Extension and Integration of the Gene Ontology (GO): Combining GO Vocabularies With External Vocabularies (original) (raw)
Abstract
Structured vocabulary development enhances the management of information in biological databases. As information grows, handling the complexity of vocabularies becomes difficult. Defined methods are needed to manipulate, expand and integrate complex vocabularies. The Gene Ontology (GO) project provides the scientific community with a set of structured vocabularies to describe domains of molecular biology. The vocabularies are used for annotation of gene products and for computational annotation of sequence data sets. The vocabularies focus on three concepts universal to living systems, biological process, molecular function and cellular component. As the vocabularies expand to incorporate terms needed by diverse annotation communities, species-specific terms become problematic. In particular, the use of species-specific anatomical concepts remains unresolved. We present a method for expansion of GO into areas outside of the three original universal concept domains. We combine concepts from two orthogonal vocabularies to generate a larger, more specific vocabulary. The example of mammalian heart development is presented because it addresses two issues that challenge GO; inclusion of organism-specific anatomical terms, and proliferation of terms and relationships. The combination of concepts from orthogonal vocabularies provides a robust representation of relevant terms and an opportunity for evaluation of hypothetical concepts.
An important goal for biological databases is the integration of different types of data. Integration connects common data elements and thus allows the combined analysis and correlation of different data sets. An important means to generate common data elements is the development and use of ontologies that represent knowledge domains. One way to represent an ontology in a biological database is to design a structured vocabulary. An ontology, as a structured vocabulary, is a hierarchy of terms in which the terms are precisely defined and the terms relate to each other in meaningful ways.
The goal of the Gene Ontology (GO) project is to design structured vocabularies to describe three universal concepts of biology: molecular function, biological process, and cellular component (The Gene Ontology Consortium 2000, 2001). As structured vocabularies, the terms have precise definitions and precise relationships to other terms. GO represents vocabulary terms in a hierarchically structured format. Vocabulary terms are linked to each other by “is a” and “part of” relationships, so that very general terms and very precise terms are both represented. The structure and format of GO are described in detail elsewhere (The Gene Ontology Consortium 2001). Each term can have one or more relationships with other terms, reflecting the complexity of the underlying biology. The structure is formally a directed acyclic graph (DAG) wherein the terms are equivalent to nodes of the graph and the relationships are equivalent to edges (Aho et al. 1983). Because GO encompasses the biology of organisms that span the phylogenetic spectrum from bacteria to vertebrates, its extensive nature has resulted in a large growth of both nodes and edges. At present the GO vocabularies contain 11,258 terms and 14,349 edges. The ontologies and related documentation for the Gene Ontology project are available at http://www.geneontology.org.
The scope and nature of the GO project provide interesting challenges for ontology construction. One problem concerns the need for species-specific terms to describe functions, processes, and components in individual groups of organisms. Conflicts in the universality of GO are apparent when curators from different organism-specific databases add species-specific terms to support the annotation of their gene products. An early example of this conflict arose in the GO project with the process of chitin metabolism, a process required for describing cell wall biosynthesis in fungi, but cuticle synthesis in insects. Making chitin metabolism a subprocess of both cell wall biosynthesis and cuticle synthesis caused errors in searches. A search for genes involved in cell wall biosynthesis resulted in genes used in insect cuticle biosynthesis that were annotated to chitin metabolism. This simple conflict was solved by creating two types of “chitin metabolism”: one involved in cell wall biosynthesis and one involved in cuticle synthesis. However, as more and more species-specific terms are added to the vocabularies, it becomes difficult to maintain both the global interspecies relationships for which GO strives and the precise terms required for intraspecies gene annotation.
Furthermore, the species-specific concepts that require integration into GO are also needed by individual organism databases to describe concepts that extend beyond the three GO concepts. For example, an anatomical term such as mouse forelimb would be useful in the developmental process section of the biological process ontology for describing pattern formation in the mouse forelimb. But, it is also useful for describing details about gene expression patterns in the mouse forelimb or phenotypes such as fused digits of the forelimb as described in the Mouse Genome Informatics databases (http://www.informatics.jax.org). Ideally, the anatomical concepts in GO would be consistent with the anatomical concepts used within a model organism database, thus allowing for integration and robust search capabilities across data sets that are pertinent to the anatomy. For example, a database user may want to search for genes that are either expressed in, function in, or show phenotypes in the heart. In addition to species-specific challenges, GO curators need to add to and refine more generic concepts than are presently represented by an already complex GO.
