Sequence-independent DNA binding and replication initiation by the human origin recognition complex (original) (raw)
Abstract
We report that a highly purified human origin recognition complex (HsORC) has intrinsic DNA-binding activity, and that this activity is modestly stimulated by ATP. HsORC binds preferentially to synthetic AT-rich polydeoxynucleotides, but does not effectively discriminate between natural DNA fragments that contain known human origins and control fragments. The complex fully restores DNA replication to ORC-depleted Xenopus egg extracts, providing strong evidence for its initiator function. Strikingly, HsORC stimulates initiation from any DNA sequence, and it does not preferentially replicate DNA containing human origin sequences. These data provide a biochemical explanation for the observation that in metazoans, initiation of DNA replication often occurs in a seemingly random pattern, and they have important implications for the nature of human origins of DNA replication.
Keywords: Origin recognition complex, DNA-binding specificity, DNA replication, origin of DNA replication
The replicon model, first proposed nearly 40 years ago, postulates that DNA replication initiates from specific chromosomal sequence elements (replicators) that are recognized by positive regulatory proteins (initiators). This model has been particularly useful for understanding the mechanisms that regulate replication of bacterial and viral chromosomes, which generally contain single sites of initiation (Jacob et al. 1963). Although the large chromosomes of eukaryotic cells contain multiple initiation sites, early genetic studies in the yeast Saccharomyces cerevisiae suggested that the replicon model also provides an accurate description of DNA replication in these more complex systems. Transformation studies resulted in the identification of short, well-defined sequence elements (autonomously replicating sequences or ARSs) that function as replicators in yeast chromosomes and plasmids. The characterization of ARS elements ultimately led to the identification of the origin recognition complex (ORC), the initiator protein that recognizes an 11-bp ARS consensus sequence (ACS) in an ATP-dependent manner (Bell and Stillman 1992).
Subsequent studies in other eukaryotes have not confirmed this simple model. Replicators in the fission yeast Schizosaccharomyces pombe are very large (500–1000 bp) and comprise several AT-rich segments that do not contain conserved consensus sequences analogous to the ACS of S. cerevisiae (Kelly and Brown 2000). Attempts to identify stable autonomously replicating sequences in human cells have been largely unsuccessful. This is because any human DNA fragment >15 kb confers on a plasmid the ability to undergo efficient and cell-cycleregulated DNA replication (Krysan et al. 1993). Nevertheless, various studies have mapped ∼20 mammalian origins of bidirectional replication (OBRs or “origins”) using a variety of approaches including 2D gels and nascent strand abundance assays. Some of these origins behave as replicators in that they support replication initiation at ectopic chromosomal locations (Todorovic et al. 1999). Two general paradigms describe known human origins (Gilbert 2001). In the first group, initiation is localized to a relatively discrete chromosomal location. This class includes the human β_-globin_ locus, where replication initiates within a <4-kb region between the adult δ_-globin_ and β_-globin_ genes, as well as the human lamin B2 locus, where the transition between continuous and discontinuous strand synthesis can be localized to a single nucleotide (Aladjem et al. 1995;Abdurashidova et al. 2000;Cimbora et al. 2000). The second class of origins consists of large zones of initiation, 10–50-kb regions where replication begins from multiple dispersed sites. The Chinese hamster ovary cell DHFR locus typifies this group, and consists of a 55-kb intergenic region that has recently been shown to contain a minimum of 20 initiation sites that are used with different efficiencies (Dijkwel et al. 2002 and references therein). Despite the discovery of these mammalian origins, no consensus DNA sequence emerges among them. Additionally, only three described human origins, including the lamin B2 origin, the PRKDC/MCM4 intergenic region, and the region upstream of the human TOP1 gene, have been shown to interact with ORC in vivo (Keller et al. 2002;Ladenburger et al. 2002). Lack of a unifying sequence or conclusive evidence for specific ORC binding at human origins illustrates the difficulty in the characterization of human replicators.
In the early embryonic cell cycles of Xenopus laevis and_Drosophila melanogaster_, DNA replication initiates in a sequence-independent manner, and this feature is retained in replication-competent Xenopus egg extracts (Blow and Laskey 1986;Newport 1987;Hyrien and Mechali1992,1993;Mahbubani et al. 1992;Walter et al. 1998;Sasaki et al. 1999). Two fundamentally distinct models can explain sequence-independent initiation. In one, metazoan ORCs have no intrinsic sequence specificity, and the observation that ORCs are not randomly distributed in somatic cells is attributable to the effects of chromatin structure, or because ORC is recruited to DNA via additional proteins that are sequence-specific. Alternatively, it may be that ORC does have intrinsic affinity for a specific replicator sequence. In this case, the lack of sequence-specificity in embryonic systems might reflect the very high concentration of the complex present in embryos (Rowles et al. 1996;Walter and Newport 1997), or an embryo-specific modification of ORC that renders it sequence-nonspecific. Importantly, recent data indicate that in_Xenopus_ egg extracts, each ORC recruits many initiation-competent MCM2–7 complexes that become widely distributed on chromatin surrounding ORC (Edwards et al. 2002). Thus, if replication initiates stochastically from these distributed MCM complexes, initiation would appear sequence-independent even if ORC were bound site-specifically. In summary, it remains unclear whether the mechanism of DNA binding by ORC in somatic and embryonic cells is fundamentally the same or different.
A better understanding of the physical properties of ORC is expected to advance our understanding of metazoan origins. Although most of what is known about ORC comes from the S. cerevisiae complex, significant progress has been made in the characterization of the Drosophila and S. pombe ORCs. As described above, S. pombe replicators are composed of asymmetric AT-rich sequence motifs. These AT-rich stretches are bound by the S. pombe origin recognition complex (SpORC), and this binding is mediated through a unique N-terminal DNA-binding domain found in SpOrc4 that contains nine copies of the HMG-I (Y)-related AT-hook motif (Chuang and Kelly 1999). SpORC interacts through this domain with multiple sites in a known S. pombe origin with similar affinity, which suggests that SpORC binding to S. pombe replicators is caused by the collective effect of multiple AT-rich binding sites (Kong and DePamphilis 2001;Lee et al. 2001;Chuang et al. 2002). This mode of replicator recognition is distinct from that of the S. cerevisiae origin recognition complex (ScORC), which recognizes one ACS element per replicator. Unlike ScORC, SpORC DNA binding is not ATP-dependent (Kong and DePamphilis 2001;Lee et al. 2001;Chuang et al. 2002;see also Takahashi and Masukata 2001). The D. melanogaster origin recognition complex (DmORC) has been shown in vivo and in vitro to bind the AT-rich chorion gene amplification control elements ACE3 and_ori-β, but no distinct ARS-like element has been determined at these sites. Additionally, DmORC appears to require ATP for binding to_ACE3 (Austin et al. 1999; Chesnokov et al. 2001).
Despite large differences in the sequence composition of eukaryotic replicators, the biochemical events underlying replication initiation are highly conserved throughout eukaryotes (for review, seeBell and Dutta 2002). The combined work in S. cerevisiae, Xenopus, S. pombe, and_Drosophila_ has shown that in G1, chromatin-bound ORC recruits the initiation factors cdc6, cdt1, and the hexameric MCM2–7 complex, the putative replicative DNA helicase (Labib and Diffley 2001). Together these proteins comprise the prereplication complex (pre-RC). As cells progress from G1 into S phase, the pre-RC is activated by CDK (cyclin-dependent kinase) and Cdc7 (Dbf4-dependent kinase), leading to origin unwinding and recruitment of DNA polymerases (Takisawa et al. 2000). Although the chromatin-binding properties of mammalian ORC and MCM2–7 in the cell cycle are consistent with the above paradigm (for review, seeFujita 1999;Mendez and Stillman 2000;Natale et al. 2000; Okuno et al. 2001;Sun et al. 2002), formal proof is lacking to show a direct role for the human origin recognition complex (HsORC) in biochemically initiating replication from cellular origins.
A biochemical analysis of HsORC will be valuable in determining how this protein selects replication initiation sites. Human ORC has been reconstituted by coinfection of insect cells with viruses encoding its subunits (Dhar et al. 2001a;Vashee et al. 2001). Using a variation of this approach, we prepared highly purified HsORC containing all six subunits. In filter binding assays, this complex bound to a DNA fragment from the human lamin B2 origin, and the binding was modestly stimulated by ATP. In competition experiments using synthetic deoxynucleotide polymers, HsORC bound better to poly(dA) · poly(dT) than to poly(dG) · poly(dC). However, HsORC did not effectively discriminate between naturally occurring DNA fragments, including AT-rich DNA derived from the lamin B2 and DHFR ori_β origins, and control DNA fragments from prokaryotic sources with significantly lower AT content. To examine whether the purified HsORC was active, we assayed its function in a soluble cell-free DNA replication system derived from_Xenopus eggs. We show that the purified HsORC fully restored DNA replication in extracts depleted of the endogenous X. laevis ORC (XlORC) complex. We further found that HsORC replicated several different plasmids of prokaryotic composition, and that it formed pre-RCs on randomly chosen prokaryotic sequences. Finally, even when HsORC was used in limiting concentrations, it did not preferentially replicate DNA templates containing known human origins. These data can explain why any DNA fragment serves as a replicator in human cells (Krysan et al. 1993), and why initiation events occur in zones at some loci. We discuss a model in which all initiation events, even those that are highly localized, are stimulated by an ORC that has little intrinsic sequence-specificity.
