A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome (original) (raw)
Abstract
A major challenge of the postgenomic era is the functional characterization of every single gene within the mammalian genome. In an effort to address this challenge, we assembled a collection of mutations in mouse embryonic stem (ES) cells, which is the largest publicly accessible collection of such mutations to date. Using four different gene-trap vectors, we generated 5,142 sequences adjacent to the gene-trap integration sites (gene-trap sequence tags;http://genetrap.de) from >11,000 ES cell clones. Although most of the gene-trap vector insertions occurred randomly throughout the genome, we found both vector-independent and vector-specific integration “hot spots.” Because >50% of the hot spots were vector-specific, we conclude that the most effective way to saturate the mouse genome with gene-trap insertions is by using a combination of gene-trap vectors. When a random sample of gene-trap integrations was passaged to the germ line, 59% (17 of 29) produced an observable phenotype in transgenic mice, a frequency similar to that achieved by conventional gene targeting. Thus, gene trapping allows a large-scale and cost-effective production of ES cell clones with mutations distributed throughout the genome, a resource likely to accelerate genome annotation and the in vivo modeling of human disease.
With the completion of sequencing of the human and mouse genomes, the interest in tools suitable for performing genome-wide mutagenesis has increased significantly. Two major mouse-mutagenesis programs have evolved: one is phenotype-driven and based on chemical (ethyl-nitroso-urea) mutagenesis (1,2), and the other is gene-driven and based on insertional mutagenesis (3,4).
Large-scale insertional mutations in mammalian cells are induced most effectively with gene traps, a class of DNA or retroviral vectors that insert a promoterless reporter gene into a large collection of chromosomal sites. By selecting for gene expression, recombinants are obtained in which the reporter gene is fused to the regulatory elements of an endogenous gene. Transcripts generated by these fusions faithfully reflect the activity of individual cellular genes and serve as molecular tags to identify and/or clone any genes linked to specific functions (3–5). Application of this technique in a genome-wide manner should allow the identification of most, if not all, active transcripts in the genome and thus is an important tool for genome annotation. More importantly, gene trapping in mouse embryonic stem (ES) cells enables the establishment of ES cell libraries with mutations in most genes, which then can be used to make mice. This opens the possibility to assign a function to each gene in the context of an entire organism.
Several smaller-sized mutagenesis screens with gene-trap vectors have been reported (4,6–9). However, the use of single gene-trap vectors in each screen, the unavailability of a complete mouse genome sequence, and a comparatively low number of analyzed insertions precluded a systematic assessment of the technology.
Based on the analysis of 5,142 sequence tags obtained from gene-trap insertions across the mouse genome, we show here that gene-trap vectors can disrupt all functional classes of genes, including disease genes, and are highly mutagenic in transgenic mice. We also show that individual gene-trap vectors complement each other in gene targeting, suggesting that the most effective way of saturating the mouse genome with mutations is by using a combination of different gene-trap vectors.
Materials and Methods
ES Cell Cultures and Gene-Trap Vectors. The E14.1 (129/Ola), CJ7 (129/Sv), R1 (129/Sv × 129/Sv-CP), and TBV-2 (129/SvP) ES cell lines were grown on irradiated (x-rays, 32 Gy) or mitomycin C (Sigma)-treated (10 μg/ml for 2.5 h) mouse embryonic fibroblast feeder layers in DMEM (GIBCO/BRL) supplemented with 10–20% (vol/vol) preselected and heat-inactivated FCS (Invitrogen), 2 mM glutamine, 1× nonessential amino acids, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol (all from Invitrogen), 1,000 units/ml leukemia inhibitory factor (Esgro, Chemicon), and optionally 5 μg/ml penicillin and streptomycin (GIBCO/BRL). ES cells were either electroporated with pT1βgeo and pT1ATGβgeo plasmid vectors or infected with U3βgeo and ROSAβgeo retroviruses as described (3,10). Gene-trap-expressing ES cell clones were selected in 200 μg/ml G418 (GIBCO/BRL), manually picked, expanded, and stored frozen in liquid nitrogen. For gene-trap sequence tag (GTST) recovery all clones were arrayed into 48-well plates, lysed, and subjected to 5′ rapid amplification of cDNA ends.