Traditionally, GO has been developed by curation-driven methods and solutions. Curators added one or a few terms at a time to the GO vocabularies and defined appropriate relationships as needed when annotating their gene products. When conflicts arose, discussion ensued, and an appropriate solution was achieved. Usually, problems concerning the logic of the directed acyclic graphs (DAGs) were addressed on a case-by-case basis and resolved by discussion. Although there is little difficulty with increased complexity of GO from a computational standpoint, the complexity becomes difficult for scientists developing the vocabularies because their expertise is limited to specific areas of the vocabulary. Ultimately, using the case-by-case method to add terms to the vocabularies and accurately and completely determine the relationships among the terms will become untenable. If GO is to remain consistent, complete, flexible, and logical, we need to develop strategies that allow for expansion of the number of terms and relationships within the ontologies that are not only biologically and logically correct, but are still manageable by human developers.
In this study, we explore a method of expanding GO by combining concepts from two vocabularies to generate a more complete vocabulary that includes specific aspects from each of the two parental vocabularies. We refer to this as the combinatorial method. We compare this method with the more conventional method of GO vocabulary construction that creates a single vocabulary de novo. The combinatorial method stems from the computational concept of creating cross products between orthogonal DAGs. A DAG cross product is generated by combining the nodes of two graphs in every possible combination. The result is a new graph in which each node has the characteristics of each of the parental graphs (Fig. 1). In a biological context, orthogonal vocabularies can be thought of as vocabularies whose terms are unrelated. In this example we use a vocabulary of generic developmental process terms and a vocabulary of anatomical terms. Similar approaches using combinations of terms have been used in other biological databases to create bins for categorizing data such as the complex annotation of patient records, but the concept has been limited to the uses of multiple root structures used for curation rather than to expand the ontologies themselves (Berman and Moore 1996). In fact, one of the major challenges facing the medical informatics community is the integration of independently controlled vocabularies (Musen 1998). We focus on creating a vocabulary to describe the development of the mammalian heart because it addresses two important issues that challenge GO: (1) the inclusion of organism-specific anatomical information and (2) the proliferation of terms and relationships.
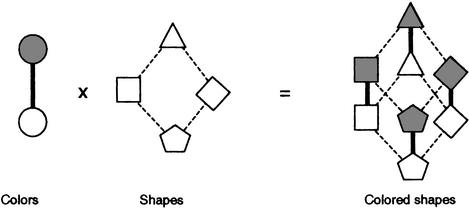
Figure 1.
DAG cross-product example. In this example, a DAG whose nodes represent colors is crossed with a DAG whose nodes represent shapes. The result is a DAG whose nodes are colored shapes. Every combination is represented, so there are eight nodes in the result. An edge connects two nodes in the cross product whenever they have the same color and their shapes are connected in the Shape DAG (8 of these), or they have the same shape and their colors are connected in the Color DAG (4 of these). In general, the number of nodes in the cross product of DAGs A and B is the number of nodes in A times the number of nodes in B. The number of edges in the cross product is the number of edges in A times the number of nodes in B, plus the number of edges in B times the number of nodes in A.
We show that the advantage of using combinatorial strategies for a project like GO becomes obvious once the vocabularies grow in size and complexity. Using defined rules to create the combinations, curators can construct new vocabularies by focusing on one subject and describing its parts and types. As long as the original vocabularies are constructed so they are compatible for generating a cross product, further extensions of the vocabularies can be generated using concepts from the novel and existing vocabularies. The importance of communication among vocabulary developers was realized early on for the development of GO. The combinatorial strategy presented here is already fostering additional interactions. The GO Web site at present hosts a repository for ontologies that will be useful in cross-product generation (http://www.geneontology.org/doc/gobo.html). The combinatorial principle emphasizes the importance of coordinating the construction of elemental vocabularies, and it illustrates the benefits of combining concepts from those vocabularies to create consistent extended vocabularies.
