Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL (original) (raw)
Abstract
HDL lowers the risk for atherosclerotic cardiovascular disease by promoting cholesterol efflux from macrophage foam cells. However, other antiatherosclerotic properties of HDL are poorly understood. To test the hypothesis that the lipoprotein carries proteins that might have novel cardioprotective activities, we used shotgun proteomics to investigate the composition of HDL isolated from healthy subjects and subjects with coronary artery disease (CAD). Unexpectedly, our analytical strategy identified multiple complement-regulatory proteins and a diverse array of distinct serpins with serine-type endopeptidase inhibitor activity. Many acute-phase response proteins were also detected, supporting the proposal that HDL is of central importance in inflammation. Mass spectrometry and biochemical analyses demonstrated that HDL3 from subjects with CAD was selectively enriched in apoE, raising the possibility that HDL carries a unique cargo of proteins in humans with clinically significant cardiovascular disease. Collectively, our observations suggest that HDL plays previously unsuspected roles in regulating the complement system and protecting tissue from proteolysis and that the protein cargo of HDL contributes to its antiinflammatory and antiatherogenic properties.
Introduction
Mortality from cardiovascular disease has decreased significantly in the United States over the past 30 years (1, 2). Many factors have contributed to this improvement, including the identification of cardiovascular risk factors, development of interventions that reduce those risk factors, and new treatments for acute coronary syndromes. Despite these advances, however, cardiovascular disease remains the leading cause of mortality in the United States (1, 2).
One important risk factor for atherosclerosis is a low level of HDL, which directly protects against atherosclerosis by removing cholesterol from artery wall macrophages (3, 4). Thus, the risk of cardiovascular disease is inversely proportional to plasma levels of HDL and its major apolipoprotein, apoA-I (5). Also, low HDL levels are particularly common in men with premature atherosclerosis (6), whereas high HDL levels associate with longevity (7).
Antiinflammatory and antioxidant properties might also contribute to the cardioprotective effects of HDL (8). Moreover, both oxidative modifications and alterations in the protein cargo of HDL may alter its biological activity, creating potentially proatherogenic particles (8, 9). We and others recently showed that HDL isolated from plasma of patients with known coronary artery disease (CAD) is oxidatively modified and that oxidation impairs reverse cholesterol transport mediated by HDL (10–12). Circulating levels of serum amyloid A (SAA), an inflammatory protein in HDL, predict the risk of heart disease in humans (13), and HDL-associated SAA has been proposed to render the lipoprotein atherogenic (14). Furthermore, alterations in the balance between pro- and antioxidative enzymes in HDL appear to play a critical role in converting the lipoprotein to a proatherogenic form (8).
We hypothesized that quantifying the protein composition of HDL might provide insights into the antiatherogenic and antiinflammatory properties of the lipoprotein. We therefore used shotgun proteomics — direct analysis of a complex mixture of proteins (15) — to study the HDL proteome. After digesting the lipoprotein with trypsin, we analyzed the resulting peptide mixture with mass spectrometry (MS) and matched the tandem mass spectra of the peptides with spectra derived from protein sequences in protein database. We found that HDL carries protein families implicated in complement activation and regulation of proteolysis, raising the possibility that these proteins contribute to the innate immune system and the cardioprotective properties of HDL.
Results
Shotgun proteomics identifies distinct protein families in HDL.
We first confirmed that liquid chromatography–electrospray ionization–tandem MS (LC-ESI-MS/MS) analysis reproducibly identifies the same HDL-associated proteins (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI26206DS1). We then used this analytical strategy to investigate the protein composition of HDL isolated by ultracentrifugation in 2 independent groups of subjects (see Methods). The first used total HDL isolated from 20 apparently healthy control subjects (group 1), and the second used HDL3 isolated from 13 subjects (group 2; 6 control subjects and 7 subjects with established CAD; additional details below). The clinical characteristics of the subjects are shown in Table 1. The following criteria were used to identify proteins: (a) a high peptide identification score (according to PeptideProphet; P > 0.90); (b) a high protein identification score (P > 0.95, ProteinProphet); and (c) at least 2 peptides unique to the protein of interest had to be detected in at least 5 subjects (total HDL) or 2 subjects (HDL3). Requiring at least 2 unique peptides with a high confidence score markedly decreases the false-positive rate of protein identification (16). Shotgun proteomics with these stringent criteria identified 48 proteins in HDL (Table 2).
Table 1 .
Clinical characteristics of study subjects
Table 2 .
Proteins detected by LC-ESI-MS/MS in HDL (group 1) and HDL3 (group 2) isolated from control and/or CAD subjects
This analytical approach detected every apolipoprotein already known to associate with HDL, except for apoA-V. Immunoblot analysis confirmed that apoA-I, apoA-II, apoC-II, apoE, clusterin (apoJ), and apoM were present in the lipoprotein. Indeed, we detected more than 90% of all proteins that have previously been identified by biochemical methods in HDL, including lecithin:cholesterol acyl transferase (LCAT), phospholipid transfer protein (PLTP), cholesteryl ester transfer protein (CETP), transferrin, all 3 human serum amyloid proteins (SAA1, SAA2, SAA4), and both paraoxonase-1 (PON1) and PON3 (17–28), strongly suggesting that our overall experimental approach was valid.
We found a number of protein families not previously known to reside in HDL, including complement pathway and regulatory proteins (complement component 4 [C4A and C4B], complement component 9 [C9], vitronectin) and endopeptidase inhibitors (α-2-antiplasmin, α-2-HS-glycoprotein, inter-α-trypsin inhibitor [α-1-microglobulin/bikunin and inter-α-trypsin inhibitor heavy chain H4], angiotensinogen, α-1-antitrypsin, serpin peptidase inhibitor [clade F, member 1], kininogen-1). We also detected plasma retinol–binding protein, which was recently implicated in the pathogenesis of type 2 diabetes (29). Interestingly, we identified the lysosomal protein prenylcysteine oxidase and hemopexin, an iron-binding protein that inhibits LDL oxidation.
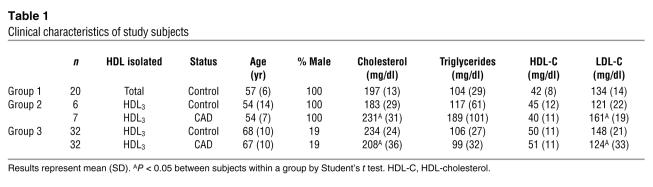
Gene Ontology (GO) analysis (Figure 1) demonstrated that 22 of the 48 HDL-associated proteins were linked to cholesterol and lipoprotein metabolism (P = 2 × 10–27). Twenty-three of the 48 were acute-phase-response proteins, whose plasma concentrations are altered markedly by acute infection and chronic inflammation (P = 1 × 10–18). Six proteins were regulators of complement activation (P = 5 × 10–5). The analysis also revealed 2 major clusters of molecular function: 8 proteins were endopeptidase inhibitors (P = 2 × 10–6), and 8 were heparin-binding proteins (SAA1, SAA2, SAA4, apoE, apoH, haptoglobin-related protein, kininogen-1, vitronectin; P = 6 × 10–5). Collectively, these observations implicate HDL in the acute-phase response and innate immunity. They also strongly support the proposal that HDL carries proteins that may play previously unsuspected roles in regulating complement activation and inhibiting proteolysis.
Figure 1. Global view of biological processes and molecular functions of HDL proteins.
