Nonsense mutation at Tyr-4046 in the DNA-dependent protein kinase catalytic subunit of severe combined immune deficiency mice (original) (raw)
Abstract
The severe combined immune deficiency (SCID) mouse was reported as an animal model for human immune deficiency. Through the course of several studies, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) gene came to be considered a candidate for the SCID-responsible gene. We isolated an ORF of the murine DNA-PKcs gene from SCID mice and their parent strain C.B-17 mice and determined the DNA sequences. The ORF of the murine DNA-PKcs gene contained 4128-aa residues and had 78.9% homology with the human DNA-PKcs gene. A particularly important finding is that a T to A transversion results in the substitution of termination codon in SCID mice for the Tyr-4046 in C.B-17 mice. No other mutation was detected in the ORF of the gene. The generality of this transversion was confirmed using four individual SCID and wild-type mice. The substitution took place in the phosphatidylinositol 3-kinase domain, and the mutated gene encodes the truncated products missing 83 residues of wild-type DNA-PKcs products. Furthermore, the quantity of DNA-PKcs transcript in wild-type and SCID cells was almost equal. These observations indicate that the DNA-PKcs gene is the SCID-responsible gene itself and that the detected mutation leads to the SCID aberration.
Severe combined immune deficiency (SCID) mice exhibit the immunodeficiency owing to a defect in their mature lymphocytes (1). Detailed analyses have disclosed an aberration of V(D)J recombination in lymphocytes and a high sensitivity to ionizing radiation in this strain (2–9). A number of researchers including ourselves used the cell fusion and microcell technique to identify the responsible gene and reached the conclusion that the region of human chromosome 8q11 had the potential to complement the SCID aberration (10–14). The scid locus was fine-mapped to 8q11 (10, 15). Meanwhile, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) gene that encodes one of the components of DNA double-strand break end-binding complex was mapped to the same locus, human chromosome 8q11 (15, 16). Furthermore, the reduction of DNA-PKcs protein and drastic reduction of kinase activity in extracts in SCID cells was observed (15, 17, 18). However, genes or their products exist downstream in the DNA-PKcs signal transduction pathway or functional homologues have a potential to exhibit phenotypes similar to those detected in complementation experiments using the DNA-PKcs gene. Indeed, we were able to establish several radioresistant transformants from SCID cell line by means of gene transfection. These transformants contain some genes other than the DNA-PKcs gene. Therefore, we considered that two facts must be established to confirm the hypothesis that the DNA-PKcs gene is the SCID-responsible gene itself: (i) the genetic locus of the murine DNA-PKcs gene resides in the centromeric region of the murine chromosome 16 in which the scid locus was assigned by cross-genetic studies (19), and (ii) the presence of the mutation(s) in the DNA-PKcs gene in SCID mice. As for the former, we confirmed the hypothesis by the fluorescence in situ hybridization method using a partial region of the murine DNA-PKcs gene as a hybridization probe (20). As for the latter, in this study, we isolated cDNAs for the whole coding region of the murine DNA-PKcs gene from SCID and C.B-17 mice and determined their DNA sequence.
MATERIALS AND METHODS
Isolation of cDNA Containing the Whole Region of ORF of Murine DNA-PKcs.
To isolate the N-terminal region of the murine DNA-PKcs gene, a cDNA library (mouse fibroblast cDNA library; CLONTECH) was used. For the C-terminal region, mouse peripheral blood leukocyte cDNA library (CLONTECH) was used. The human DNA-PKcs genes clone 13 and 1 were used for hybridization probes, respectively (21). Plaque hybridizations were carried out according to a procedures described previously (22). The probes were labeled with [α-32P]dCTP using a nick translation kit (Nippon Gene, Toyama, Japan).
Cell Lines.