METHODS
The Narrative Approach
To judge the utility of the combinatorial method, we first constructed an ontology for heart development using the conventional method of GO development. Using three textbooks and 15 references, we were able to construct a version of GO to describe mammalian heart development (Rugh 1990; Sucov et al. 1994; Kern et al. 1995; Lyons 1996; Biben and Harvey 1997; Kirby 1997; Gimond et al. 1998; Huang et al. 1998; King et al. 1998; Abdelwahid et al. 1999; Eferl et al. 1999; Kaufman and Bard 1999; Pereira et al. 1999; Swiderski et al. 1999; Tanaka et al. 1999; Woldeyesus et al. 1999; Jaspard et al. 2000; Tevosian et al. 2000). The vocabulary shown in Figure 2 describes heart development much like the overview that would be given at a seminar or in a review article, and it will be referred to as the “narrative” approach to vocabulary development. An important aspect of the narrative approach is that it creates a single vocabulary that describes both simple and, in this case, complex concepts. The vocabulary is itself a stand-alone entity and does not have any formal relationships with any other vocabulary. The structure of the vocabulary and all other vocabularies in this manuscript is represented in the common GO format. Each term of the vocabulary is placed on a separate line. The number of ‖ characters preceding the terms indicates a relationship to the term above it. The “is a” and “part of” relationships are denoted by % and < symbols, respectively. The vocabulary in Figure 2 shows that heart development begins with an endoderm/mesoderm inductive event, resulting in the thickening of the cardiac regions and the formation of the primitive heart tube. The cells of the heart tube undergo differentiation into the primitive layers of the heart, and the tube undergoes folding and remodeling to form the primitive heart. The vocabulary describes the development of the various structures of the heart and the cellular processes that occur during their development. The vocabulary contains 58 specific terms and 80 relationships. However, close inspection reveals that the vocabulary is incomplete. For example, “breakdown of the dorsal mesocardium” may be described as a type of “apoptosis” and should contain all of the cellular processes that are characteristic of apoptosis. Once completed, it would suffice to add the mammalian heart development tree shown in Figure 2 to the existing biological process graph as a type of developmental process, but the simple addition would probably be deficient in its relationships and integration with other areas in the existing GO. Additionally, according to GO rules, each of the new unique terms in Figure 2 requires a novel precise definition.
Figure 2.
A vocabulary describing heart development constructed using literature references. The format of the vocabulary and other vocabularies in this manuscript is as follows: Indentation reflects parent–child relationships; the < symbol indicates that the child is a part of its parent; and the % symbol indicates that the child is a type of its parent. Multiple parentage is indicated by two terms on the same line, where the first term is a child of the second term. The colored portion of the graph corresponds to the similarly colored diagram in the schematic shown in Figure 6.
The Combinatorial Approach
In the experimental method for building an ontology to describe mouse heart development, we constructed an ontology by combining terms and relationships from two existing ontologies representing two orthogonal concepts involved in heart development: anatomy and developmental processes. An important distinction between this and the narrative approach is the use of more than one elemental vocabulary to describe complex terms. As we discuss below, the result of this is the creation of a relationship between the combinatorial vocabulary and each of the vocabularies that was used in its construction. The rationale behind this approach is that a mouse heart anatomical vocabulary should completely describe the anatomy of the heart, and a developmental process vocabulary should completely describe all of the general biological processes involved in development. Therefore, we should be able to combine the concepts from the two vocabularies to describe all of the processes involved in the development of all of the anatomical parts of the heart. For the anatomical vocabulary, we chose the mouse anatomical dictionary used by the Mouse Gene Expression Database (Bard et al. 1998; Ringwald et al. 2001). The mouse anatomical dictionary lists the anatomical structures for every stage of development. The structures are represented hierarchically, with “part of” relationships defined between structures and substructures. To simplify the approach for illustrative purposes, we condensed the anatomical dictionary to eliminate the time component, and we defined “is a” and “part of” relationships between the structures of the developing heart to be consistent with the relationships in the GO vocabularies (Fig. 3). We also modified the organogenesis portion of the GO biological process ontology by eliminating all terms from the vocabulary that refer to specific anatomical structures (Fig. 4). This step ensured that there would not be conflicting conceptual information in the two vocabularies, that is, it ensures orthogonality between the two vocabularies.
Figure 3.
A consolidated version of the mouse anatomical dictionary. The time component has been removed, and primitive structures have been defined as “types of” the more mature structure. This vocabulary can be combined with developmental processes to describe the processes underlying the development of the structures.
Figure 4.
A modified developmental process ontology. The ontology describes processes, but does not refer to anatomical structures that would be included in an anatomical dictionary. This vocabulary can be combined with anatomical concepts to describe developmental processes occurring in specific structures.
Once the individual vocabularies were generated, we generated two top-level cross products of the anatomical vocabulary and the developmental process vocabulary by combining the most general term of each respective vocabulary with all of the terms from the other vocabulary. First, if the anatomical dictionary completely describes all of the parts of the heart, then the product of the heart anatomy and the general concept of development should describe all aspects of heart development from an anatomical perspective. In the generation of this cross product, we retained the relationships between the terms in the anatomical dictionary for the relationships between the combined terms. For example, if the “myocardium” is a part of the “primitive heart tube,” then “development of the myocardium” is a part of the “development of the primitive heart tube.” The cross product is generated with all of the terms because all anatomical structures in the heart develop. The cross product contains the same number of terms that are in the anatomical dictionary, and the terms and their relationships accurately describe heart development from an anatomical perspective (Fig. 5A). For example, “development of the cardiac muscle of the right common atrial chamber” is a part of “common atrial chamber development,” which is a type of “atrial chamber development,” which is a part of “heart development.” Second, if the “organogenesis” portion of the developmental process ontology describes all aspects of organogenesis, then the combination of the top-level anatomical term “heart” with all of the terms from the “organogenesis” vocabulary should describe all of the processes that can possibly be involved in heart development (Fig. 5B). Of course, when we use the term heart in this context, we refer to the mouse heart, because the term originates from the mouse anatomical dictionary. Furthermore, all of these processes are more specific types of the general developmental processes in the original vocabulary. For example “formation of an epithelial tube during heart development” is a type of “formation of an epithelial tube.” Note that although the two top-level cross products are illustrated as two separate graphs, they are in fact part of the same DAG. For example, the term “heart development” in Figure 5, A and B, can be thought of as the common root for two paths through the graph.