Proteins in total HDL and HDL3 were identified using LC-ESI-MS/MS (Table 2). Proteins detected in HDL were associated with biological functions using GO process annotations. This approach demonstrated significant overrepresentation of proteins involved in several categories, including lipid metabolism (P = 2 × 10–27), the acute-phase response (P = 1 × 10–18), protease inhibitor activity (P = 2 × 10–6), and complement regulation (P = 5 × 10–5). apoH, β-2-glycoprotein I; AGT, angiotensinogen; AHSG, α-2-HS-glycoprotein; AMP, bikunin; FGA, fibrinogen; HRP, haptoglobin-related protein; HPX, hemopexin; ITIH4, inter-α-trypsin inhibitor heavy chain H4; KNG1, kininogen-1; LCAT, lecithin-cholesterol acyltransferase; ORM2, α-1-acid glycoprotein 2; PLTP, phospholipid transfer protein; RBP4, retinol binding protein; SERA1, α-1-antitrypsin; SERF1, serpin peptidase inhibitor (clade F, member 1); SERF2, α-2-antiplasmin; TF, transferrin; TTR, transthyretin; VTN, vitronectin.
apoA-I particles and HDL carry members of overlapping families of proteins.
To further investigate the protein composition of HDL, we used affinity chromatography to isolate lipoprotein particles containing apoA-I, the major HDL protein. The particles were then analyzed by LC-ESI-MS/MS. We used this approach because it is well established that proteins can dissociate from lipoproteins during ultracentrifugation (30, 31), eliminating some proteins that HDL would carry under physiological conditions. Additionally, isolation by our orthogonal techniques and detection of the same proteins in both apoA-I particles and HDL would strongly support the proposal that those proteins actually associate with HDL and are not just abundant plasma components.
apoA-I particles prepared from plasma of healthy subjects was digested with trypsin and analyzed by LC-ESI-MS/MS. Control studies used columns containing beads cross-linked to albumin or nonspecific immunoglobulin. This approach identified 11 of the 13 proteins in the apoA-I particles that were not previously known to reside in HDL and/or HDL3. Importantly, this included all of the HDL-associated complement-related proteins, the thiol protease inhibitor kininogen-1, and 5 of the 6 serine-type endopeptidase inhibitors. In contrast, we failed to detect plasma retinol–binding protein or serpin peptidase inhibitor (clade F, member 1), suggesting that apoA-I particles may not contain these proteins. We also found peptides derived from the α, β, and γ chains of fibrinogen and complement factor H–related protein, which were previously isolated from a minor HDL fraction (32).
apoB-100 (the major LDL protein) and albumin were detected in HDL and HDL3. These proteins are abundant in plasma and therefore might have been contaminants. The dense fraction of LDL can also be coisolated with HDL by ultracentrifugation. However, we detected both of these proteins in apoA-I isolated by affinity chromatography. Only low levels of apoB-100 and albumin were detected in material eluted from control affinity columns (matrix alone or cross-linked to nonspecific immunoglobulin), which argues against nonspecific binding. It is noteworthy that albumin binds free fatty acids and other hydrophobic compounds with high affinity and that other investigators have proposed that HDL may interact with albumin (20). Moreover, apoA-I has recently been found associated with albumin isolated biochemically or serum albumin by affinity chromatography, suggesting that the apolipoprotein can associate with albumin (33).
Complement component 3 complexes with HDL.
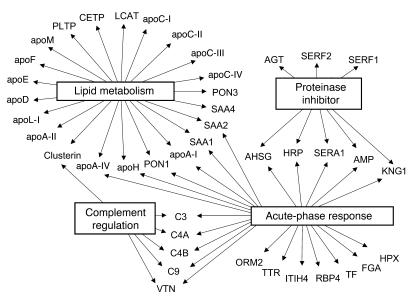
GO analysis revealed a highly significant association of HDL with complement factors (C3, C4, C9) as well as the complement-regulatory proteins vitronectin and clusterin. A major effector of the complement system, C3, has been implicated in atherogenesis (34). All 3 unique peptides identified by MS/MS in HDL3 are between residues 954 and 1,303 of C3dg (Figure 2, A and B). This region contains a thioester bond that can cross-link C3dg to target proteins (Figure 2C), raising the possibility that C3dg forms a covalent complex with HDL proteins.
Figure 2. Mass spectrometric and immunological detection of C3 in HDL3.
HDL3 or apoA-I complexes were isolated from plasma of control subjects. In parallel studies, plasma was passed over columns containing albumin or nonspecific immunoglobulin covalently cross-linked to beads. Isolated proteins were separated under denaturing conditions by gel electrophoresis, transferred to nitrocellulose, and analyzed through immunoblotting. (A and B) MS/MS spectra of 2 peptides unique to C3 in HDL3. (C) Sequence of the C3dg domain of human complement C3. The 3 tryptic digest peptides detected selectively in HDL3 of CAD subjects are underlined. (D) Detection of proteins reactive with antibody to C3. Lane 1: plasma. Lanes 2 and 3: eluate from control columns (albumin and IgG, respectively). Lane 4: apoA-I complexes. Lanes 5–7: 3 independent preparations of HDL3 isolated from pooled human plasma.
To biochemically confirm that C3 is a component of HDL, we isolated HDL3 from pooled plasma and immunoblotted it with a polyclonal antibody against C3. Three independent HDL3 preparations exhibited major bands of immunoreactive material with apparent MWs of approximately 40 kDa, 45 kDa, and 65 kDa (Figure 2D, lanes 5–7). We also detected immunoreactive C3 in apoA-I complexes isolated by affinity chromatography (Figure 2D, lane 4) but not in the material eluted from control columns (Figure 2D, lanes 2 and 3). The patterns of immunoreactive C3 in HDL3 and immunoisolated apoA-I differed, suggesting that the different preparations of HDL contained distinct species of C3. These observations strongly suggest that proteolytic fragments derived from C3 associate with HDL.
Endopeptidase inhibitor and complement-regulatory proteins associate with HDL.
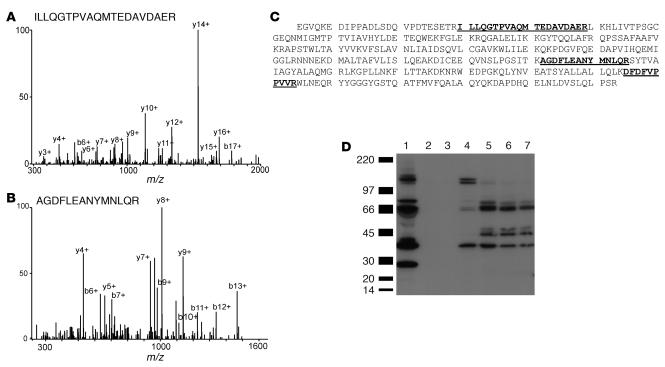
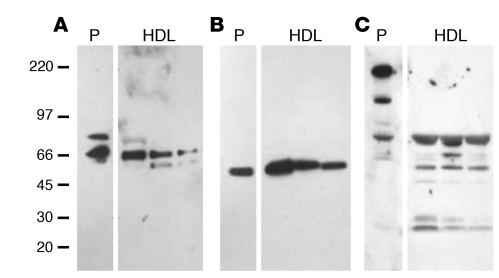
Immunoblot analysis of proteins separated by SDS-PAGE confirmed biochemically that vitronectin and 2 endopeptidase inhibitors, inter-α-trypsin inhibitor and α-2-HS glycoprotein (Figure 3), were associated with HDL3.