5′ Rapid Amplification of cDNA Ends and Sequencing. cDNAs were prepared from the polyadenylated RNA by using a RoboAmp robotic device (MWG Biotec, Ebersberg, Germany) with a processing capacity of 96 samples per day. Samples of 2 × 105 cells were lysed in 1 ml of lysis buffer containing 100 mM Tris·HCl, pH 8.0/500 mM LiCl/10 mM EDTA/1% lithium-dodecyl sulfate (LiDS)/5 mM DTT. Polyadenylated RNA was captured from the lysates by biotin-labeled oligo(dT) primers according to manufacturer instructions (Roche Diagnostics, Indianapolis) and placed on streptavidin-coated 96-well plates (AB Gene, Surrey, U.K.). After washing, solid-phase cDNA synthesis was performed in situ by using random hexamers and SuperScript II reverse transcriptase (Invitrogen). To remove excess primers the cDNAs were filtered through multiscreen PCR plates (Millipore). The 5′ ends of the purified cDNAs were tailed with dCTPs by using terminal transferase, terminal deoxynucleotidyl transferase (Invitrogen), following manufacturer instructions.
For PCR amplification of GTSTs, the following vector-specific primers were used: (i) pT1βgeo and pT1ATGβgeo: 5′-CTA CTA CTA CTA GGC CAC GCG TCG ACT AGT ACG GGI IGG GII GGG IIG-3′ and 5′-GCC AGG GTT TTC CCA GTC ACG A-3′; and 5′-CTA CTA CTA CTA GGC CAC GCG TCG ACT AGT AC-3′ and 5′-TGT AAA ACG ACG GCC AGT GTG AAG GCT GTG CGA GGC CG-3′ (nested); and (ii) U3βgeo and the ROSAβgeo: 5′-GCC ATT CAG GCT GCG CAA-3′; and 5′-CAA GGC GAT TAA GTT GGG TAA TG-3′ (nested). Amplification products were directly sequenced by using AB377 or ABI3700 sequencing machines (Applied Biosystems).
GTST Analysis. After filtering sequences against repeats and removing all vector sequences from the GTSTs, a PHRED score was assigned to each individual nucleotide. GTSTs qualified as informative if they were at least 50 nt long and exhibited a minimum mean PHRED score of 20 (Fig. 4, which is published as supporting information on the PNAS web site,www.pnas.org). Homology searches were performed by using the publicly available sequence databases and the blastn algorithm. Databases included GenBank, UniGene, Online Mendelian Inheritance in Man (OMIM) (all atwww.ncbi.nlm.nih.gov),ensembl (www. ensembl.org), RIKEN (www.rarf.riken.go.jp), and GeneOntology (www.geneontology.org).
ES Cell Injections, Breeding, and Genotyping. 129Sv/J (TBV-2, R1, and E14.1) ES cell-derived chimeras were generated by injecting C57BL/6 blastocysts. The resulting male chimeras were bred to C57BL/6 females, and agouti offspring were tested for transgene transmission by tail blotting. Animals heterozygous for gene-trap insertions were backcrossed to C57BL/6 mice, and phenotypes were assessed in homozygous F2 offspring.
Results and Discussion
We used the gene-trap vectors pT1βgeo, pT1ATGβgeo, ROSAβgeo, and U3βgeo to transduce a promoterless β-galactosidase-neomycin phosphotransferase (βgeo) reporter gene into mouse ES cells. In pT1βgeo, pT1ATGβgeo, and ROSAβgeo, βgeo is flanked by an upstream 3′ splice consensus sequence (splice acceptor) and a downstream polyadenylation site to ensure its activation from integrations into introns (“intron trap”) (11–13). U3βgeo lacks a splice acceptor sequence and therefore is activated mostly from integrations into exons (“exon trap”) (10,14). Because all these gene-trap vectors require a cellular promoter for activation, the maximum number of genomic targets equals the number of expressed genes. The vectors pT1βgeo and pT1ATGβgeo were transduced as DNA into ES cells by electroporation. The vectors U3βgeo and ROSAβgeo were transduced as retroviruses into ES cells by infection.
