Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control (original) (raw)
Abstract
The Hippo pathway plays a key role in organ size control by regulating cell proliferation and apoptosis in Drosophila. Although recent genetic studies have shown that the Hippo pathway is regulated by the NF2 and Fat tumor suppressors, the physiological regulations of this pathway are unknown. Here we show that in mammalian cells, the transcription coactivator YAP (Yes-associated protein), is inhibited by cell density via the Hippo pathway. Phosphorylation by the Lats tumor suppressor kinase leads to cytoplasmic translocation and inactivation of the YAP oncoprotein. Furthermore, attenuation of this phosphorylation of YAP or Yorkie (Yki), the Drosophila homolog of YAP, potentiates their growth-promoting function in vivo. Moreover, YAP overexpression regulates gene expression in a manner opposite to cell density, and is able to overcome cell contact inhibition. Inhibition of YAP function restores contact inhibition in a human cancer cell line bearing deletion of Salvador (Sav), a Hippo pathway component. Interestingly, we observed that YAP protein is elevated and nuclear localized in some human liver and prostate cancers. Our observations demonstrate that YAP plays a key role in the Hippo pathway to control cell proliferation in response to cell contact.
[Keywords: YAP, Lats, Mst, contact inhibition, NF2, Hippo]
Body and organ size of metazoans is determined by cell number and cell size. The opposing action of cell proliferation and apoptosis controls cell number in particular tissue and organs (Conlon and Raff 1999). Recent genetic studies in Drosophila have shown that the Hippo signaling pathway plays a key role in restricting organ size by controlling both cell proliferation and apoptosis (Edgar 2006; Harvey and Tapon 2007; Pan 2007). Hippo (Hpo) is a Ste20 family protein kinase that complexes with a regulatory scaffold protein Salvador (Sav) (Kango-Singh et al. 2002; Tapon et al. 2002; Harvey et al. 2003; Pantalacci et al. 2003; Udan et al. 2003; Wu et al. 2003). The Hpo/Sav complex phosphorylates and activates Warts (Wts), a NDR (nuclear Dbf2-related) family protein kinase. Wts has an activating subunit Mats (Mob as tumor suppressor) (Lai et al. 2005; Wei et al. 2007). The Wts/Mats complex inhibits Yorkie (Yki), a transcription coactivator (Huang et al. 2005), possibly via direct phosphorylation, although the precise mechanism has yet to be determined. In Drosophila, key downstream targets of Yki include cyclin E, diap-1; and the bantam microRNA (Huang et al. 2005; Nolo et al. 2006; Thompson and Cohen 2006).
Although elusive for several years, the signals upstream of Hpo are now emerging. The NF2 tumor suppressor, also known as Merlin (Mer), and Expanded (Ex), two ezrin/radixin/moesin (ERM) family actin-binding proteins (McClatchey and Giovannini 2005; Okada et al. 2007), have been shown to positively regulate the Hippo pathway in Drosophila (Hamaratoglu et al. 2006). Interestingly, genetic data indicate that Fat, a protocadherin tumor suppressor, also functions upstream of Hpo (Bennett and Harvey 2006; Cho et al. 2006; Hariharan 2006; Silva et al. 2006; Willecke et al. 2006; Tyler and Baker 2007; Yin and Pan 2007). The fact that Fat may interact with another protocadherin, Dachsous, at the cell surface (Matakatsu and Blair 2004; Halbleib and Nelson 2006) suggests an exciting possibility that the Hippo pathway may be involved in cell growth regulation in response to cell–cell contact.
Components of the Hippo pathway are highly conserved in mammals, including YAP (Yes-associated protein), Lats1/2, Mob, Mst1/2, Sav, Merlin, Ex1/2, and Fat4 (Yki, Wts, Mats, Hpo, Sav, Mer, Expanded, and Fat homologs, respectively). Human YAP, Lats1, Mst2, and Mob1 can functionally rescue the respective Drosophila mutants, suggesting the functional conservation of these proteins in mammals (Edgar 2006). Interestingly, YAP has been shown recently to be a candidate oncogene in the human chromosome 11q22 amplicon (Overholtzer et al. 2006; Zender et al. 2006). In addition, mutations of Lats1/2, Sav, and Mob have been implicated in tumorigenesis (St John et al. 1999; Tapon et al. 2002; Lai et al. 2005; Takahashi et al. 2005; Harvey and Tapon 2007). In spite of their conservation and intimate relationship with cancer, the Hippo pathway has not been systematically studied in mammalian cells.
