Arginase Modulates NF-κB Activity via a Nitric Oxide–Dependent Mechanism (original) (raw)
Abstract
NF-κB is a versatile transcription factor that regulates a wide array of processes, including inflammation and survival, and plays a critical role in the etiology of inflammatory lung diseases. Nitric oxide (NO) has been suggested to play an antiinflammatory role through S-nitrosation of components of NF-κB pathway. NO production can be modulated by changing the availability of its substrate, L-arginine. Arginases compete with NO synthases (NOSs) for their common substrate, L-arginine, and thereby have the potential to alter the signaling function of NO. The goal of the present study was to determine the impact of arginase manipulation on NO, and subsequent effects on NF-κB activation, in lung epithelial cells. Our results demonstrate that reduction of arginase activity enhanced cellular content of NO and S-nitrosated proteins, and resulted in decreases in TNF-α– or LPS-stimulated NF-κB DNA binding and transcriptional activity, in association with enhanced S-nitrosation of p50. The effects of arginase inhibition on NF-κB were reversed by the generic NOS inhibitor, N-ω-nitro-l-arginine methyl ester (l-NAME), suggesting a causal role for NO in the attenuation of NF-κB induced by arginase suppression. Conversely, overexpression of arginase I decreased cellular S-nitrosothiol content and enhanced IκB kinase activity and NF-κB DNA binding, and decreased S-nitrosation of p50. Collectively, our data point to a regulatory mechanism wherein NF-κB is controlled through arginase-dependent regulation of NO levels, which may impact on chronic inflammatory diseases that are accompanied by NF-κB activation and upregulation of arginases.
Keywords: arginase, lung epithelial cells, nitric oxide, NF-κB, S-nitrosation
CLINICAL RELEVANCE
Arginase was recently presented as a new player in asthma. This article suggests that arginase regulates transcription factor NF-κB via NO-dependent redox changes. This would provide insights into the role of arginase in inflammatory disease.
NF-κB is a transcription factor that regulates a multitude of cellular processes, ranging from inflammation to survival and proliferation. NF-κB activity is tightly controlled by its inhibitory components, IκBs, which reside in the cytosol complexed with NF-κB dimers, thereby preventing the nuclear localization of NF-κB, and minimizing transcriptional activity. In response to most stimuli, the activation of NF-κB requires the activation of IκB kinases (IKKs), which phosphorylate IκB, RelA, and histone H3 (1–3), and promote NF-κB nuclear retention and transcriptional activity, leading to the activation of diverse target genes that encode for inflammatory cytokines, chemokines, acute phase proteins, immunoreceptors, and so forth (4).
Inflammatory diseases are commonly associated with enhanced local production of nitric oxide (NO) and other oxidants (5, 6). As a signaling molecule, NO regulates a wide range of physiologic responses, ranging from vessel relaxation to gene transcription. The ability of NO to modify specific protein cysteine residues, referred to as S-nitrosylation or S-nitrosation, is now well recognized, and has emerged as a prototype redox-based post-translational modification (7, 8). Indeed, NO has been demonstrated to regulate NF-κB via S-nitrosation, and S-nitrosation of cysteines 62 and 179 of p50 and IKKβ, respectively, have been shown to inhibit NF-κB (9–11). It should be highlighted that considerable debate exists regarding the proper nomenclature to describe NO binding to sulfhydryl groups of protein. Nitrosation is the correct chemical term to define attachment of NO to nucleophilic centers, including sulfhydryl groups. However, it is common in the signal transduction field to refer to this NO-dependent modification as S-nitrosylation instead S-nitrosation, as an analogy to other post-translational modifications, such as phosphorylation, glycosylation, prenylation, and so forth.
NO is generated by NO synthases (NOSs) by oxidation of its substrate, L-arginine. In addition to NOS, L-arginine is also metabolized by arginases, which hydrolyze L-arginine to urea and ornithine (12). Two isoforms of arginase exist, arginase I (AI) and -II (AII), which are expressed in several cell types, including immune cells, fibroblasts, and epithelial cells (13). Several studies, mostly conducted in myeloid cells, have illuminated a competitive balance in the regulation of both arginase and inducible NOS (iNOS), and demonstrated that arginase can impact NOS function by depleting the bioavailability of L-arginine needed for NO biosynthesis (14–17).
Although the aforementioned studies demonstrated that NO inhibits NF-κB through S-nitrosation of IKKβ and p50, those studies were conducted with high levels of administered S-nitrosothiols. Consequently, the physiologic relevance of NO in the regulation of NF-κB in lung epithelium remains elusive. Because of recent studies demonstrating arginase upregulation in allergic lung disease (18, 19), and the documented functional significance of epithelial-dependent NF-κB activation in lung inflammation and allergic lung disease (20–22), the goal of the present study was to determine whether manipulation of arginase affects the NF-κB pathway in lung epithelial cells, and whether this occurs in an NO-dependent manner.
MATERIALS AND METHODS
Cell Culture and Reagents
A line of spontaneously transformed mouse alveolar type II epithelial cells (C10) was propagated in CRML-1066 medium, containing 50 U/ml penicillin–50 μg/ml streptomycin (P/S), 2 mM L-glutamine, and 10% FBS, all from GIBCO-BRL (Carlsbad, CA). Murine recombinant TNF-α was purchased from Calbiochem (San Diego, CA). Ultrapure LPS (Escherichia coli, 0111:B4) was purchased from List Biological Laboratories (Campbell, CA). Primary mouse tracheal epithelial (MTE) cells were isolated and propagated according Wu et al. (23) and used within three passages.
The specific competitive inhibitor of AI and -II, a boronic acid–based arginine analog, _S_-(2-boronoethyl)-L-cysteine (BEC; Calbiochem), was used to inhibit overall arginase activity. Cells were incubated with 1 mM BEC, or general NOS inhibitor, N-ω-nitro-l-arginine methyl ester (l-NAME; 1 mM), for 20–24 h. To knock down AI protein expression, cells were transfected with AI siRNA (Ambion, Austin, TX), using siPORTamine, according to the manufacturer's instructions, and the assays were performed 24–48 h later. A control siRNA was used as a reagent control.
Mouse AI in pCMV-SPORT 6 plasmid was purchased from Invitrogen (Carlsbad, CA), and subcloned in pcDNA3 expression vector. Cells were transfected, and the assays were performed 24 h later.
ELISA
CCL20 (macrophage inflammatory protein-3α) and KC (neutrophil attractant protein, member of the α CXCL1 chemokine family) concentrations were determined in MTE cell culture media using a Quantikine ELISA Kit (R&D Systems, Minneapolis, MN), according to the manufacturer's instructions. Values were calculated using a standard curve.
Immunoprecipitation and Western Blot Analysis
AI or endothelial NOS (eNOS) were immunoprecipitated from cell lysates using specific antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C, and incubated with protein A agarose for 1 h at 4°C. Matching isotype IgG was used as a reagent control. Immunoprecipitates or cell lysates were mixed with 2× Laemmli buffer, boiled, and loaded on polyacrylamide gels. Proteins were transferred to nitrocellulose membrane, which was blocked with 5% skim milk overnight at 4°C. Antibodies directed against AI or eNOS (Santa Cruz Biotechnology) were used to evaluate protein expression.