From 11,266 ES cell clones containing gene-trap insertions in expressed genes, we isolated 8,423 sequences adjacent to the gene-trap integration sites (GTSTs). As summarized in Table 1, 5,142 of these sequences provided useful GTSTs. The other sequences were either of low quality or were too short (<50 nucleotides) to be informative (see Materials and Methods and Fig. 4).
Table 1. Summary of GTST results and homology analysis in GenBank (release 133).
| Gene-trap vector | pT1βgeo | pT1ATGβgeo | U3βgeo | ROSAβgeo | Total |
|---|---|---|---|---|---|
| No. of insertions sequenced | 3,866 | 1,581 | 2,100 | 876 | 8,423 |
| No. of GTSTs | 2,526 | 771 | 1,111 | 734 | 5,142† |
| NR* homology | 2,093 (82.9%) | 579 (75.1%) | 627 (56.4%) | 451 (61.4%) | 3,750 (72.9%) |
| EST homology | 190 (7.5%) | 103 (13.4%) | 192 (17.3%) | 138 (18.8%) | 623 (12.1%) |
| No homology | 243 (9.6%) | 89 (11.5%) | 292 (26.3%) | 145 (19.8%) | 769 (15.0%) |
GenBank (NCBI) homology analysis revealed that 3,750 (72.9%) of the GTSTs belonged to known genes, 623 (12.1%) were ESTs, and 769 (15%) had no match in the database (Table 1). In comparison to our previous analysis (7), the number of matches to known genes increased by 26%, clearly reflecting the sustained progress in sequencing of the human and mouse genomes. Moreover, when nonmatching “novel” (previously uncharacterized) sequences (769) were aligned to the ensembl database, 41% (389) produced a match (Table 2). However, despite the availability of a nearly complete mouse genome sequence, 7.4% (380 of 5,142; Tables 1 and2) failed to produce a match in any database. Although this could be the result of some strain-specific variations between mouse genomes, it may also reflect the fact that some sequences are not yet available from the genome sequence, which still contains gaps.
Table 2. Genome matches of “novel” GTSTs.
| Gene-trap vector | Novel GTSTs | Genome matches | In annotated genes | In predicted exons | In predicted introns |
|---|---|---|---|---|---|
| All | 769 | 389 | 214 | 84 | 130 |
| Without 3′ splice site (U3βgeo) | 292 | 189 | 104 | 24 | 80 |
| With 3′ splice site (pT1βgeo, pT1ATGβgeo, and ROSAβgeo) | 477 | 200 | 110 | 60 | 50 |
Fifty-five percent of the genome-matching GTSTs were in annotated genes. Interestingly, the frequency of U3βgeo insertions into predicted introns was almost twice as high as that obtained with all the other vectors (Table 2), confirming previous studies showing that the U3-type exon-trap vectors can be activated also from integrations into the introns of expressed genes (9,15). Unexpectedly, 50 of 110 GTSTs obtained with the other vectors were also part of predicted introns (Table 2), although intronic sequences should have been removed by splicing (3,4). Although in nine instances the intron-matching GTSTs resulted from aberrant splicing, we assumed that the other 41 GTSTs are actually part of exons annotated incorrectly by the current gene-prediction programs. To substantiate this hypothesis, we selected 10 annotated genes for additional expression studies. By using RT-PCR and primers complementary to the intron-annotated GTSTs and to the corresponding downstream exons (Fig. 5_A_, which is published as supporting information on the PNAS web site), we obtained amplification products in five instances. Direct sequencing of these products revealed splicing of the GTSTs to the downstream exons (Fig. 5_B_), indicating that a significant proportion of intron-matching GTSTs indeed are part of mispredicted exons.