A fundamental property of a normal cell is to cease proliferation upon reaching confluence, a phenomenon referred to as cell contact inhibition (Eagle and Levine 1967). In contrast, cancer cells are able to escape cell contact inhibition, which enhances their ability to invade host tissues and metastasize (Hanahan and Weinberg 2000). This is also one of the most commonly used criteria for cellular transformation in vitro (Abercrombie 1979). Although activation of oncogenes and inactivation of tumor suppressor genes can prevent contact inhibition, the precise molecular mechanism is not clear.
In this report we show that YAP is regulated by the Hippo pathway and may play an important role in mediating cell contact inhibition. YAP is phosphorylated and inhibited by the Lats tumor suppressor, and this phosphorylation results in its association with 14–3–3 and cytoplasmic localization. This regulatory mechanism is utilized in YAP regulation by cell density and is likely conserved in Drosophila. Furthermore, overexpression of YAP antagonizes density-dependent gene regulation and contact inhibition, whereas expression of dominant-negative YAP restores contact inhibition in a human cancer cell line bearing a deletion of Sav. Moreover, we showed that YAP expression levels and nuclear localization are strongly elevated in some human cancers.
Results
YAP localization and phosphorylation are regulated by cell density
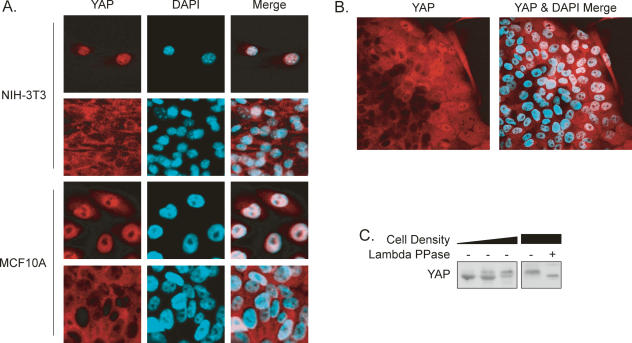
YAP is a transcription coactivator and a candidate oncogene, but neither its function in cancers nor its physiological regulation has been established. Interestingly, we found that YAP localization was regulated by cell density (Fig. 1A). At low density, YAP was predominantly localized in the nuclei of NIH-3T3 cells. In contrast, YAP translocated to the cytoplasm at high density. Similar observations were made in the MCF10A human breast epithelial cell line (Fig. 1A). This translocation was unlikely due to differential medium conditions, because in cell colonies YAP was preferentially localized to nuclei in cells at the edge but displayed cytoplasmic localization in cells toward the center (Fig. 1B). Given the fact that YAP is a transcription coactivator acting in the cell nucleus (Yagi et al. 1999), our results indicate that YAP may be inhibited by high cell density.
Figure 1.
YAP localization and phosphorylation are regulated by cell density. (A) YAP localization is affected by cell density. NIH-3T3 and MCF10A cells were cultured sparsely or to confluence. YAP was stained with anti-YAP antibody. (B) MCF10A cells at the edge of a large colony have high nuclear YAP. YAP was stained with anti-YAP antibody. (C) High cell density induces YAP phosphorylation. NIH-3T3 cell lysates from cells at different densities were probed with anti-YAP antibody. λ phosphatase treatment is indicated.
Besides translocation, YAP from high-density cultures displayed a slower electrophoretic migration (Fig. 1C). This density-dependent mobility shift was due to phosphorylation because phosphatase treatment converted YAP to the fast migrating form, suggesting that YAP phosphorylation is regulated by cell density. Together, the above observations indicate a possible relationship between YAP phosphorylation and cytoplasmic localization upon high cell density.