Results
Purification of recombinant human ORC
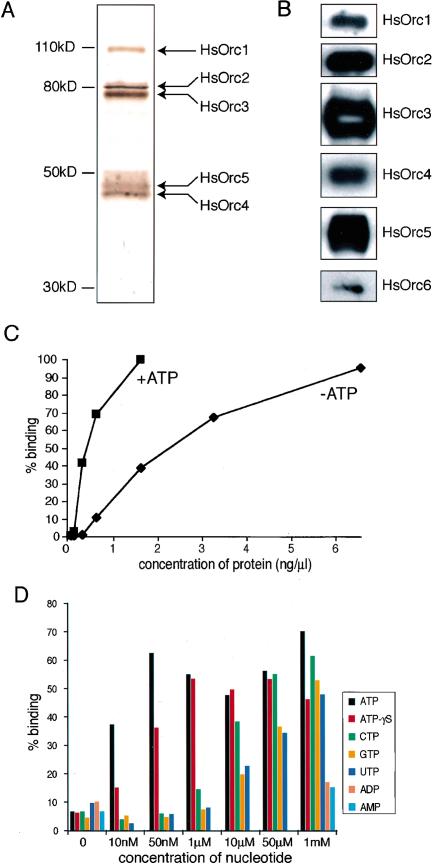
In an effort to better characterize human ORC, we first sought to purify the complex to homogeneity. To do this, we expressed the HsOrc2 subunit, epitope-tagged with the hemagluttanin (HA) and hexahistidine (His6) moieties, and the other five HsORC subunits in Sf9 insect cells using the baculovirus system (Vashee et al. 2001). Recombinant HsORC was solubilized from a chromatin-enriched fraction using 0.5 M NaCl and purified by sequential affinity chromatography on Ni-agarose and F7 anti-HA antibody-agarose. The eluate from the F7 anti-HA antibody-agarose was analyzed by SDS/PAGE and silver staining (Fig. 1A). The purified preparation contained six proteins with mobilities consistent with the molecular weights (MW) of the HsOrc1–HsOrc6 subunits (Fig. 1A). The HsOrc2–HsOrc5 subunits stained with similar intensities, suggesting that they were present at comparable levels in this preparation. The HsOrc1 subunit stained with lower intensity, suggesting that it was present at substoichiometric amounts. Analysis of a number of independent preparations of purified HsORC revealed that the relative amount of HsOrc1 varied from preparation to preparation and was consistently less than the amounts of HsOrc2–HsOrc5. HsOrc6 also stained weakly and was only observed when the other subunits were overstained (Fig. 1A;see also Fig. 4A, below), suggesting that HsOrc6, like HsOrc1, was present in substoichiometric amounts in our purified recombinant HsORC. The presence of HsOrc6, as well as the other HsORC subunits, was confirmed by Western blot analysis (Fig. 1B). In previous studies, we and others have shown that a number of stable subcomplexes can be formed from HsOrc2–HsOrc5 subunits expressed in insect cells (Dhar et al. 2001a;Vashee et al. 2001). Therefore, it is likely that our preparations of purified HsORC contain such subcomplexes, in addition to the holocomplex comprising all six subunits.
Figure 1.
Purification of recombinant HsORC and DNA binding by HsORC. (A) Composition of purified recombinant HsORC. HsORC was purified from a chromatin-enriched extract by Ni-agarose and anti-HA antibody affinity chromatography. The purified HsORC was analyzed by SDS-PAGE, and the individual subunits were detected by silver staining. (B) Purified HsORC was analyzed by SDS-PAGE and Western blotting with subunit-specific antibodies. (C) Titration of protein. Recombinant HsORC at the indicated concentrations was incubated with 0.5 nM radiolabeled lamin B2 ori II probe in the presence or absence of 1 mM ATP. The amount of radioactive lamin B2 ori II DNA bound by HsORC was determined by the nitrocellulose filter-binding assay. (D) Recombinant HsORC prefers ATP but does not require hydrolysis for DNA-binding activity. Nitrocellulose assays were performed as described in A except that 0.75 ng/μL recombinant HsORC and the indicated concentrations of various nucleotides were used.
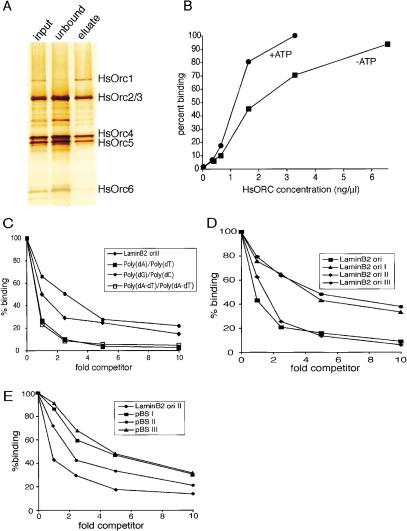
Figure 4.
Properties of DNA-affinity-purified HsORC. (A) DsDNA affinity purification of HsORC. HsORC prepared as inFigure 1A was further purified by adsorption to magnetic beads containing immobilized DNA followed by elution with 500 mM NaCl as in Figure 3B. SDS-PAGE was used to analyze 1 μL of input, 2 μL of unbound, and 1 μL of eluted fractions, and the individual subunits were detected by silver staining. (B) Titration of protein. DNA-affinity-purified HsORC at the indicated concentrations was incubated with 0.5 nM radiolabeled lamin B2 ori II probe in the presence or absence of 1 mM ATP. The amount of radioactive lamin B2 ori II DNA bound by HsORC was determined by the nitrocellulose filter-binding assay. (C_–_E) Competition experiments. DNA-affinity-purified HsORC was incubated with a fixed quantity of radiolabeled lamin B2 ori II probe and various amounts of different competitor DNAs in the absence of ATP. Similar results were obtained in the presence of ATP. (C) Competition assays with synthetic polynucleotides were performed as described forFigure 2A. (D) Competition assays with human lamin B2 origin DNA fragments were performed as in Figure 3A. (E) Competition assays with nonorigin DNAs were performed as inFigure 3C.
Recombinant human ORC has intrinsic DNA-binding activity
To examine whether recombinant HsORC is able to bind DNA, we carried out a series of nitrocellulose-filter binding assays with a 450-bp fragment of duplex DNA containing the human lamin B2 origin region (lamin B2 II). This fragment encompasses potential start sites for leading-strand DNA synthesis as recently described by Falaschi and coworkers (Abdurashidova et al. 2000).Figure 1C shows an example of one such assay in which various concentrations of purified recombinant HsORC were incubated with 0.5 nM32P-labeled lamin B2 II DNA probe in the presence or absence of ATP. In the presence of ATP, half-maximal binding to the labeled probe was observed at 0.4 ng/μL HsORC. Although the precise concentration of active complexes in this preparation of HsORC is unknown, the data indicate that HsORC binds to lamin B2 II with at least nanomolar affinity (see also Fig. 4, below). HsORC also bound tightly to the probe in the absence of ATP, but its apparent affinity for DNA was approximately one-fifth of that observed in the presence of ATP. The stimulation of the DNA-binding activity of HsORC by ATP was observed in multiple experiments with a variety of DNA molecules. The magnitude of the stimulatory effect ranged from three- to fivefold. This behavior is unlike that of the S. cerevisiae or S. pombe ORCs; the former is completely dependent on ATP for DNA binding (Klemm et al. 1997), whereas the latter exhibits no dependence on ATP (Kong and DePamphilis 2001;Lee et al. 2001;Chuang et al. 2002).
We tested several different nucleotides at a wide range of concentrations for their ability to stimulate the binding of recombinant HsORC to the32P-labeled lamin B2 II DNA probe in nitrocellulose-filter binding assays (Fig. 1D). Under the conditions of our experiment, ATP stimulated binding at all concentrations tested, even as low as 10 nM. In contrast, CTP, GTP, and UTP stimulated DNA binding only at concentrations of 10 μM and higher. Neither ADP nor AMP stimulated the ability of HsORC to bind the probe, even at concentrations as high as 1 mM. These data indicate that recombinant HsORC prefers ATP at least several hundredfold more than other nucleotides. We also observed that a nonhydrolyzable analog of ATP, ATPγS, was able to stimulate the DNA-binding activity of HsORC at concentrations of 50 nM and higher, suggesting that ATP hydrolysis is not required for stimulation (Fig. 1D). These results are in agreement with those previously reported for other ORCs that are dependent on ATP for DNA-binding activity (Klemm et al. 1997;Chesnokov et al. 2001).
Recombinant HsORC preferentially binds to AT-rich sequences
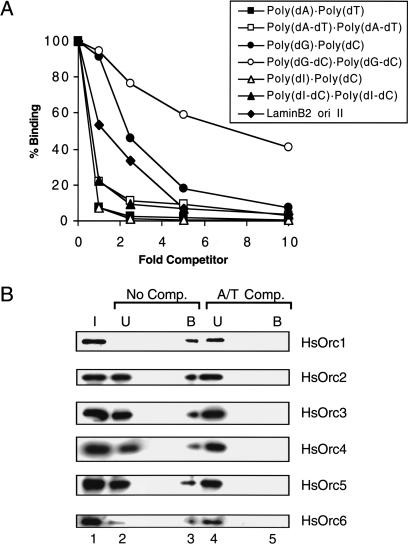
To better understand the DNA-binding properties of recombinant HsORC, we determined the ability of different synthetic polynucleotides to compete for binding of HsORC to the32P-labeled lamin B2 II DNA probe. The relative binding affinity of HsORC for each competitor DNA was estimated from the DNA concentration required to reduce binding of the labeled probe by 50% in the nitrocellulose filter-binding assay. The results of representative competition experiments are shown in Figure 2A. When unlabeled lamin B2 II DNA was used as competitor, we observed the expected 50% reduction of HsORC binding to the labeled probe at a competitor-:probe ratio of ∼1:1 (Fig. 2A, diamonds). Polynucleotides containing exclusively A and T residues, such as poly(dA) · poly(dT) or poly(dA-dT) · poly(dA-dT), were more effective competitors than lamin B2 II DNA (Fig. 2A, closed and open squares). Based on the results of multiple assays, we estimated that the affinities of recombinant HsORC for poly(dA) · poly(dT) and poly(dA-dT) · poly(dA-dT) are ∼45- and 5-fold greater, respectively, than for the lamin B2 II DNA probe. In contrast, the relative binding affinities of HsORC for poly(dG) · poly(dC) and poly(dG-dC) · poly(dG-dC) were approximately three- and eightfold lower, respectively, than for the lamin B2 II DNA probe (Fig. 2A, closed and open circles). Interestingly, the competition data for poly(dI) · poly(dC) and poly(dI-dC) · poly(dI-dC) were very similar to those obtained for poly(dA) · poly(dT) and poly(dA-dT) · poly(dA-dT), respectively (Fig. 2A, cf. triangles and squares). Because the minor grooves of poly(dI) · poly(dC) and poly(dA) · poly(dT) are structurally similar, this finding suggests that minor groove contacts play an important role in determining the DNA-binding properties of recombinant HsORC.