Fibroblast cell lines SCNT3A and CBN2 were established from the tail of C.B-17 scid/scid and C.B-17 +/+ mice, respectively, by spontaneous transformation in long-term culture. 46–6 (+/+), S41, S55 (scid/scid), and SN-3 (+/scid) are Abelson murine leukemia virus-transformed pre-B cell line (23, 24). Culture conditions are DMEM (GIBCO/BRL) supplemented with 10% fetal calf serum for fibroblast cell lines, and RPMI 1640 medium (GIBCO/BRL) supplemented with 5% fetal calf serum and 50 μM 2-mercaptoethanol for pre-B cell line.
Reverse Transcriptase–PCR (RT-PCR).
Total RNAs were prepared from 1 × 107 cells with RNeasy total RNA Kit (Qiagen, Hilden, Germany), and 1 μg of total RNA fraction was used for the first strand synthesis. The synthesis was performed by Superscript II (GIBCO/BRL) with primer 5′-CCTTTATCTGACCATCTCGCCAGCACCATA-3′ (Fig. 1, primer a) according to the method recommended by the supplier. It was important in the analysis not to misread the DNA sequence through misincorporation at PCR steps. Therefore, the samples used for DNA sequence analyses were prepared by combining PCR products amplified with different sets of primers: 5′-TGTGCAACTTCACTAAGTCCATGGAAGAAG-3′ (Fig. 1, primer c) and 5′-AGGAGGTCCTCTCGGAGACAGAATGCTTTA-3′ (Fig. 1, primer b) or 5′-GCCTACGATTACTTACCCTGCATGCATCTC-3′ (Fig. 1, primer d) and 5′-AGGAGGTCCTCTCGGAGACAGAATGCTTTA-3′ (Fig. 1, primer b). We used LA PCR Kit version 2 (Takara Shuzo, Kyoto). The conditions of RT-PCR were as follows: 94°C, 1 min, 1 cycle; 98°C, 20 sec; 68°C, 15 min, 30 cycles; 72°C, 10 min, 1 cycle. The combined amplified samples were completely digested with either _Sau_3AI or _Pst_I restriction enzymes and recloned into M13 mp18 or pBluescript II SK(−). Contiguous deletion clones were prepared from the clones more than 400 bp long (double-stranded nested deletion kit; Pharmacia Biotech). These samples were used for DNA sequence analysis.
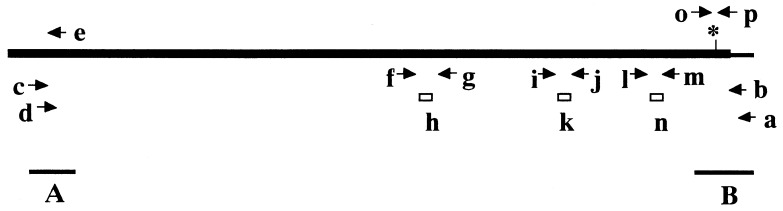
Figure 1.
Location of the primers used for the sequencing and quantitative assay of DNA-PKcs RNA. The DNA-PKcs genes were obtained from C.B-17 and SCID cell lines by long and accurate PCR methods with the indicated primers b, c, and d after the first strand synthesis using the primer a. The 5′ rapid amplification of cDNA ends (RACE) was accomplished using primer e. These primers (a–d) were designed based on the DNA sequence data from the clones (A and B) isolated from the mouse cDNA library. A pair of primers (o and p) were used to verify the generality of the SCID mutation (indicated by an asterisk). Three pairs of primers (f and g, i and j, and l and m) were used with the respective Taqman probes (h, k, and n) for the quantitation of the DNA-PKcs gene expression.
With primer 5′-TCTGGGGATCTTCTTCCATGGACTTAGTGA-3′ (Fig. 1, primer e), 5′-RACE reaction was performed as reported (25). RACE and RT-PCR were carried out with RNA prepared from a C.B-17-derived cell line (CBN2) and a SCID-derived cell line (SCNT3A). Finally, the remaining gap regions that could not be isolated by either method were amplified by PCR with appropriate primers.