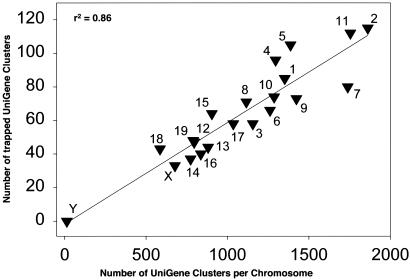
To localize the GTSTs cytogenetically, we screened the UniGene database using the GenBank accession number as an identifier. Allowing for an_e_ value ≤10–20, we identified 1,349 GTSTs in mapped UniGene clusters that were distributed among all chromosomes except the Y chromosome (Table 3). There was a direct correlation between the number of GTSTs on a given chromosome and the number of UniGene clusters on that chromosome, indicating that gene-trap insertions are dispersed throughout the genome and occur more frequently in chromosomes with a high density of genes (Fig. 1).
Table 3. Distribution of gene-trap insertions among chromosomes.
| Chromosome | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | X | Y | Total | |
| No. of gene-trap insertions | 85 | 115 | 58 | 96 | 105 | 66 | 80 | 71 | 73 | 74 | 112 | 47 | 44 | 37 | 64 | 40 | 58 | 43 | 48 | 33 | 0 | 1,349 |
Fig. 1.
Correlation between gene-trap insertions and the number of UniGene clusters per chromosome.
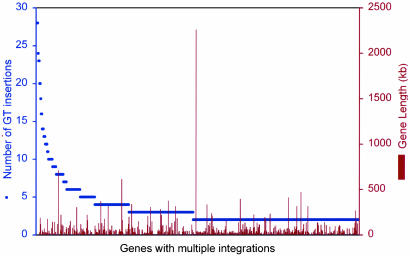
Several preferred integration sites or “hot spots” were observed, some of which were hit >20 times. Examples include the UniGene clusters 38,186 and 36,541, the growth-arrest gene Gas5, the C-terminal-binding protein 2, and the Jumonji (mouse) homolog (Table 8, which is published as supporting information on the PNAS web site). We identified a total of 441 UniGene clusters containing two or more gene-trap insertions, which corresponds to 25% of the recovered UniGene clusters and suggests that 75% of all genes are randomly accessible for gene-trap insertions. Forty-five percent of the hot spots contained multiple (more than two) insertions of more than one of the vectors and thus were vector-independent. Of the remaining vector-specific hot spots, 12% were recognized only by pT1βgeo, 10% by pT1ATGβgeo, 16% by U3βgeo, and 17% by ROSAβgeo vectors. Moreover, the gene-trap hot spots were not sequence-specific and were not related to gene size (Fig. 2), suggesting that they are most likely defined by secondary chromatin structure. Considering that over half of all the hot spots are vector-specific, we believe that the most effective way to saturate the genome with gene-trap insertions is with gene-trap vector combinations.
Fig. 2.
Number of gene-trap (GT) insertions into annotated hot spots. All genes with two or more insertions were classified as hot spots. Gene lengths were derived from GenBank.
