNA binding protein involved in either plasmid segregation or regulation of the alfAB operon.
Taken together, our results suggest that AlfA is part of a DNA segregation machinery responsible for pBET131 stability, and that AlfA polymers represent a new type of bacterial cytoskeletal filament composed of a protein distinct in sequence from the bacterial actins previously implicated in DNA segregation. As has been observed with other plasmid segregation systems, the dynamic properties of the cytoskeletal protein are critical for its function in segregation (Moller-Jensen et al, 2002; Lim et al, 2005).
Sporulation poses a unique challenge for plasmid segregation, since the sporulation septum is synthesized at the extreme cell pole, thereby making it unlikely that a plasmid lacking efficient mechanisms for segregation and polar anchoring will enter the future spore. We have developed new Cre-loxP based assays to determine if plasmids enter the forespore before polar septation or after septation (e.g. by an SpoIIIE-based mechanism). In this assay, expression of GFP in the forespore only occurs after the Cre mediated excision of a kan cassette flanked by loxP sites. By expressing the Cre recombinase only in the mother cell, we could then determine if DNA that first resided in the mother cell was then later translocated into forespores. This assay, unlike other approaches for determining the subcellular location of DNA, does not require that the cells be fixed or that the DNA be tagged with a large protein complex that could alter the plasmid's segregation properties. The Cre-loxP assay is also extraordinarily sensitive, since in theory only four molecules of Cre could result in full activation of the reporter. Using this assay we found that pBET131 enters the forespore before polar septation.
In agreement with our Cre-loxP assay, pBET131 segregation into the forespore is not affected by the absence of SpoIIIE but instead depends upon both AlfA and RacA. Although the effect during sporulation of deleting either one alone was only two- to three-fold, each sporulation assay represents a single segregation event. Therefore, even a small (two fold) decrease in stability would result in greater than 99% plasmid loss from the population after just seven cycles of sporulation. We propose a model in which pBET131 segregation is mediated by AlfA and that the dynamic polymerization of this actin-like protein is essential to its ability to separate plasmids and move them to the cell poles at the onset of sporulation. Once at the poles, the plasmids are anchored there by host factors such as RacA. This model for plasmid segregation into the forespore is similar to that for the chromosomal origin of replication oriC, whose segregation toward the sporangium cell pole requires both RacA and a partitioning system (Soj/Spo0J) (Ireton et al, 1994; Sharpe and Errington, 1996; Ben-Yehuda et al, 2003; Wu and Errington, 2003).
Natural low copy number plasmids are generally extraordinarily stable and rely upon multiple mechanisms (including partitioning systems, post-segregational killing systems, and conjugation) to ensure that plasmid free cells are rarely produced. Here, we have studied a small but actively partitioned mini-pLS32 derivative, pBET131, that provides a minimal system for studying plasmid segregation in B. subtilis. Since pBET131 is not completely stable during either vegetative growth or sporulation, additional stability factors are likely present on the larger and more stable 70 kb pLS32 plasmid. This large plasmid therefore provides a model for understanding the segregation of large DNA molecules that are likely to depend on overlapping segregation machineries.