Figure 5.
(A) The global development concept has been combined with the anatomical concepts from the anatomical dictionary. This figure only illustrates the first 21 lines of the complete vocabulary. The complete vocabulary would include all of the anatomical terms in Figure 3. In the combinatorial terms presented here, the concept of development taken from the process ontology is shown in boldface. This ontology provides an “anatomical” view of heart development. (B) The developmental process ontology has been combined with the anatomical concept of the heart. In this case a simple rule of adding the phrase “during heart development” was added to the developmental process ontology. This new ontology gives a low-resolution “embryological” picture of heart development. This figure only illustrates the first 29 lines of the vocabulary. The complete vocabulary would include all of the terms shown in Figure 4.
RESULTS
Examination of the vocabularies in Figure 5 shows that generating the two top-level combinations gives a complete, albeit low-resolution, picture of heart development. There are at least two options to generate a higher resolution picture of the process. We could generate the entire cross product between the two original graphs, that is, combine every term from each vocabulary with every term from the other vocabulary. This would result in a large number of terms and would result in all possible combinations of anatomical terms with developmental process terms. An advantage to this approach is that it generates all possibilities of processes that can occur in combination with an anatomical structure, including some that may not have been discovered experimentally or some that may not exist. For example, a complete cross product would generate the term “pattern formation/primitive heart tube.” This term describes the pattern-formation taking place in the development of the primitive heart tube. From the cross product we can generate a hypothesis that there might be an axis set up during heart tube development or that there may be segmentation occurring during heart tube development because “axial development” and “segmentation” are both types of “pattern formation.” Thus, another advantage to generating cross products is that they may broaden our thinking about how processes occur.
Alternatively, we could use our biological knowledge to pick and choose subelements of each vocabulary to generate subgraphs. This limits the new ontology to processes that are known, but provides an accurate description of a process as it is presently understood. It is important to note that although the “pick and choose” approach might be used for the initial vocabulary, all of the combinations of terms from both vocabularies are still possible, and therefore the combinatorial vocabulary becomes easily extendible as our knowledge about a process improves. Although the cross-product terms are restricted by our present knowledge, the creation of the terms must not result in incorrect information. If an expert biological curator deems a cross-product ontology incorrect, this indicates that the original ontologies are incorrect.
We will illustrate the “pick and choose” approach using primitive “heart tube formation.” From a narrative standpoint, the primitive heart tube forms in three steps: (1) A mesenchymal-to-epithelial transition forms the cardiogenic plate (Fig. 6B). (2) This epithelial sheet then forms an epithelial tube (Fig. 6C). (3) The epithelial tube differentiates into three layers: endocardium, myocardium, and cardiac jelly (Fig. 6D). Using this very general narrative approach, we are able to identify the areas of the top-level vocabularies that need to be expanded and combined to describe these initial stages of development (Fig. 7). From the anatomical perspective, we need expansion of the “cardiogenic plate development” and “primitive heart tube development” sections (Fig. 7A). From the process perspective, expansion of both the “morphogenesis of an epithelium” section and the “cell differentiation” section is required (Fig. 7B). When we create the combined terms, they should relate to the existing terms using the rules we described for the initial top-level vocabularies. First, when an anatomical term is combined with a process term and is inserted into the process-based vocabulary, the new process becomes a type of the generic process. Second, when an anatomical term is combined with a process term and is inserted into the anatomy-based vocabulary, the new term retains its relationship with the parental structure. Four areas of the top-level vocabularies were expanded. Figure 8 shows the final product after the processes of epithelial tube formation and cell differentiation are included into the appropriate sections of the vocabulary. Figure 8, A and B, is essentially the same vocabulary presented from the process perspective and the anatomical perspective. A comparison of the vocabulary in Figure 8 with the vocabularies shown in Figures 3 and 4 illustrates that the terms and the relationships in the final graph were actually defined in the original parental vocabularies. For example, we already knew that the differentiation of any cell type would include commitment, specification, and determination, even if we were not expert developmental biologists. Therefore, if the “formation of endocardium” requires “cell differentiation,” then we can learn from the graph in Figure 4 that part of the process of this cell differentiation will be cell commitment as is illustrated in the term “cell commitment during endocardium development” (Fig. 8B).