Figure 2.
Recombinant HsORC preferentially binds to AT-rich sequences. (A) Competition with synthetic polynucleotide duplex DNA. Fixed amounts of recombinant HsORC and radioactive lamin B2 ori II DNA were incubated with various amounts of competitor polynucleotide duplex DNA in the presence of ATP. The fraction of input radiolabeled lamin B2 ori II DNA bound to HsORC was determined by the nitrocellulose filter-binding assay. The fold competitor is in terms of concentration of nucleotides. (B) Recombinant HsORC binds to poly(dA) · poly(dT). Recombinant HsORC was incubated with oriP DNA conjugated to magnetic beads in the absence (No Comp.) or presence (A/T Comp.) of poly(dA) · poly(dT). The bound proteins were analyzed by SDS-PAGE and detected by Western blotting with antibodies specific for each HsORC subunit. Input material (I), unbound fraction (U), and bound fraction (B) are indicated.
To confirm that the observed DNA-binding activity was caused by recombinant HsORC and not to a contaminating protein in our preparation, we carried out a competition experiment in which we monitored the distribution of protein, rather than DNA, in the bound and unbound fractions. For this purpose, DNA was immobilized on magnetic beads and incubated with recombinant HsORC in the presence or absence of a 10-fold excess of poly(dA) · poly(dT) as competitor. After removal of the unbound protein, the HsORC subunits bound to the immobilized DNA were recovered in SDS and detected by Western blotting (Fig. 2B, lane 3). In the absence of competitor DNA, all of the HsOrc1 and most of the HsOrc6 in the purified HsORC preparation was recovered in the DNA bound fraction (Fig. 2B, cf. lanes 1,2,3). Significant amounts of the HsOrc2–HsOrc5 subunits were also recovered in the bound fraction, but a large proportion of each of these subunits remained in the unbound fraction (Fig. 2B, cf. lanes 2 and 3). This observation is consistent with the possibility, discussed above, that our purified HsORC preparations contain subcomplexes of HsOrc2–HsOrc5, as well as holocomplexes of all six subunits. The data suggest that holocomplexes bind preferentially to DNA. When an excess of poly(dA) · poly(dT) was added to the binding reaction, all six HsORC subunits were recovered exclusively in the unbound fraction (Fig. 2B, cf. lanes 4 and 5). This finding demonstrates conclusively that recombinant HsORC is capable of binding to poly(dA) · poly(dT) with high affinity and also provides confidence in the validity of the results obtained by nitrocellulose-filter binding assays.
HsORC does not effectively discriminate between origin DNA and control DNA
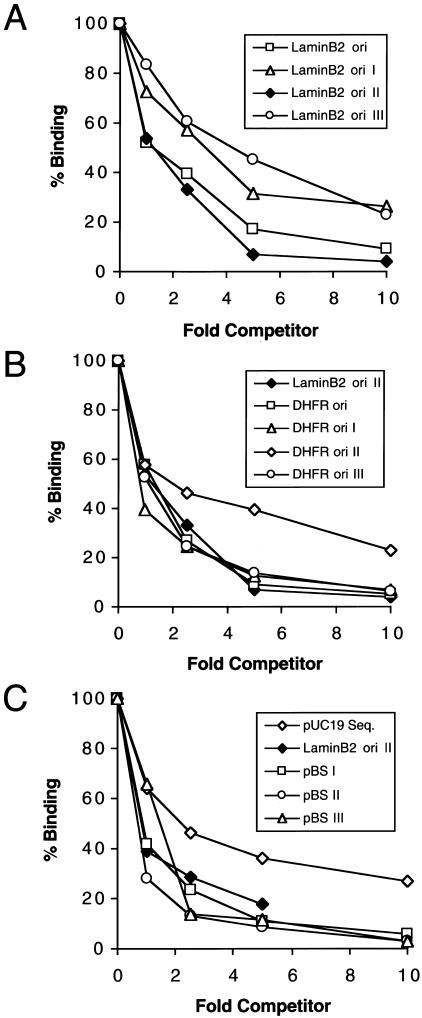
Several characterized human origins are AT-rich and contain long stretches of As and Ts (AT tracts). Because HsORC exhibited a preference for synthetic polynucleotides that were AT-rich, we wondered whether HsORC interacted preferentially with the AT-rich sequences present in origins. We therefore compared the abilities of various DNA fragments derived from mammalian origins to compete for binding of HsORC to the 32P-labeled lamin B2 II DNA probe. In the first set of experiments, we studied a 2-kb region surrounding the start site of DNA replication near the lamin B2 gene (lamin B2 ori). The complete 2-kb lamin B2 origin fragment, as well as three subfragments, were tested as competitors (Fig. 3A). One subfragment (lamin B2 II) bound HsORC with approximately the same affinity as the complete lamin B2 origin (Fig. 3A, cf. squares and diamonds). The other two subfragments (lamin B2 I and lamin B2 III) exhibited slightly lower (threefold to fourfold) affinities for HsORC (Fig. 3A, triangles and circles). The relative binding affinities of the subfragments were roughly correlated with their respective AT contents (51% AT for lamin B2 II, 41% AT for lamin B2 I, and 32% AT for lamin B2 III). Similar experiments were performed with the Chinese hamster DHFR oriβ. The DNA fragments derived from the DHFR origin had relatively high AT contents (59%–63%). All were excellent competitors (Fig. 3B), and their relative affinities for HsORC fell within a narrow (twofold) range. One of these fragments, fragment DHFR ori I (61% AT), contains four long A tracts, but it did not compete better than fragment DHFR ori III (63% AT) which has no A tracts (Fig. 3B, cf. triangles and circles). Based on these results, it appears that AT tracts do not confer preferential binding, whereas the overall AT content may have a modest effect.
Figure 3.
Recombinant HsORC binds DNA with little sequence specificity. Recombinant HsORC was incubated with a fixed quantity of radiolabeled lamin B2 ori II probe and various amounts of different competitor DNAs in the presence of ATP as indicated. The extent of binding of the radioactive lamin B2 ori II DNA was measured by the nitrocellulose filter-binding assay. As inFigure 2, the fold competitor is in terms of concentration of nucleotides. (A) Competition with human lamin B2 origin DNA. Overlapping fragments that constitute the human lamin B2 origin were generated by PCR and used as competitors. (B) Competition with Chinese hamster DHFR origin DNA. Fragments that span DHFR oriβ were synthesized by PCR and used as competitors. (C) Competition with nonorigin DNA. A fragment of pUC19 and three fragments of pBS (I, II, and III) were used as competitors.
To extend this analysis, we also tested the ability of prokaryotic sequences to compete for HsORC binding.Figure 3C shows the results obtained with several fragments derived from the plasmids pBluescript II KS+ and pUC19. The relative affinities of these fragments for HsORC fell within a fourfold range. Interestingly, two of the prokaryotic DNA fragments, pBS I (45% AT) and pBS II (52% AT), competed for HsORC binding as well or better than the lamin B2 II origin fragment (51% AT;Fig. 3C, cf. squares, circles, filled diamonds). In addition, using lamin B2 ori II as the internal standard, it is evident that pBS I, which has low AT content and lacks A tracts, binds HsORC equally as well as DHFR ori I, which has a high AT content and four long A tracts (Fig. 3, cf. B and C). Collectively, the data presented inFigure 3 show that our purified HsORC binds to all naturally occurring DNA sequences with affinities that fall into a narrow range. We conclude that recombinant HsORC does not have a strong preference for DNA sequences found in the vicinity of mammalian origins, even when they contain several AT tracts and their AT content is above average.
Properties of DNA affinity-purified HsORC
As noted above, the preparation of HsORC described inFigure 1A, although highly purified, likely contained substoichiometric amounts of Orc1 and Orc6. Given that Orc1 is known to be required for ATP-dependent DNA binding in yeast, we were concerned about its low abundance in our preparations. We therefore further purified the complex using dsDNA affinity chromatography. A large amount of HsORC subunits 2–6 flowed through the column (Fig. 4A, lane 2). In contrast, all the HsORC1 was retained on the column (Fig. 4A, lane 3). Therefore, this affinity-purification step resulted in significant enrichment of HsORC1-containing complexes (“holocomplexes”), and it suggests that HsOrc1 is required for DNA binding. HsORC holocomplexes had a similar affinity for DNA as the original preparation (cf. Figs.1C and4B). Assuming this more purified HsORC contains few if any subcomplexes, and that most of the molecules are active in DNA binding, the apparent dissociation constant of HsORC for lamin B2 II DNA was estimated at ∼3 nM in the presence of ATP. Like the original HsORC preparation, the DNA-binding activity of the holocomplex preparation was stimulated only moderately by ATP (Fig. 4B, cf. squares and circles). Similarly, HsORC holocomplexes bound to AT-rich sequences with higher affinity than to GC-rich sequences (Fig. 4C) and bound to multiple DNA fragments in the lamin B2 origin region with similar affinities (Fig. 4D). Importantly, HsORC holocomplexes did not discriminate significantly between origin and control DNA fragments (Fig. 4E). We conclude that the biochemical properties of the original HsORC preparation accurately reflect those of ORCs that contain more stoichiometric amounts of the HsOrc1 subunit.