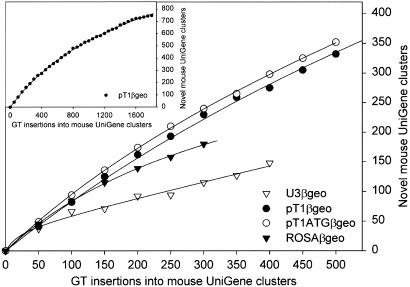
To estimate how effectively the various vectors trap genes that had not been trapped before, we determined the number of insertions required by each vector to trap a novel UniGene cluster.Fig. 3 shows that the vectors with a splice acceptor site (pT1βgeo, pT1ATGβgeo, and ROSAβgeo) trapped a different gene with almost every insertion. However, results from pT1βgeo, for which more insertions are available, suggest that the trapping efficiency decreases with an increasing number of insertions, presumably because of a gradual reduction of the pool of trappable genes (Fig. 3 Insert). In contrast, U3βgeo, which does not contain a splice acceptor, consistently required two or more insertions to hit a novel UniGene cluster (Fig. 3). The inferior gene-trapping efficiency of U3βgeo reflects its comparatively small pool of genomic integration targets, consisting mainly of the exons of expressed genes. As a result, U3βgeo integrated more frequently into a given genomic hot spot than any of the other vectors. With an average insertion frequency of 4.1 insertions per hot spot, U3βgeo exceeded the average hot-spot insertion frequency of the other vectors by almost 2-fold (Table 8).
Fig. 3.
Frequency of gene-trap (GT) insertions into unique UniGene clusters. Data points represent the number of novel UniGene clusters accumulating with every 50 insertions. For further explanation see text.
Because gene inactivations induced by gene-trap vectors with a splice acceptor sequence partly depend on effective splicing, the frequency of aberrant splicing events was determined by analyzing the splice junctions induced by each individual vector. Because the frequency of aberrant splicing was essentially similar for all gene-trap vectors (pT1βgeo = 3.5%; pT1ATGβgeo = 5.5%; ROSAβgeo = 4.0%), we conclude that the splice acceptor sequences used in this analysis are equally efficient [i.e., engrailed splice acceptor sequence for pT1βgeo and pT1ATGβgeo (11,12) and adenovirus major late transcript splice acceptor sequence for ROSAβgeo (13)]. Interestingly, >80% of the aberrantly spliced integrations into annotated genes were atypically in exons, suggesting that ectopic splice sites inside exons are recognized ineffectively by the splicing enzymes.
Because the relative mutagenicity of the gene-trap vectors likely depends on their position within a gene, we looked at the insertion site of each gene-trap vector with regard to its location within the full-length cDNA.Table 4 shows that the vast majority of retroviral gene-trap insertions involved the 5′ half of genes, confirming a reported preference of retroviral integrations (9,16). Interestingly, >50% of the U3βgeo insertions were in 5′ untranslated regions (Table 4), presumably due to a relatively high stringency of selection that requires gene-trap vectors without a splice acceptor to insert close to an active cellular promoter. Although plasmid vectors also exhibited a slight preference for the 5′ ends of genes, insertions were distributed more evenly over the coding region of a gene, indicating that even longer fusion proteins are stable (Table 4). Finally, one U3βgeo integration was recovered from an intronless gene (glutathione peroxidase 4/ENSMUSG00000038809). Although this was a unique event, it demonstrates that U3βgeo vectors can also disrupt single exon genes.
Table 4. Gene-trap vector insertion site preference in full-length cDNAs (according to RefSeq).
| Gene trap vector | Total no. of insertions | Insertions in 5′ UTR (%) | Insertions in 5′ CDS (%) | Insertions in 3′ CDS (%) | Insertions in 3′ UTR (%) |
|---|---|---|---|---|---|
| pT1βgeo | 1,385 | 222 (16.0) | 589 (42.5) | 511 (36.9) | 63 (4.6) |
| pT1ATGβgeo | 395 | 97 (24.6) | 160 (40.5) | 125 (31.6) | 13 (3.3) |
| U3βgeo | 324 | 176 (54.3) | 101 (31.2) | 31 (9.6) | 16 (4.9) |
| ROSAβgeo | 302 | 100 (33.1) | 152 (50.3) | 37 (12.3) | 13 (4.3) |
| Total | 2,406 | 595 (24.7) | 1,002 (41.6) | 704 (29.3) | 105 (4.4) |
To analyze the functional spectrum of the genes represented in the GTST library, we classified the trapped UniGene clusters based on their known or putative function by using the GeneOntology database.Table 5 shows that the vectors used in this study inserted into all functional classes of mammalian genes, although with different frequencies, which suggests that the effective trapping of some specific classes of genes may require more specialized gene-trap vectors (17,18).
