Targeted high-throughput sequencing of tagged nucleic acid samples (original) (raw)
Abstract
High-throughput 454 DNA sequencing technology allows much faster and more cost-effective sequencing than traditional Sanger sequencing. However, the technology imposes inherent limitations on the number of samples that can be processed in parallel. Here we introduce parallel tagged sequencing (PTS), a simple, inexpensive and flexible barcoding technique that can be used for parallel sequencing any number and type of double-stranded nucleic acid samples. We demonstrate that PTS is particularly powerful for sequencing contiguous DNA fragments such as mtDNA genomes: in theory as many as 250 mammalian mtDNA genomes can be sequenced in a single GS FLX run. PTS dramatically increases the sequencing throughput of samples in parallel and thus fully mobilizes the resources of the 454 technology for targeted sequencing.
INTRODUCTION
Since its introduction, the benefits of 454 sequencing (1) have been exploited for an increasing number of applications, including genomic sequencing (2), cDNA sequencing (3) and ultra-deep amplicon sequencing (4). Despite its power in generating a great number of sequences from few samples, 454 sequencing is at present unsuitable for studies requiring targeted sequence data from many different samples. This limitation is particularly pertinent to population and medical genetics studies. The 454 sequencing plate can be physically divided into a maximum of sixteen regions, each of which yields an average of 0.63 and 2.88 Mb per run for the GS20 and the new GS FLX sequencing platforms, respectively. If shotgun libraries from 16 human mtDNA genomes are sequenced in a single run, each genome will on average be covered 70- and 350-fold for GS20 and GS FLX, respectively. Despite the low cost per sequenced nucleotide, the high coverage and cost per run make this approach impractical. Furthermore, the physical separation of the plate reduces the total number of sequences retrieved from one run to roughly half and consumes more time and material, as each sample must be processed separately. Thus, even further physical separation of the plate would not solve these inherent problems.
One approach to overcoming these limitations is to barcode samples with sample-specific sequence tags. Samples are pooled prior to 454 sequencing and are identified after sequencing by their unique sequence tags. This approach has been used to barcode cDNA libraries with tagged primers for reverse transcription (5). Recently, Binladen et al. (6) used 5′-tagged PCR primers to distinguish amplicon sequences derived from different sources. However, this method requires the synthesis of sample-specific primers for each target under study, which is time-consuming and cost-prohibitive when dealing with large sample sizes. Currently, no method exists for barcoding small genomic libraries or amplicons derived from untagged PCR primers. The resources of 454 sequencing therefore cannot be fully exploited for applications that require low coverage sequencing of many different samples.
Despite the potential benefits a barcoding method for 454 sequencing offers, any proposed technique must ensure efficient use of sequencing resources and high data reliability. Incomplete reactions and sequencing errors can result in background sequences without a sequence tag, heterogeneous sequence representation among samples and false-assignment of sequences to their sample origin. We have developed a method called parallel tagged sequencing (PTS) that largely alleviates these problems.
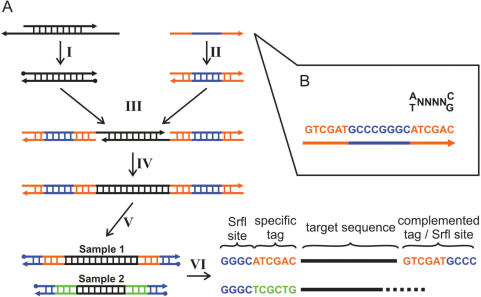
PTS is based on a ligation strategy analogous to the one utilized in the standard 454 library preparation procedure (1), the use of barcoding adapters and a restriction system that excludes background sequences. An outline of the method is displayed in Figure 1A. In separate reactions DNA molecules from different samples are blunt end repaired and phosphorylated. Subsequently, sample-specific barcoding adapters are ligated to both ends of the molecules, and the resulting nicks are removed by a strand-displacing polymerase. Each barcoding adapter is comprised of a single self-hybridized palindromic oligonucleotide containing an SrfI restriction site flanked by complementary sequence tags (Figure 1B). This setup requires only a single oligonucleotide to be synthesized in order to barcode each sample. The barcoded samples are then quantified, pooled in a ratio reflecting the proportion of sequences desired from each sample and treated with phosphatase to remove residual 5′ phosphates of unligated ends. Such unligated ends may arise during the adapter fill-in step in molecules containing single-strand nicks caused by nebulization. Half of the adapter is then cut off by SrfI, which is a rare cutter in mammalian genomes (7). It leaves blunt ends with 5′ phosphates and allows the pooled samples to be directly processed using the standard 454 library preparation. Phosphatase treatment in conjunction with SrfI digestion effectively reduces background sequences when starting from nebulized DNA; untagged molecule ends are prevented from being ligated to the universal 454 adapters during library preparation.