RT-PCR and Semiquantitative PCR
Total RNA from cell culture was prepared by using RNeasy Qiagen mini kit (Qiagen Inc., Valencia, CA), DNase treated, and reverse transcribed into cDNA. Semiquantitative TaqMan PCR was performed using Assay On Demand for AI or AII from Applied Biosystems (Framingham, MA). Values were normalized to hypoxanthine phosphoribosyltransferase (HPRT) housekeeping gene. RT-PCR was performed with specific AI forward 5′-GGA ACC CAG AGA GAG CAT GA-3′ and reverse 5′-AAG GCG TTT GCT TAG CTG TG-3′ and AII forward 5′-CTG CCA ATC ATG TTC CTG AG-3′ and reverse 5′-TGA TCC AGA CAG CCA TTT CA-3′ primers (IDT, Coralville, IA).
Arginase Activity
Arginase activity was evaluated in cell cultures as described previously (24). Urea production by arginase was determined spectrophotometrically at 540 nm using a standard curve generated with urea.
Measurement of Nitrite and Nitroso/Nitrosyl Content by Chemiluminescence
The cellular content of nitrite and nitroso/nitrosyl (NOx; RSNO: S-nitrosothiols; RNNO: _N_-nitroso adducts, including _N-_nitrosoamines, and metal nitrosyl other than NO-heme) was determined using a group-specific reductive denitrosation by iodine-iodide with subsequent detection of NO liberated by gas-phase ozone chemiluminescence (25, 26), using an NO analyzer (Ionics, Boulder, CO). Cell lysates were centrifuged, and 50 μl of supernatants injected into a purge vessel containing 5 ml of 45 mM potassium iodide and 10 mM iodine (I2) in glacial acetic acid, at 60°C, purged continuously with nitrogen (27). For the purpose of clarity, we will refer to these measurements hereafter as cellular NOx content.
Assessment of S-Nitrosation Using Chemical Derivatization Coupled to Immunoprecipitation and Western Blotting
The biotin labeling of S-nitrosated proteins in lysates was based on previously described procedures (11, 28, 29). For immunoprecipitation of p50, the biotinylated lysates were incubated with specific polyclonal antibody for 1.5 h at 4°C. Biotinylation of p50 was detected using streptavidin–horseradish peroxidase conjugate. To avoid potential interference of ascorbate in the reduction of disulfide bonds (30), we lowered its concentration to 1 mM. Control experiments were performed in which sodium ascorbate was omitted, preventing the reduction of S-nitrosothiols (29). Lastly, to assess the contribution of endogenous biotinylated proteins, _N_α-(3-maleimidylpropionyl)biocytin (MPB) was omitted in some samples. Samples were protected from light during all procedures before electrophoresis.
Assessment of S-Nitrosation in Intact Cells Using Biotin Derivatization and Analysis by Confocal Microscopy
To evaluate overall S-nitrosation in intact cells, a biotin derivatization method was used in combination with fluorophore labeling and visualization by confocal microscopy (31). Cells were analyzed by confocal microscopy (magnification: ×40) using an Olympus BX50 microscope (Olympus America, Inc., Melville, NY) coupled to a Bio-Rad (Hercules, CA) MRC 1024 confocal scanning laser microscope system. Control experiments were performed in which blocking of SH groups with _N_-ethylmaleimide or S-nitrosated protein reduction by ascorbate were omitted. To assess the contribution of endogenous biotinylated proteins, the biotin label was omitted in some control coverslips.
In Vitro IKK Assay
C10 cells were exposed to test agents, transferred to ice, washed twice with PBS, and lysates were prepared and the kinase assay performed as described previously (32). Data were expressed as fold increase of kinase activity compared with sham controls. IKKγ (BD Pharmingen, San Jose, CA). Western blots were performed as a loading control.
Electrophoretic Mobility Shift Assay
To determine the binding activity of protein complexes to canonical NF-κB DNA consensus sequence, nuclear extracts were prepared and electrophoretic mobility shift assay was performed, as described previously (32). Previously conducted supershift assays (32) using antibodies directed against RelA or p50 ascertained that DNA binding complexes contain RelA and p50 proteins (data not shown).
NF-κB Luciferase Reporter Gene Assay
A C10 cell line stably expressing an NF-κB–luciferase reporter construct was used to measure NF-κB activity, as previously described (32). Luciferase units were expressed relative to micrograms of protein in the lysates (relative Luciferase units).
Statistical Analysis
All experiments were performed in triplicate and/or repeated at least twice. Results are expressed as means (± SD). Significant statistical differences between the groups were evaluated using Student's t test or ANOVA with the Student-Newman-Keuls test to adjust for multiple pair-wise comparisons. In all analyses, the level of significance used was P < 0.05.
RESULTS
Airway Epithelial Cells Express Functional Arginase
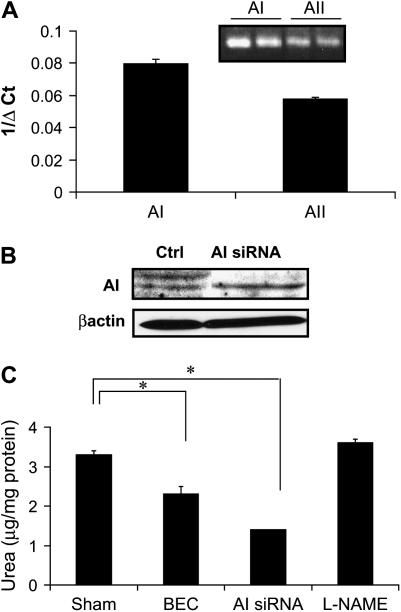
It is well known that myeloid (33) and endothelial cells express arginases, and their activity can control NO production and consequent vasoactivity (34). Because this has not been clearly established in airway epithelial cells, we first evaluated whether arginase was expressed in these cells. Indeed, results in Figure 1A demonstrate that mouse lung epithelial cells (C10 cells) express AI and AII, as detected by PCR analysis. Because AI is cytosolic, has been shown to affect NO metabolism (35), and its mRNA is most abundant in C10 cells (Figure 1A), we chose to use AI siRNA to knock down AI. Results in Figure 1B demonstrate that arginase expression was markedly decreased with AI siRNA, whereas control siRNA did not affect expression of AI (Figure 1B and data not shown). Moreover, incubation of C10 cells with the arginase inhibitor, BEC, resulted in an ∼ 30% decrease in arginase activity, whereas AI siRNA resulted in a 50% reduction of arginase activity. The generic NOS inhibitor, l-NAME, did not affect the activity of arginase (Figure 1C). Collectively, these data demonstrate that arginase expression and activity are detected in C10 mouse lung epithelial cells, and can be suppressed with siRNA or pharmacologic approaches.
Figure 1.