Table 5. Functional gene classes targeted by gene-trap insertions.
| Class* | Annotated genes | Trapped genes (%) |
|---|---|---|
| Ligand binding or carrier | 2,002 | 240 (12.0) |
| Enzyme/enzyme regulator | 1,491 | 138 (9.3) |
| Transcription factors/cofactors | 416 | 45 (10.8) |
| Transporter | 437 | 25 (5.7) |
| Signal transducer | 779 | 24 (3.1) |
| Structural protein | 193 | 11 (5.7) |
| Chaperone | 49 | 11 (22.5) |
| Translation regulator | 27 | 8 (30.0) |
| Motor | 39 | 7 (17.9) |
| Defense/immunity | 44 | 1 (2.3) |
| Cell adhesion molecule | 78 | 1 (1.3) |
| Apoptosis regulator | 30 | 1 (0.3) |
Because the development of mouse models for human disease is a major goal of the human genome project, we also searched our library for integrations into genes involved in human disease. Using the Online Mendelian Inheritance in Man (OMIM) database, we found 204 GTSTs that corresponded to 90 previously characterized disease genes (Table 9, which is published as supporting information on the PNAS web site). ES cell clones with these insertions can be used to produce mouse mutant strains that may replicate the genetic defects and the symptomology of specific human disorders, and that may be useful for testing therapeutic methods. For example, we recently characterized a mouse strain with a phenotype closely resembling congenital nephrotic syndrome (19,20).
To analyze the frequency of obvious phenotypes developing after gene-trap insertions, we injected 29 randomly selected ES cell clones into blastocysts and produced mutant mice from them. As shown inTable 6, 59% of the mice developed an obvious phenotype when bred to homozygosity, a frequency comparable with conventional gene targeting and to reported gene-trap screens (6,13). Interestingly, over half of the observed phenotypes were embryonic or perinatal-lethal (Table 7), suggesting that a significant proportion of the genes expressed in ES cells are required for embryonic development.
Table 6. Frequency of phenotypes obtained with gene-trap vectors.
| Gene-trap vector | Phenotypes/mutant strains | Frequency, % |
|---|---|---|
| pT1βgeo | 6/11 | 55 |
| pT1ATGβgeo | 5/9 | 56 |
| U3βgeo | 7/9 | 78 |
| All | 17/29 | 59 |
Table 7. Mutant phenotypes induced by gene-trap insertions.
| Line | Gene name | Symbol | Phenotype |
|---|---|---|---|
| A006B04 | Sprouty homolog 4 (Drosophila) | Spry4 | Limb deformation |
| A20010 | Novel | Pigmentation defects | |
| M004D05 | Selenocysteine tRNA gene transcription-activating factor | Staf | Sterility |
| M016A06 | PHD finger protein 2 | Phf2 | Dwarfism |
| W027B02 (20) | Nephrin 1 | Nphs1 | Nephrotic syndrome |
| W036C08 | Baculoviral IAP repeat-containing 6 | Birc6 | Placenta defects |
| W044B06 | Neurochondrin | Ncdn-pending | Lacrimal gland hypertrophy |
| W052E02 | Synaptojanin 2 | Synj2 | Perinatal-lethal |
| 3C7 (21) | Latent transforming growth factor β-binding protein 4 | Ltbp4 | Pulmonary emphysema, colorectal cancer |
| F053A01 | ITSN | Itsn | Embryonic-lethal |
| F045D05 | Ect2 oncogene | Ect2 | Embryonic-lethal |
| M016E07 | Splicing factor (CC1.3) | CC1.3 | Embryonic-lethal |