Materials and methods
Strains, growth conditions and genetic manipulations
Strains were induced to sporulate by resuspension in minimal salts (Sterlini and Mandelstam, 1969). All strains (Table V) are derivatives of PY79 (Youngman et al, 1984) or JH642 (Gilman and Chamberlin, 1983) and were constructed using standard methods (Hoch, 1991). Plasmids are listed in Table VI and oligonucleotides are listed in Table VII and Supplementary data. pBET131 (Tanaka and Ogura, 1998) was kindly provided by T Tanaka. pGAP100 and pGAP101 were constructed by amplifying the P_spoIIQ_-gfp expression cassette (for pGAP100) from pMDS13 or the P spoIIQ ∷loxP-kan-loxP∷gfp cassette from pEB19 (for pGAP101) using primers 2QGFP1 and 2QGFP4 and ligating the _Bgl_II digested fragment into pBET131 restricted with _Bam_HI (Sharp and Pogliano, 2002). A point mutation (D168A) in the alfA gene of pBET131 was created using the λ red system with oligonucleotide NHP17 to generate pNCH113 (Costantino and Court, 2003). The alfB gene of pBET131 was replaced with the kan gene from pUB110 using the λ red system and a PCR product obtained with oligonucleotides orf6/kanFWD-1 and orf6/RVRS that were designed to leave expression of the potential downstream orf7 intact. pNCH103 (alfA-GFP) and pNCH129 (alfAD168A-GFP) were made by amplifying the alfA gene with primers NHP5 and NHP7, digesting with KpnI and NheI, and then ligating into the _Kpn_I/_Nhe_I sites of pMutin-GFP (Kaltwasser et al, 2002). pNCH103 and pNCH129 were integrated into pBET131 by a single recombination event in B. subtilis to create pNCH106 and pNCH135, respectively. pJSA1 was made by amplifying the chloramphenicol acetyltransferase gene from pMDS13 (Sharp and Pogliano, 2002) using primers cat1 & cat2, digesting with _Eco_RI and _Hin_dIII and ligating into similarly digested pBR322. The 1.5 kb oriN region from pBET131 was amplified using PCR and primers oriN1b and oriN2, digested with _Eco_R1 and ligated into the _Eco_R1 site of pJSA1 yielding pJSA3 (oriented in the same direction as cat). To make pJSA5, a fragment containing the spoIIQ promoter was amplified by PCR from pMDS13 with primers N2Qsal and C2Qbam, digested with _Sal_I and _Bam_HI, and ligated into similarly digested pJSA3. Plasmid pJSA6 was constructed by amplifying gfp from pMDS13 using NgfpBgl and 2QGFP5, digesting the fragment with _Bgl_II and _Bam_HI and ligating it with BamHI digested pJSA5. Plasmid pEB255 containing the 3 kb alfAB region was constructed by using primers EBP208 and EBP209 to amplify the region, digesting the fragment with _Bam_HI, and ligating into similarly digested pJSA3.
Table 5.
Strains and pLS32 derivatives used in this study
| Strain | Relevant genotypea | Source, construction or referenceb |
|---|---|---|
| EBS21 | sacA∷PspoIID creΩspc | pEB15 → PY79 (SpcR) |
| EBS38 | amyE∷PspoIIQ∷loxP-kan-loxP∷gfpΩcat | pEB19 → PY79 (CmR) |
| EBS40 | sacA∷PspoIID creΩspc | EBS38 → EBS21 (CmR) |
| amyE∷PspoIIQ∷loxP-kan-loxP∷gfpΩcat | ||
| EBS42 | sacA∷PspoIIR−creΩspc | pEB27 → PY79 (SpcR) |
| EBS52 | cotC∷cat | RL52 → PY79 (CmR) |
| EBS87 | Δ_racA∷kan_ | pEB57 → PY79 (KanR) |
| EBS102 | Δ_racA∷kan_ (pGAP100) | pGAP100 → EBS87 (KanR) |
| EBS109 | Wild type (pBET131) | pBET131 → PY79 (CmR) |
| EBS111 | Wild type (pGAP100) | pGAP100 → PY79 (CmR) |
| EBS114 | Wild type (pJSA6) | pJSA6 → PY79 (CmR) |
| EBS125 | Δ_(soj-spo0J)_ (pGAP100) | pGAP100 → KP527 (CmR) |
| EBS143 | Δ_racA∷kan, amyE∷PspoIIQ-gfpΩcat_ | KP646 → EBS87 (CmR) |
| EBS280 | lacA∷spc | 1HA01(Hartl et al, 2001) → PY79(spcR) |
| EBS375 | spoIIIE73–11 (pGAP100) | pGAP100 → KP541 (CmR) |