Figure 1.
Workflow for barcoding and sequencing double-stranded DNA samples. (A) The DNA is blunt end repaired (I) and sample-specific self-complementary oligos are self-hybridized to form double-stranded barcoding adapters (II), which are subsequently ligated to both ends of the DNA molecules (III). Resulting nicks are removed by strand-displacement (IV). Individually tagged samples are combined into a single master library and treated with phosphatase. Half of the adapter is removed by SrfI digestion (V) leaving ligatable ends for the standard 454 library production. After sequencing (VI), the tags are used to identify the sample origin and removed prior to further analysis. (B) Barcoding tags begin with either an A or T, followed by four freely chosen nucleotides and end with C or G with no homopolymers allowed. From 324 possible tags, 72 can be chosen to differ by at least two substitutions.
The sequence output from barcoded libraries begins with the sequence key TCAG, which originates from the universal 454 adapters, followed by four non-informative nucleotides GGGC from the SrfI site. Since the 454 technique uses sequencing-by-synthesis, all G's are incorporated in the same nucleotide flow as the last key base, and therefore do not decrease the read length. The origin of the sequence is identified through the adjacent sequence tag. We developed a tag design that is particularly robust to sequencing errors associated with homopolymers, a well-known problem in 454 sequencing (1,8,9). At a length of 6 bp, it allows the pooling of a maximum of 72 samples into a single sequencing library (Figure 1B). Since the 454 sequencing plate is always split into at least two regions, this configuration allows for the parallel processing of up to 144 samples in a single run.
We demonstrate the power of parallel tagged 454 sequencing by shotgun sequencing six human mtDNA genomes on two 16th plate regions of the GS20 platform. Published Sanger sequences for these samples allow a comparison of the two sequencing approaches. Additionally, we verified the reproducibility of the method by independently sequencing a second set of six human mtDNA genomes.
MATERIALS AND METHODS
Construction of shotgun libraries
Six human genomic DNA samples and their respective mtDNA genome sequences were obtained from a previous study and the CEPH panel (10,11). For the replicate experiment, six additional samples were kindly provided by Mark Stoneking. The mtDNA genomes were amplified in two overlapping fragments using the Expand Long Range dNTPack kit (Roche) and equimolar mixtures of 5′ blocked and unmodified PCR primers in order to avoid sequence overrepresentation within the two amplicon overlaps (oligonucleotide sequences listed in Supplementary Table 2). Amplicons were purified using the PCR purification kit (Qiagen) and quantified using an ND-1000 spectrophotometer (Nanodrop Technologies). Amplicons from each sample were combined in equal molar ratios to obtain roughly one microgram of DNA per sample. Shotgun DNA libraries with a mean fragment size of ∼500 bp were prepared using nebulizers and chemicals from the GS20 library preparation kit (Roche) according to the manual.