| M017A08 | Heterogeneous nuclear ribonucleoprotein C (C1/C2) | Hnrpc | Embryonic-lethal |
| M019E03 | KIAA0240 | KIAA0240 | Embryonic-lethal |
| M020A01 | Plectin 1, intermediate filament-binding protein, 500 kDa | Plec1 | Embryonic-lethal |
| W023D11 | Novel | Embryonic-lethal | |
| W078F01 | Peroxisomal biogenesis factor 14 | Pex14 | Embryonic-lethal |
| F035B07 | Nuclear factor of κ light chain gene enhancer in B cells 1, p 105 | Nfkb1 | Not obvious |
| F041B05 | HSPC063 protein ESTs, weakly similar to I53869 zinc finger protein—mouse (Mus musculus) | EST | Not obvious |
| M017C03 | EST | EST | Not obvious |
| M019D01 | Chromobox homolog 1 (Drosophila HP1 β) | Cbx1 | Not obvious |
| W008G09 | ESTs, highly similar to T34020 zinc finger protein—rat (Rattus norvegicus) | EST | Not obvious |
| W024F10 | Homo sapiens cDNA FLJ30453 fis, clone BRACE2009307, weakly similar to P120 PROTEIN | Pkp4 | Not obvious |
| W027F01 | Msx-interacting zinc finger | Miz1 | Not obvious |
| W047A01 | Baculoviral IAP repeat-containing 6 | Birc6 | Not obvious |
| W056E05 | Dystrophin, muscular dystrophy | Dmd | Not obvious |
| W063E06: | Fibroblast growth factor-inducible 13 | Fin13 | Not obvious |
| W073D02 | Dentin matrix protein 1 | Dmp1 | Not obvious |
| Aquarius | Aquarius | Aqr | Not obvious |
We conclude that gene-trap mutagenesis is an efficient approach for annotating and dissecting the function of mammalian genes. Its large-scale implementation has already enabled the worldwide establishment of several databases containing GTSTs from hundreds of mouse genes (4,6–9). Collectively, these databases provide an unprecedented resource for the scientific community in the postgenomic era, because clones from the corresponding ES cell libraries can be used immediately to cost-effectively generate mouse models of human disease. Clearly, the goal of understanding the function of every gene in the genome could be attained more quickly with the establishment of ES cell libraries with mutations in every single gene. Because each gene-trap vector seems to have its own set of specific hot spots, we conclude that the most effective generation of an ES cell library saturated with mutations should involve a collection of different gene-trap vectors. The ongoing collaboration within the international mouse-mutagenesis consortium (22) is likely to achieve complete saturation of the mouse genome within the next few years.
Supplementary Material
Supporting Information
Acknowledgments
We thank Drs. William. C. Skarnes for the pT1βgeo vectors, H. Earl Ruley for the U3βgeo, and Philippe Soriano for the ROSAβgeo vectors. We acknowledge Susanne Bourier, Franziska Köhler, Katharina Kuhlmeier, Sava Michailidou, Ines Peiser, Armin Reffelmann, Irina Rodionova, Cordula Schulz, Beata Thalke, Sandra Schwarzmeier, Beate Walther, and Carsta Werner for excellent technical assistance. This work was supported by grants from the Bundesministerium für Bildung und Forschung to the German Gene Trap Consortium.
Abbreviations: ES, embryonic stem; GTST, gene-trap sequence tag.