| EBS397 | cotC∷cat∷PspoIIQ∷loxP-kan-loxP∷gfp | pEB171 → EBS52 |
| EBS400 | sacA∷PspoIIR creΩspc, cotC∷cat∷PspoIIQ∷loxP-kan-loxP∷gfp | EBS400 → EBS397 (SpcR) |
| EBS402 | sacA∷PspoIID−creΩspcR, cotC∷cat∷PspoIIQ∷loxP-kan-loxP∷gfp | EBS21 → EBS397 (SpcR) |
| EBS443 | Δ_racA: kan_ (pBET131) | pBET131 → EBS 87 (CmR) |
| EBS646 | Δ_racA∷kan_ (pJSA5) | pJSA5 → EBS 87 (CmR) |
| EBS650 | Δ_racA∷kan_ (pNCH113) | pNCH113 → EBS 87 (CmR) |
| JP947 | sacA∷PspoIID−creΩspc (pGAP101) | pGAP101 → EBS21 (CmR) |
| JP948 | sacA∷PspoIIR−creΩspc (pGAP101) | pGAP101 → EBS42 (CmR) |
| JP969 | sacA∷PspoIIR creΩspc, amyE∷PspoIIQ∷loxP-kan-loxP∷gfpΩcat | EBS42 → EBS38 (KanR) |
| JP3666 | PY79 (pFG6001) | pFG6001 → PY79 (CmR) |
| KP527 | Δ_(soj-spo0J)_ | Sharp and Pogliano (1999) |
| KP541 | spoIIIE73–11 | Sharp and Pogliano (1999) |
| KP646 | amyE∷PspoIIQ-gfpΩcat | Sharp and Pogliano (2002) |
| NHB10 | PY79 (pNCH106) | pNCH106 → PY79 (CmR) |
| NHB11 | PY79 (pJSA5) | pJSA5 → PY79 (CmR) |
| NHB22 | PY79 (pNCH113) | pNCH113 → PY79 (CmR) |
| NHB55 | PY79 (pNCH135) | pNCH135 → PY79 (CmR) |
| NHB91 | JH642 (pJSA5) | pJSA5 → JH642 (CmR) |
| NHB92 | JH642 (pBET131) | pBET131 → JH642 (CmR) |
| NHB93 | JH642 (pNCH113) | pNCH113 → JH642 (CmR) |
| PY79 | wt | Youngman et al (1984) |
| JH642 | trpC2, phe-1 | Gilman and Chamberlin (1983) |
| RL52 | trpC2, cotC∷cat | Donovan et al (1987) |
| a_Pn_, promoter; loxP, recombination site for Cre; plasmids indicated in parenthesis. | ||
| bPlasmid or strain DNA used to transform a given strain is indicated to the left of the arrow. The arrow points to the strain that was transformed and the drug used to select for transformants is indicated in parentheses. |
Table 6.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| pBET131 | pBR322 derivative containing 7.2 kb of pLS32 including the oriN region | Tanaka and Ogura (1998) |
| pDJ1730 | Source of spectinomycin resistance cassette (spc) | Guerout-Fleury et al (1996) |
| pEB14 | pEB7 with PspoIID-cre | This study |
| pEB15 | pEB14 with PspoIID-creΩspc | This study |
| pEB171 | pJSA1 with _PspoIIQ_-loxP-kan-loxP-gfp cloned into the _Nco_I site in cat | This study |
| pEB19 | pMDS13 with _PspoIIQ_-loxP-kan-loxP-gfpΩcat | This study |
| pEB24 | pSac-Cm with PspoIIR | This study |
| pEB27 | pEB24 with PspoIIR_-cre_Ωspc | This study |
| pEB53 | pSU18 with kan | This study |
| pEB57 | pEB53 with kan flanked by 527 bp upstream and 752 bp downstream of the racA gene | This study |
| pEB255 | pJSA3 containing 3 kb alfAB region | This study |
| pEB7 | pSac-Cm with PspoIID | This study |
| pFG6001 | pBET131 alfB∷kan | This study |
| pGAP100 | pBET131 with PspoIIQ -gfp reporter | This study |
| pGAP101 | pBET131 with PspoIIQ∷loxP-kan-loxP∷gfp reporter | This study |
| pJSA1 | pBR322 with cat | This study |
| pJSA3 | pJSA1 with oriN from pBET131 | This study |
| pJSA5 | pJSA3 with PspoIIQ | This study |
| pJSA6 | pJSA5 with PspoIIQ -gfp reporter | This study |
| pMDS13 | amyE integrating _PspoIIQ_-gfpΩcat | Sharp and Pogliano (2002) |
| pNCH103 | pMutin-GFP with alfA-gfp | This study |
| pNCH106 | pBET131 with alfA-gfp | This study |
| pNCH113 | PBET131 with D168A mutation in alfA | This study |
| pNCH129 | pMutin-GFP with alfAD168A-gfp | This study |
| pNCH135 | PBET131 with alfAD168A-gfp | This study |
| pSac-Cm | sacA integrating vector | Middleton and Hofmeister (2004) |
| pSU18 | Bartolome et al (1991) | |
| pUK19 | Source of kan for Cre/loxP experiments | Ju et al (1998) |
| pUB110 | Source of kan for alfB∷kan knockout | Bacillus genetic stock center |
| aDetails of plasmid constructions are provided in the Materials and methods and in the Supplementary data. |
Table 7.