Figure 3. Immunodetection of vitronectin (A), α-2-HS glycoprotein (B) and α-1–intertrypsin inhibitor (C) in HDL3.
HDL3 was isolated from the plasma of 3 subjects by ultracentrifugation. HDL-associated proteins were separated under denaturing conditions by gel electrophoresis, transferred to a PVDF membrane, and subjected to immunoblot analysis. P, plasma.
Recent studies have demonstrated that HDL carries a wide range of low-MW peptides (35). However, the apparent MWs of vitronectin and α-2-HS glycoprotein in plasma and HDL3 were indistinguishable, strongly suggesting a lack of proteolytic processing of the protein forms identified in HDL. The major form of immunoreactive inter-α-trypsin inhibitor detected in plasma was consistent with its reported MW of 240 kDa, which is thought to represent a complex of 1 light and 2 heavy chains (36). The material in HDL3 that immunoreacted with the polyclonal antibody to inter-α-trypsin inhibitor differed from that in plasma. Their apparent MWs were consistent with those anticipated for the uncomplexed heavy and light chains. The light chain of this protein, also termed bikunin, contains 2 Kunitz-type protease inhibitor domains, which can complex with proteins. It is therefore of interest that an immunoreactive protein with an apparent MW somewhat greater than that of the light chain of inter-α-trypsin inhibitor was detected in HDL.
Collectively, these observations indicate that HDL3 contains immunoreactive vitronectin, α-2-HS glycoprotein, and the heavy and light chains of inter-α-trypsin inhibitor. They also raise the possibility that 2 of the proteins we detected, inter-α-trypsin inhibitor and complement factor C3, may form complexes with other HDL-associated proteins.
HDL isolated from healthy subjects and subjects with established CAD carry different protein cargoes.
To begin to investigate the protein composition of HDL from CAD subjects, we focused on HDL3, the lipoprotein’s dense subfraction. We selected HDL3 to minimize potential contamination with LDL because our analyses detected the LDL protein apoB100 in total HDL of control subjects.
We isolated HDL3 from the plasma of 7 men with CAD and 6 apparently healthy age-matched men (group 2 in Table 1; age 54 ± 7 [mean ± SD] and 54 ± 14 years, respectively). The CAD patients were recently diagnosed, as documented by clinical symptoms consistent with angina and q waves on their EKG or by the presence of at least 1 stenotic lesion (>50%) on coronary angiography. However, they were clinically stable, and at least 3 months had elapsed since their acute coronary syndrome. None of the subjects smoked cigarettes, had liver or renal disease, or had received lipid-lowering medications for at least 4 weeks before their blood was collected. The healthy controls had no known history of CAD, were not hyperlipidemic or diabetic, had no family history of premature CAD, and were not receiving lipid-lowering therapy. The patients with CAD had higher levels of total cholesterol, LDL, and triglycerides in their plasma than the healthy controls (Table 1; group 2). Importantly, there were no major differences in levels of HDL-cholesterol in this small cohort of CAD patients and controls (40 ± 11 [mean ± SD] and 45 ± 12 mg/dl, respectively).
Quantifying proteins by MS remains a formidable technical challenge. However, recent studies of model systems support the proposal that peptide counting — summing all the peptides derived from a single protein in an LC-ESI-MS/MS analysis — can assess relative protein abundance (37–39). To assess the relative abundance of HDL-associated proteins, we developed the following strategy. Peptides in the tryptic digest of HDL samples were separated using a fully automated 2-dimensional LC-MS system with 10-step cation-exchange fractionation and 90 minutes reverse-phase elution. This approach facilitated extensive data-dependent MS/MS sampling and the generation of an adequate number of peptide counts to reproducibly reflect relative peptide abundance (38, 39). We then developed an empiric test based on the peptide count, termed the “peptide index,” to provide a semiquantitative measure of relative protein abundance in different groups of subjects (see Methods). Last, we determined whether the peptide index derived from each protein in healthy controls differed significantly from that in patients with CAD.
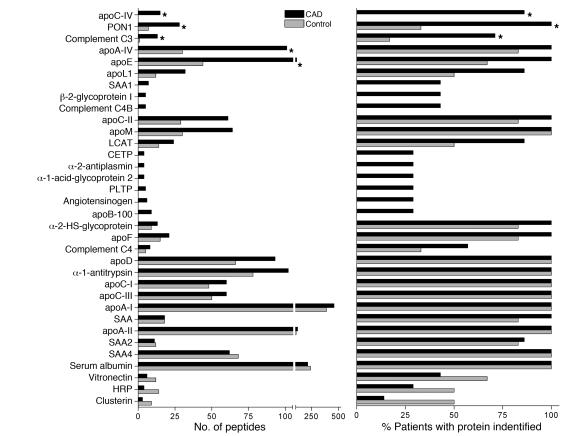
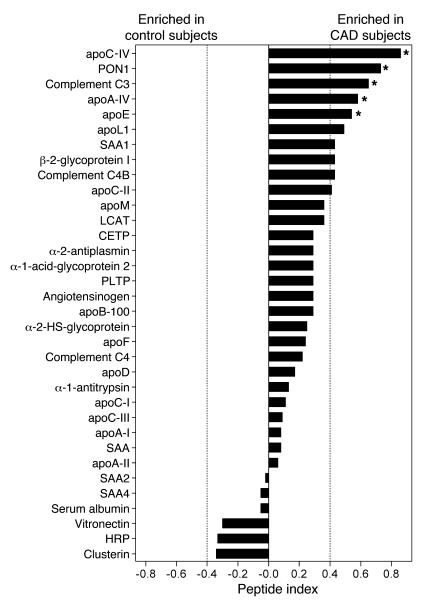
The number of peptides detected for each protein, the number of subjects in each group with detectable peptides, and the peptide index for each protein are shown in Figures 4 and 5. Permutation analysis suggested that a peptide index of less than –0.40 or greater than 0.40 identifies selective enrichment of proteins in either control subjects or CAD subjects, respectively (see Methods). We therefore used a peptide index of greater than 0.40 as the initial criterion for classifying proteins as being potentially more abundant in CAD subjects (Figure 5). This approach identified 8 proteins in HDL3 from the CAD patient population. Six of the 8 apparently more abundant proteins — PON1, apoC-II, apoA-IV, apoC-IV, apoL-1, and apoE — had previously been identified in HDL. Interestingly, peptides derived from C3 and C4 also appeared to be enriched in patients with CAD. In contrast, none of the HDL-associated proteins were more abundant in the control subjects. Statistical analysis of peptide abundance indicated that 5 of the 8 proteins in HDL3 were enriched significantly in the CAD patient population (Figure 5; P = 0.0003 to 0.03). Collectively, these observations suggest that HDL3 of CAD subjects is relatively enriched in specific proteins (Supplemental Table 1).
Figure 4. Peptide number and number of subjects with detectable peptides in HDL3 isolated from control subjects and CAD subjects.
HDL3 was isolated from the plasma of 6 control subjects and 7 subjects with established CAD (group 2; Table 1). HDL-associated proteins were subjected to LC-ESI-MS/MS analysis. All protein identifications required detection of at least 2 unique peptides from each protein from at least 2 individuals (Table 1). The P value was assessed by Student’s t test (peptide number) or Fisher’s exact test (subject number). *P < 0.05.
Figure 5. Relative abundance of proteins isolated from HDL3 of control and CAD subjects.