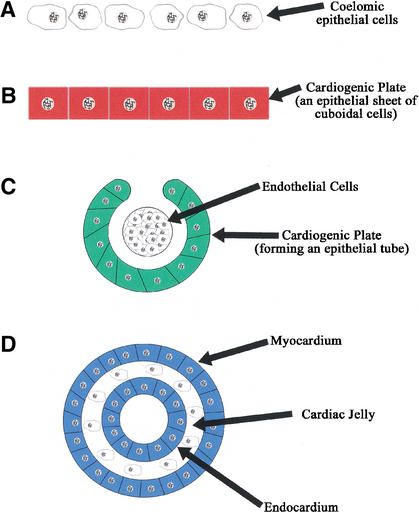
Figure 6.
A schematic representation of the processes that occur during the formation of the primitive heart tube. The schematic is modeled after the description given by Kaufman and Bard (1999). (A) The coelomic epithelial cells that are destined to form the heart tube. (B) The coelomic epithelial cells have formed the cuboidal cells of the cardiogenic plate. (C) The plate is undergoing morphogenesis to form the primitive heart tube. (D) The heart tube is complete, and cells have differentiated to give rise to the endocardium, cells forming the cardiac jelly, and the myocardium. For illustrative purposes, the colors of this figure correspond to the colors in each of the text-based graphs of Figures 2 and 8.
Figure 7.
The sections of the initial combinatorial ontologies that require expanding to describe formation of the primitive heart tube. (A) This shows that we are dealing with the anatomical concepts of heart, cardiogenic plate, and primitive heart tube. (B) This shows that we need to describe the events that occur during the development of an epithelial sheet. (C) This illustrates that we need to include the processes involved in cell differentiation. The colored portion of the graph corresponds to the similarly colored diagram in the schematic shown in Figure 6.
Figure 8.
The complete graph generated by successive combination of terms. In the first stage, terms describing the formation of an epithelial tube during the development of the primitive heart tube were inserted into the graph shown in Figure 7. In the second stage, terms that describe the differentiation of cells in the appropriate tissues were inserted. (A) The graph shown from an anatomical perspective. (B) The graph shown from a developmental process perspective. The portion of each term that was derived from the “anatomical” view of development generated in Figure 5A is shown in boldface.
DISCUSSION
The combinatorial approach has several advantages over the narrative approach for building complex vocabularies, as comparison of the ontology generated in Figure 8 with the ontology generated in Figure 2 demonstrates. From a graph perspective, the narrative approach describes cardiogenic plate and primitive heart tube formation using 9 nodes and 13 edges. In the combinatorial approach, 28 nodes and 71 edges are generated. When inspecting the graphs from a biological perspective, it is clear that the processes are described more accurately and completely in the combinatorial approach. For example, many of the details of the formation and morphogenesis of the primitive heart tube, such as movements and division of cells, are omitted in the narrative approach. It is quite conceivable that gene products that are involved in the oriented division of cells in the heart tube would need to be annotated at that level of specificity. To annotate to that level of specificity in the narrative vocabulary in Figure 2, we would need to manually expand the “formation of heart tube” node and create relationships and definitions for all of the new terms describing movements and division of cells. It is important to note that using this trial-and-error approach, or through a completely extensive review of the literature, the narrative vocabulary can eventually attain the level of complexity/accuracy of a vocabulary generated by the combinatorial approach. However, the added levels of complexity will undoubtedly make the vocabulary very difficult to expand and modify from the perspective of a human curator.
One might argue that the vocabulary in Figure 8 is still insufficient. In particular, the graph does not give us a sense of “when” processes are taking place. Part of this problem is an artifact of our simplification of the anatomical dictionary to remove temporal information. If the anatomical dictionary were to contain temporal information or relationships such as one structure being derived from another, then this information could be directly incorporated into the combinatorial graph. A potential problem with the combinatorial approach is that there is a critical dependency on the parental vocabularies. For example, the concept of tissue remodeling is missing from the developmental process vocabulary but is certainly taking place during heart organogenesis. This points out that each combinatorial vocabulary is entirely dependent on its parents, and problems with the parental vocabulary will be propagated through every combinatorial vocabulary that uses it. Thus, the potential problem actually leads to an advantage because the combinatorial approach can reveal deficiencies in the parental vocabularies and lead to their improvement. Of course, once the problem is solved in the parental vocabulary, it would also be solved in all of the combinatorial vocabularies as well, alleviating the need for the same problem being solved over and over again. Therefore, the quality and completeness of the original vocabularies are of critical importance. In this example, “tissue remodeling” and all of its children would need to be added to the original “developmental process” vocabulary at the appropriate places. It is also of critical importance that the parental vocabularies contain independent concepts, that is, they are orthogonal. If a process such as “alveolar branching morphogenesis” were inadvertently placed in the developmental process vocabulary, as opposed to “branching morphogenesis” being placed in the process vocabulary and “alveolae” in the anatomy vocabulary, this would create problems when combining the process with the heart vocabulary because the alveolae are components of the lung.