Oligonucleotides used in this study
| Oligonucleotide designation | Oligonucleotide sequencea,b,c |
|---|---|
| EBP7 | 5′ggactag_t_aatttactgaccgtacacc |
| EBP8 | 5′ccagatctatcatttacgcgttaatggctaat |
| EBP9 | 5′ccagatctgcagccctggcgaatggcga |
| EBP10 | 5′tctgtcgaccccctatgcaagggttta |
| EBP12 | 5′cgggatccgtgagcggatgccaagatt |
| EBP14 | 5′ttgtcgacagatctactagtcattgtttcatcacctca |
| EBP15 | 5′cctactagtaaataacttcgtatagcatacattatacgaagttatgcgctcgggacccctatcta |
| EBP16 | 5′acggatccataacttcgtataatgtatgctatacgaagttatcccagcgaaccatttgaggtga |
| EBP38 | 5′ttgtcgacagatctactagtcatcggtccccaccgt |
| EBP39 | 5′cgggatccatatgcgctctgattcat |
| EBP53 | 5′ttgcagatctccatgataacg |
| EBP54 | 5′tttactagtctgcaccgtctttgcggag |
| EBP55 | 5′tttctcgagctcattcgatccccgag |
| EBP56 | 5′tttgcatgcaatcctcctggcgccgat |
| EBP82 | 5′ttttagatctggcgccgcacaccatc |
| EBP83 | 5′ttttactagtgcctacttcactgtctgaaag |
| EBP126 | 5′tttacatgtagaattcggatctgctag |
| EBP151 | 5′aaaaacatgtctattacggccgtttgtata |
| EBP174 | 5′cccagatctcatttccgagcttcgtgaggccg |
| EBP175 | 5′cccggatccgaaagccgtgttgatggcgcctg |
| EBP208 | 5′-ggggatccgtcgacaggaatgtttagcaaggaggta-3′ |
| EBP209 | 5′-gggggatccgccctctttcttagtagttg-3′ |
| cat1 | 5′atccgaattctcatgtttgac |
| cat2 | 5′agcaagcttcggggcaggttagtgaca |
| 2QGFP1 | 5′tcaagatctattaaaaactggtctgat |
| 2QGFP4 | 5′atgagatctagaattcggatctgctag |
| N2QSal | 5′gcgtcgacagaattcggatctgctag |
| 2QGFP5 | 5′tcaggatccattaaaaactggtctgat |
| oriN1b | 5′ttagaattccagaagaatatgtagaag |
| oriN2 | 5′agcgaattcagcttcctcctaaaatc |
| oriNint1 | 5′ggatggttcaatttgacctctc |
| oriNint2 | 5′gttcttctgatcggagacaag |
| orf6/kanFWD-1 | 5′cagttggattgcttctaaaatacggtaggaagtttgaggagatgtttgcgtgacaaacgggccagtttgttgaag |
| orf6/kanRVRS | 5′cttgaatctcgagtaagtctttttctttaatttcaattttcataagaaccctcctggttaatgcgccatgacagccatg |
| NgfpBgl | 5′ggaagatctaagcttactagtagtaaagga |
| NHP5 | 5′gcgcggtaccccttgctgccaacaagg |
| NHP7 | 5′ggtaccgctagccgcaaacatctcc |
| NHP17 | 5′cttctcagattcattaaactgcgtcattgtagctgctggttctaagacattaaacgtcctttatttaatc |
| aUnderlined sequence indicates restriction site. | |
| bBold lettering indicates the site of mutation on a mutagenic oligonucleotide. | |
| cUse of EBP oligonucleotides is described in Supplementary data. |
A detailed description of the Cre/loxP system for detecting postseptational plasmid partitioning is described in the Supplementary data. In brief, cell-specific expression of Cre recombinase was accomplished by fusing the coding sequence of cre to either the spoIID (mother cell) or the spoIIR (forespore) promoters and translational start sites, and integrating the hybrid genes at the sacA locus using derivatives of pSac-Cm. The loxP-kan-loxP∷gfp reporter was constructed by inserting a kanamycin resistance cassette (Ju et al, 1998) flanked by loxP sites (Sternberg et al, 1981) at the beginning of GFP, thereby disrupting the reading frame of this gene. This disrupted reporter gene was fused to the spoIIQ promoter (_P_FS) (creating plasmid pEB19) and then subcloned into pBET131 or integrated into the chromosome at amyE using a derivative of pMDS13 or into the chromosomal cat gene of EBS52 using a derivative of pJSA1. Cre-mediated recombination between loxP sites excises the kanamycin resistance cassette and restores the reading frame of GFP.