Human ORC supports initiation of DNA replication in a cell-free system
We wished to determine whether our preparation of purified recombinant HsORC was active for DNA replication. To this end, we turned to a cell-cycle-regulated in vitro DNA replication system derived from X. laevis egg extracts (Walter et al. 1998). The system uses two extracts, egg cytosol (EC) and NucleoPlasmic Extract (NPE). EC supports the events that occur in the G1 phase of the cell cycle, including the formation of the prereplication complex (pre-RC) by the ordered assembly of ORC, cdc6, cdt1, and the MCM2–7 complex onto added DNA. The pre-RC is activated for initiation by the subsequent addition of NPE, which contains high levels of the protein kinases cdk2 and cdc7, and therefore mimics the S phase (Walter et al. 1998;Walter 2000).
To test if the purified human ORC was functional, we asked whether the endogenous Xenopus ORC could be replaced with purified HsORC. Anti-XlOrc2 antibodies were used to deplete XlOrc2 from egg cytosol, and the depleted extract was supplemented with HsORC. Subsequently a eukaryotic DNA template, demembranated Xenopus sperm chromatin, was added and after 30 min, NPE was supplied to initiate DNA replication. It was not necessary to deplete XlORC from NPE because NPE contains potent inhibitors of de novo pre-RC formation (Walter et al. 1998). As expected, extracts that were mock-depleted supported DNA replication (Fig. 5A, lanes 2,5,8), whereas extracts depleted of XlOrc2 were inactive for DNA replication (Fig. 5A, lanes 3,6,9). Addition of the recombinant HsORC restored DNA replication of the XlOrc2-depleted extracts to the level seen in mock-depleted extract. In this experiment, 73% of total input DNA was replicated after 45 min (seeFig. 5A legend). Similar replication efficiencies were achieved in all subsequent replication assays (data not shown). The results in Figure 5A were quantitated using a PhosphorImager, the maximum amount of DNA replication achieved was arbitrarily assigned a value of 100, and the results were graphed (Fig. 5B). Rescue of DNA replication by HsORC was not simply caused by readdition of HsOrc2 because a complex containing Orc2–Orc6 was unable to rescue DNA replication (C. Cvetic, S. Vashee, T.J. Kelly, and J.C. Walter, unpubl.). Indeed, we found that XlOrc2 depletion also removed XlOrc1 (data not shown), as previously reported (Romanowski et al. 1996). Finally, we found that all six subunits of HsORC load onto chromatin in egg extracts (Fig. 6;data not shown), indicating that the entire complex is functioning to stimulate DNA replication. We conclude that our preparation of recombinant HsORC is active.
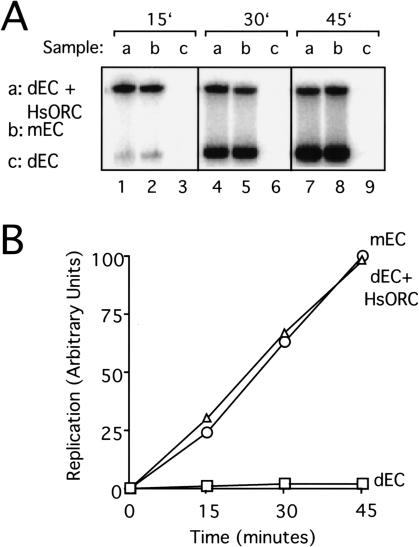
Figure 5.
Purified HsORC supports DNA replication. (A) Sperm chromatin (9000/μL final concentration) was incubated with XlOrc2-depleted egg cytosol (EC; sample c), XlOrc2-depleted EC supplemented with 13.5 ng/μL final concentration HsORC (sample a), or mock-depleted EC (sample b) for 30 min, followed by incubation with NPE. Replication products were separated on a 0.8% agarose gel and visualized by autoradiography. Incorporation of [α-32P]dATP was measured at 15, 30, and 45 min after addition of NPE. The fraction of input DNA replicated by 45 min (73%) was calculated from the fraction of the endogenous dNTP pool consumed (Blow and Laskey 1986), assuming an endogenous concentration of dATP of 50 μM in egg cytosol and NPE. (B) The radioactivity in each lane of the experiment shown in_A_ was measured using a PhosphorImager, normalized such that the highest value was equal to 100, and graphed.
Figure 6.
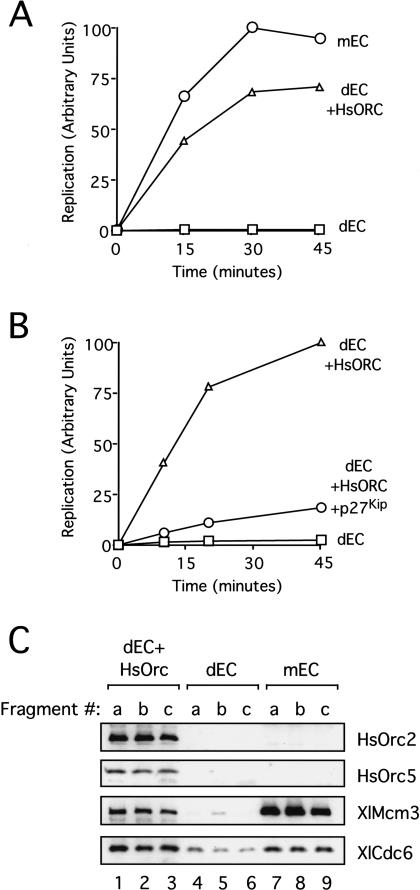
HsORC can initiate DNA replication from any DNA sequence. (A) HsORC can replicate a plasmid of prokaryotic composition. pBS (9 ng/μL final concentration) was incubated with XlOrc2-depleted EC (squares), XlOrc2-depleted EC supplemented with 14.5 ng/μL final concentration HsORC (triangles), or mock-depleted EC (circles) for 30 min, followed by incubation with NPE. DNA replication was measured at 15, 30, and 45 min after addition of NPE. (B) HsORC-dependent replication is inhibited by p27Kip. pBluescript (pBS;9 ng/μL final concentration) was incubated with XlOrc2-depleted EC (squares) or XlOrc2-depleted extract supplemented with 45 ng/μL HsORC for 30 min (circles, triangles). NPE (triangles, squares) or NPE preincubated with 85 ng/μL final concentration p27Kip (circles) was added to each reaction, and DNA replication was measured 15, 30, and 45 min after addition of NPE. (C) Three nonoverlapping 200-bp DNA fragments (termed a, b, and c) were PCR-amplified from pBluescript and coupled separately to magnetic beads. The beads were incubated for 20 min in XlOrc2-depleted EC supplemented with 10 ng/μL HsORC (lanes 1_–_3), XlOrc2-depleted EC (lanes_4_–6), or mock-depleted EC (lanes_7_–9). The beads were then isolated and washed. Bound proteins were separated by SDS-PAGE and transferred to PVDF membrane that was cut into strips and probed with antibodies against HsOrc2, HsOrc5, XlMcm3, and XlCdc6.
Sequence-independent initiation of DNA replication by HsORC
It has been shown both in vivo and in vitro in Xenopus egg extracts that embryonic XlORC does not require a eukaryotic origin sequence for initiation of DNA replication (Hyrien and Mechali 1992;Mahbubani et al. 1992; Walter et al. 1998). To determine whether human ORC requires a specific DNA sequence for initiation, we tested whether HsORC would be able to support DNA replication of pBluescript (pBS), a DNA template of exclusively prokaryotic composition. Our system is well suited to ask such a question because, unlike conventional nuclear assembly egg extracts (Blow and Laskey 1988), the nucleus-free replication system supports highly efficient and XlORC-dependent DNA replication of simple DNA substrates such as pBS (Walter et al. 1998). Endogenous XlORC was removed from the extract using XlOrc2 antibodies, and the extracts were supplemented with HsORC. We found that HsORC was able to replicate pBS with an efficiency close to that of endogenous XlORC (Fig. 6A, cf. circles and triangles). Therefore, human ORC does not appear to require a eukaryotic origin sequence to initiate DNA replication. To confirm that HsORC-dependent replication on pBS used a normal initiation mechanism, we asked whether it was Cdk2-dependent. As shown in Figure 6B, p27Kip, a Cdk2 inhibitor, severely reduced replication in this reaction, demonstrating that replication stimulated by HsORC follows a physiological mechanism.