Arginase is expressed in epithelial cells, and its activity can be inhibited after incubation with the boronic acid–based arginine analog, BEC, or knocked down by siRNA. (A) Arginase I (AI) and arginase II (AII) mRNA expression in an alveolar epithelial cell line (C10) was analyzed by TaqMan-PCR. Relative expression is represented as 1/ΔCT, where higher values indicate higher levels of expression (inset: RT-PCR). (B) ArgI protein expression evaluated via Western blot analysis after transfection with control siRNA, or AI siRNA. (C) Attenuation of arginase activity in C10 cells after exposure to the arginase inhibitor, BEC (1 mM), for 24 or 48 h post-transfection with AI siRNA. As a control, cells were exposed to 1 mM l-NAME for 24 h. *P < 0.05, compared with sham by ANOVA. All experiments were repeated at least twice.
Inhibition of Arginase Increases Cellular NO Content
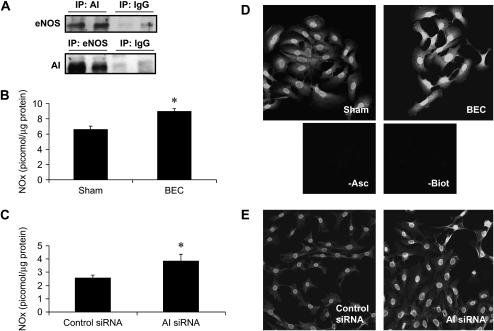
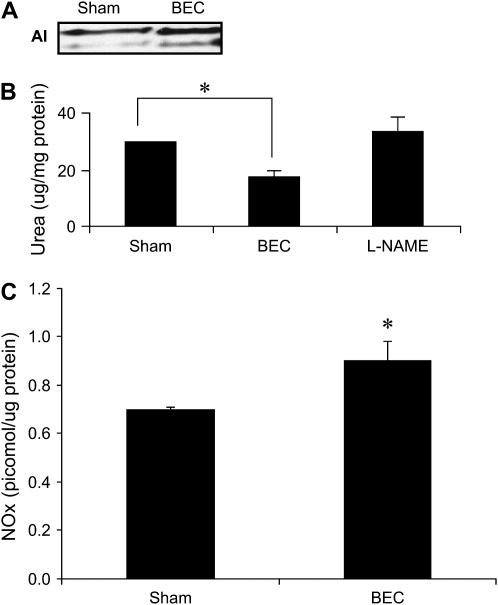
As mentioned previously here, several studies have demonstrated a competitive balance in the regulation of both arginase and NOS, and suggested an involvement of arginase in repression of NOS activity (14, 15, 17). One important aspect of this competition is the colocalization of both arginase and NOS isoforms (36). We therefore investigated whether AI was colocalized with NOS isoforms in C10 cells. After immunoprecipitation of AI, but not control IgG, eNOS protein was detected after immunoblotting of immunoprecipitated proteins (Figure 2A, top panel), whereas neither neuronal NOS (nNOS) nor iNOS were detected (data not shown). In addition, immunoprecipitation of eNOS also resulted in detection of AI via Western blotting (Figure 2A, bottom panel), providing some evidence for colocalization of AI and eNOS in lung epithelial cells. We next examined whether inhibition of arginase activity affected the cellular NO content through measurement of NOx in cells. Results in Figures 2B and 2C demonstrate that inhibition of arginase activity by BEC, or knockdown by AI siRNA increased cellular NOx content, which corresponded to increased levels of S-nitrosated proteins in intact cells (Figures 2D and 2E). In agreement with our previous observations (31), omission of ascorbate, which reduces SNO bonds, before the addition of the biotin-MPB label, markedly decreased the observed reactivity. Furthermore, fluorescence was not apparent when the biotin-MPB label was omitted, indicating that the observed fluorescence was not due to endogenous biotin (Figures 2D and 2E). To corroborate the relevance of these observations, which suggest that AI controls NOx levels in lung epithelial cells, we extended our studies to primary MTE cells. Results in Figure 3 demonstrate that AI is expressed in MTE cells (Figure 3A), and that arginase activity can be attenuated with BEC (Figure 3B). Identical to our results in C10 cells, inhibition of AI in primary MTE cells led to increases in the intracellular NOx content (Figure 3C).
Figure 2.
AI is colocalized with eNOS and its inhibition or knockdown increases cellular content of NOx and total S-nitrosated proteins. (A) AI (AI; top) or eNOS (bottom) were immunoprecipitated from C10 cells, and immunoprecipitates were submitted to SDS-PAGE for detection of eNOS (top) or AI (bottom) using specific antibodies, followed by incubation with a horseradish peroxidase (HRP)–conjugated secondary antibody. As a control, preimmune IgG was used in the immunoprecipitations. All immunoprecipitations were done in two independent samples. C10 cells were exposed to BEC (B) or transfected with control or AI-specific siRNA (C) for 24 or 48 h, respectively, and lysed in HEPES buffer containing 0.1 mM EDTA, 0.01 mM neocuproine, 20 mM NEM, 0.5% CHAPS, and 0.1% SDS. NOx were detected in lysates by chemiluminescence, using a purge vessel containing 5 ml of potassium iodide, 45 mM/iodine (I2), 10 mM in glacial acetic acid, at 60°C. Results are expressed as pmol NO/μg protein using a standard curve generated with S-nitrosoglutathione (GSNO). *P < 0.05 (Student's t test). All experiments were repeated at least three times. C10 cells were exposed to BEC for 24 h (D) or transfected with control of AI siRNA (E) for 48 h, fixed, and S-nitrosylated proteins visualized after biotin derivatization, incubation with streptavidin-AlexaFluor 568, and detection via confocal microscopy. As reagent controls, the biotin label (−Biot), or ascorbate (−asc) were omitted. Image analysis was performed by evaluating pixel intensity of at least 10 individual cells (IMAGEJ software; http://rsb.info.nih.gov/ij/index.html), and the values (mean ± SD) in arbitrary units were as follows: sham, 43.8 ± 4.5 versus BEC, 79.8 ± 13.7 (P < 0.001), Student's t test; and control siRNA, 44.6 ± 2.7 versus A1 siRNA, 56.6 ± 9.6 (P < 0.001), Student's t test. All experiments were repeated at least three times.
Figure 3.
Arginase is expressed in MTE cells, and its activity can be inhibited after incubation with the boronic acid–based arginine analog, BEC. (A) AI expression in control MTE cells, or after incubation with 1 mM BEC, for 24 h. (B) Attenuation of arginase activity in MTE cells after exposure to the arginase inhibitor, BEC, for 24 h. As a reagent control, cells were exposed to 1 mM l-NAME for 24 h (C). MTE cells were exposed to BEC for 24 h and NOx were detected in lysates as described previously. Results are expressed as pmol NO/ug protein using a standard curve generated with GSNO. *P < 0.05 compared with sham: ANOVA (B); Student's t test (C). All experiments were repeated at least three times.