The DNA sequence was determined in each region at least five times on both strands. The sequence analysis was carried out with an automatic DNA sequencer using an Applied Biosystems PRISM Dye Primer Cycle Sequencing FS Ready Reaction Kit (Perkin–Elmer).
Test for Generality of the Mutation.
The following primers were used for the PCR: m6-DNA-PK(+), 5′-GGAAAAGAATTGGTATCCAC-3′ (Fig. 1, primer o); and m8-DNA-PK(−), 5′-GTTGGCCCCTGCTAACTTTC-3′ (Fig. 1, primer p). The template DNAs were prepared from the spleen of SCID and C.B-17 mice purchased from Nippon Clea (Tokyo) by using SepaGene Kit (Sanko-jyunyaku, Tokyo). The subjects were 4-week-old female mice. One microgram of cellular DNA fractions was used for template. The amplification reaction was carried out in a 50 μl volume in the presence of 200 μM dNTPs, 0.4 μM primer, 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris·HCl (pH 8.3), and 1.25 units of Taq DNA polymerase (Takara Shuzo). Five microliters of each sample was digested with restriction enzyme _Alu_I (Takara Shuzo).
Quantitative RT-PCR.
Total RNA preparation from 1 × 107 was performed as mentioned above. Quantitative RT-PCR to measure the DNA-PKcs transcripts of SCID and normal cells was used with three independent primer sets and oligonucleotide probes using an Applied Biosystems PRISM 7700 Sequence Detector (Perkin–Elmer) (26, 27). The primer sets and probes are described in Fig. 1: primer f, 5′-AACACGTGATAGCAGAGTTGG-3′; primer g, 5′-CTCTACATGTTGGAGAAGGATG-3′; primer h, 5′-CGAGAACTTCTGAATCCTGTTGTGG-3′; primer i, 5′-AGACTCGCCAAGTTACAGTCTG-3′; primer j, 5′-CTGAAGCATGCAACGAACA-3′; primer k, 5′-TGGATGAGGATGAAGAGTCCATTGAC-3′; primer l, 5′-TGTAAGATCACTGGCTCGCT-3′; primer m, 5′-TGGCCTCGAATAACAATGC-3′; and primer n, 5′-TCACGTGCGGATCTCTGGATT-3′.
For internal control of the RNA, expression of β-actin was examined by RT-PCR as described above. For the first-strand synthesis, 5′-CAGCTCAGTAACAGTCCGCCTAGAAGCACT-3′ was used as a primer. The following primers are used for the PCR: 5′-GAACCCTAAGGCCAACCGTGAAAAGATGAC-3′, and 5′-TGATCTTCATGGTGCTAGGAGCCAGAGCAG-3′.
Amplified products were separated by agarose gel electrophoresis and visualized with ethidium bromide. The intensity of the bands was quantified using the Bio-Profil system (Vilber-Lourmat, Marne La Vallee, France).
RESULTS AND DISCUSSION
We used PCR to isolate the full-length murine DNA-PKcs cDNA. Initially, the N-terminal and C-terminal regions of the cDNA were isolated from the murine cDNA library by plaque hybridization. Human DNA-PKcs gene fragments were used as the probes. Four independent clones containing the C-terminal region were isolated from 500,000 plaques (20), and only 1 clone containing the N-terminal region was isolated by screening 600,000 plaques (Fig. 1, clones A and B). These results suggest the extremely low expression of the DNA-PKcs gene in the mouse cell line used for the preparation of the cDNA library (28). RT-PCR was performed with the primers designed in the isolated N- and C-terminal regions. Single bands of 12 kb were amplified using the total RNA fraction prepared from both SCID-derived and C.B-17-derived mice cell lines. It is noteworthy that the length of the amplified fragments was identical in the two strains and the intensity of the fragments was nearly equal. More of the 5′ region which could not be isolated by plaque hybridization was obtained by the 5′-RACE technique using the primer designed in the sequence on the isolated N-terminal clone.