HDL3 was isolated from plasma of 6 control subjects and 7 subjects with established CAD. The relative abundance of proteins in the CAD subjects versus the control subjects was assessed by the peptide index, as described in Methods. *P < 0.05 by Student’s t test (peptide number) or Fisher’s exact test (subject number).
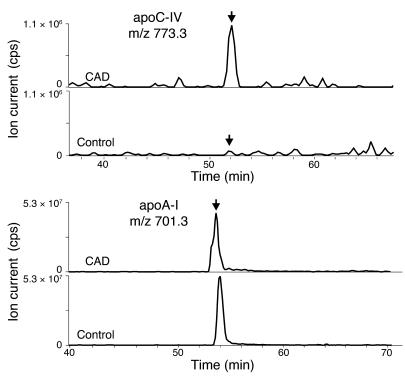
Because peptide counting is only semiquantitative, we used extracted ion chromatograms to quantify the relative abundance of peptides unique to the proteins that seemed to be more abundant in the CAD patients (39). In this technique, the ion current for a given peptide and charge state are extracted from the full-scan mass spectrum and used to construct a chromatogram. This approach demonstrated that peptides derived from apoC-IV were more abundant in CAD patients than in control subjects (Figure 6A). In contrast, there was no apparent difference in the relative abundance of peptides derived from apoA-I (Figure 6B).
Figure 6. Reconstructed ion chromatograms of peptides derived from apoC-IV and apoA-I in HDL3.
Tryptic digests of HDL3 isolated from CAD subjects and control subjects were subjected to LC-ESI-MS/MS analysis as described in the legend to Figure 1. Reconstructed ion chromatograms of peptides (arrows) unique to apoC-IV and apoA-I were extracted from the full-scan mass survey spectra of each analysis. cps, counts per second.
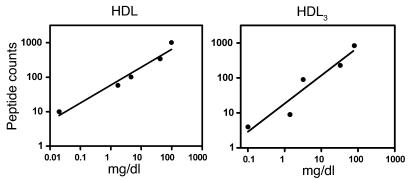
To provide additional evidence that peptide counting monitors protein abundance, we examined the relationship among the number of unique peptides we detected in HDL and HDL3 (Table 2); the estimated concentration of CETP in HDL3 (40); and the reported concentrations for apoA-I, apoA-II, apoC-II, apoB, and transferrin in HDL and HDL3 isolated by ultracentrifugation (41). Both the number of unique peptides detected by MS/MS analysis and the protein concentrations varied over 3 orders of magnitude. Linear regression analysis (Figure 7) demonstrated a strong correlation between protein concentration and the number of unique peptides for the proteins in HDL and HDL3 (_r_2 = 0.98 and 0.88, respectively). This approach also suggested that under our experimental conditions, the limit of detection for MS/MS analysis for HDL3 was approximately 0.1 mg/ml, as we detected CETP (4 unique peptides, approximately 0.1 mg/dl; ref. 40) but failed to detect apoA-V (estimated plasma concentration approximately 0.02 mg/dl; ref. 42). These results support the proposal that the peptide count can quantify proteins in HDL (38, 39).
Figure 7. Relationship between unique peptides and protein concentration in HDL and HDL3.
The total number of unique peptides detected by MS in HDL and HDL3 is as shown in Table 2. The protein concentrations of apoA-I (98 mg/dl), apoA-II (42 mg/dl), apoC-II (4.6 mg/dl), apoB (1.7 mg/dl), and transferrin (0.02 mg/dl) in HDL3 and HDL isolated by ultracentrifugation are those reported by McPherson et al. (where HDL equals the sum of HDL2 and HDL3; ref. 41). The concentration of CETP in HDL3 (0.1 mg/dl) was estimated as 50% that of plasma CETP (40). Linear regression analysis revealed a strong correlation (_r_2 = 0.98 and 0.88) between protein concentration and unique peptides in HDL and HDL3, respectively.
apoE is enriched in HDL3 in subjects with established CAD.
To extend our observations to more subjects and confirm our observations biochemically, we used an antibody-based approach to quantify levels of apoE in HDL3 isolated from 32 healthy control subjects and 32 age-, sex-, HDL-, and triglyceride-matched CAD subjects enrolled in the Pravastatin Inflammation/CRP Evaluation (PRINCE) study (43). The clinical characteristics of the subjects (group 3) are shown in Table 1. Briefly, control subjects were part of a primary prevention cohort and had no symptoms of clinically significant CAD and no family history of premature CAD. Subjects with established CAD had symptoms consistent with angina and q waves on their EKGs or at least 1 stenotic lesion (>50%) detectable by coronary angiography. The CAD subjects had experienced their acute coronary syndrome at least 3 months previously and were not taking lipid-lowering drugs. The control subjects were apparently healthy, had no underlying medical conditions, were nonsmokers, and were not receiving any lipid-modulating therapy.
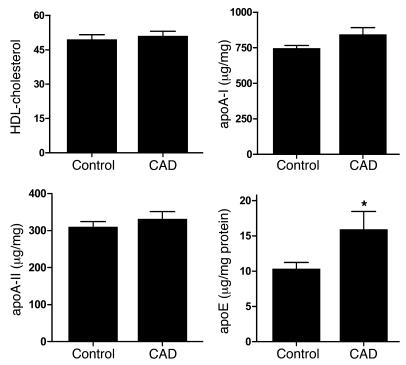
The levels of HDL-cholesterol, apoA-I, apoA-II, and apoE of the 64 subjects are shown in Figure 8. Due to the matching criteria, the 2 groups had virtually identical HDL-cholesterol levels. In contrast, the level of apoE in HDL3 isolated from the control subjects and CAD subjects were 10 ± 5 (mean ± SD) and 16 ± 4 μg/mg protein, respectively (P = 0.02). It is important to note that both apoA-I and apoA-II were quantified simultaneously with apoE in HDL3 and that no significant differences were observed in either apolipoprotein. Covariant analysis indicated that sex, total cholesterol, and LDL cholesterol did not contribute significantly to the variation in apoE levels.
Figure 8. Quantification of HDL-cholesterol, apoA-I, apoA-II, and apoE in HDL3 isolated from control and CAD subjects matched for plasma levels of triglycerides.
Plasma was obtained from 32 apparently healthy individuals and 32 individuals with established coronary artery disease (group 3; Table 1). HDL3 was isolated from plasma by sequential ultracentrifugation. apoA-I, apoA-II, and apoE were quantified by ELISA. Results are normalized to the protein content of HDL3. *P = 0.02 by a 2-tailed Student’s t test.
The subjects enrolled in the PRINCE study (group 3; Table 1) differed from those used for our proteomics analysis (group 2; Table 1). The latter were male, and LDL levels were 30% higher in the CAD subjects than in the control subjects (P = 0.005). In contrast, 81% of the PRINCE subjects were female, and levels of LDL were slightly but significantly lower (16%; P = 0.01) in the CAD subjects. Collectively, these results indicate that HDL3 of CAD subjects is enriched in apoE and that hyperlipidemia and sex are unlikely to account for the observed differences. Our observations may be clinically relevant, given that we detected elevated levels of apoE in HDL3 isolated from 2 very different populations of CAD subjects.
HDL isolated from atherosclerotic lesions and HDL from CAD patients carries a subset of the same proteins.
We and others have previously shown that HDL isolated from atherosclerotic lesions contains higher levels of oxidized amino acids than HDL isolated from plasma (10–12). This observation raises the possibility that proteins found in HDL from CAD patients might come in part from the artery wall.