The Hippo pathway regulates YAP phosphorylation, activity, and localization
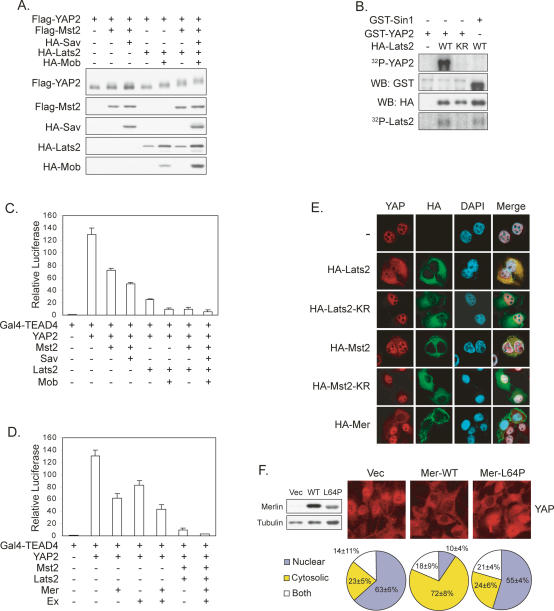
In Drosophila, it has been reported that Yki, the YAP homolog, is inhibited by the Hippo pathway, possibly via phosphorylation (Huang et al. 2005). Therefore, we tested effects of the Hippo pathway on YAP phosphorylation. All cDNAs used in the cell culture studies are of human or mouse origin. YAP2, one of the two alternatively spliced forms of human yap, was coexpressed with the Hippo pathway kinases Mst2 or Lats2. We found that expression of Mst2 or Lats2 caused a modest mobility shift of YAP2 that was further enhanced by Sav and Mob, the respective regulatory subunits of Mst2 and Lats2 (Fig. 2A). Moreover, coexpression of both Mst2 and Lats2 resulted in a dramatic mobility shift of YAP2. These results indicate that ectopic expression of Mst2 and Lats2 induces YAP2 phosphorylation.
Figure 2.
The Hippo pathway regulates YAP phosphorylation, activity, and localization. (A) Coexpression of Mst2 and Lats2 decreases YAP2 mobility. Flag-YAP2 was cotransfected with indicated plasmids into HEK293 cells. Western blot was performed as indicated. (B) In vitro phosphorylation of YAP2 by Lats2. HA-Lats2 was immunoprecipitated from transfected HEK293 cells. In vitro kinase assay was performed using purified GST-YAP2 as a substrate in the presence of [32P]ATP. GST-Sin1 was used as a negative control. (KR) Kinase-inactive mutant. (C) YAP2 activity is inhibited by Mst2 and Lats2. Indicated plasmids were cotransfected with a 5× UAS-luciferase reporter and a CMV-β-gal construct into 293T cells. Luciferase activity was measured and normalized to β-galactosidase activity. (D) YAP2 activity is inhibited by Merlin and Expanded. Experiments are similar to those in C. The Ex used is human FRMD6. (E) Activation of the Hippo pathway causes YAP cytoplasmic localization. HeLa cells were transfected with indicated plasmids. Endogenous YAP2 was stained to visualize the localization. (F) Cell density-induced YAP translocation is Merlin dependent. RT4-D6-P2T Schwannoma cell lines with empty vector, inducible wild-type Merlin, or a Merlin-L64P mutant were cultured to confluence. Merlin expression was induced by doxycycline for 2 d. (Left panel) Expression of Merlin was determined by Western blot. Endogenous YAP was stained and YAP localization was quantified.
In order to test the possibility of direct phosphorylation of YAP2 by Lats2, we performed an in vitro kinase assay using purified GST-YAP2 and immunoprecipitated Lats2. As shown in Figure 2B, Lats2, but not the kinase-inactive Lats2-KR, phosphorylated YAP2. In contrast, Mst2 poorly phosphorylated GST-YAP2, even though it had much stronger autophosphorylation than that of Lats2 (Supplementary Fig. S1A). These data demonstrate that Lats2 directly phosphorylates YAP2, while Mst2 stimulates YAP2 phosphorylation indirectly in vivo, perhaps by activating Lats2.