Parallel tagged sequencing
Roughly 500 ng of nebulized DNA were blunt end repaired in 40 μl reactions containing 1× buffer Tango, 100 μM each dNTP, 1 mM ATP, 40U T4 polynucleotide kinase, 12U T4 DNA polymerase (all Fermentas) and 0.125 μg/μl BSA (Sigma). After incubation for 15 min at 12°C and 15 min at 25°C, the reactions were purified using the MinElute PCR Purification kit (Qiagen), which was also used for all subsequent purification steps. Double-stranded barcoding adapters were prepared by briefly heating 100 μM solutions of palindromic oligos to 95°C and cooling them to room temperature at a rate of 0.1°C/s. Adapters were ligated to the blunt end DNA in separate 40 μl reactions, containing 1× T4 ligase buffer, 5% PEG-4000, 5U T4 ligase (all Fermentas) and 8 μl barcoding adapter. After incubation at 22°C for 1 h, the reactions were purified. The DNA was then incubated for 20 min at 37°C in 30 μl reactions containing 1× Thermopol buffer, 250 μM of each dNTP and 8U Bst polymerase (NEB), followed by reaction purification. According to the nanodrop, purified yields were between 25 and 43 ng/μl, but since we observed intrasample measurement variation of ±20%, triplicate measurements were carried out to estimate mean concentrations.
One hundred nanogram of adapter-ligated DNA from each of the six samples were pooled and treated with calf intestine phosphatase in a single 40 μl reaction containing 1× NEBuffer 3 and 30U calf intestinal phosphatase (NEB) for 1 h at 37°C. After purification, the DNA was digested with SrfI in a 30 μl reaction containing 1× universal buffer and 10U SrfI (Stratagene) for 3 h at 37°C and purified again. A single sequencing library was produced using the standard GS20 library preparation procedure, but starting at the adapter ligation step. According to the standard GS20 sequencing procedure, the library was directly sequenced on two 16th regions of a full GS20 sequencing plate without a prior titration run.
Sequence data processing
Raw 454 reads from the standard flow files (SFF) begin with the artificial sequence TCAGGGGC followed by the 6 bp barcode. To compensate for homopolymer miscalls and carry forward effects, two to six Gs, followed by one or two Cs and an additional G were accepted while scanning for barcode sequences. If the following 6 bp had the general pattern of a barcode, starting with A or T, ending with C or G with adjacent bases differing, the ‘clip_adapter_left’ entry in the SFF file was overwritten to the point after the barcode, causing the 454 software tools to ignore this part of the sequence. The 3′ ends of the reads were then scanned for the reverse-complement of the barcode sequence and, if found, the ‘clip_adapter_right’ entry was set to also remove this part. The modified SFF files were then separated according to the barcode sequences using ‘sfffile’ and directly submitted into runMapper v1.0.53.17 for assembly. The consensus sequences are available as Supplementary Data.
RESULTS
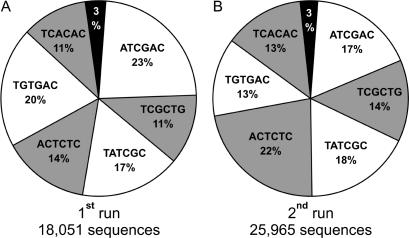
Nebulized DNA fragments from six human mtDNA genomes were barcoded, pooled into a single sequencing library and sequenced on a small region of the 454 picotitre plate. An independent replicate experiment was carried out using six different samples. Only a limited amount of variation was observed for the sequence representation among samples: between 1995 and 4154 sequences were obtained from each sample in the first experiment (Figure 2A); the second experiment yielded between 3267 and 5731 sequences from each sample (Figure 2B). The variation was observed both within and between runs, indicating that there is no underlying ligation bias leading to differential efficiencies in the tagging reactions. In fact, we could not detect incomplete tagging with any of the barcoding adapters when optimizing the protocol using a PCR product as template. Therefore, we think that a more accurate quantification of the number of molecules contained in each barcoded sample is likely to further increase the homogeneity of sequence representation among samples. This could be achieved by DNA quantification methods more sensitive than UV absorption, such as fluorescence-based picogreen quantification assays (12). In addition, for each barcoded sample the mean fragment size could be determined by capillary electrophoresis (13), which allows for inferring the actual molar concentration rather than the simple mass concentration, thereby accounting for differences in fragment size distributions after nebulization.
Figure 2.
Sequence representation of barcoded shotgun libraries obtained from two independently constructed sequencing libraries (A and B, respectively).