To test this proposal, we isolated HDL by ultracentrifugation from human carotid atherosclerotic tissue recovered from 6 patients during surgery. Because arterial tissue contains relatively low levels of apoA-I, total HDL was isolated by ultracentrifugation to maximize the yield. Lesion HDL analyzed by immunoblotting with a rabbit polyclonal antibody monospecific for human apoA-I contained a predominant protein with the predicted molecular mass of apoA-I (data not shown). Quantifying apoA-I by immunoblotting in lesions, extracted lesions, and lesion HDL showed that this procedure recovered approximately 80% of the immunoreactive protein originally present in the atherosclerotic tissue. HDL isolated from 2 individuals was combined, digested with trypsin, and analyzed with LC-ESI-MS/MS. Using the criterion of 2 unique peptides with high confidence identifications, we identified more than 100 proteins in lesion HDL in 3 independent analyses. Importantly, 3 of the 5 proteins that appeared enriched in the HDL3 samples from the CAD patients were also present in lesion HDL, and apoE was the most abundant of these proteins (Supplemental Table 2).
Discussion
We employed shotgun proteomics to test the hypothesis that HDL might carry proteins that make a previously unsuspected contribution to its cardioprotective and antiinflammatory activities. Forty-eight proteins were identified in HDL isolated by ultracentrifugation from healthy controls and/or CAD subjects. Twenty-two proteins with well-characterized roles in lipid metabolism and the antioxidant properties of HDL were detected, which validates our experimental approach. Importantly, we found 13 proteins not previously known to reside in HDL.
We used annotations by the GO Consortium to connect the complex array of proteins we identified in HDL to biological processes. Twenty-two of the proteins were linked to cholesterol and lipoprotein metabolism. Remarkably, there were more acute-phase-response proteins (23 of 48), whose plasma concentrations are altered markedly by acute and chronic inflammation. A number of proteins not previously known to reside in HDL, including C4A/C4B, C9, and vitronectin, were linked to the complement pathway and its regulation. Our detection of the extracellular matrix protein vitronectin and the lysosomal protein prenylcysteine oxidase suggests that HDL components can be derived from cells other than those that synthesize the lipoprotein in the liver and intestine. The detection of multiple proteins with roles in complement activation, together with the identification of specific apoA-I complexes that help kill pathogens (44), is consistent with the suggestion that HDL serves as a platform for the assembly of proteins involved in the innate immune response.
Proteolysis of artery wall proteins is thought to play a critical role in plaque rupture, the major cause of myocardial infarction and sudden death in subjects with CAD (45). It is noteworthy that we found α-2-antiplasmin, α-2-HS-glycoprotein, angiotensinogen, inter-α-trypsin inhibitor, α-2-macroglobulin, and serpin peptidase inhibitor (clade F, member 1) in HDL. All of these proteins have serine proteinase inhibitor domains. Serine proteinase inhibitors, termed serpins, are key regulators of numerous biological pathways that initiate inflammation, coagulation, apoptosis, angiogenesis, and matrix degradation (46). Moreover, we also detected the thiol proteinase inhibitor kininogen-1 and haptoglobin-related protein. The latter contains a trypsin-like serine proteinase domain with a substitution in 1 of its catalytic triad residues that may allow it to act as a decoy substrate to prevent proteolysis. Collectively, these observations raise the possibility that HDL plays a previously unsuspected role in preventing plaque rupture, perhaps by protecting tissue from promiscuous proteolysis.
Complement activation is implicated in tissue damage during acute myocardial infarction (47, 48). It may therefore be significant that we found multiple complement components and complement-regulatory proteins in HDL. Moreover, in vitro studies indicate that HDL blocks the assembly of the terminal complement attack complex on endothelial cells (49). Inhibition of complement deposition may limit injury to cardiac cells and prevent activation of the procoagulant response in endothelium, macrophages, and platelets, 2 critical events in acute thrombosis.
When we extended our analysis to HDL3 from men with established CAD, we found the lipoprotein to be selectively enriched in 5 proteins. One protein was C3, suggesting that this HDL-associated immune system protein may contribute to vascular disease. It is noteworthy that C3 is produced by human monocyte-derived macrophages (50) and that 3 of the other enriched proteins — apoE, apoC-IV, and apoA-IV — are involved in lipid metabolism and cholesterol export from macrophages. apoE and apoC-IV are part of a gene cluster that is induced in macrophages by cholesterol-sensing nuclear receptors that protect against atherosclerosis in mice (51). apoA-IV, along with apoA-I and apoC-III, belongs to another gene cluster that also protects against atherosclerosis when expressed in mice (52). Taken together, our observations suggest that HDL3 from CAD patients is selectively enriched in proteins that play critical roles in macrophage biology, lipid metabolism, and the inflammatory response. Several of these proteins are produced by cholesterol-loaded macrophages, suggesting that a fraction of the proteins that are overproduced in CAD patients may be derived from macrophages in atherosclerotic lesions. Alternatively, conditions associated with CAD may enhance their production by the liver, promote their association with circulating HDL, or remodel HDL particles so that they redistribute to denser HDL subfractions (3).
Our proteomics analyses suggested that apoE was the most significantly enriched protein in CAD HDL3. To confirm that this approach can estimate protein abundance semiquantitatively, we measured apoE levels immunochemically in HDL3 isolated from a second set of 64 subjects: 32 with established CAD and 32 age-matched controls. Levels of apoE were significantly higher in HDL3 isolated from the CAD subjects. In striking contrast, the 2 groups had similar levels of HDL-cholesterol, apoA-I, apoA-II, and plasma triglycerides. Importantly, relative protein abundance, as assessed by MS and biochemical assays, was similar, supporting the proposal that peptide counts and the peptide index can assess relative protein abundance in biological material.
Many lines of evidence indicate that apoE plays a role in modulating atherogenesis. apoE is produced by cholesterol-loaded macrophages, where it can promote cholesterol efflux during its secretion. Circulating and macrophage-produced apoE removes macrophage cholesterol through its interaction with the ATP-binding cassette transporter (ABC) A1. apoE-rich HDL2 particles are efficient acceptors for macrophage cholesterol exported by another ABC transporter termed ABCG1 (53). apoE may be important for the expansion of HDL particles required for driving net cholesterol transport from macrophage foam cells (54). Epidemiological and clinical studies suggest that elevated levels of HDL2 are cardioprotective. Moreover, apoE levels are lower in HDL2 (the less dense subfraction of HDL) isolated from subjects with CAD (55), and apoE-enriched HDL2 isolated from CETP-deficient subjects removes potentially toxic cholesterol from macrophages much more efficiently than control HDL2 (53). Our demonstration of elevated levels of apoE in HDL3 isolated from subjects with established CAD raises the possibility that redistribution of apoE from HDL2 to HDL3 impairs cholesterol efflux and promotes the formation of macrophage foam cells in vivo.
In summary, our results indicate that HDL carries a population of proteins that function in lipid metabolism, proteinase inhibition, complement activation, and the acute-phase response. It will be of interest to study a larger number of subjects to determine whether levels of the proteins we detected in HDL3 from CAD subjects might be useful indicators of cardiovascular risk and to investigate the proposal that chronic inflammation alters the protein composition of HDL, making it atherogenic (3, 8, 13).
Methods
Isolation of HDL.