Arginase Activity Modulates NF-κB Activity
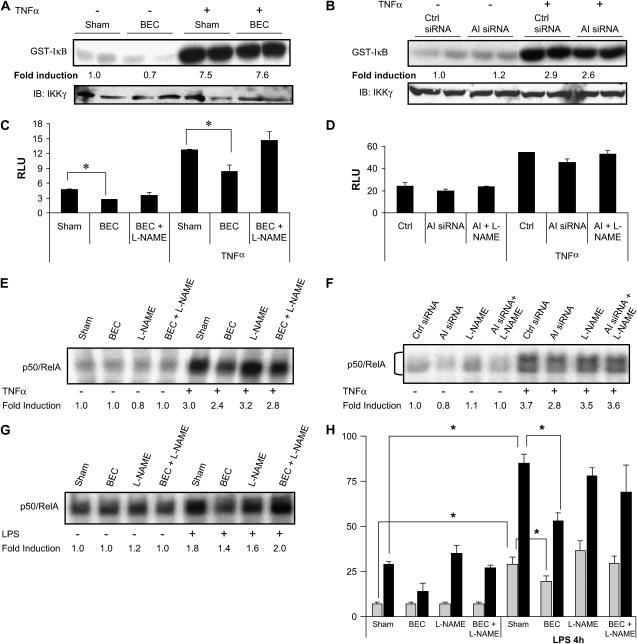
After demonstrating that arginase inhibition or knockdown increased the cellular content of NOx in lung epithelial cells, we next investigated the implications for regulation of NF-κB, as this transcription factor has been demonstrated to be sensitive to inhibition by NO (9–11). Initially, we investigated whether manipulation of arginase interfered with the activity of IKK, the enzymatic complex responsible for NF-κB activation (37). In contrast to our previous observations demonstrating inhibition of IKK after administration of exogenous S-nitrosothiols (11), arginase inhibition or knockdown did not affect baseline or TNF-α–stimulated IKK activity (Figures 4A and 4B). However, arginase inhibition or knockdown affected NF-κB downstream of IKK, based upon analysis of NF-κB transcriptional activity and DNA binding. Results in Figures 4C and 4D demonstrate that inhibition or knockdown of arginase caused a small but consistent attenuation of both basal and TNF-α–stimulated NF-κB transcriptional activity, which was restored by the NOS inhibitor, l-NAME, suggesting an involvement of NO in the inhibition of NF-κB. Because NO has been shown to inhibit the ability of NF-κB to bind to its DNA consensus sequence, we next evaluated the impact of arginase suppression on NF-κB–DNA binding activity. Results in Figures 4E and 4F demonstrate that inhibition or knockdown of arginase decreased the binding of NF-κB to DNA in cells stimulated with TNF-α, consistent with repression of NF-κB reporter gene activity. Similar effects of arginase inhibition on NF-κB–DNA binding were also observed in MTE cells stimulated with LPS (Figure 4G). In addition, incubation of MTE cells with BEC also caused an attenuation of LPS-induced expression of the NF-κB–regulated proinflammatory mediators, KC and CCL20 (Figure 4H).
Figure 4.
Arginase inhibition or knockdown does not alter activation of IKK, but attenuates NF-κB DNA binding and NF-κB transcriptional activity. C10 cells were incubated in the presence of BEC for 24 h (A) or transfected with AI siRNA for 48 h (B), stimulated with TNF-α (10 ng/ml, 5 min), and lysed in kinase buffer. IKK complex was immunopreciptated, and in vitro IKK assay was performed using GST-IκB as the substrate (top panels). Bottom panels are Western blots for IKKγ. Results represent two independent samples for each condition. C10 cells stably transfected with an NF-κB–luciferase reporter vector were incubated with 1 mM BEC or 1 mM l-NAME for 20–24 h (C), or transfected with AI siRNA for 48 h in the presence or absence of l-NAME (D), and subsequently stimulated with 10 ng/ml TNF-α for 6 h. Cells were lysed for determination of luciferase activity. Results are expressed as relative luciferase units (RLU) after normalization to protein content. C10 cells were treated with BEC in the presence or absence of l-NAME for 24 h (E), or transfected with control or AI siRNA (F), as described previously, before stimulation with TNF-α (10 ng/ml) for 4 h, after which nuclear extracts were prepared for assessment of NF-κB–DNA binding, via electrophoretic mobility shift assay (EMSA). DNA-binding complexes containing both RelA and p50 are shown. Note that the EMSA shown in (E) was not resolved sufficiently to visualize individual RelA/p50 complexes. MTE cells were treated with BEC or l-NAME in the presence or absence of 1 μg/ml LPS for 4 h for evaluation of NF-κB–DNA binding by EMSA (G) or assessment of the NF-κB–driven genes, KC (ng/ml; shaded bars) or CCL20 (pg/ml; solid bars), by ELISA (H). *P < 0.05 compared with sham; ANOVA. All experiments were repeated at least three times.
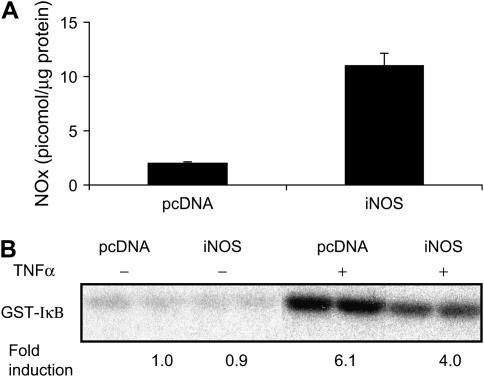
Because the inhibitory effects of NF-κB after suppression of arginase in MTE cells were reversed in the presence of l-NAME, they point to an important role for NO in the attenuation of NF-κB or mediator production in lung epithelial cells. As AII is also expressed in C10 cells (Figure 1A), we conducted experiments in which we simultaneously knocked down AI and AII. Intriguingly, this strategy did not lead to further increases in NOx content or NF-κB inhibition, and results were indistinguishable from those observed after knockdown of AI alone (data not shown). Our current data, showing no effect of increased NOx content and S-nitrosated proteins after suppression of arginase on IKKβ activity, are in apparent contrast with our previous observations demonstrating that exogenously added S-nitrosothiols effectively inhibited IKKβ in C10 cells. However, it is likely that cellular SNO levels were higher in the previous study compared with the mild increase in NOx content achieved after arginase inhibition. Indeed, overexpression of iNOS in C10 cells caused a marked increase in cellular content of NOx (Figure 5A), and resulted in a marked attenuation of TNF-α–stimulated IKKβ activity (Figure 5B), consistent with our previous data (11).
Figure 5.
Overexpression of iNOS causes robust increases in NOx content and inhibits the ability of TNF-α to stimulate IKK activity in C10 lung epithelial cells. (A) C10 cells were transiently transfected with iNOS cDNA for 24 h before assessment of cellular NOx, as described previously. (B) C10 cells were transiently transfected with iNOS plasmid for 24 h, and stimulated with TNF-α (10 ng/ml) for 5 min before the assessment of IKK via an in vitro kinase assay, as described in Figure 4. All experiments were repeated at least twice.
Arginase Overexpression Decreases NO Content and Increases NF-KB Activity
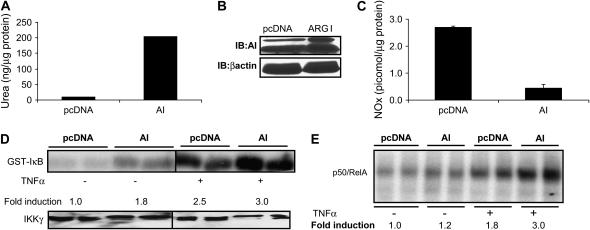
We next investigated whether augmentation of arginase activity would diminish the cellular content of NOx, and, consequently, the ramifications for activation of the NF-κB pathway. Overexpression of AI markedly increased ArgI activity, and expression (Figures 6A and 6B), which was sufficient to decrease steady-state levels of NOx in cells (Figure 6C). Importantly, arginase overexpression increased both basal and TNF-α–stimulated IKK activity (Figure 6D), and enhanced NF-κB binding to its consensus DNA element (Figure 6E).