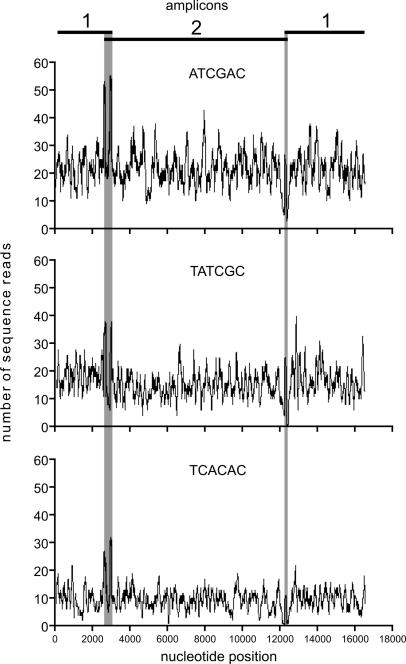
As little as ∼3% of the sequences obtained from both runs could not be assigned to a correct sequence tag. Whereas most of these sequences did not begin with a tag, 260 of the 44 016 sequences generated in both runs carried tag sequences that differed by at least one substitution from the tags that were used. In 154 of these cases, the tag sequences differed by at least two substitutions and could therefore possibly be assigned to a false sample. Depending on the number of tags used, this represents a maximum false assignment rate of 0.35%. High coverage makes it unlikely that single misidentified reads will affect consensus sequence accuracy, but if necessary, the false assignment rate could be reduced by extending the tag length. The average coverage for each genome ranged from 10- to 22-fold and from 19- to 32-fold in the first and second experiment, respectively (see Supplementary Table 1). The coverage distribution along the mtDNA genome shows only moderate variation, with the exception of under-representation of reads close to the amplicon ends (Figure 3 and Supplementary Figure 1). This arises due to reduced breakage of molecules at their ends. Thus, apart from the regions adjacent to the shorter amplicon overlap, all positions of the mtDNA genomes were covered by at least one sequence read, except for 3 bp from one of the 12 sequences.
Figure 3.
Three coverage plots representing the spectrum of coverage obtained for the six human mtDNA genomes with known Sanger sequence. The overlapping regions of the long-range amplicons are indicated by gray shading.
To assess the resulting sequence quality, the consensus sequences of six mtDNA genomes were compared to their previously published Sanger sequences. In 99 kb of sequence four substitutions were detected between the Sanger and the 454 consensus sequences, corresponding to more than 99.99% agreement. However, we observed single base-pair insertions or deletions at a frequency of 0.27% on average (see Supplementary Table 1). Indel rate estimates derived from previous 454 genome sequencing projects are lower (1,8,9), but in-depth comparisons are difficult due to the different natures of the sequenced genomes and the limited amount of data available in this study. The frequency of indels decreases as coverage increases (see Supplementary Figure 2), but higher coverage sequencing would be required to determine whether the indel rate decreases to previously determined rates at >30-fold coverage.
DISCUSSION
PTS allows the parallel generation of DNA sequences from a large number of samples using 454 high-throughput sequencing. It provides several advantages over simple physical separation of the sequencing plate or previously published tagging techniques. First, it allows highly parallel sequencing of small shotgun libraries derived, for example, from long-range amplicons or plasmids. Second, PTS can be applied to the sequencing of single or pooled PCR products. It thereby facilitates a simple switch from classical Sanger sequencing to 454 sequencing without the need to change the experimental design of existing PCR applications (e.g. by ordering new primers). This is particularly useful for applications that require microbial subcloning, since 454 sequences derive from single molecules. The oligonucleotide tags can be reused for any PCR system and on any type of double-stranded DNA. Third, the number of samples that can be sequenced in parallel using PTS is increased by an order of magnitude compared to the use of a plate's 16 subunits. Moreover, since physical separation decreases the overall number of sequences obtained, PTS enables targeted DNA sequencing at a lower price per nucleotide. In fact, optimal use of this technique can drastically reduce sequencing costs compared to traditional Sanger sequencing. Using PTS, a single run of the GS20 platform, which usually yields at least 20 Mb, can currently provide sequences for more than 50 mtDNA genomes at ∼20-fold coverage. With an output of 100 Mb per run it should be possible to obtain more than 250 mtDNA genome sequences at this coverage on the higher throughput GS FLX platform. Our data indicate that aiming for 20-fold average coverage is sufficient to obtain complete consensus sequences for all or the vast majority of the samples, as we expect each genome to be covered between 10- and 30-fold. Even at 10-fold average coverage, the resulting sequence quality is sufficient to accurately identify substitutional differences. However, the frequency of single base-pair insertion and deletion errors in and around homopolymers increases with lower coverage. These errors can be easily identified and eliminated when closely related sequences are available for comparison, but may represent a more serious problem when this is not the case. Therefore, the optimal coverage range should be determined according to each study's particular requirements. For nuclear sequences, higher average coverage will be necessary if polymorphic positions must be reliably detected.