While building the ontologies, there are also several advantages to the combinatorial approach. A curator can focus on one domain of biology at a time to build a primary graph that will be combined with other graphs generated by curators with different areas of expertise. In this example, the graphs were “heart anatomy” and “developmental process.” The heart anatomy vocabulary was built by expert anatomists, and the developmental process vocabulary was built by developmental biologists. The two graphs could have been “dysmorphogenesis” and “anatomy” and could have been built by an expert pathologist and an anatomist, respectively. The combinatorial approach allows for a better allocation of resources in which experts share their results through the combination of their graphs. When modifications to the graphs are required, they will be made to the parental graphs so that they can be shared by all of the graphs that result from combination. The approach will not only maintain consistency throughout the ontology but will improve on accuracy because experts will be working on the parental graphs.
Another key advantage to the combinatorial approach is that required definitions for the combined terms can be automatically generated. In the GO project, the biological concepts that are represented by nodes of the graph are actually represented in the definition of the terms (Ashburner and Lewis 2001). If the terms are well defined, then the combinatorial terms can be defined by a manipulation of the definitions of the two parental terms. An example of this is the insertion of the words “during” and “development” into the combinatorial terms so that they make semantic sense. The same approach can be used with definitions.
One potential problem with the combinatorial approach in a project like GO is that as vocabularies are generated for various organisms, the new combinatorial terms may not be appropriate for use in future combinations because the two graphs may no longer be orthogonal. To choose a trivial example, it does not make sense to combine a developmental process term already containing a mouse anatomical component with a fly anatomical component. Thus, we need to satisfy a rule that never allows a combinatorial term containing an anatomical component to be combined with another anatomical component term. There are at least three solutions to this problem: First, we could generate combinatorial terms one at a time and insert them by hand into the parental vocabularies. Although this approach would be very accurate, it would also be time-consuming and would not take advantage of the inherent relationships that exist in any automated way.
Second, we could flag nodes in GO that are generated by combinatorial terms based on the vocabularies used in their creation. Then when GO is used to generate new combinatorial graphs, curators will have a choice of eliminating the existing combinatorial terms if either of the original terms contained a concept from a graph that is nonorthogonal to the new graph, such as anatomical structures from two different organisms. GO already uses a similar mechanism by flagging organism-specific terms with the string “sensu organism.” These terms and their children are then “set aside” for use and further development by organism-specific curators. This approach is appropriate, but it has a disadvantage inherent in the narrative approach; entire subgraphs are inserted into the parental graph with a single “sensu” term at the root. Thus, there is a potential for missing relationships between the subgraphs and the parental graph and between different subgraphs.
Third, and probably most appropriate, we could keep the combinatorial graphs separate from the parent GO vocabularies, but record the relationships of the combinatorial terms with the GO parents. In this approach, the parental vocabularies remain as “pure” ontologies, whereas the combinatorial vocabularies would be “working” ontologies that would relate back to their parents. Terms from separate combinatorial graphs would relate to each other through their common “pure” GO parent terms. Thus both GO and, in this case, the anatomical dictionary could change independently, and the combinatorial “working” graph would simply be recalculated to reflect the changes. An advantage to this is that as the “pure” graphs are corrected and refined, the “working” graphs would automatically be updated. A direct requirement of this type of updating scheme would be to design logical rules used for the generation of combinatorial graphs. The rules would be used in a semiautomated way for both the construction of the graphs and the update of the graphs as the parents change. One example of this type of rule would be:
Graph1 (Process)
‖Node1 (apoptosis)
∥Node2 (induction of apoptosis) is a part of Node1 (apoptosis)
∥Node3 (killing of inflammatory cells) is a type of Node1 (apoptosis)
∥‖Node4 (terminal differentiation of inflammatory cells) is a part of Node3 (killing of inflammatory cells)
∥‖Node5 (induction of apoptosis of inflammatory cells) is a part of Node3 (killing of inflammatory cells)
Graph2 (Anatomy)
NodeA (limb)
‖NodeB (handplate) is a part of NodeA
∥NodeC (interdigital mesenchyme) is a part of NodeB
Graph3 (New combinatorial concept)