Figure 6.
Overexpression of AI decreases cellular NOx content and increases NF-κB activity. C10 cells were transiently transfected with ArgI plasmid for 24 h and arginase activity (A), protein expression (B), and NOx content (C) were analyzed in cell lysates, as described in Figures 1 and 2. (D) C10 cells were transfected with AI plasmid and treated with TNF-α (10 ng/ml 5 min), and IKK activity was evaluated as described previously. Bottom panel shows Western blot for IKKγ. (E) C10 cells were transfected with pcDNA or ArgI cDNA and treated with TNF-α (10 ng/ml) for 4 h, at which time, nuclear extracts were prepared for evaluation of NF-κB–DNA binding via EMSA, as described previously. DNA-binding complexes containing both RelA and p50 are shown. Results represent two independent samples for each condition. All experiments were repeated at least three times.
Modulation of Arginase Affects S-Nitrosation of p50
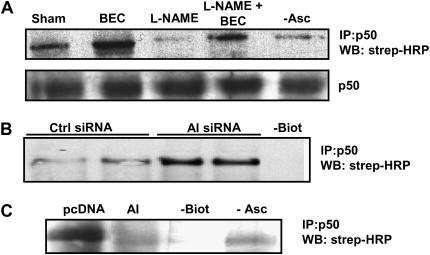
Previous studies demonstrated that S-nitrosation of cysteine 62 within the p50 subunit of NF-κB prevents DNA binding activity (10). We therefore investigated whether modulation of NF-κB activity by manipulation of arginase was associated with altered S-nitrosation of p50, based upon the indirect biotin switch method. This method employs ascorbate-dependent reduction, and subsequent biotin labeling of S-nitrosated proteins. In the present experiments, we used 1 mM ascorbate to avoid possible reduction of disulfide bonds (30, 31). Results in Figure 7A demonstrate baseline S-nitrosation of p50. Inhibition of arginase by BEC enhanced S-nitrosation of p50, whereas the NOS inhibitor, l-NAME, resulted in decreased S-nitrosation of p50. Importantly, l-NAME prevented the BEC-induced increases in S-nitrosation of p50, consistent with the previously demonstrated effect of l-NAME on restoring NF-κB transcriptional activity (Figures 4C and 4D). AI knockdown via siRNA led to similar increase of p50 S-nitrosation (Figure 7B), whereas overexpression of AI markedly decreased p50 S-nitrosation (Figure 7C). In aggregate, these data demonstrate that arginase regulates the NF-κB pathway, and suggests an important physiologic role of NO herein, potentially through the control of S-nitrosation of p50 and, consequently, interference with the ability of NF-κB to bind DNA.
Figure 7.
Arginase activity controls S-nitrosation of p50. (A) C10 cells were treated with 1 mM BEC in the presence or absence of l-NAME (1 mM) for 24 h, (B) transfected with AI siRNA for 48 h, or (C) transfected with AI plasmid. The biotin switch method was performed to assess S-nitrosation of p50; p50 was immunopreciptated (IP) and biotinylated p50 was detected by Western blot using streptavidin- conjugated HRP (strep-HRP). In (A), lower panel shows Western blot loading control for p50 in whole cell lysates. As reagent controls, the biotin label (−Biot), or ascorbate (−Asc) were omitted. All experiments were repeated at least three times.
DISCUSSION
Investigation into the functional significance of NF-κB activation in airway epithelia has illuminated its importance in the pathophysiology of inflammatory lung disease (20–22). Therefore, understanding the molecular signals and mediators that control the strength of activation of NF-κB will be a critical endeavor toward the identification of strategies aimed at limiting inflammation. NO has been shown to exert an antiinflammatory role in lung epithelial cells through inhibition of the NF-κB pathway, and attenuation of NOS function has the potential to augment NF-κB activation (10, 11). Competition of arginase and NOS for the same substrate, L-arginine, is a well known scenario in myeloid cells (16, 38) and raises the intriguing possibility that arginase can affect NO function (39, 40) and, consequently, inflammation. Arginase competition with NOS may be particularly relevant in allergic airways diseases, where prominent arginase upregulation and chronic inflammation occur. In support of a regulatory role of arginase in the signaling function of NO, arginase activity has been demonstrated to increase airway hyperresponsiveness and attenuate airway or vessel relaxation by inhibition of NO (34, 41, 42) and competition with nNOS (43). Furthermore, in experimental autoimmune encephalitis in which AI was upregulated, its inhibition resulted in delayed onset of disease, reduced disease severity, and expedited recovery in association with enhanced NO production and reduced secretion of cytokines (44).
In the present study, we demonstrate that cellular arginase activity is critical in the control of the NF-κB pathway, and have provided evidence that this may occur through alterations in the NO content produced by the basal activity of constitutive NOS. Our results demonstrate that cellular increases in NOx after arginase suppression inhibit the NF-κB pathway downstream of IKK, likely by inhibiting the ability of NF-κB to bind to DNA, as a result of S-nitrosation of p50. These findings are in line with those of another report in epithelial cells, demonstrating that S-nitrosothiols did not interfere with degradation of IκBα or nuclear accumulation of NF-κB, but inhibited the ability of NF-κB to bind DNA (45, 46). S-nitrosation of cysteine 62 of the p50 subunit of NF-κB was previously shown to inhibit the ability of NF-κB to bind DNA (45, 47), and provides a likely explanation for the attenuation of NF-κB seen in our study. We cannot, however, rule out the possibility that other components of the NF-κB pathway are targets for S-nitrosation, or that oxidative modifications other than S-nitrosation are caused by the changes in NOx content, and thus account for the modulation of NF-κB shown in the present study. We recently reported, using the same cell line that was used in the present study, that exogenous S-nitrosothiols cause S-nitrosation of IKKβ, inhibiting NF-κB proximally in its pathway of activation (11). In contrast, our present study demonstrates that increases in S-nitrosothiols after arginase inhibition do not affect the extent of IKK activation, but attenuate the NF-κB pathway downstream of IKK. This apparent inconsistency may stem from potentially different chemical forms of NO, concentration, or subcellular localization that arose after arginase suppression in the present study compared with extracellularly delivered S-nitrosothiols, which require uptake through the L-Cys amino acid transporter (48). Furthermore, NO reacts with O2 within the hydrophobic environment ∼ 300-times faster than in an aqueous medium. Therefore, biological membranes and other hydrophobic compartments are important sites for formation of NO-derived reactive molecules (49). In this context, it is noteworthy that the IKK signalsome is recruited from the cytoplasm to the membrane in response to TNF-α stimulation (50), which could promote S-nitrosation of IKK complex by NO encountered via the extracellular milieu. Nonetheless, additional experiments will be needed to formally test the notion that distinct concentrations and chemical forms of NO, and their compartmentalization (51), govern the exact targets for NO critical to inhibition of the NF-κB pathway.