Through this study we took great care to prevent the missequencing caused by the PCR step owing to the low fidelity of Taq DNA polymerase. We used LA Taq DNA polymerase, which shows a fidelity (3.2 × 10−6 errors/bp) in DNA polymerization step more than 6-fold higher than that of Taq DNA polymerase (2.0 × 10−5 errors/bp) (29, 30). In addition, the templates for sequencing were independently amplified with different sets of primers before being combined. Sequencing was conducted at least five times on both strands in each region divided with utilized restriction enzymes. The full length of the transcript is more than 12.5 kbp.
The amino acid sequence of the DNA-PKcs of C.B-17 mice is shown in Fig. 2. The murine DNA-PKcs gene has a huge ORF containing 4128 aa and it shows 78.9% homology with the human DNA-PKcs ORF. We found that there are some highly conserved regions, as shown in Fig. 3. The regions are expected to encode a domain possessing an important function.
Figure 2.
Deduced amino acid sequence from the nucleotide sequence of murine DNA-PKcs gene. The phosphatidylinositol 3-kinase (PI3K) motifs are indicated with solid bars. Tyr-4046 is indicated by an arrow, which is terminated in the SCID DNA-PKcs gene (see also Fig. 4). The leucin zipper motif is conserved (asterisks). The box indicates the potential recognition sites of CPP32 (apopine) (31, 32).
Figure 3.
Comparison of the amino acid sequence of the DNA-PKcs between human and mouse. All amino acid residues are indicated by three symbols; vertical bars (∥) are for the identical residues and small horizontal bars (−) are for the residues with similar biochemical features; spaces are for the residues with no similarity between the human and mouse counterparts. There are some extremely conserved regions, which are surrounded by open squares. Solid bars (▪) are for the consensus sequence of PI3K.
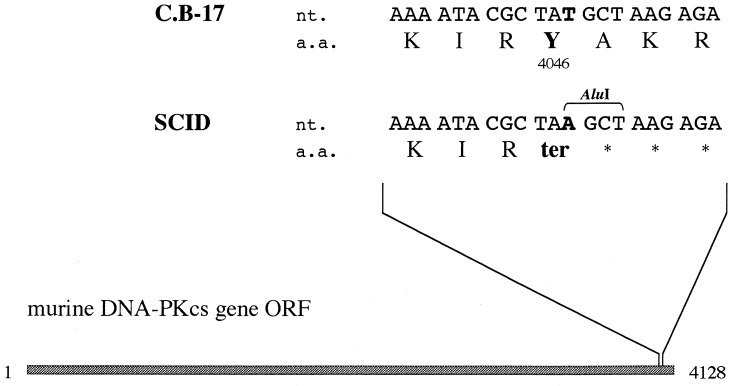
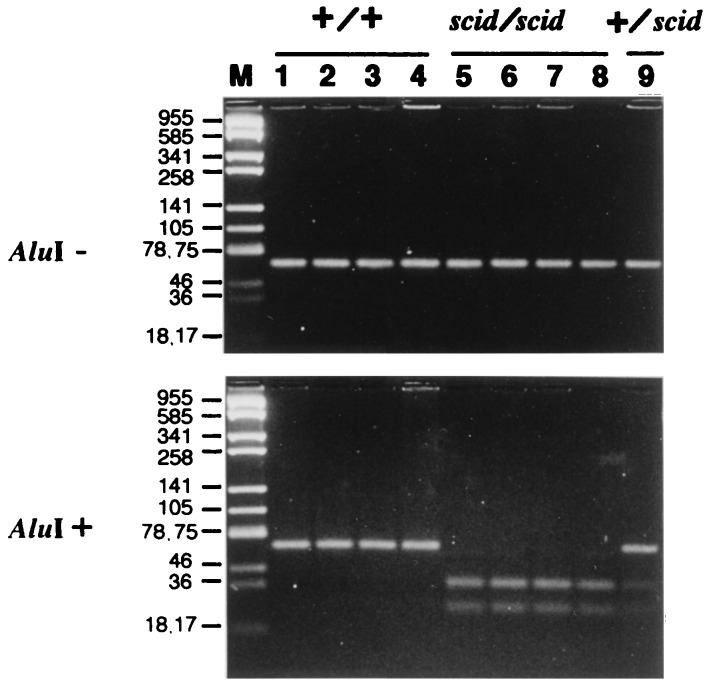
We detected the nonsense mutation on Tyr-4046 residue in SCID DNA-PKcs ORF. As shown in Fig. 4, substitution of nucleotide A for nucleotide T truncates the gene product, resulting in 83-aa removal from the C terminus. No mutation other than the nonsense mutation was detected in the ORF of SCID mice. We tested whether the nonsense mutation could be verified with SCID and C.B-17 mice. As indicated in Fig. 4, since the substitution occurring in SCID mice creates a restriction enzyme _Alu_I recognition site, we amplified the regions of interest with appropriate primers and digested them with restriction enzyme _Alu_I. The amplified DNA sample without the mutation should be 64 bp long, and the sample with the mutation should be divided in lengths of 38 and 26 bp after _Alu_I digestion. The results clearly demonstrated that the mutation occurred only in SCID individuals (Fig. 5). The mutation of the regions was also confirmed by sequence analyses.