To determine the fraction of spores containing plasmids, spores were harvested after 24 h of sporulation, heated at 80°C for 30 min and then germinated on LB plates. The resulting colonies were screened for chloramphenicol resistance by patching on LB and LB chloramphenicol (5 μg/ml) plates. The fraction of plasmid containing spores that contain loxP-kan-loxP∷gfp were scored by patching colonies from germinated spores onto LB chloramphenicol and LB kanamycin (10 μg/ml) plates.
Assaying plasmid stability during growth
To determine the stability of the pBET131 plasmid during vegetative growth, cells were grown at 37°C in LB media containing chloramphenicol (5 μg/ml) to mid-logarithmic phase, washed twice with LB, diluted 1:106 in LB, and grown at 37°C without selection for 15 generations. At _t_=0 and 15 generations, 200–300 cells were spread onto LB plates, incubated overnight, and the resulting colonies tested by patching onto LB plates with and without chloramphenicol.
Microscopy
Cells were grown to the desired stage of sporulation in the presence of 0.5 μg/ml. FM 4-64, stained with 2 μg/ml DAPI and images collected as described (Pogliano et al, 1999).
Supplementary Material
Supplementary Figure 1
Acknowledgments
We are grateful to T Tanaka for plasmid pBET131, Jim Sawitzke and Don Court for the lambda red recombination strains, Marc Sharp for helpful discussions and Kit Pogliano for critical reading of the manuscript. This work was supported by grants from the NSF (MCB-0215752) and the NIH (R01GM073898) to JP, by an NIH Postdoctoral Fellowship (GM65692-03) to ECB, and by an HHMI predoctoral fellowship to ALD.
References
- Austin S, Ziese M, Sternberg N (1981) A novel role for site-specific recombination in maintenance of bacterial replicons. Cell 25: 729–736 [DOI] [PubMed] [Google Scholar]
- Bartolome B, Jubete Y, Martinez E, de la Cruz F (1991) Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102: 75–78 [DOI] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, Robinson C (2000) Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290: 995–997 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Fujita M, Liu XS, Gorbatyuk B, Skoko D, Yan J, Marko JF, Liu JS, Eichenberger P, Rudner DZ, Losick R (2005) Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol Cell 17: 773–782 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Losick R (2002) Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109: 257–266 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R (2003) RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536 [DOI] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA 89: 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-Lopez R, Errington J (2003a) The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell 4: 19–28 [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R, Errington J (2003b) A dynamic bacterial cytoskeleton. Trends Cell Biol 13: 577–583 [DOI] [PubMed] [Google Scholar]
- Costantino N, Court DL (2003) Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci USA 100: 15748–15753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan W, Zheng L, Sandman K, Losick R (1987) Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol 196: 1–10 [DOI] [PubMed] [Google Scholar]
- Errington J (2003a) Dynamic proteins and a cytoskeleton in bacteria. Nat Cell Biol 5: 175–178 [DOI] [PubMed] [Google Scholar]
- Errington J (2003b) Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1: 117–126 [DOI] [PubMed] [Google Scholar]
- Errington J, Bath J, Wu LJ (2001) DNA transport in bacteria. Nat Rev Mol Cell Biol 2: 538–545 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Garner EC, Campbell CS, Mullins RD (2004) Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306: 1021–1025 [DOI] [PubMed] [Google Scholar]
- Gerdes K, Moller-Jensen J, Ebersbach G, Kruse T, Nordstrom K (2004) Bacterial mitotic machineries. Cell 116: 359–366 [DOI] [PubMed] [Google Scholar]
- Gilman M, Chamberlin MJ (1983) Developmental and genetic regulation of Bacillus subtilis genes transcribed by σ28-RNA polymerase. Cell 35: 285–293 [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P (1996) Plasmids for ectopic integration in Bacillus subtilis. Gene 180: 57–61 [DOI] [PubMed] [Google Scholar]
- Hartl B, Wehrl W, Wiegert T, Homuth G, Schumann W (2001) Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J Bacteriol 183: 2696–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JA (1991) Genetic analysis in Bacillus subtilis. Methods Enzymol 204: 305–320 [DOI] [PubMed] [Google Scholar]
- Holmes KC, Sander C, Valencia A (1993) A new ATP-binding fold in actin, hexokinase and Hsc70. Trends Cell Biol 3: 53–59 [DOI] [PubMed] [Google Scholar]
- Ireton K, Gunther NW, Grossman AD (1994) spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol 176: 5320–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Gerdes K (1997) Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR–parC complex. J Mol Biol 269: 505–513 [DOI] [PubMed] [Google Scholar]