An important aspect of substrate competition between arginases and NOS is their subcellular localization. In the present study, we provided some evidence that AI is associated with eNOS in mouse lung epithelial cell line, indicating that the competition between these enzymes for their common substrate possibly occurs in a specific subcellular domain, although this possibility needs to be formally tested. In agreement with our observations, recent findings demonstrated that mitochondrial AII is colocalized with nNOS, limiting its substrate availability and leading to increasing basal contractility of isolated myocytes (36). In addition to direct substrate competition, the interregulation of NOS and arginase pathways may also occur through the action of downstream products. Ornithine, the direct product of arginase activity, is the substrate for ornithine decarboxylase (ODC), which produces putrescine, the precursor of the polyamines, spermine and spermidine (52). Spermine has been shown to attenuate iNOS protein levels and NO production in response to Helicobacter pylori. In the same report, siRNA-mediated knockdown of ODC increased _H. pylori_–induced iNOS protein expression and NO production (53), indicating the functional significance of the ODC–polyamine pathway in the control of NO. Conversely, evidence that NO affects the arginase/polyamine axis is also apparent based upon a report demonstrating that NO inhibits ODC via S-nitrosation of cysteine 360 in the active site of the enzyme (54). In addition, cytokines can also influence the balance between NOS and arginase in inflammation. Whereas the T helper cell (Th) type 1 cytokine, IFN-γ, induces iNOS, the Th2 cytokines IL-4 and IL-10 induce arginase (14, 33, 55–57). In aggregate, these data demonstrate a number of cross-regulatory events between NOS and arginase pathways that regulate the strength of activation of either pathway. Although the functional ramification of arginase and NOS induction and their reciprocal regulation likely depend on the disease process, cytokine milieu, and cell type, these studies collectively support the concept that the balance of their activation may be critical in the pathophysiology of inflammatory diseases, and suggest a role of NO therein.
In contrast to the lack of effect on IKK activity observed after arginase suppression, arginase overexpression was sufficient to activate IKK and increase NF-κB DNA binding. Arginase overexpression caused substantial decreases in the cellular NOx content, which might affect basal S-nitrosation of IKK sufficiently to promote activation, analogous to our previous observations in Jurkat T cells, where inhibition of NOS was sufficient to mediate IKK activation (11). In contrast, arginase inhibition or knockdown resulted in less robust increases in cellular NOx content, in agreement with previous studies demonstrating that intracellular concentrations of arginine are already above the _K_m for NOS activity (58). Additional increases of intracellular L-arginine after arginase suppression would therefore be anticipated to cause only mild increases in NOS activity, explaining the lack of effects on IKK. To illuminate these discrepancies, it will be critical to implement strategies to determine subcellular NO levels and chemical forms and quantify their changes after arginase or NOS manipulation, as mentioned previously here.
Another consideration is that consumption of L-arginine after arginase overexpression could uncouple NOS, leading to production of superoxide in addition to NO, and, consequently, the generation of highly reactive species, such as peroxynitrite (15, 41, 59), which may affect the NF-κB pathway in certain cell types (60, 61). Finally, the possibility has to be considered that the robust expression of functional arginase leads to the formation of its products, urea and ornithine (62), which may contribute to the augmentation of NF-κB activity, independent of NO. In support of this possibility, recent reports have demonstrated that polyamines increase NF-κB binding and activity in breast cancer cells (63, 64), although effects of the products of arginase catalytic activity on IKKβ remain to be determined. Based upon our data demonstrating increases in IKK and decreases in S-nitrosation of p50 after arginase overexpression, it is clear that multiple mechanisms are operative to enhance activation of NF-κB.
In conclusion, our study demonstrates that indirect manipulation of NO content, through modulation of arginase activity, represents an important biochemical mechanism to regulate the activity of NF-κB. These findings have implications for the pathophysiology of chronic inflammatory conditions, such as asthma (19), rheumatoid arthritis (65), psoriasis (66), and cardiovascular disease (34), where NF-κB and arginase activation have been documented.
Acknowledgments
The authors thank Pamela Vacek for statistical analysis.
This work was supported by National Institutes of Health grants RO1 HL60014 and RO1 HL074295 (Y.J.H.); NCRR COBRE P20 RR 15557.
Originally Published in Press as DOI: 10.1164/rccm.2006-0329SM on January 11, 2007
Conflict of Interest Statement: Y.J.H. received $1,500 from Sepracor for a scientific presentation in 2005. Y.J.H., K.C., and A.V. are listed as inventors on a patent application to detect S-nitrosylated proteins.
References
- 1.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol 2000;18:621–663. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature 2003;423:655–659. [DOI] [PubMed] [Google Scholar]
- 3.Hall G, Singh IS, Hester L, Hasday JD, Rogers TB. Inhibitor-κB kinase-β regulates LPS-induced TNF-α production in cardiac myocytes through modulation of NF-κB p65 subunit phosphorylation. Am J Physiol Heart Circ Physiol 2005;289:H2103–H2111. [DOI] [PubMed] [Google Scholar]
- 4.Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR. Identification of direct genomic targets downstream of the nuclear factor-κB transcription factor mediating tumor necrosis factor signaling. J Biol Chem 2005;280:17435–17448. [DOI] [PubMed] [Google Scholar]
- 5.Conner EM, Grisham MB. Inflammation, free radicals, and antioxidants. Nutrition 1996;12:274–277. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Chen F. Reactive oxygen species (ROS), troublemakers between nuclear factor-κB (NF-κB) and c-Jun NH(2)-terminal kinase (JNK). Cancer Res 2004;64:1902–1905. [DOI] [PubMed] [Google Scholar]
- 7.Stamler JS, Lamas S, Fang FC. Nitrosylation: the prototypic redox-based signaling mechanism. Cell 2001;106:675–683. [DOI] [PubMed] [Google Scholar]
- 8.Mannick JB, Schonhoff CM. Nitrosylation: the next phosphorylation? Arch Biochem Biophys 2002;408:1–6. [DOI] [PubMed] [Google Scholar]