Figure 4.
Nonsense mutation in DNA-PKcs gene in SCID mice. The dark box indicates the ORF (1-4128). Only the mutated region is indicated above. nt. and a.a. indicate the nucleotide sequence and amino acid sequence, respectively. The replaced nucleotides and amino acid residues are shown in boldface type. The one-letter symbols are used to specify amino acids. _Alu_I indicates the recognition sequence of restriction enzyme _Alu_I.
Figure 5.
Confirmation for generality of the mutation. The single bands of 64 bp were amplified by PCR (Upper) followed by _Alu_I digestion (Lower). The products containing the mutation were digested into 38- and 26-bp bands. Four individuals of C.B-17 (+/+) (lanes 1–4), scid/scid (lanes 5–8), and +/scid (lane 9) were analyzed. (Lane M) pUC19 digested with _Sau_3AI was used for molecular weight marker and the molecular weight of each band is indicated (bp). Samples were resolved by electrophoresis with 5% Nu Sieve agarose (FMC).
In addition, to confirm that the nonsense mutation was not caused spontaneously during maintenance, we tested the mutation with DNA samples derived from SCID mice purchased from Nippon Clea (Fig. 5) and donated by Melvin Bosma (Fox Chase Cancer Center, Philadelphia) (data not shown), and SCID cell lines donated by Melvin Bosma (Fig. 5). Then we detected the mutation in all samples. These observations indicated that the nonsense mutation is not caused and fixed during maintenance.
Homology searches performed by other groups disclosed that the DNA-PKcs belong to the PI3K family (21, 33, 34). However the association of putative PI3K domain region with DNA-PKcs activity has not been clarified. Our finding indicates this relationship. The Tyr-4046 residue is conserved between mouse and human but not among the PI3K family. The putative kinase region with highest homology ranges from amino acid 3779 to 4098 in mice. The motifs (DXXXXN and DFG) implicated in catalytic activity are at 3941–3943 and 3922–3927. Therefore, the identified nonsense mutation (4046) resides just 3′ to the motifs. This fact might indicate the direct involvement of the Tyr residue in the active center. On the other hand, it is also possible that the nonsense mutation affects the stability and/or folding of the DNA-PKcs protein. The fact that the transcripts of the DNA-PKcs were similar in SCID and normal cells while the protein level decreased suggests this possibility (15, 17, 18). In any case, the C-terminal region from the Tyr residue is critical for DNA-PKcs activity. Indeed, in addition to the active center of PI3K, there is homologous regions among the PI3K family at the C terminus (4098–4128). The ataxia telangiectasia gene (ATM) is a member of the PI3K family. In ATM, two types of mutation were reported to take place in the region, which correspond to the SCID mutation reported (91RD90, AT103LO) (35).