Node6 (apoptosis of interdigital mesenchyme)
‖Node7 (signaling from condensing mesenchyme cells) is a part of Node6 (apoptosis of interdigital mesenchyme)
If a curator sees a need for Node6, a combinatorial term that is a type of Node1 involving NodeC, then:
If Node2 is a part of Node1, then a type of Node2 can be described that is also a part of Node6. Node7 is always a part of Node6, but is not always a part of Node1 (it is a specific process that occurs only as a part of Node6). So, Graph1 × 3 can also be represented as (where, e.g., 2/6 means a Node2 involving the anatomical structure in Node6):
Node1 (apoptosis)
‖Node2 (induction of apoptosis) is a part of Node1 (apoptosis)
∥Node2/6 (induction of apoptosis of interdigital mesenchyme) is a type of Node2 (induction of apoptosis)
‖Node3 (killing of inflammatory cells) is a type of Node1 (apoptosis)
∥Node4 (terminal differentiation of inflammatory cells) is a part of Node3 (killing of inflammatory cells)
∥Node5 (induction of apoptosis of inflammatory cells) is a part of Node3 (killing of inflammatory cells)
‖Node6 (apoptosis of interdigital mesenchyme) is a type of Node1 (apoptosis)
∥Node2/6 (induction of apoptosis of interdigital mesenchyme) is a part of Node6 (apoptosis of interdigital mesenchyme)
∥Node7 (signaling from condensing mesenchyme cells) is a part of Node6 (apoptosis of interdigital mesenchyme)
This example gives an illustration of how a combinatorial graph can be generated and maintained. Note that in this case, we have inserted the combinatorial terms into the parental graph to show the relationships between the combinatorial and parental terms. In the case of graphs representing anatomy and developmental process, the development of a specific anatomical structure will be composed of a type of general developmental process that is specified by the structure. Node6 represents the term describing both anatomy and development of which the development term is Node1. Node2/6 and its relationships would be calculated. Node7 would be a specific process that only applies to the development of the anatomical structure represented in Node6 and would be added by a curator.
The advantage of using combinatorial strategies for a project like GO becomes obvious once the vocabularies grow in size and complexity. Using rules like the one illustrated above, curators can construct vocabularies by focusing on one subject and describing its parts and types. Other curators who are using the concepts of a well-established vocabulary are not required to reconstruct that vocabulary to fit their specific needs, but instead can use the well-established vocabulary. For example, a curator describing the “morphogenesis of an epithelium” in the heart and a curator describing the “morphogenesis of an epithelium” in the kidney would both use the same parental vocabulary for the “morphogenesis of an epithelium” and would add their specific appropriate anatomical terms. The two parts of the vocabulary are then guaranteed to be internally consistent. Furthermore, the definitions of the new terms can be directly derived from the definitions of the parental terms and will be inherently consistent as well. Using the cross-product approach, ambitious projects like GO can remain manageable for human construction and curation.
This study examined two approaches for the construction of structured vocabularies to describe developmental processes that are involved in the development of species-specific anatomical structures. The analysis compares the present narrative approach to vocabulary development with a new approach taking advantage of combining terms from two existing vocabularies. Our work shows that the combinatorial approach provides a method of vocabulary construction and maintenance that allows for the evolving complexities of extensive vocabularies like those used in the Gene Ontology. It is clear from our practical experience with heart development, that the most biologically relevant method for creating graphs will combine aspects of both manual inspection and computational creation of terms. Expert curators will still be required to generate the original vocabularies, then curators will use the “pick and choose” approach and computational tools to generate new combinatorial terms for a complex vocabulary. The combination of both expert biological knowledge and tools for creating cross-product vocabularies should allow for a robust expansion of vocabularies such as GO to include other knowledge domains.
WEB SITE REFERENCES
http://www.geneontology.org; The Gene Ontology Consortium.
http://www.geneontology.org/doc/gobo.html; ontologies that will be useful in cross-product generation.
http://www.informatics.jax.org; Mouse Genome Informatics.
Acknowledgments
This work was funded by NIH Grant HG02273 to the Gene Ontology Consortium and Grant HD33745 to the Gene Expression Database (GXD). The authors would like to thank Michael Ashburner, Harold Drabkin, Monica McAndrews-Hill, and Constance Smith for their critical reading of the manuscript and W. John Boddy for his help with the figures. This paper does not describe software or specific programs.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dph@informatics.jax.org; FAX: (207) 288-6132.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.580102.