- 9.Marshall HE, Merchant K, Stamler JS. Nitrosation and oxidation in the regulation of gene expression. FASEB J 2000;14:1889–1900. [DOI] [PubMed] [Google Scholar]
- 10.Marshall HE, Stamler JS. Inhibition of NF-κ B by S-nitrosylation. Biochemistry 2001;40:1688–1693. [DOI] [PubMed] [Google Scholar]
- 11.Reynaert NL, Ckless K, Korn SH, Vos N, Guala AS, Wouters EF, van der Vliet A, Janssen-Heininger YM. Nitric oxide represses inhibitory κB kinase through S-nitrosylation. Proc Natl Acad Sci USA 2004;101:8945–8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ash DE. Structure and function of arginases. J Nutr 2004;134:2760S–2764S. (discussion 2765S–2767S). [DOI] [PubMed] [Google Scholar]
- 13.Yu H, Yoo PK, Aguirre CC, Tsoa RW, Kern RM, Grody WW, Cederbaum SD, Iyer RK. Widespread expression of arginase I in mouse tissues: biochemical and physiological implications. J Histochem Cytochem 2003;51:1151–1160. [DOI] [PubMed] [Google Scholar]
- 14.Modolell M, Corraliza IM, Link F, Soler G, Eichmann K. Reciprocal regulation of the nitric oxide synthase/arginase balance in mouse bone marrow–derived macrophages by Th1 and Th2 cytokines. Eur J Immunol 1995;25:1101–1104. [DOI] [PubMed] [Google Scholar]
- 15.Bronte V, Serafini P, Mazzoni A, Segal DM, Zanovello P. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol 2003;24:302–306. [DOI] [PubMed] [Google Scholar]
- 16.Chicoine LG, Paffett ML, Young TL, Nelin LD. Arginase inhibition increases nitric oxide production in bovine pulmonary arterial endothelial cells. Am J Physiol Lung Cell Mol Physiol 2004;287:L60–L68. [DOI] [PubMed] [Google Scholar]
- 17.Barksdale AR, Bernard AC, Maley ME, Gellin GL, Kearney PA, Boulanger BR, Tsuei BJ, Ochoa JB. Regulation of arginase expression by T-helper II cytokines and isoproterenol. Surgery 2004;135:527–535. [DOI] [PubMed] [Google Scholar]
- 18.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest 2003;111:1863–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King NE, Rothenberg ME, Zimmermann N. Arginine in asthma and lung inflammation. J Nutr 2004;134:2830S–2836S. (discussion 2853S). [DOI] [PubMed] [Google Scholar]
- 20.Poynter ME, Irvin CG, Janssen-Heininger YM. A prominent role for airway epithelial NF-κ B activation in lipopolysaccharide-induced airway inflammation. J Immunol 2003;170:6257–6265. [DOI] [PubMed] [Google Scholar]
- 21.Poynter ME, Cloots R, van Woerkom T, Butnor KJ, Vacek P, Taatjes DJ, Irvin CG, Janssen-Heininger YM. NF-κ B activation in airways modulates allergic inflammation but not hyperresponsiveness. J Immunol 2004;173:7003–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broide DH, Lawrence T, Doherty T, Cho JY, Miller M, McElwain K, McElwain S, Karin M. Allergen-induced peribronchial fibrosis and mucus production mediated by IκB kinase β–dependent genes in airway epithelium. Proc Natl Acad Sci USA 2005;102:17723–17728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu R, Smith D. Continuous multiplication of rabbit tracheal epithelial cells in a defined, hormone-supplemented medium. In Vitro 1982;18:800–812. [DOI] [PubMed] [Google Scholar]
- 24.Corraliza IM, Campo ML, Soler G, Modolell M. Determination of arginase activity in macrophages: a micromethod. J Immunol Methods 1994;174:231–235. [DOI] [PubMed] [Google Scholar]
- 25.Janero DR, Bryan NS, Saijo F, Dhawan V, Schwalb DJ, Warren MC, Feelisch M. Differential nitros(yl)ation of blood and tissue constituents during glyceryl trinitrate biotransformation in vivo. Proc Natl Acad Sci USA 2004;101:16958–16963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhawan V, Schwalb DJ, Shumway MJ, Warren MC, Wexler RS, Zemtseva IS, Zifcak BM, Janero DR. Selective nitros(yl)ation induced in vivo by a nitric oxide-donating cyclooxygenase-2 inhibitor: a NObonomic analysis. Free Radic Biol Med 2005;39:1191–1207. [DOI] [PubMed] [Google Scholar]
- 27.Feelisch M, Rassaf T, Mnaimneh S, Singh N, Bryan NS, Jourd'Heuil D, Kelm M. Concomitant S-, N-, and heme-nitros(yl)ation in biological tissues and fluids: implications for the fate of NO in vivo. FASEB J 2002;16:1775–1785. [DOI] [PubMed] [Google Scholar]
- 28.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001;2001:PL1. (http://stke.sciencemag.org/cgi/content/full/sigtrans;2001/86/pl1, accessed on December 4, 2001.) [DOI] [PubMed]
- 29.Li S, Whorton AR. Regulation of protein tyrosine phosphatase 1B in intact cells by S-nitrosothiols. Arch Biochem Biophys 2003;410:269–279. [DOI] [PubMed] [Google Scholar]
- 30.Huang B, Chen C. An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic Biol Med 2006;41:562–567. [DOI] [PubMed] [Google Scholar]
- 31.Ckless K, Reynaert NL, Taatjes DJ, Lounsbury KM, van der Vliet A, Janssen-Heininger Y. In situ detection and visualization of S-nitrosylated proteins following chemical derivatization: identification of Ran GTPase as a target for S-nitrosylation. Nitric Oxide 2004;11:216–227. [DOI] [PubMed] [Google Scholar]
- 32.Korn SH, Wouters EF, Vos N, Janssen-Heininger YM. Cytokine-induced activation of nuclear factor-κ B is inhibited by hydrogen peroxide through oxidative inactivation of IκB kinase. J Biol Chem 2001;276:35693–35700. [DOI] [PubMed] [Google Scholar]
- 33.Munder M, Eichmann K, Moran JM, Centeno F, Soler G, Modolell M. Th1/Th2-regulated expression of arginase isoforms in murine macrophages and dendritic cells. J Immunol 1999;163:3771–3777. [PubMed] [Google Scholar]
- 34.Berkowitz DE, White R, Li D, Minhas KM, Cernetich A, Kim S, Burke S, Shoukas AA, Nyhan D, Champion HC, et al. Arginase reciprocally regulates nitric oxide synthase activity and contributes to endothelial dysfunction in aging blood vessels. Circulation 2003;108:2000–2006. [DOI] [PubMed] [Google Scholar]
- 35.Scaglia F, Brunetti-Pierri N, Kleppe S, Marini J, Carter S, Garlick P, Jahoor F, O'Brien W, Lee B. Clinical consequences of urea cycle enzyme deficiencies and potential links to arginine and nitric oxide metabolism. J Nutr 2004;134:2775S–2782S. (discussion 2796S–2797S). [DOI] [PubMed] [Google Scholar]
- 36.Steppan J, Ryoo S, Schuleri KH, Gregg C, Hasan RK, White AR, Bugaj LJ, Khan M, Santhanam L, Nyhan D, et al. Arginase modulates myocardial contractility by a nitric oxide synthase 1–dependent mechanism. Proc Natl Acad Sci USA 2006;103:4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziegelbauer K, Gantner F, Lukacs NW, Berlin A, Fuchikami K, Niki T, Sakai K, Inbe H, Takeshita K, Ishimori M, et al. A selective novel low-molecular-weight inhibitor of IκB kinase-β (IKK-β) prevents pulmonary inflammation and shows broad anti-inflammatory activity. Br J Pharmacol 2005;145:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klasen S, Hammermann R, Fuhrmann M, Lindemann D, Beck KF, Pfeilschifter J, Racke K. Glucocorticoids inhibit lipopolysaccharide-induced up-regulation of arginase in rat alveolar macrophages. Br J Pharmacol 2001;132:1349–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cook HT, Jansen A, Lewis S, Largen P, O'Donnell M, Reaveley D, Cattell V. Arginine metabolism in experimental glomerulonephritis: interaction between nitric oxide synthase and arginase. Am J Physiol 1994;267:F646–F653. [DOI] [PubMed] [Google Scholar]