Three groups of subjects were used to study the protein composition of HDL (Table 1). The first group (20 control male subjects from Seattle) was used for proteomics analysis of total HDL. The second group (6 control subjects and 7 CAD subjects from Seattle) was used for proteomics analysis of HDL3. The third group (64 subjects enrolled in the PRINCE study (43) was used for quantification of apoE in HDL3. All protocols involving human material were approved by the Human Studies Committees at the University of Washington, Brigham and Women’s Hospital, Harvard Medical School, and Wake Forest University. All subjects provided informed consent.
Blood anticoagulated with EDTA was collected from subjects recruited at the University of Washington or Brigham and Women’s Hospital. The subjects had fasted overnight. Atherosclerotic tissue was harvested at carotid endarterectomy at Wake Forest University, snap-frozen, and stored at –80°C until analysis. Lesions from a single individual (approximately 0.5 g wet weight) were mixed with dry ice and pulverized with a pestle in a stainless steel mortar. HDL was extracted from the tissue powder (10). HDL (d = 1.063–1.210 g/ml) and HDL3 (d = 1.110–1.210 g/ml) were isolated by ultracentrifugation (56). apoA-I complexes were isolated from human plasma using apoA-I–specific goat polyclonal antibody covalently coupled to HiTrap NHS-activated HP columns (GE HealthCare) (57). As a control, parallel analyses were run using material eluted from washed beads coupled to albumin or nonspecific immunoglobulin. Protein was determined using the Bradford assay, with albumin as the standard.
Tryptic digest.
HDL was precipitated with 10% trichloroacetic acid and resolubilized with 6 M urea in 25 mM ammonium bicarbonate. Reduced, alkylated proteins were suspended in 0.6 M urea in 25 mM ammonium bicarbonate, digested overnight at 37°C with trypsin (1:20, wt/wt, trypsin/HDL protein), acidified with acetic acid, dried under vacuum, and resuspended in 0.1% formic acid. Tryptic digests were desalted with a C18 ZipTip (Millipore) prior to MS analysis.
LC-ESI-MS/MS.
Tryptic digests of HDL3 were separated using 2-dimensional LC with strong cation exchange and reverse-phase capillary LC columns. The LC system was interfaced with a Finnigan LCQ Deca ProteomeX ion trap mass spectrometer (Thermo Electron) equipped with an orthogonal electrospray interface. Tryptic digests of total HDL and apoA-I particles were analyzed by reverse-phase capillary LC coupled with electrospray ionization and a Thermo Electron linear ion trap mass spectrometer. Details are provided in the Supplemental Methods.
Peptide and protein identification.
MS/MS spectra were searched against the human International Protein Index (IPI) database v.3.01 (http://www.ebi.ac.uk/IPI/IPIhelp.html) (58), using the SEQUEST search engine (Thermo Electron) with fixed Cys alkylation and variable Met oxidation modifications (58). One incomplete cleavage site was allowed in peptides for trypsin-restricted searches. The SEQUEST results were further validated using PeptideProphet and ProteinProphet (http://tools.proteomecenter.org/TPP.php) (59, 60), using an adjusted probability of greater than 0.90 for peptides and greater than 0.95 for proteins. Each charge state of a peptide was considered a unique identification. MS/MS spectra from proteins identified with less than 6 peptides were confirmed by manual inspection.
Immunoblotting and apolipoprotein.
Reduced proteins separated by SDS-PAGE were transferred to a PVDF membrane and probed with a rabbit polyclonal anti-C3 antibody (Quidel Corp.), a sheep polyclonal anti-α-2-HS glycoprotein antibody (BioDesign), a rabbit polyclonal anti-vitronectin antibody (BIODESIGN International), or a sheep polyclonal antibody to the inter-α-trypsin inhibitor complex (Nordic Immunological Laboratories). A multiplex ELISA (Linco) was used to quantify levels of apoE, apoA-I, and apoA-II (61).
Statistics.
For each protein identified by MS/MS, the peptide index was calculated as: [(peptides in CAD subjects/total peptides) × (% of CAD subjects with ≥1 peptide)] – [(peptides in control subjects/total peptides) × (% of control subjects with ≥1 peptide)]. To validate the peptide index, we performed a permutation analysis. The cutoffs for statistical significance at 95% confidence interval (based on 1,000 random permutations) were peptide indices of –0.41 and +0.44 for relative enrichment in control subjects and CAD subjects, respectively. Therefore, we chose a peptide index of less than –0.4 or greater than 0.4, respectively, to identify selective enrichment of proteins in control subjects or CAD subjects.
For proteins that appeared enriched by the peptide index, Student’s unpaired 2-tailed t test was used to compare the number of unique peptides identified in CAD patients versus healthy subjects. For proteins found in only 1 group of subjects, a 1-sample t test was used to compare the number of unique peptides to a theoretical mean of 0. Fisher’s exact test was used to compare the number of CAD patients versus healthy subjects from which each protein was identified. The Mann-Whitney U test for nonparametric data was used to compare levels of HDL, apoA–I, apoA–II, and apoE in control and CAD subjects. For all statistical analyses, P < 0.05 was considered significant.
GO analysis.
Functional annotation of proteins detected in HDL3 was obtained from the GO database (http://www.geneontology.org; ref. 62). Functional categories within HDL3 proteins that were significantly enriched relative to 3,020 human plasma proteins (63) were determined using the GOTree Machine’s algorithm (http://genereg.ornl.gov/gotm; ref. 64). For a given GO category, the relative enrichment of genes encoding the proteins detected in HDL3 relative to all reference genes in that category were calculated (http://david.abcc.ncifcrf.gov/home.jsp). A cutoff value of P < 0.0001 was used to report a functional category as significantly overrepresented. Results were confirmed by repeating the analysis using the Expression Analysis Systematic Explorer algorithm (EASE; http://david.niaid.nih.gov/david/ease.htm). EASE uses a variant of the 1-tailed Fisher exact probability test to determine an EASE score, which ranks overrepresented GO processes (65). To address the multiple comparisons problem that arises when many processes are evaluated simultaneously, we performed a permutation analysis (n = 1,000 permutations) to calculate the global false discovery rate (FDR). To improve statistical confidence in our results, all enriched functional categories were required to be significant using both methods (P < 0.0001 and FDR < 0.001).
Supplementary Material
Supplemental data
Acknowledgments
This research was supported by grants from the NIH (HL086798, DK017047, DK02456, HL57557, HL074223, P30ES07033, P30DK017047, HL078527, and PO1HL030086) and the Donald W. Reynolds Foundation. T. Vaisar and S. Pennathur were supported by a Pilot and Feasibility Award from the Diabetes and Endocrinology Research Center and by the Juvenile Diabetes Research Foundation, respectively. MS experiments were performed by the Mass Spectrometry Resource, Department of Medicine, and the Mass Spectrometry Core, Diabetes and Endocrinology Research Center at the University of Washington.
Footnotes
Nonstandard abbreviations used: CAD, coronary artery disease; CETP, cholesteryl ester transfer protein; GO, Gene Ontology; LC-ESI-MS/MS, liquid chromatography–electrospray ionization–tandem MS; MS, mass spectrometry; PON1, paraoxonase-1; PRINCE study, Pravastatin Inflammation/CRP Evaluation study; SAA, serum amyloid A.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:746–756 (2007). doi:10.1172/JCI26206
Subramaniam Pennathur and Jaeman Byun’s present address is: Department of Medicine, University of Michigan, Ann Arbor, Michigan, USA.
Tomas Vaisar, Subramaniam Pennathur, and Pattie S. Green contributed equally to this work.