For sequencing continuous stretches of DNA, the shotgun sequencing of long-range PCR fragments using PTS provides several advantages over traditional Sanger sequencing. First, it is much less time-consuming, as it reduces the work load for setting up PCRs and sequencing reactions. Second, it decreases the risk of allelic dropout due to primer template mismatches, which is especially important when working with species for which no genomic sequence information exists. Third, apart from the primer sequences, no prior information is needed for sequence assembly. This feature is especially useful for reliably obtaining unknown exon–intron structures of genes when primers are designed to anneal to conserved exonic sequences. The use of SrfI, a restriction enzyme with restrictions sites approximately every 150 kb in the human genome, guarantees a minimal number of dropouts due to restriction occurring within a sequence fragment. Even if a sequence fragment contains an SrfI site, due to the random fragmentation of DNA by nebulization, this would only prevent coverage immediately adjacent to the SrfI site itself.
In summary, we have developed a system that allows for high-throughput sequencing of targeted DNA sequences using the 454 system or any other parallel sequencing system (14). Our technique substantially increases the throughput for targeted sequencing compared to traditional Sanger sequencing, while at the same time decreasing the cost per nucleotide. PTS can be readily adapted to any existing experimental design and enables sequencing any type of double-stranded DNA from multiple samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are indebted to Barbara Höffner, Antje Weihmann and Barbara Höber for running the 454 sequencer, Mark Stoneking for providing DNA samples and Christine B. Green for comments on the manuscript. We thank Richard E. Green, Jr., Adrian Briggs, Holger Römpler, Nadin Rohland and Janet Kelso for helpful discussions, Stratagene for freely providing SrfI and the Max Planck Society for financial support. Funding to pay the Open Access publication charges for this article was provided by the Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gowda M, Li H, Alessi J, Chen F, Pratt R, Wang GL. Robust analysis of 5′-transcript ends (5′-RATE): a novel technique for transcriptome analysis and genome annotation. Nucleic Acids Res. 2006;34:e126. doi: 10.1093/nar/gkl522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat. Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen KL, Hogh AL, Emmersen J. DeepSAGE – digital transcriptomics with high sensitivity, simple experimental protocol and multiplexing of samples. Nucleic Acids Res. 2006;34:e133. doi: 10.1093/nar/gkl714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simcox TG, Marsh SJ, Gross EA, Lernhardt W, Davis S, Simcox ME. SrfI, a new type-II restriction endonuclease that recognizes the octanucleotide sequence, [sequence: see text] Gene. 1991;109:121–123. doi: 10.1016/0378-1119(91)90596-4. [DOI] [PubMed] [Google Scholar]
- 8.Moore MJ, Dhingra A, Soltis PS, Shaw R, Farmerie WG, Folta KM, Soltis DE. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingman M, Gyllensten U. Mitochondrial genome variation and evolutionary history of Australian and New Guinean aborigines. Genome Res. 2003;13:1600–1606. doi: 10.1101/gr.686603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol. Biol. Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- 12.Ahn SJ, Costa J, Emanuel JR. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 1996;24:2623–2625. doi: 10.1093/nar/24.13.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panaro NJ, Yuen PK, Sakazume T, Fortina P, Kricka LJ, Wilding P. Evaluation of DNA fragment sizing and quantification by the agilent 2100 bioanalyzer. Clin. Chem. 2000;46:1851–1853. [PubMed] [Google Scholar]
- 14.Bentley DR. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]