A possible explanation for the efficient replication of pBS by HsORC is that pBS contains a “cryptic origin” that resembles human origin sequences. In theory, a simple way to test this idea would be to map where initiation events occur on pBS. However, we have recently shown that in_Xenopus_ egg extracts, ORC recruits many initiation competent MCM2–7 complexes that become widely distributed on chromatin (Edwards et al. 2002). Consequently, even if ORC binding were localized, the pattern of initiation events may well not be. Therefore, we used a different strategy to address the possibility of a cryptic origin. We reasoned that if a cryptic origin exists, most DNA sequences chosen at random from pBS would not be expected to support replication initiation. We therefore asked whether HsORC could load onto three different 200-bp fragments of pBS chosen at random. All three fragments have a similar AT content of ∼45%. The fragments were PCR-amplified using a biotinylated primer and then immobilized on streptavidin-coated magnetic beads to allow for their removal from the extracts. Using this system, we previously showed that the Xenopus ORC can assemble pre-RCs on linearized pBS (Edwards et al. 2002). When HsORC was added to XlOrc2-depleted egg cytosol, pre-RC formation occurred on all three immobilized DNA fragments (Fig. 6C, lanes 1–3). Although these experiments measure MCM2–7 binding rather than DNA replication initiation, they fully survey ORC's role in replication initiation, as it has been shown in Xenopus extracts that ORC becomes dispensable once MCM2–7 has been loaded onto the chromatin (Hua and Newport 1998;Rowles et al. 1999). In summary, the data inFigure 6 demonstrate that HsORC supports DNA replication of prokaryotic DNA, and that this likely occurs because it can stimulate pre-RC formation on any DNA sequence.
Human ORC replicates origin-containing and “originless” plasmids with the same efficiency
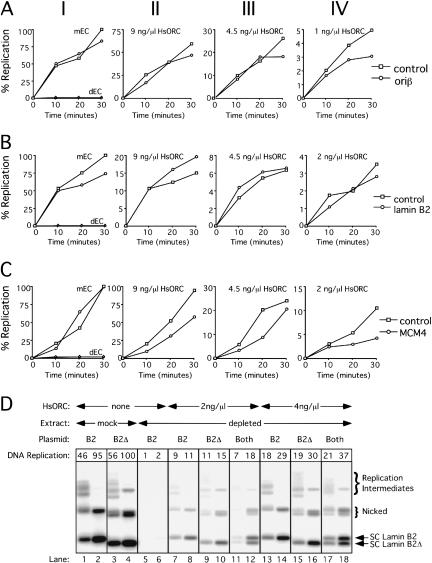
In the above experiments, it is possible that HsORC was added at saturating concentrations, thereby allowing HsORC to functionally interact with any DNA sequence. To address this issue, we performed replication assays using HsORC at limiting concentrations on plasmids containing or lacking known human origins. As shown in Figure 7A, panel I, the replication of a plasmid containing 5.8 kb of the DHFR ori_β locus was essentially the same as the replication of a similarly sized prokaryotic plasmid (pET24gp4-63) in mock-depleted extracts. The oriβ DNA fragment is known to contain a replicator element as it is sufficient to induce DNA replication at ectopic chromosomal locations in mammalian cells (Altman and Fanning 2001). When extracts were depleted of XlORC and supplemented with HsORC, the two plasmids also replicated at similar efficiency with all concentrations of HsORC used, including the lowest concentration, where replication was 5% of the control (Fig. 7A, panels II–IV). Similar results were obtained when comparing a lamin B2 ori plasmid (containing 2 kb of DNA encompassing the start site of DNA replication) with a different prokaryotic plasmid (φpRI10), as shown in Figure 7B. Finally, we performed this experiment on a third plasmid containing the intergenic region between the human MCM4 and_PRKDC genes, the first characterized human origin that is also an HsORC-binding site in vivo (Ladenburger et al. 2002). In this case, the same general result was observed, except that replication of the control plasmid (φpRI10) was slightly better than that of the ori plasmid when ORC was limiting (Fig. 7C). These results suggest that even when human ORC is limiting, it has the same propensity to initiate synthesis on origin and nonorigin DNA in Xenopus egg extracts.
Figure 7.
HsORC replicates origin-containing and “originless” plasmids with the same efficiency. (A_–_C) DNA replication of an origin-containing plasmid (circles) and a control plasmid containing only prokaryotic DNA sequences (squares) was compared separately in mock-depleted egg cytosol (panel I), or XlOrc2-depleted egg cytosol supplemented with different amounts of HsORC (panels II–IV). In panel_I_, DNA replication of the control plasmid (triangles) and the ori-containing plasmid (diamonds) was also measured using XlOrc2-depleted egg cytosol that was not supplemented with HsORC. (A) Control plasmid, pET24gp4–63;ori plasmid, contains the DHFR oriβ origin of replication. (B) Control plasmid, φpRI10;ori plasmid, contains the lamin B2 origin of replication. (C) Control plasmid, φpRI10;oriplasmid, contains the intergenic region between MCM4 and PRKDC. Plasmid concentration was 9 ng/μL in egg cytosol. (D) DNA replication of the lamin B2 plasmid (B2), an originless control plasmid (B2Δ), or both was examined in mock-depleted (lanes_1_–4), XlORC-depleted (lanes 5,6), or XlORC-depleted extract supplemented with HsORC (lanes_7_–18) as above. In each case, total plasmid concentration was 10 ng/μL in egg cytosol. At 15 and 45 min, reaction products were separated on an agarose gel, and replication intermediates and products (Walter and Newport 2000), are indicated to the right. The total radioactivity in each lane was determined with a PhosphorImager (see boxes).
To examine more directly whether HsORC preferentially replicates substrates that contain a human origin, we added a plasmid containing the lamin B2 origin (B2), as well as a control plasmid (B2Δ) to the same extract. We created the control plasmid by removing from the lamin B2 ori plasmid a 450-bp fragment containing the ORC-binding site and the replication initiation start site (see Materials and Methods). When tested individually, both plasmids replicated with similar efficiency in mock-depleted (Fig. 7D, lanes 1–4) or XlORC-depleted extracts supplemented with 2 ng/μL or 4 ng/μL HsORC (Fig. 7D, lanes 7–10,13–16). When added together, the two plasmids also replicated with similar efficiency by HsORC (Fig. 7D, lanes 11,12,17,18). This comparison is best made by examining the supercoiled forms of the plasmids, as these are easily distinguishable (see arrows). At both concentrations of HsORC, the amount of DNA replication in the supercoiled forms of B2 and B2Δ differed by no more than 20% (data not shown). HsORC was limiting in these experiments because DNA replication was only 18% and 37% of the level seen in mockdepleted extract, respectively (Fig. 7D, cf. lanes 4,12,18). This experiment further demonstrates that in Xenopus egg extracts, HsORC does not show a marked preference for DNA replication of origin-containing DNAs.
Discussion
The problem of how replication initiation sites are selected on the chromosomes of somatic cells has been difficult to resolve. Development of a unified theory is impeded by the fact that initiation events appear to be specific at some loci and random at others. Moreover, whereas some experiments indicate that only particular DNA sequences possess replicator activity, other experiments indicate that any DNA sequence of sufficient length can support initiation. To further address the biochemical mechanism underlying initiation site selection in humans, we have characterized human ORC. We show that HsORC supports efficient DNA replication in a cell-free system, lending further strong support to the idea that HsORC is the mammalian initiator protein. Remarkably, HsORC is able to initiate DNA replication from any DNA sequence. In addition, we find that purified human ORC does not effectively discriminate between origin DNA and random DNA in filter binding and DNA replication assays. Below, we propose a model for initiation site selection in metazoans.
DNA-binding properties of HsORC
Efficient occupancy of a specific chromosomal DNA sequence by a DNA-binding protein depends on its ability to distinguish between its cognate binding site and large amounts of unspecific competitor DNA (von Hippel et al. 1974). In mammalian cells, where replication initiation events are spaced ∼100–200 kb apart, HsORC would need to prefer origin DNA sequences to other sequences by several orders of magnitude. We found that HsORC bound ∼45-fold better to poly(dA) · poly(dT) than to naturally occurring DNA fragments. However, among natural DNAs, HsORC had almost the same affinity for human origins (lamin B2 and DHFR ori-β) and multiple control DNA fragments consisting of prokaryotic DNA. In these experiments, no significant preference was observed for DNA fragments that had a high AT content or fragments that contained long AT tracts. Therefore, the DNA-binding properties of HsORC purified from insect cells do not explain its targeting to origins of DNA replication in vivo (see also below).
The preferential binding of HsORC to synthetic rich-AT DNA is shared by the_S. pombe_ ORC (Kong and DePamphilis 2001;Lee et al. 2001;Chuang et al. 2002). DNA binding by SpORC to AT-rich DNA is mediated by a unique N-terminal domain in SpOrc4, which contains nine AT-hook motifs known to make minor groove contacts (Huth et al. 1997;Chuang and Kelly 1999). Interestingly, the similar affinity of HsORC for poly(dI) · poly(dC) and the AT-rich polynucleotides (Fig. 2A) suggests that some of its preference for AT-rich DNA depends on minor groove interactions, because the minor grooves of poly(dA) · poly(dT) and poly(dI) · poly(dC) are functionally similar. However, none of the subunits of HsORC contains a canonical AT-hook motif, so any minor groove interactions must be mediated by other unidentified motifs. It is interesting to speculate that HsORC may sometimes be recruited to DNA via interaction with an AT-hook motif-containing protein that became fused to Orc4 in the S. pombe complex. Importantly, although HsORC and SpORC share a preference for AT-rich-containing DNA, SpORC differs from HsORC in that it is found to bind with greater affinity to origin DNA than to control DNA in different assays (Chuang and Kelly 1999;Lee et al. 2001;Chuang et al. 2002). This observation implies that SpORC binds more selectively than HsORC to AT-rich DNA. Indeed, AT tracts have been shown to be important for the function of some origins in S. pombe (Kim and Huberman 1998;Okuno et al. 1999), whereas no such requirement has been demonstrated for human origins.