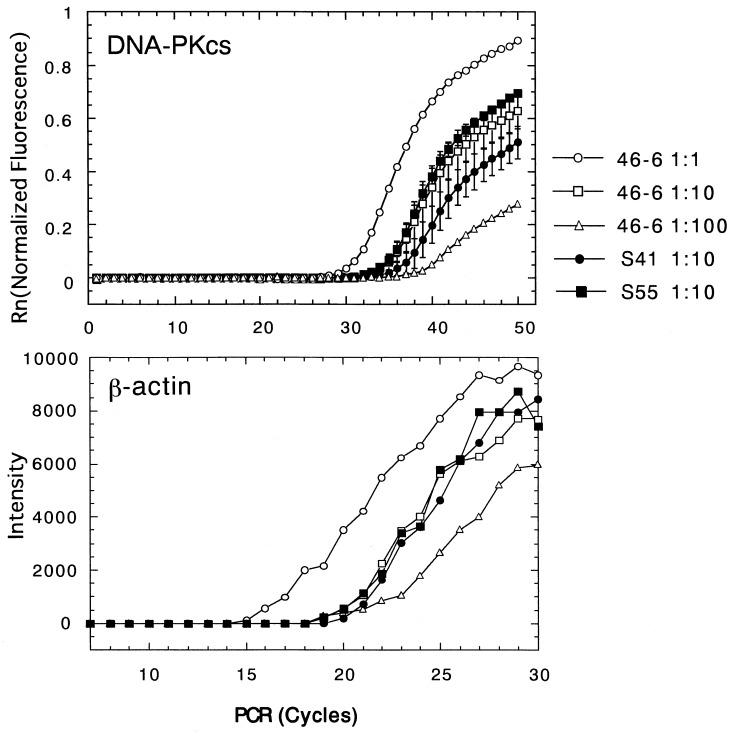
To gain further information on the SCID defect, we measured the DNA-PKcs transcript in SCID cells. It is very difficult to accurately examine the expression rate of the DNA-PKcs gene coding more than 12-kbp mRNA with Northern blot hybridization. Therefore, we used quantitative RT-PCR by amplifying different regions but did not detect a significant difference in mRNA of DNA-PKcs between SCID and normal cells (Fig. 6 and Table 1).
Figure 6.
Measurement of the DNA-PKcs gene expression between wild-type and SCID cell lines. DNA-PKcs gene expression was measured by the RT-PCR method using the Applied Biosystems Prism 7700 (Perkin–Elmar) as described. The representative fluorescence intensity (Rn) was indicated as the function of the frequency of the PCRs with the probe n (Upper). The wild-type 46–6 was tested with RNA samples of different dilutions; 1:1 (○), 1:10 (□), and 1:100 (▵). For the SCID cell lines, the samples from S41 (•) and S55 (▪) were both tested with 1:10 diluted samples. Standard deviation is indicated as a vertical bar. The initial RNA samples were normalized by the number of the exponentially growing culture cells. (Lower) For reference, β-actin gene expression was measured from the identical RNA samples.
Table 1.
Quantitation of RT-PCR products
| Probe* | Cell line | ||
|---|---|---|---|
| 46-6 | S41 | S55 | |
| n | 10,000† 17,000 | 4,400 5,800 | 12,000 19,000 |
| k | 10,000† 16,000 | 5,800 3,200 | 12,000 15,000 |
| h | 10,000† 17,000 | 3,400 4,200 | 9,700 23,000 |
Our observations indicate that the DNA-PKcs gene is the gene responsible for scid mutation. To confirm the role of the gene in SCID aberration, reverse genetics under in vivo and in vitro conditions should be conducted. Unfortunately, however, cloning of the full-length DNA-PKcs cDNA into the plasmid-expression vector is currently extremely difficult. In particular, the 5′ flanking region of the PI3K encoding region proved impossible to be cloned into available vectors. The analyses will be facilitated by the mice in which the mutation in DNA-PKcs gene is introduced or removed.