- Ju J, Luo T, Haldenwang WG (1998) Forespore expression and processing of the SigE transcription factor in wild-type and mutant Bacillus subtilis. J Bacteriol 180: 1673–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltwasser M, Wiegert T, Schumann W (2002) Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl Environ Microbiol 68: 2624–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GE, Derman AI, Pogliano J (2005) Bacterial DNA segregation by dynamic SopA polymers. Proc Natl Acad Sci USA 102: 17658–17663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NJ, Dutton RJ, Pogliano K (2006) Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol Microbiol 59: 1097–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer WJ, Castilla-Llorente V, Villar L, Murray H, Errington J, Salas M (2005) Molecular basis for the exploitation of spore formation as survival mechanism by virulent phage phi29. EMBO J 24: 3647–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton R, Hofmeister A (2004) New shuttle vectors for ectopic insertion of genes into Bacillus subtilis. Plasmid 51: 238–245 [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K (2003) Bacterial mitosis: parM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol Cell 12: 1477–1487 [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J, Jensen R, Lowe J, K G (2002) Prokaryotic DNA segregation by an actin-like filament. EMBO J 21: 3119–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Jensen J, Lowe J (2005) Increasing complexity of the bacterial cytoskeleton. Curr Opin Cell Biol 17: 75–81 [DOI] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-DeMello A, Perez A, Sun Y-L, Pogliano K (1999) A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol 31: 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Sharp MD, Pogliano K (2002) Partitioning of chromosomal DNA during establishment of cellular asymmetry in Bacillus subtilis. J Bacteriol 184: 1743–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R (2001) Morphological coupling in development: lessons from prokaryotes. Dev Cell 1: 733–742 [DOI] [PubMed] [Google Scholar]
- Ryter A (1965) Etude morphologie de la sporulation de Bacillus subtilis. Ann Inst Pasteur (Paris) 108: 40–60 [PubMed] [Google Scholar]
- Sauer B (1993) Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol 225: 890–900 [DOI] [PubMed] [Google Scholar]
- Setlow B, Magill N, Febbroriello P, Nakhimousky L, Koppel DE, Setlow P (1991) Condensation of the forespore nucleoid early in sporulation of Bacillus species. J Bacteriol 173: 6270–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K (1999) An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA 96: 14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K (2002) Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295: 137–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe ME, Errington J (1996) The Bacillus subtili soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol Microbiol 21: 501–509 [DOI] [PubMed] [Google Scholar]
- Sherratt DJ (2003) Bacterial chromosome dynamics. Science 301: 780–785 [DOI] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J (1969) Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J 113: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Hamilton D, Austin S, Yarmolinsky M, Hoess RH (1981) Site-specific recombination and its role in the life cycle of bacteriophage P1. Cold Spring Harbor Symp Quant Biol 1: 297–309 [DOI] [PubMed] [Google Scholar]
- Stragier P, Losick R (1996) Molecular genetics of sporulation in Bacillus subtilis. Ann Rev Genetics 30: 297–341 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Ishida H, Maehara T (2005) Characterization of the replication region of plasmid pLS32 from the Natto strain of Bacillus subtilis. J Bacteriol 187: 4315–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ogura M (1998) A novel Bacillus natto plasmid pLS32 capable of replication in Bacillus subtilis. FEBS Lett 422: 243–246 [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Wang SC, Shapiro L (2005) The bacterial nucleoid: A highly organized and dynamic structure. J Cell Biochem 96 (3): 506–521 [DOI] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ (1991) Actin microfilament dynamics in locomoting cells. Nature 352: 126–131 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Amos L, Lowe J (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413: 39–44 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J (2002) F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J 21: 6935–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne GD (2001) A structural view of Cre-loxP site-specific recombination. Annu Rev Biophys Biomol Struct 30: 87–104 [DOI] [PubMed] [Google Scholar]
- Webb CD, Decatur A, Teleman A, Losick R (1995) Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol 177: 5906–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC-H, Grossman AD, Wright A, Losick R (1997) Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88: 667–674 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (1994) Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264: 572–575 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (1998) Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol 27: 777–786 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (2003) RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol 49: 1463–1475 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J (1995) A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev 9: 1316–1326 [DOI] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Sandman K (eds.) (1984) New Genetic Methods, Molecular Cloning Strategies, and Gene Fusion Techniques for Bacillus Subtilis Which Take Advantage of Tn917 Insertional Mutagenesis. Academic Press: NewWork [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1