Of the three ORCs that have been biochemically characterized in some detail, the properties of HsORC may resemble most closely those of the_Drosophila_ ORC (Austin et al. 1999;Chesnokov et al. 2001). Our studies show that HsORC can bind to DNA in the absence of ATP, although DNA binding is stimulated twofold to fivefold by ATP. Similarly, the binding of DmORC to DNA is largely nonspecific and independent of ATP, but stimulation of binding by ATP can be observed under special experimental conditions (e.g., excess competing DNA). Although it is clear that DmORC, like HsORC, can bind nonspecifically to many different DNA sequences in vitro, it has also been reported that the protein exhibits some preference for AT-rich sites within ACE3 and _ori_β, specialized DNA elements that direct gene amplification in Drosophila follicle cells (Austin et al. 1999). Determination of the extent of this preference awaits quantitative binding studies, and it will be interesting to determine whether DmORC is able to initiate replication in a sequence-independent manner.
Our purified preparation of HsORC containing six subunits was similar in composition to the ORCs of the budding and fission yeasts, D. melanogaster, and X. laevis (Bell and Stillman 1992;Gossen et al. 1995;Rowles et al. 1996;Moon et al. 1999). However, there were subtle differences—the HsOrc1 and HsOrc6 subunits were consistently underrepresented compared with the other subunits, but HsORC1-containing complexes could be enriched using DNA affinity chromatography, and their properties were the same as the less homogeneous ORC preparation. Because we have also observed similar underrepresentation of HsOrc1 and HsOrc6 in ORCs purified from human cells (Vashee et al. 2001), we suspect that these subunits may be less tightly associated with the complex than the other subunits. It is worth noting that several recent studies have suggested that the regulation of expression or localization of HsOrc1 and HsOrc6 during the cell cycle may differ from that of the other HsORC subunits (Mendez and Stillman 2000;Natale et al. 2000;Chesnokov et al. 2001;Kreitz et al. 2001; Prasanth et al. 2002).
DNA sequence requirements for replication activity of HsORC
HsORC supported efficient DNA replication of three different plasmids of exclusively prokaryotic composition. Replication of these plasmids was not likely because of cryptic sites that resemble eukaryotic replicators because HsORC also nucleated prereplication complex formation on three short, randomly chosen prokaryotic DNA fragments. Together, these data support the argument that HsORC is able to initiate DNA replication from essentially any DNA sequence. These results notwithstanding, it was possible that HsORC might preferentially replicate certain sequences over others. This does not appear to be the case because even when HsORC was severely limiting, replication on plasmids containing various human origins was no more efficient than replication of prokaryotic plasmids, or on control plasmids with the origin removed, in the case of lamin B2. These data demonstrate that the lack of specificity is not due to saturating concentrations of HsORC. It was conceivable that sequence-independent initiation by HsORC in Xenopus egg extracts was due to an embryospecific factor that binds DNA cooperatively with HsORC, rendering it sequence nonspecific. However, the fact that the purified complex, isolated from a eukaryotic expression system, binds without sequence specificity in filter binding assays does not support this model. Additionally, we find that to achieve efficient DNA replication, significantly higher concentrations of HsORC are required than efficient filter binding (cf. Figs. 1C,4A and7). This finding suggests that DNA-binding conditions in extracts are more stringent than in a purified system, and it supports the argument against a cooperativity model in which an embryonic factor increases the affinity of HsORC for nonspecific DNA.
A model for origin targeting of HsORC
Our data help to account for many observations that have been difficult to explain using the conventional replicon model. First, it has been shown that essentially any DNA fragment greater than ∼15 kb, including fragments derived from prokaryotic sources, can replicate autonomously when introduced into human embryonic kidney cells (Krysan et al. 1993). These results are exactly what is expected if the initiator protein has low sequence specificity. Second, at several chromosomal loci, initiation occurs at many sites within 10–50-kb “initiation zones” (Little et al. 1993;Dijkwel and Hamlin 1995; Dijkwel et al. 2000;Ina et al. 2001). At one of these loci, DHFR, there is evidence that before the “origin decision point” in mid-G1, potential replication start sites (pre-RCs) are even more widely distributed than the initiation events detected in S phase (Gilbert et al. 1995;Wu and Gilbert 1996;Okuno et al. 2001), and this observation applies to many chromosomal locations (F. Li and D. Gilbert, pers. comm.). Although these observations could be explained by the recruitment of many MCM2–7 complexes to chromatin via a highly localized ORC (Edwards et al. 2002), they are also consistent with the idea that HsORC itself binds to many sites within the initiation zone.
The foregoing arguments notwithstanding, initiation of DNA replication in metazoans is not completely random, and in some cases, it may be highly specific. How can a model in which ORC binds sequence nonspecifically account for these data? To explain the absence of completely random initiation, we propose, as have others (Gilbert 2001), that HsORC binding is affected by chromatin structure. There is some indirect evidence for this idea. First, positioning of a nucleosome directly over the ACS region in S. cerevisiae abrogates replicator activity, presumably by blocking ORC binding (Simpson 1990). Second, ORC binding and initiation events are frequently localized to CpG islands, undermethylated regions of open chromatin structure (Antequera and Bird 1999;Ladenburger et al. 2002). A restrictive effect of chromatin would explain the completely random replication initiation seen on plasmids (Krysan et al. 1993), if chromatin structure is not fully formed on these DNA templates. In those cases in which replication initiation appears to be highly localized (Aladjem et al. 1995;Abdurashidova et al. 2000;Cimbora et al. 2000), HsORC might bind DNA cooperatively with a chromosomal protein(s) that is sequence-specific. Precedents for this idea come from the observation that DmORC binding to the chorion locus is dependent on E2F (Royzman et al. 1999) and by experiments suggesting that HsORC might be recruited to the EBV origin of DNA replication directly or indirectly by EBNA-1, a highly sequence-specific viral initiator protein (Chaudhuri et al. 2001;Dhar et al. 2001b;Schepers et al. 2001).
An alternative view of origin site selection is that HsORC is highly sequence-specific. In this case, endogenous HsORC would have to be posttranslationally modified in a way that is not recapitulated in our preparations of HsORC purified from insect cells. The modification would have to affect ORC binding such that it prefers origin DNA over random DNA by several orders of magnitude. A highly specific ORC would provide a satisfactory explanation for the initiation event seen at the lamin B2 locus. However, the same ORC that binds specifically at one locus should not bind randomly at another, and so it would be difficult to explain why any DNA can support DNA replication in vivo (Krysan et al. 1993). At present, there is no way to conclusively determine the intrinsic DNA-binding properties of endogenous HsORC. However, our data indicate that the affinity of HsORC for origin and nonorigin DNA molecules is quite high, with_K_ds in the low nanomolar range. Given this high affinity, it is hard to image the existence of other, more “specific” recognition sequences on chromosomal DNA, as such sites would have binding constants in the picomolar range to effectively compete for the available HsORC. Given all the data, in our view, the more parsimonious model is one in which ORC is sequence nonspecific. Whatever the ultimate answer, in this study we have shown conclusively that HsORC is able to initiate DNA replication from any DNA sequence, explaining the extraordinary flexibility with respect to where DNA replication initiation can occur in cells. One advantage of this flexible mechanism is that it allows great plasticity of genome organization during evolution (Gilbert 2001). Chromosome rearrangements and other changes could more easily be tolerated because they would not create large regions in the genome that lack origins of replication.
Materials and methods
Purification of recombinant HsORC
For purification of recombinant HsORC, a baculovirus expression vector containing an N-terminal hemagglutinin (HA) and hexahistidine (His6)-tagged HsORC2 gene was constructed as follows: the full-length cDNA was amplified by PCR using oligonucleotides, 5′-GAAGGCCTATGAGTAAACCAGAATTA AAGG-3′ and 5′-GGGGTACCTCAAGCCTCCTCTTCTTCC TTTTC-3′ as primers and a HeLa MATCHMAKER cDNA library (Clontech) as template. The PCR product was digested with _Stu_I and Kpn_I and cloned into the corresponding sites in a pFastBac1 plasmid (Invitrogen, Inc.) containing the_HA-His 6 –Sporc4 fusion gene (Chuang et al. 2002) and confirmed by sequencing. The recombinant virus was produced and amplified according to the manufacturer's instructions (Bac-to-Bac Baculovirus Expression Systems, Invitrogen, Inc.). Recombinant baculoviruses expressing the other five HsORC subunits have been described previously (Vashee et al. 2001).
Sf9 insect cells were cultured at 27°C in Grace's medium supplemented with 10% fetal bovine serum, penicillin (50 units/mL), and streptomycin (50 μg/mL;Invitrogen, Inc.). For expression of HsORC, Sf9 insect cells (2 × 106 cells/mL) were coinfected with six recombinant baculoviruses expressing all six subunits at a multiplicity of infection of 4. After 48 h, cells from 300-mL cultures were harvested and washed once with ice-cold hypotonic buffer (20 mM HEPES at pH 7.5, 5 mM KCl, 0.5 mM MgCl2, 0.5 mM dithiothreitol) containing 0.2 M sucrose and collected by centrifugation. Nuclei were prepared as described (Challberg and Kelly 1979) and lysed by incubation with ISLB (20 mM Tris-HCl at pH 6.8, 0.4 M sorbitol, 150 mM potassium acetate, 5 mM MgCl2, 5 mM MgSO4, 1% Triton X-100) on ice for 10 min. The suspension was centrifuged at 21,000_g_ at 4°C for 15 min, and the resulting pellet was extracted with OPB2 (50 mM HEPES at pH 7.5, 5 mM MgCl2,0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, 10% glycerol, 0.01% Tween 20) plus 500 mM NaCl on ice for 1 h. The supernatant collected after centrifugation at 140,000_g_ at 4°C for 45 min was incubated with 1 mL of Ni-agarose (QIAGEN) at 4°C for 2 h. The resin was washed with 15 mL of OPB2 plus 500 mM NaCl, and bound proteins were eluted with OPB2 plus 500 mM NaCl, containing 250 mM imidazole. The eluate from the Niagarose step was incubated with 0.25 mL of anti-HA antibody-conjugated agarose (F7-agarose;Santa Cruz Biotech) at 4°C for 2 h. The beads were washed once with OPB2 plus 500 mM NaCl, once with OPB2 plus 250 mM NaCl, and twice with OPB2 plus 100 mM NaCl. The F7-agarose-bound proteins were eluted by incubation with OPB2 plus 100 mM NaCl, containing 2 mg/mL HA peptide, at 4°C overnight. This step yielded a nearly homogeneous HsORC, which was used for in vitro experiments.