REFERENCES
- Abdelwahid E, Pelliniemi LJ, Niinikoski H, Simell O, Tuominen J, Rahkonen O, Jokinen E. Apoptosis in the pattern formation of the ventricular wall during mouse heart organogenesis. Anat Rec. 1999;256:208–217. doi: 10.1002/(SICI)1097-0185(19991001)256:2<208::AID-AR12>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Aho AV, Hopcroft JE, Ullman JD. Data structures and algorithms. Reading, MA: Addison-Wesley; 1983. Directed graphs; pp. 219–221. [Google Scholar]
- Ashburner M, Lewis S. In silico biology. Novartis Symposium. 2002. On ontologies for biologists: The Gene Ontology—Uncoupling the web. (in press). [PubMed] [Google Scholar]
- Bard JBL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock R, Davidson DR. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech Dev. 1998;74:111–120. doi: 10.1016/s0925-4773(98)00069-0. [DOI] [PubMed] [Google Scholar]
- Berman JJ, Moore GW. SNOMED-encoded surgical pathology databases: A tool for epidemiologic investigation. Mod Pathol. 1996;9:944–950. [PubMed] [Google Scholar]
- Biben C, Harvey RP. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes & Dev. 1997;11:1357–1369. doi: 10.1101/gad.11.11.1357. [DOI] [PubMed] [Google Scholar]
- Eferl R, Sibilia M, Hilberg F, Fuchsbichler A, Kufferath I, Guertl B, Zenz R, Wagner EF, Zatloukal K. Functions of c-Jun in liver and heart development. J Cell Biol. 1999;145:1049–1061. doi: 10.1083/jcb.145.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Creating the gene ontology resource: Design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimond C, Baudoin C, van der Neut R, Kramer D, Calafat J, Sonnenberg A. Cre-loxP-mediated inactivation of the α6A integrin splice variant in vivo: Evidence for a specific functional role of α6A in lymphocyte migration but not in heart development. J Cell Biol. 1998;143:253–266. doi: 10.1083/jcb.143.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GY, Wessels A, Smith BR, Linask KK, Ewart JL, Lo CW. Alteration in connexin 43 gap junction gene dosage impairs conotruncal heart development. Dev Biol. 1998;198:32–44. doi: 10.1006/dbio.1998.8891. [DOI] [PubMed] [Google Scholar]
- Jaspard B, Couffinhal T, Dufourcq P, Moreau C, Duplaa C. Expression pattern of mouse sFRP-1 and mWnt-8 gene during heart morphogenesis. Mech Dev. 2000;90:263–267. doi: 10.1016/s0925-4773(99)00236-1. [DOI] [PubMed] [Google Scholar]
- Kaufman MH, Bard JBL. The anatomical basis of mouse development. London: Academic Press; 1999. The heart and its associated vascular system; pp. 77–92. [Google Scholar]
- Kern MJ, Argao EA, Potter SS. Homeobox genes and heart development. Trends Cardiovasc Med. 1995;5:47–54. doi: 10.1016/1050-1738(94)00033-6. [DOI] [PubMed] [Google Scholar]
- King T, Beddington RS, Brown NA. The role of the brachyury gene in heart development and left–right specification in the mouse. Mech Dev. 1998;79:29–37. doi: 10.1016/s0925-4773(98)00166-x. [DOI] [PubMed] [Google Scholar]
- Kirby M. The heart. In: Thorogood Peter., editor. Embryos, genes and birth defects. Chichester: J. Wiley; 1997. pp. 231–250. [Google Scholar]
- Lyons GE. Vertebrate heart development. Curr Opin Genet Dev. 1996;6:454–460. doi: 10.1016/s0959-437x(96)80067-0. [DOI] [PubMed] [Google Scholar]
- Musen MA. Domain ontologies in software engineering: Use of Protege with the EON architecture. Methods Inf Med. 1998;37:540–550. [PubMed] [Google Scholar]
- Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes & Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald M, Eppig JT, Begley DA, Corradi JP, McCright IJ, Hayamizu TF, Hill DP, Kadin JA, Richardson JE. The mouse gene expression database. Nucleic Acids Res. 2001;29:98–101. doi: 10.1093/nar/29.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugh R. The mouse: Its reproduction and development. New York: Oxford University Press; 1990. Circulatory system; pp. 268–272. [Google Scholar]
- Sucov HM, Dyson E, Gumeringer CL, Price J, Chien KR, Evans RM. RXR α mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes & Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- Swiderski RE, Reiter RS, Nishimura DY, Alward WL, Kalenak JW, Searby CS, Stone EM, Sheffield VC, Lin JJ. Expression of the Mf1 gene in developing mouse hearts: Implication in the development of human congenital heart defects. Dev Dyn. 1999;216:16–27. doi: 10.1002/(SICI)1097-0177(199909)216:1<16::AID-DVDY4>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Woldeyesus MT, Britsch S, Riethmacher D, Xu L, Sonnenberg-Riethmacher E, Abou-Rebyeh F, Harvey R, Caroni P, Birchmeier C. Peripheral nervous system defects in erbB2 mutants following genetic rescue of heart development. Genes & Dev. 1999;13:2538–2548. doi: 10.1101/gad.13.19.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]