Acknowledgments
We thank Drs. S. Abrahamson and K. Yokoro for encouragement and useful suggestions. Dr. S. Jackson generously provided human DNA-PKcs hybridization probes; Dr. M. Bosma provided cell lines S41, S55, and SN-3; and Dr. T. Takemori provided cell line 46-6. We thank Dr. H. Ishihara, Mr. S. Fukushima, and Dr. O. Sakatsume for technical advice. We also thank Mr. and Ms. M. Edo, Y. Inada, K. Tagawa, N. Kai, H. Miki, Y. Mizuta, E. Kubo, S. Sasanuma, J. Nohata, T. Kimura, and T. Hayao for technical assistance; and Mr. B. F. Burke-Gaffney and Ms. M. Hirose for editing the English manuscript. This publication is partly supported by the Radiation Effects Research Foundation, Hiroshima and Nagasaki, Japan. The Radiation Effects Research Foundation is a private nonprofit foundation funded equally by the Japanese Ministry of Health and Welfare and the U.S. Department of Energy through the National Academy of Sciences.
ABBREVIATIONS
DNA-PKcs
DNA-dependent protein kinase catalytic subunit
SCID
severe combined immune deficiency
RACE
rapid amplification of cDNA ends
PI3K
phosphatidylinositol 3-kinase
RT-PCR
reverse transcriptase–PCR
Note Added in Proof:
Since this manuscript was submitted, two papers have appeared (36, 37) identifying the same mutation as we have as present in the SCID mouse. Neither of these papers, unlike the present paper, excluded the possibility of other relevant mutations in the SCID gene.
Footnotes
The sequence reported in this paper has been deposited in the GenBank database (accession no. D87521D87521).
References
- 1.Bosma G C, Custer R P, Bosma M J. Nature (London) 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 2.Schuler W, Weiler I J, Schuler A, Phillips R A, Rosenberg N, Mak T W, Kearney J F, Perry R P, Bosma M. Cell. 1986;46:963–972. doi: 10.1016/0092-8674(86)90695-1. [DOI] [PubMed] [Google Scholar]
- 3.Hendrickson E A, Schatz D G, Weaver D T. Genes Dev. 1988;2:817–829. doi: 10.1101/gad.2.7.817. [DOI] [PubMed] [Google Scholar]
- 4.Kim M G, Schuler W, Bosma M J, Marcu K B. J Immunol. 1988;141:1341–1347. [PubMed] [Google Scholar]
- 5.Malynn B A, Blackwell T K, Fulop G M, Rathbun O A, Furley A J W, Ferrier P, Heinke L B, Phillips R A, Yancopoulos G D, Alt F W. Cell. 1988;54:453–460. doi: 10.1016/0092-8674(88)90066-9. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki K, Nishikawa S, Sakano H. J Immunol. 1988;141:1348–1352. [PubMed] [Google Scholar]
- 7.Blackwell T K, Malynn B A, Pollack R R, Ferrier P, Covey L R, Fulop G M, Phillips R A, Yancopoulos G D, Alt F W. EMBO J. 1989;8:735–742. doi: 10.1002/j.1460-2075.1989.tb03433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulop G M, Phillips R A. Nature (London) 1990;347:479–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- 9.Biedermann K A, Sun J, Giaccia A J, Tosto L M, Brown J M. Proc Natl Acad Sci USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh M, Hamatani K, Komatsu K, Araki R, Takayama K, Abe M. Radiat Res. 1993;134:364–368. [PubMed] [Google Scholar]
- 11.Kirchgessner C U, Tosto L M, Biedermann K A, Kovacs M, Araujo D, Stanbridge E J, Brown J M. Cancer Res. 1993;53:6011–6016. [PubMed] [Google Scholar]