DNA-binding assays
For nitrocellulose filter-binding assays, a 450-bp fragment (lamin B2 ori II) that corresponds to positions 3750–4200 bp from the human lamin B2 origin (GenBank accession no. M94363) was synthesized by PCR using the following oligonucleotides: 5′-GGAATTCCTAGCGTCGCCGCCTGCAGCCTCT-3′ and 5′-GGAATTCATCACGTGACGAAGAGTCAGCTTG-3′,as the primers. The PCR DNA fragment was labeled at terminal restriction sites with [α-32P]dATP (Applied Bioscience) using the Klenow fragment of DNA polymerase I. The 15-μL binding reactions contained 50 mM HEPES (pH 7.5), 75 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, 10% glycerol, 0.01% Tween 20, 120 μg/mL BSA, 0.5 nM radiolabeled lamin B2 II, and the indicated concentrations of HsORC. Where indicated, ATP was added at a final concentration of 1 mM. After incubation at 25°C for 30 min, the reaction mixtures were passed through HA nitrocellulose filters (MF-Membrane Filters, Millipore Corp.), washed with 2 mL of OPB2, dried, and quantitated in a scintillation spectrometer.
For DNA-binding assays testing the effects of various nucleotides, the binding reactions were performed as described above except that 0.75 ng/μL HsORC was used and the concentrations of nucleotides were as indicated inFigure 1D.
For competition assays, synthetic deoxyribonucleotide polymers were purchased from Amersham Biosciences. Lamin B2 origin competitors, lamin B2 ori (2500–4500 bp), lamin B2 ori I (2500–3800 bp), lamin B2 ori II (3750–4200 bp), and lamin B2 ori III (4150–4500 bp) were synthesized by PCR. The numbers correspond to those of the humlamb sequence (GenBank accession no. M94363). DHFR oriβ competitors, DHFR ori (1991–3120 bp), DHFR ori I (1991–2451 bp), DHFR ori II (2450–2800 bp), and DHFR ori III (2801–3120 bp) were synthesized by PCR. The numbers correspond to those of GenBank accession no. Y09885. The pUC19 sequence was amplified by PCR using the following oligonucleotides: 5′-CGTAATCATGGTCATAGCTGT-3′ and 5′-CACGCTTCCCGAAGGGAGAAAGG-3′, as primers and the plasmid pMCD (Caddle et al. 1990) as template. The pBS I, II, and III competitors were generated by cleaving the plasmid pBluescript II SK+ with the restriction enzyme_Ear_I. The assays were performed as described above with the following modifications: 5 nM of radioactive lamin B2 II (∼25 ng), indicated concentrations of competitors, and 10–20 nM of HsORC were used.
For the DNA-binding experiments ofFigure 2B, a 2.3-kb DNA fragment containing the Epstein-Barr virus oriP was amplified by PCR using 5′-biotinylated oligonucleotide, Bio-5′-TCCAAC CAAACCGACTCTGACGG-3′ and 5′-CCAATACGCAAACC GCCTCTCCC-3′ as primers, and pGEMoriP (Frappier and O'Donnell 1991) as template. The PCR product was then coupled to streptavidin-conjugated magnetic beads (Dynalbeads M-280 Streptavidin; Dynal Corp.) according to the manufacturer's instructions. The binding assays were performed by incubating 150 fmole of HsORC with beads containing 75 fmole of oriP DNA in 15 μL of OPB2 buffer with 120 μg/mL BSA and 75 mM NaCl at 25°C for 30 min. Where indicated, a 10-fold excess of poly(dA) · poly(dT) was included in the reaction mixture. Beads were washed three times with 200 μL of OPB2 plus 75 mM NaCl. The bound proteins were released by addition of 1× SDS loading dye and separated by 10% SDS-PAGE followed by Western blotting analysis with antibodies against HsORC subunits 1 through 6 (Vashee et al. 2001).
DNA replication and immobilized DNA-fragment-binding assays
Egg cytosol preparation, NPE preparation, and DNA replication assays were carried out as described (Walter et al. 1998). Plasmid templates for replication included pMCD (Caddle et al. 1990), p449 (Connelly et al. 1998), pET24gp463 (Lee and Richardson 2001), φ10pRI10 (Ikeda and Richardson 1986), pBS (Stratagene), and pUC19laminB2, which was created by PCR amplification of the region surrounding the lamin B2 origin (2500–4500 bp) from human genomic DNA (Clontech) using the following primers: 5′-CGGGATCCTGCAGCTCAAGTCTTAAAGAC-3′ and 5′-GGGGTACCGGACTACAACTCCCACACGAC-3′. The PCR product was cloned into pUC19 using the _Bam_HI and _Kpn_I restriction sites. The lamin B2Δ plasmid was created by excising the region between the_Pml_I and _Bgl_II sites on pUC19laminB2 (nucleotides 3728–4197 of human lamin B2, accession no. M94363).
To calculate DNA replication efficiency inFigure 7A–C, we needed to account for the fact that DNA replication of the origin and control plasmids in undepleted Xenopus egg extracts sometimes differed (on average by 30%). Therefore, for each plasmid, the results of DNA replication under each condition were normalized to the maximum amount of replication possible on each template in undepleted extracts.
Generation of immobilized linear templates was as described (Edwards et al. 2002) except that different PCR primers were used. Using the _Sca_I site as a reference site on pBS KS- (position 2524), we used primers starting at positions 156 (biotinylated) and 345 to amplify pBSa, 1087 (biotinylated) and 1282 to amplify pBSb, and 1453 (biotinylated) and 1643 to amplify pBSc. Chromatin binding to immobilized DNA templates was as described (Edwards et al. 2002).
Acknowledgments
We thank Ellen Fanning for providing the pMCD plasmid, Charles Richardson for the pET24gp463 and φ10pRI10 plasmids, and Carl Anderson for the p449 plasmid. We thank Tatsuro Takahashi and Gernot Walter for critical reading of the manuscript and all of the members of the Kelly and Walter labs for helpful discussions. This work was supported by NIH grants to J.W. (GM62267) and T.K. The work was also supported by a Burroughs Wellcome Career Award in the Biomedical Sciences (J.W.) and a Stuart Trust Pilot Project (J.W.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding authors.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1084203.