See the related Commentary beginning on page 595.
References
- 1.Braunwald E. Cardiovascular medicine at the turn of the millennium: triumphs, concerns, and opportunities. N. Engl. J. Med. 1997;337:1360–1369. doi: 10.1056/NEJM199711063371906. [DOI] [PubMed] [Google Scholar]
- 2.Hoyert D.L., Kung H.-C., Smith B.L. Deaths: preliminary data for 2003. Natl. Vital Stat. Rep. 2005;53:1–48. [PubMed] [Google Scholar]
- 3.Rader D.J. Molecular regulation of HDL metabolism and function: implications for novel therapies. J. Clin. Invest. 2006;116:3090–3100. doi: 10.1172/JCI30163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tall A.R., Costet P., Wang N. Regulation and mechanisms of macrophage cholesterol efflux. J. Clin. Invest. 2002;110:899–904. doi: 10.1172/JCI200216391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon D.J., Rifkind B.M. High-density lipoprotein — the clinical implications of recent studies. N. Engl. J. Med. 1989;321:1311–1316. doi: 10.1056/NEJM198911093211907. [DOI] [PubMed] [Google Scholar]
- 6.Wilson P.W., Abbott R.D., Castelli W.P. High density lipoprotein cholesterol and mortality. The Framingham Heart Study. Arteriosclerosis. 1988;8:737–741. doi: 10.1161/01.atv.8.6.737. [DOI] [PubMed] [Google Scholar]
- 7.Barzilai N., et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- 8.Barter P.J., et al. Antiinflammatory properties of HDL. Circ. Res. 2004;95:764–772. doi: 10.1161/01.RES.0000146094.59640.13. [DOI] [PubMed] [Google Scholar]
- 9.Shao B., Oda M.N., Oram J.F., Heinecke J.W. Myeloperoxidase: an inflammatory enzyme for generating dysfunctional high density lipoprotein. Curr. Opin. Cardiol. 2006;21:322–328. doi: 10.1097/01.hco.0000231402.87232.aa. [DOI] [PubMed] [Google Scholar]
- 10.Bergt C., et al. The myeloperoxidase product hypochlorous acid oxidizes HDL in the human artery wall and impairs ABCA1-dependent cholesterol transport. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13032–13037. doi: 10.1073/pnas.0405292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennathur S., et al. Human atherosclerotic intima and blood of patients with established coronary artery disease contain high density lipoprotein damaged by reactive nitrogen species. J. Biol. Chem. 2004;279:42977–42983. doi: 10.1074/jbc.M406762200. [DOI] [PubMed] [Google Scholar]
- 12.Zheng L., et al. Apolipoprotein A-I is a selective target for myeloperoxidase-catalyzed oxidation and functional impairment in subjects with cardiovascular disease. J. Clin. Invest. 2004;114:529–541. doi: 10.1172/JCI200421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chait A., Han C.Y., Oram J.F., Heinecke J.W. Thematic review series: The immune system and atherogenesis. Lipoprotein-associated inflammatory proteins: markers or mediators of cardiovascular disease? J. Lipid. Res. 2005;46:389–403. doi: 10.1194/jlr.R400017-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Olin K.L., Potter-Perigo S., Barrett P.H., Wight T.N., Chait A. Biglycan, a vascular proteoglycan, binds differently to HDL2 and HDL3: role of apoE. Arterioscler. Thromb. Vasc. Biol. 2001;21:129–135. doi: 10.1161/01.atv.21.1.129. [DOI] [PubMed] [Google Scholar]
- 15.Link A.J., et al. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 16.Resing K.A., Ahn N.G. Proteomics strategies for protein identification. FEBS Lett. 2005;579:885–889. doi: 10.1016/j.febslet.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Draganov D.I., Stetson P.L., Watson C.E., Billecke S.S., La Du B.N. Rabbit serum paraoxonase 3 (PON3) is a high density lipoprotein-associated lactonase and protects low density lipoprotein against oxidation. J. Biol. Chem. 2000;275:33435–33442. doi: 10.1074/jbc.M004543200. [DOI] [PubMed] [Google Scholar]
- 18.Duchateau P.N., et al. Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas. Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J. Biol. Chem. 1997;272:25576–25582. doi: 10.1074/jbc.272.41.25576. [DOI] [PubMed] [Google Scholar]
- 19.Getz G.S., Reardon C.A. Paraoxonase, a cardioprotective enzyme: continuing issues. Curr. Opin. Lipidol. 2004;15:261–267. doi: 10.1097/00041433-200406000-00005. [DOI] [PubMed] [Google Scholar]
- 20.Heller M., et al. Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins. Proteomics. 2005;5:2619–2630. doi: 10.1002/pmic.200401233. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson H., Leanderson P., Tagesson C., Lindahl M. Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005;5:1431–1445. doi: 10.1002/pmic.200401010. [DOI] [PubMed] [Google Scholar]
- 22.Kotite L., Zhang L.H., Yu Z., Burlingame A.L., Havel R.J. Human apoC-IV: isolation, characterization, and immunochemical quantification in plasma and plasma lipoproteins. J. Lipid Res. 2003;44:1387–1394. doi: 10.1194/jlr.M300087-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Kunitake S.T., et al. Identification of proteins associated with apolipoprotein A-I-containing lipoproteins purified by selected-affinity immunosorption. Biochemistry. 1994;33:1988–1993. doi: 10.1021/bi00174a003. [DOI] [PubMed] [Google Scholar]
- 24.McVicar J.P., Kunitake S.T., Hamilton R.L., Kane J.P. Characteristics of human lipoproteins isolated by selected-affinity immunosorption of apolipoprotein A-I. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1356–1360. doi: 10.1073/pnas.81.5.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogorzalek Loo R.R., Yam L., Loo J.A., Schumaker V.N. Virtual two-dimensional gel electrophoresis of high-density lipoproteins. Electrophoresis. 2004;25:2384–2391. doi: 10.1002/elps.200405955. [DOI] [PubMed] [Google Scholar]
- 26.Rezaee F., Casetta B., Levels J.H., Speijer D., Meijers J.C. Proteomic analysis of high-density lipoprotein. Proteomics. 2006;6:721–730. doi: 10.1002/pmic.200500191. [DOI] [PubMed] [Google Scholar]
- 27.Xu N., Dahlback B. A novel human apolipoprotein (apoM). J. Biol. Chem. 1999;274:31286–31290. doi: 10.1074/jbc.274.44.31286. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L.H., Kotite L., Havel R.J. Identification, characterization, cloning, and expression of apolipoprotein C-IV, a novel sialoglycoprotein of rabbit plasma lipoproteins. J. Biol. Chem. 1996;271:1776–1783. doi: 10.1074/jbc.271.3.1776. [DOI] [PubMed] [Google Scholar]
- 29.Graham T.E., et al. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N. Engl. J. Med. 2006;354:2552–2563. doi: 10.1056/NEJMoa054862. [DOI] [PubMed] [Google Scholar]
- 30.Kunitake S.T., Jarvis M.R., Hamilton R.L., Kane J.P. Binding of transition metals by apolipoprotein A-I-containing plasma lipoproteins: inhibition of oxidation of low density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6993–6997. doi: 10.1073/pnas.89.15.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung M.C., Wolf A.C. Differential effect of ultracentrifugation on apolipoprotein A-I-containing lipoprotein subpopulations. J. Lipid Res. 1988;29:15–25. [PubMed] [Google Scholar]