References
- 1.Brown, S. D. & Balling, R. (2001) Curr. Opin. Genet. Dev. 11**,** 268–273. [DOI] [PubMed] [Google Scholar]
- 2.Beier, D. R. (2000) Mamm. Genome 11**,** 594–597. [DOI] [PubMed] [Google Scholar]
- 3.Floss, T. & Wurst, W. (2002) Methods Mol. Biol. 185**,** 347–379. [DOI] [PubMed] [Google Scholar]
- 4.Stanford, W. L., Cohn, J. B. & Cordes, S. P. (2001) Nat. Rev. Genet. 2**,** 756–768. [DOI] [PubMed] [Google Scholar]
- 5.Hicks, G. G., Shi, E. G., Chen, J., Roshon, M., Williamson, D., Scherer, C. & Ruley, H. E. (1995) Methods Enzymol. 254**,** 263–275. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell, K. J., Pinson, K. I., Kelly, O. G., Brennan, J., Zupicich, J., Scherz, P., Leighton, P. A., Goodrich, L. V., Lu, X., Avery, B. J., et al. (2001) Nat. Genet. 28**,** 241–249. [DOI] [PubMed] [Google Scholar]
- 7.Wiles, M. V., Vauti, F., Otte, J., Fuchtbauer, E. M., Ruiz, P., Fuchtbauer, A., Arnold, H. H., Lehrach, H., Metz, T., von Melchner, H. & Wurst, W. (2000) Nat. Genet. 24**,** 13–14. [DOI] [PubMed] [Google Scholar]
- 8.Zambrowicz, B., Friedrich, G., Buxton, E., Lilleberg, S., Person, C. & Sands, A. (1998) Nature 392**,** 608–611. [DOI] [PubMed] [Google Scholar]
- 9.Hicks, G. G., Shi, E., Li, X. M., Li, C. H., Pawlak, M. & Ruley, H. E. (1997) Nat. Genet. 16**,** 338–344. [DOI] [PubMed] [Google Scholar]
- 10.von Melchner, H., DeGregori, J. V., Rayburn, H., Reddy, S., Friedel, C. & Ruley, H. E. (1992) Genes Dev. 6**,** 919–927. [DOI] [PubMed] [Google Scholar]
- 11.Gossler, A., Joyner, A. L., Rossant, J. & Skarnes, W. C. (1989) Science 244**,** 463–465. [DOI] [PubMed] [Google Scholar]
- 12.Skarnes, W. C., Auerbach, B. A. & Joyner, A. L. (1992) Genes Dev. 6**,** 903–918. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich, G. & Soriano, P. (1991) Genes Dev. 5**,** 1513–1523. [DOI] [PubMed] [Google Scholar]
- 14.Scherer, C. A., Chen, J., Nachabeh, A., Hopkins, N. & Ruley, H. E. (1996) Cell Growth Differ. 7**,** 1393–1401. [PubMed] [Google Scholar]
- 15.Wempe, F., Yang, J. Y., Hammann, J. & von Melchner, H. (2001) Genome Biol. 2**,** research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohdewohld, H., Weiher, H., Reik, W., Jaenisch, R. & Breindl, M. (1987) J. Virol. 61**,** 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skarnes, W., Moss, J., Hurtley, S. & Beddington, R. (1995) Proc. Natl. Acad. Sci. USA 92**,** 6592–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebauer, M., von Melchner, H. & Beckers, T. (2001) Genome Res. 11**,** 1871–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patrakka, J., Martin, P., Salonen, R., Kestila, M., Ruotsalainen, V., Mannikko, M., Ryynanen, M., Rapola, J., Holmberg, C., Tryggvason, K. & Jalanko, H. (2002) Lancet 359**,** 1575–1577. [DOI] [PubMed] [Google Scholar]
- 20.Rantanen, M., Palmen, T., Patari, A., Ahola, H., Lehtonen, S., Astrom, E., Floss, T., Vauti, F., Wurst, W., Ruiz, P., et al. (2002) J. Am. Soc. Nephrol. 13**,** 1586–1594. [DOI] [PubMed] [Google Scholar]
- 21.Sterner-Kock, A., Thorey, I. S., Koli, K., Wempe, F., Otte, J., Bangsow, T., Kuhlmeier, K., Kirchner, T., Jin, S., Keski-Oja, J. & von Melchner, H. (2002) Genes Dev. 16**,** 2264–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadeau, J. H., Balling, R., Barsh, G., Beier, D., Brown, S. D., Bucan, M., Camper, S., Carlson, G., Copeland, N., Eppig, J., et al. (2001) Science 291**,** 1251–1255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information