References
- Abdurashidova G., Deganuto, M., Klima, R., Riva, S., Biamonti, G., Giacca, M., and Falaschi, A. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287**:** 2023-2026. [DOI] [PubMed] [Google Scholar]
- Aladjem M.I., Groudine, M., Brody, L.L., Dieken, E.S., Fournier, R.E., Wahl, G.M., and Epner, E.M. 1995. Participation of the human β-globin locus control region in initiation of DNA replication. Science 270**:** 815-819. [DOI] [PubMed] [Google Scholar]
- Altman A.L. and Fanning, E. 2001. The Chinese hamster dihydrofolate reductase replication origin β is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21**:** 1098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F. and Bird, A. 1999. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr. Biol. 9**:** R661-R667. [DOI] [PubMed] [Google Scholar]
- Austin R.J., Orr-Weaver, T.L., and Bell, S.P. 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes & Dev. 13**:** 2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P. and Dutta, A. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71**:** 333-374. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Stillman, B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357**:** 128-134. [DOI] [PubMed] [Google Scholar]
- Blow J.J. and Laskey, R.A. 1986. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 47**:** 577-587. [DOI] [PubMed] [Google Scholar]
- ____. 1988. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature 332**:** 546-548. [DOI] [PubMed] [Google Scholar]
- Caddle M.S., Lussier, R.H., and Heintz, N.H. 1990. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J. Mol. Biol. 211**:** 19-33. [DOI] [PubMed] [Google Scholar]
- Challberg M.D. and Kelly Jr., T.J. 1979. Adenovirus DNA replication in vitro. Proc. Natl. Acad. Sci. 76**:** 655-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri B., Xu, H., Todorov, I., Dutta, A., and Yates, J.L. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. 98**:** 10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I., Remus, D., and Botchan, M. 2001. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. 98**:** 11997-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang R.Y. and Kelly, T.J. 1999. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AThooks. Proc. Natl. Acad. Sci. 96**:** 2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang R.Y., Chretien, L., Dai, J., and Kelly, T.J. 2002. Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: Interaction with origin DNA and Cdc18 protein. J. Biol. Chem. 277**:** 16920-16927. [DOI] [PubMed] [Google Scholar]
- Cimbora D.M., Schubeler, D., Reik, A., Hamilton, J., Francastel, C., Epner, E.M., and Groudine, M. 2000. Long-distance control of origin choice and replication timing in the human β-globin locus are independent of the locus control region. Mol. Cell. Biol. 20**:** 5581-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly M.A., Zhang, H., Kieleczawa, J., and Anderson, C.W. 1998. The promoters for human DNA-PKcs (PRKDC) and MCM4: Divergently transcribed genes located at chromosome 8 band q11. Genomics 47**:** 71-83. [DOI] [PubMed] [Google Scholar]
- Dhar S.K., Delmolino, L., and Dutta, A. 2001a. Architecture of the human origin recognition complex. J. Biol. Chem. 276**:** 29067-29071. [DOI] [PubMed] [Google Scholar]
- Dhar S.K., Yoshida, K., Machida, Y., Khaira, P., Chaudhuri, B., Wohlschlegel, J.A., Leffak, M., Yates, J., and Dutta, A. 2001b. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106**:** 287-296. [DOI] [PubMed] [Google Scholar]
- Dijkwel P.A. and Hamlin, J.L. 1995. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell. Biol. 15**:** 3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P.A., Mesner, L.D., Levenson, V.V., d'Anna, J., and Hamlin, J.L. 2000. Dispersive initiation of replication in the Chinese hamster rhodopsin locus. Exp. Cell Res. 256**:** 150-157. [DOI] [PubMed] [Google Scholar]
- Dijkwel P.A., Wang, S., and Hamlin, J.L. 2002. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell. Biol. 22**:** 3053-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.C., Tutter, A.V., Cvetic, C., Gilbert, C.H., Prokhorova, T.A., and Walter, J.C. 2002. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 277**:** 33049-33057. [DOI] [PubMed] [Google Scholar]
- Frappier L. and O'Donnell, M. 1991. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J. Biol. Chem. 266**:** 7819-7826. [PubMed] [Google Scholar]
- Fujita M. 1999. Cell cycle regulation of DNA replication initiation proteins in mammalian cells. Front. Biosci. 4**:** D816-D823. [DOI] [PubMed] [Google Scholar]
- Gilbert D.M. 2001. Making sense of eukaryotic DNA replication origins. Science 294**:** 96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D.M., Miyazawa, H., and DePamphilis, M.L. 1995. Sitespecific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol. Cell. Biol. 15**:** 2942-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M., Pak, D.T., Hansen, S.K., Acharya, J.K., and Botchan, M.R. 1995. A_Drosophila_ homolog of the yeast origin recognition complex. Science 270**:** 1674-1677. [DOI] [PubMed] [Google Scholar]
- Hua X.H. and Newport, J. 1998. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol. 140**:** 271-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth J.R., Bewley, C.A., Nissen, M.S., Evans, J.N., Reeves, R., Gronenborn, A.M., and Clore, G.M. 1997. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4**:** 657-665. [DOI] [PubMed] [Google Scholar]
- Hyrien O. and Mechali, M. 1992. Plasmid replication in Xenopus eggs and egg extracts: A 2D gel electrophoretic analysis. Nucleic Acids Res. 20**:** 1463-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1993. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 12**:** 4511-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R.A. and Richardson, C.C. 1986. Interactions of the RNA polymerase of bacteriophage T7 with its promoter during binding and initiation of transcription. Proc. Natl. Acad. Sci. 83**:** 3614-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ina S., Sasaki, T., Yokota, Y., and Shinomiya, T. 2001. A broad replication origin of Drosophila melanogaster, oriDα, consists of AT-rich multiple discrete initiation sites. Chromosoma 109**:** 551-564. [DOI] [PubMed] [Google Scholar]
- Jacob F., Brenner, J., and Cuzin, F. 1963. On the regulation of DNA replication in bacteria. Cold Spring Harb. Symp. Quant. Biol. 28**:** 329-348. [Google Scholar]
- Keller C., Ladenburger, E.M., Kremer, M., and Knippers, R. 2002. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 277**:** 31430-31440. [DOI] [PubMed] [Google Scholar]
- Kelly T.J. and Brown, G.W. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69**:** 829-880. [DOI] [PubMed] [Google Scholar]
- Kim S.M. and Huberman, J.A. 1998. Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol. Cell. Biol. 18**:** 7294-7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm R.D., Austin, R.J., and Bell, S.P. 1997. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell 88**:** 493-502. [DOI] [PubMed] [Google Scholar]
- Kong D. and DePamphilis, M.L. 2001. Site-specific DNA binding of the_Schizosaccharomyces pombe_ origin recognition complex is determined by the Orc4 subunit. Mol. Cell. Biol. 21**:** 8095-8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz S., Ritzi, M., Baack, M., and Knippers, R. 2001. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 276**:** 6337-6342. [DOI] [PubMed] [Google Scholar]
- Krysan P.J., Smith, J.G., and Calos, M.P. 1993. Autonomous replication in human cells of multimers of specific human and bacterial DNA sequences. Mol. Cell. Biol. 13**:** 2688-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. and Diffley, J.F. 2001. Is the MCM2-7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev. 11**:** 64-70. [DOI] [PubMed] [Google Scholar]
- Ladenburger E.M., Keller, C., and Knippers, R. 2002. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 22**:** 1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J. and Richardson, C.C. 2001. Essential lysine residues in the RNA polymerase domain of the gene 4 primase-helicase of bacteriophage T7. J. Biol. Chem. 276**:** 49419-49426. [DOI] [PubMed] [Google Scholar]
- Lee J.K., Moon, K.Y., Jiang, Y., and Hurwitz, J. 2001. The_Schizosaccharomyces pombe_ origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc. Natl. Acad. Sci. 98**:** 13589-13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R.D., Platt, T.H., and Schildkraut, C.L. 1993. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 13**:** 6600-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbubani H.M., Paull, T., Elder, J.K., and Blow, J.J. 1992. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 20**:** 1457-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J. and Stillman, B. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20**:** 8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K.Y., Kong, D., Lee, J.K., Raychaudhuri, S., and Hurwitz, J. 1999. Identification and reconstitution of the origin recognition complex from_Schizosaccharomyces pombe_. Proc. Natl. Acad. Sci. 96**:** 12367-12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale D.A., Li, C.J., Sun, W.H., and DePamphilis, M.L. 2000. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G1 transition in mammals. EMBO J. 19**:** 2728-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J. 1987. Nuclear reconstitution in vitro: Stages of assembly around protein-free DNA. Cell 48**:** 205-217. [DOI] [PubMed] [Google Scholar]
- Okuno Y., Satoh, H., Sekiguchi, M., and Masukata, H. 1999. Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol. Cell. Biol. 19**:** 6699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., McNairn, A.J., den Elzen, N., Pines, J., and Gilbert, D.M. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20**:** 4263-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth S.G., Prasanth, K.V., and Stillman, B. 2002. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science 297**:** 1026-1031. [DOI] [PubMed] [Google Scholar]
- Romanowski P., Madine, M.A., Rowles, A., Blow, J.J., and Laskey, R.A. 1996. The_Xenopus_ origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 6**:** 1416-1425. [DOI] [PubMed] [Google Scholar]
- Rowles A., Chong, J.P., Brown, L., Howell, M., Evan, G.I., and Blow, J.J. 1996. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell 87**:** 287-296. [DOI] [PubMed] [Google Scholar]
- Rowles A., Tada, S., and Blow, J.J. 1999. Changes in association of the_Xenopus_ origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 112**:** 2011-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I., Austin, R.J., Bosco, G., Bell, S.P., and Orr-Weaver, T.L. 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes & Dev. 13**:** 827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Sawado, T., Yamaguchi, M., and Shinomiya, T. 1999. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 19**:** 547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A., Ritzi, M., Bousset, K., Kremmer, E., Yates, J.L., Harwood, J., Diffley, J.F., and Hammerschmidt, W. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20**:** 4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R.T. 1990. Nucleosome positioning can affect the function of a_cis_-acting DNA element in vivo. Nature 343**:** 387-389. [DOI] [PubMed] [Google Scholar]
- Sun W.H., Coleman, T.R., and DePamphilis, M.L. 2002. Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin. EMBO J. 21**:** 1437-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T. and Masukata, H. 2001. Interaction of fission yeast ORC with essential adenine/thymine stretches in replication origins. Genes Cells 6**:** 837-849. [DOI] [PubMed] [Google Scholar]
- Takisawa H., Mimura, S., and Kubota, Y. 2000. Eukaryotic DNA replication: From pre-replication complex to initiation complex. Curr. Opin. Cell Biol. 12**:** 690-696. [DOI] [PubMed] [Google Scholar]
- Todorovic V., Falaschi, A., and Giacca, M. 1999. Replication origins of mammalian chromosomes: The happy few. Front. Biosci. 4**:** D859-D868. [DOI] [PubMed] [Google Scholar]
- Vashee S., Simancek, P., Challberg, M.D., and Kelly, T.J. 2001. Assembly of the human origin recognition complex. J. Biol. Chem. 276**:** 26666-26673. [DOI] [PubMed] [Google Scholar]
- von Hippel P.H., Revzin, A., Gross, C.A., and Wang, A.C. 1974. Non-specific DNA binding of genome regulating proteins as a biological control mechanism: I. The lac operon: Equilibrium aspects. Proc. Natl. Acad. Sci. 71**:** 4808-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J.C. 2000. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 275**:** 39773-39778. [DOI] [PubMed] [Google Scholar]
- Walter J. and Newport, J.W. 1997. Regulation of replicon size in_Xenopus_ egg extracts. Science 275**:** 993-995. [DOI] [PubMed] [Google Scholar]
- ____. 2000. Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell 5**:** 617-627. [DOI] [PubMed] [Google Scholar]
- Walter J., Sun, L., and Newport, J. 1998. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell 1**:** 519-529. [DOI] [PubMed] [Google Scholar]
- Wu J.R. and Gilbert, D.M. 1996. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science 271**:** 1270-1272. [DOI] [PubMed] [Google Scholar]