- 32.Park C.T., Wright S.D. Fibrinogen is a component of a novel lipoprotein particle: factor H-related protein (FHRP)-associated lipoprotein particle (FALP). Blood. 2000;95:198–204. [PubMed] [Google Scholar]
- 33.Gundry R.L., Fu Q., Jelinek C.A., VanEyk J.E., Cotter R.J. Investigation of an albumin-enriched fraction of human serum and its albuminome. J. Clin. Proteomics. 2007 doi: 10.1002/prca.200600276. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oksjoki R., Kovanen P.T., Pentikainen M.O. Role of complement activation in atherosclerosis. Curr. Opin. Lipidol. 2003;14:477–482. doi: 10.1097/00041433-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 35.Hortin G.L., Shen R.F., Martin B.M., Remaley A.T. Diverse range of small peptides associated with high-density lipoprotein. Biochem. Biophys. Res. Commun. 2006;340:909–915. doi: 10.1016/j.bbrc.2005.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuo L., Hascall V.C., Kimata K. Inter-alpha-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J. Biol. Chem. 2004;279:38079–38082. doi: 10.1074/jbc.R300039200. [DOI] [PubMed] [Google Scholar]
- 37.Washburn M.P., Ulaszek R.R., Yates J.R., 3rd. Reproducibility of quantitative proteomic analyses of complex biological mixtures by multidimensional protein identification technology. Anal. Chem. 2003;75:5054–5061. doi: 10.1021/ac034120b. [DOI] [PubMed] [Google Scholar]
- 38.Liu H., Sadygov R.G., Yates J.R., 3rd. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 39.Old W.M., et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Marcel Y.L., et al. Distribution and concentration of cholesteryl ester transfer protein in plasma of normolipemic subjects. J. Clin. Invest. 1990;85:10–17. doi: 10.1172/JCI114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McPherson P.A., Young I.S., McKibben B., McEneny J. High density lipoprotein subfractions: isolation, composition and their duplicitous role in oxidation. J. Lipid Res. 2007;48:86–95. doi: 10.1194/jlr.M600094-JLR200. [DOI] [PubMed] [Google Scholar]
- 42.O’Brien P.J., et al. The novel apolipoprotein A5 is present in human serum, is associated with VLDL, HDL, and chylomicrons, and circulates at very low concentrations compared with other apolipoproteins. Clin. Chem. 2005;51:351–359. doi: 10.1373/clinchem.2004.040824. [DOI] [PubMed] [Google Scholar]
- 43.Albert M.A., Danielson E., Rifai N., Ridker P.M. Effect of statin therapy on C-reactive protein levels: the pravastatin inflammation/CRP evaluation (PRINCE): a randomized trial and cohort study. JAMA. 2001;286:64–70. doi: 10.1001/jama.286.1.64. [DOI] [PubMed] [Google Scholar]
- 44.Shiflett A.M., Bishop J.R., Pahwa A., Hajduk S.L. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J. Biol. Chem. 2005;280:32578–32585. doi: 10.1074/jbc.M503510200. [DOI] [PubMed] [Google Scholar]
- 45.Davies M.J. A macro and micro view of coronary vascular insult in ischemic heart disease. Circulation. 1990;82(Suppl.):II38–II46. [PubMed] [Google Scholar]
- 46.Richardson J., Viswanathan K., Lucas A. Serpins, the vasculature, and viral therapeutics. Front. Biosci. 2006;11:1042–1056. doi: 10.2741/1862. [DOI] [PubMed] [Google Scholar]
- 47.Niculescu F., Rus H. The role of complement activation in atherosclerosis. Immunol. Res. 2004;30:73–80. doi: 10.1385/IR:30:1:073. [DOI] [PubMed] [Google Scholar]
- 48.Pepys M.B., et al. Targeting C-reactive protein for the treatment of cardiovascular disease. Nature. 2006;440:1217–1221. doi: 10.1038/nature04672. [DOI] [PubMed] [Google Scholar]
- 49.Hamilton K.K., Zhao J., Sims P.J. Interaction between apolipoproteins A-I and A-II and the membrane attack complex of complement. Affinity of the apoproteins for polymeric C9. J. Biol. Chem. 1993;268:3632–3638. [PubMed] [Google Scholar]
- 50.Strunk R.C., Kunke K.S., Giclas P.C. Human peripheral blood monocyte-derived macrophages produce haemolytically active C3 in vitro. Immunology. 1983;49:169–174. [PMC free article] [PubMed] [Google Scholar]
- 51.Mak P.A., et al. Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors alpha and beta. J. Biol. Chem. 2002;277:31900–31908. doi: 10.1074/jbc.M202993200. [DOI] [PubMed] [Google Scholar]
- 52.Vergnes L., et al. Expression of human apolipoprotein A-I/C-III/A-IV gene cluster in mice induces hyperlipidemia but reduces atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2000;20:2267–2274. doi: 10.1161/01.atv.20.10.2267. [DOI] [PubMed] [Google Scholar]
- 53.Matsuura F., Wang N., Chen W., Jiang X.C., Tall A.R. HDL from CETP-deficient subjects shows enhanced ability to promote cholesterol efflux from macrophages in an apoE- and ABCG1-dependent pathway. J. Clin. Invest. 2006;116:1435–1442. doi: 10.1172/JCI27602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahley R.W., Huang Y., Weisgraber K.H. Putting cholesterol in its place: apoE and reverse cholesterol transport. J. Clin. Invest. 2006;116:1226–1229. doi: 10.1172/JCI28632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson H.M., Patel J.C., Russell D., Skinner E.R. Alterations in the concentration of an apolipoprotein E-containing subfraction of plasma high density lipoprotein in coronary heart disease. Clin. Chim. Acta. 1993;220:175–187. doi: 10.1016/0009-8981(93)90046-7. [DOI] [PubMed] [Google Scholar]
- 56.Mendez A.J., Oram J.F., Bierman E.L. Protein kinase C as a mediator of high density lipoprotein receptor-dependent efflux of intracellular cholesterol. J. Biol. Chem. 1991;266:10104–10111. [PubMed] [Google Scholar]
- 57.Cheung M.C., et al. Characterization of high density lipoprotein subspecies: structural studies by single vertical spin ultracentrifugation and immunoaffinity chromatography. J. Lipid Res. 1987;28:913–929. [PubMed] [Google Scholar]
- 58.Kersey P.J., et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 59.Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 60.Yan W., et al. A dataset of human liver proteins identified by protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry. Mol. Cell. Proteomics. 2004;3:1039–1041. doi: 10.1074/mcp.D400001-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Vuletic S., et al. Reduced CSF PLTP activity in Alzheimer’s disease and other neurologic diseases; PLTP induces ApoE secretion in primary human astrocytes in vitro. J. Neurosci. Res. 2005;80:406–413. doi: 10.1002/jnr.20458. [DOI] [PubMed] [Google Scholar]
- 62.[Anonymous]. . Creating the gene ontology resource: design and implementation. Gene Ontology Consortium. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Omenn G.S., et al. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 64.Zhang B., Schmoyer D., Kirov S., Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hosack D.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A.2003Identifying biological themes within lists of genes with EASE. Genome Biol. 4R70. 10.1186/gb-2003-4-10-r70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Navab M., et al. The oxidation hypothesis of atherogenesis: the role of oxidized phospholipids and HDL. J. Lipid Res. 2004;45:993–1007.. doi: 10.1194/jlr.R400001-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data