- 12.Komatsu K, Ohta T, Jinno Y, Niikawa N, Okumura Y. Hum Mol Genet. 1993;2:1031–1034. doi: 10.1093/hmg/2.7.1031. [DOI] [PubMed] [Google Scholar]
- 13.Banga S S, Hall K T, Sandhu A K, Weaver D T, Athwal R S. Mutat Res. 1994;315:239–247. doi: 10.1016/0921-8777(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 14.Kurimasa A, Nagata Y, Shimizu M, Emi M, Nakamura Y, Oshimura M. Hum Genet. 1994;93:21–26. doi: 10.1007/BF00218907. [DOI] [PubMed] [Google Scholar]
- 15.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 16.Sipley J D, Menninger J C, Hartley K O, Ward D C, Jackson S P, Anderson C W. Proc Natl Acad Sci USA. 1995;92:7515–7519. doi: 10.1073/pnas.92.16.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 18.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosma G C, Davisson M T, Reutsch N R, Sweet H O, Shultz L D, Bosma M J. Immunogenetics. 1989;29:54–57. doi: 10.1007/BF02341614. [DOI] [PubMed] [Google Scholar]
- 20.Hamatani K, Matsuda Y, Araki R, Itoh M, Abe M. Immunogenetics. 1996;45:1–5. doi: 10.1007/s002510050159. [DOI] [PubMed] [Google Scholar]
- 21.Hartley K O, Gell D, Smith G C M, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 22.Benton W D, Davis R W. Science. 1977;196:180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- 23.Lieber M R, Hesse J E, Lewis S, Bosma G C, Rosenberg N, Mizuuchi K, Bosma M J, Gellert M. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 24.Takemori T, Mizuguchi J, Miyazoe I, Nakanishi M, Shigemoto K, Kimoto H, Shirasawa T, Maruyama N, Taniguchi M. EMBO J. 1990;9:2493–2500. doi: 10.1002/j.1460-2075.1990.tb07428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson U E M, Heid C A, Williams P M. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 27.Heid C A, Stevens J, Livak K J, Williams P M. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 28.Miller R D, Hogg J, Ozaki J H, Gell D, Jackson S P, Riblat R. Proc Natl Acad Sci USA. 1995;92:10782–10795. doi: 10.1073/pnas.92.23.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes W E. Gene. 1992;112:29–35. doi: 10.1016/0378-1119(92)90299-5. [DOI] [PubMed] [Google Scholar]
- 30.Barnes W M. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Q, Lees-Miller S P, Kumar S, Zhang N, Chan D W, Smith G C M, Jackson S P, Alnemri E S, Litwack G, Khanna K K, Lavin M F. EMBO J. 1996;15:3238–3246. [PMC free article] [PubMed] [Google Scholar]
- 32.Teraoka H, Yumoto Y, Watanabe F, Tsukada K, Suwa A, Enari M, Nagata S. FEBS Lett. 1996;393:1–6. doi: 10.1016/0014-5793(96)00842-3. [DOI] [PubMed] [Google Scholar]
- 33.Hunter T. Cell. 1995;83:1–4. doi: 10.1016/0092-8674(95)90225-2. [DOI] [PubMed] [Google Scholar]
- 34.Poltoratsky V P, Shi X, York J D, Lieber M R, Carter T H. J Immunol. 1995;155:4529–4533. [PubMed] [Google Scholar]
- 35.Gilad S, Khosravi R, Shkedy D, Uziel T, Ziv Y, et al. Hum Mol Genet. 1996;5:433–439. doi: 10.1093/hmg/5.4.433. [DOI] [PubMed] [Google Scholar]
- 36.Blunt T, Gell D, Fox M, Taccioli G E, Lehmann A R, Jackson S P, Jeggo P A. Proc Natl Acad Sci USA. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danska J S, Holland D P, Mariathasan S, Williams K M, Guidos C. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]