- 40.Rutschman R, Lang R, Hesse M, Ihle JN, Wynn TA, Murray PJ. Cutting edge: Stat6-dependent substrate depletion regulates nitric oxide production. J Immunol 2001;166:2173–2177. [DOI] [PubMed] [Google Scholar]
- 41.Meurs H, McKay S, Maarsingh H, Hamer MA, Macic L, Molendijk N, Zaagsma J. Increased arginase activity underlies allergen-induced deficiency of cNOS-derived nitric oxide and airway hyperresponsiveness. Br J Pharmacol 2002;136:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maarsingh H, Tio MA, Zaagsma J, Meurs H. Arginase attenuates inhibitory nonadrenergic noncholinergic nerve-induced nitric oxide generation and airway smooth muscle relaxation. Respir Res 2005;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maarsingh H, Leusink J, Bos IS, Zaagsma J, Meurs H. Arginase strongly impairs neuronal nitric oxide–mediated airway smooth muscle relaxation in allergic asthma. Respir Res 2006;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu L, Hilliard B, Carmody RJ, Tsabary G, Shin H, Christianson DW, Chen YH. Arginase and autoimmune inflammation in the central nervous system. Immunology 2003;110:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthews JR, Botting CH, Panico M, Morris HR, Hay RT. Inhibition of NF-κB DNA binding by nitric oxide. Nucleic Acids Res 1996;24:2236–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marshall HE, Stamler JS. Nitrosative stress–induced apoptosis through inhibition of NF-κ B. J Biol Chem 2002;277:34223–34228. [DOI] [PubMed] [Google Scholar]
- 47.Matthews JR, Wakasugi N, Virelizier JL, Yodoi J, Hay RT. Thioredoxin regulates the DNA binding activity of NF-κ B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res 1992;20:3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Hogg N. The mechanism of transmembrane S-nitrosothiol transport. Proc Natl Acad Sci USA 2004;101:7891–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Miller MJ, Joshi MS, Thomas DD, Lancaster JR Jr. Accelerated reaction of nitric oxide with O2 within the hydrophobic interior of biological membranes. Proc Natl Acad Sci USA 1998;95:2175–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen G, Cao P, Goeddel DV. TNF-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol Cell 2002;9:401–410. [DOI] [PubMed] [Google Scholar]
- 51.Espey MG, Miranda KM, Thomas DD, Wink DA. Distinction between nitrosating mechanisms within human cells and aqueous solution. J Biol Chem 2001;276:30085–30091. [DOI] [PubMed] [Google Scholar]
- 52.Wu G, Morris SM Jr. Arginine metabolism: nitric oxide and beyond. Biochem J 1998;336:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bussiere FI, Chaturvedi R, Cheng Y, Gobert AP, Asim M, Blumberg DR, Xu H, Kim PY, Hacker A, Casero RA Jr, et al. Spermine causes loss of innate immune response to Helicobacter pylori by inhibition of inducible nitric-oxide synthase translation. J Biol Chem 2005;280:2409–2412. [DOI] [PubMed] [Google Scholar]
- 54.Bauer PM, Buga GM, Fukuto JM, Pegg AE, Ignarro LJ. Nitric oxide inhibits ornithine decarboxylase via S-nitrosylation of cysteine 360 in the active site of the enzyme. J Biol Chem 2001;276:34458–34464. [DOI] [PubMed] [Google Scholar]
- 55.Corraliza IM, Soler G, Eichmann K, Modolell M. Arginase induction by suppressors of nitric oxide synthesis (IL-4, IL-10 and PGE2) in murine bone-marrow–derived macrophages. Biochem Biophys Res Commun 1995;206:667–673. [DOI] [PubMed] [Google Scholar]
- 56.Hesse M, Modolell M, La Flamme AC, Schito M, Fuentes JM, Cheever AW, Pearce EJ, Wynn TA. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J Immunol 2001;167:6533–6544. [DOI] [PubMed] [Google Scholar]
- 57.Iniesta V, Gomez-Nieto LC, Molano I, Mohedano A, Carcelen J, Miron C, Alonso C, Corraliza I. Arginase I induction in macrophages, triggered by Th2-type cytokines, supports the growth of intracellular Leishmania parasites. Parasite Immunol 2002;24:113–118. [DOI] [PubMed] [Google Scholar]
- 58.Lee J, Ryu H, Ferrante RJ, Morris SM Jr, Ratan RR. Translational control of inducible nitric oxide synthase expression by arginine can explain the arginine paradox. Proc Natl Acad Sci USA 2003;100:4843–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noris M, Todeschini M, Cassis P, Pasta F, Cappellini A, Bonazzola S, Macconi D, Maucci R, Porrati F, Benigni A, et al. L-arginine depletion in preeclampsia orients nitric oxide synthase toward oxidant species. Hypertension 2004;43:614–622. [DOI] [PubMed] [Google Scholar]
- 60.Janssen YM, Soultanakis R, Steece K, Heerdt E, Singh RJ, Joseph J, Kalyanaraman B. Depletion of nitric oxide causes cell cycle alterations, apoptosis, and oxidative stress in pulmonary cells. Am J Physiol 1998;275:L1100–L1109. [DOI] [PubMed] [Google Scholar]
- 61.Matata BM, Galinanes M. Peroxynitrite is an essential component of cytokines production mechanism in human monocytes through modulation of nuclear factor-κ B DNA binding activity. J Biol Chem 2002;277:2330–2335. [DOI] [PubMed] [Google Scholar]
- 62.Wei LH, Wu G, Morris SM Jr, Ignarro LJ. Elevated arginase I expression in rat aortic smooth muscle cells increases cell proliferation. Proc Natl Acad Sci USA 2001;98:9260–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shah N, Thomas TJ, Lewis JS, Klinge CM, Shirahata A, Gelinas C, Thomas T. Regulation of estrogenic and nuclear factor κ B functions by polyamines and their role in polyamine analog–induced apoptosis of breast cancer cells. Oncogene 2001;20:1715–1729. [DOI] [PubMed] [Google Scholar]
- 64.Zaletok S, Alexandrova N, Berdynskykh N, Ignatenko N, Gogol S, Orlovsky O, Tregubova N, Gerner E, Chekhun V. Role of polyamines in the function of nuclear transcription factor NF-κB in breast cancer cells. Exp Oncol 2004;26:221–225. [PubMed] [Google Scholar]
- 65.Corraliza I, Moncada S. Increased expression of arginase II in patients with different forms of arthritis: implications of the regulation of nitric oxide. J Rheumatol 2002;29:2261–2265. [PubMed] [Google Scholar]
- 66.Bruch-Gerharz D, Schnorr O, Suschek C, Beck KF, Pfeilschifter J, Ruzicka T, Kolb-Bachofen V. Arginase 1 overexpression in psoriasis: limitation of inducible nitric oxide synthase activity as a molecular mechanism for keratinocyte hyperproliferation. Am J Pathol 2003;162:203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]