YAP has been shown to interact with and activate the TEAD family transcription factors, which have four highly conserved members (Vassilev et al. 2001). To assess the effect of phosphorylation on YAP activity, we utilized a reporter system consisting of a 5× UAS-luciferase reporter and a Gal4 DNA-binding domain fused to TEAD4 (Gal4-TEAD4). In the absence of YAP, Gal4-TEAD4 had low basal activity. However, when YAP2 was cotransfected, the reporter was strongly activated (Fig. 2C). Coexpression of Lats2 or Mst2, but not the kinase-inactive mutants, resulted in a dose-dependent inhibition of the reporter (Supplementary Fig. S1B,C). Reminiscent of the effect seen on phosphorylation, YAP2 activity was further inhibited by coexpressing Mst2/Sav or Lats2/Mob, and even more dramatically inhibited by a combination of all four proteins (Fig. 2C). This inhibition of YAP2 activity was also observed in COS7 and HeLa cells (data not shown). We also tested the effect of Merlin and Expanded on YAP2 activity. Coexpression of either caused a modest but reproducible inhibition of YAP2 activity (Fig. 2D). Furthermore, Merlin and Expanded enhanced the inhibition of YAP2 by Mst2 and Lats2. Consistently, Merlin also caused a mobility shift of YAP2 (Supplementary Fig. S1D).
Next, we addressed whether Mst2 and Lats2 affected YAP localization. In HeLa cells, endogenous YAP was localized in the nucleus at low cell density (Fig. 2E). However, expression of Lats2, but not the kinase-inactive mutant, caused a dramatic redistribution of YAP to the cytoplasm (Fig. 2E). Similarly, expression of Mst2 but not the kinase-inactive mutant increased cytoplasmic YAP, although less dramatically. Expression of Merlin also resulted in YAP cytoplasmic translocation, supporting the role of Merlin in the Hippo pathway in mammalian cells. These results suggest that activation of the Hippo pathway may cause cytoplasmic translocation of YAP through phosphorylation by Lats.
Schwannoma is the major tumor type associated with Merlin mutation. We examined YAP localization in the RT4-D6-P2T rat Schwannoma cell line, which is incapable of inducing Merlin expression at high cell density as normal Schwann cells do (Morrison et al. 2001). We observed that the majority of Schwannoma cells showed nuclear YAP localization even under high density (Fig. 2F). Interestingly, expression of Merlin wild-type, but not a cancer-derived L64P mutant, restored YAP cytoplasmic translocation. Together, these results further support the involvement of Merlin and the Hippo pathway in the regulation of YAP translocation in response to cell density.
Lats inhibits YAP by phosphorylating HXRXXS motifs
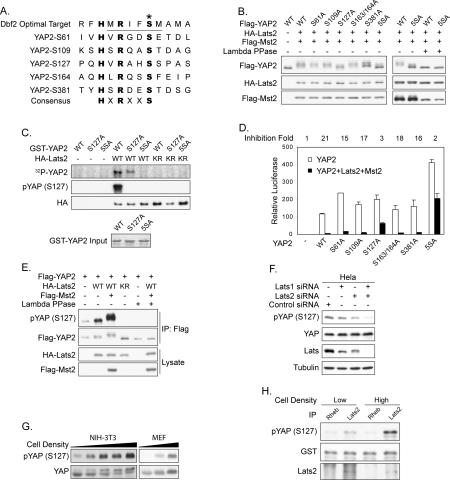
Lats belongs to the NDR family of protein kinases (Hergovich et al. 2006). Previous biochemical studies have shown that the yeast Dbf2 kinase recognizes an RXXS motif in its substrates (Mah et al. 2005). Interestingly, in search of such a consensus, we noticed that YAP2 contains five HXRXXS motifs (Fig. 3A), of which three are conserved in Drosophila. It is worth noting that the peptides utilized in elucidating the Dbf2 recognition motif also had a histidine at position −5 (Mah et al. 2005).
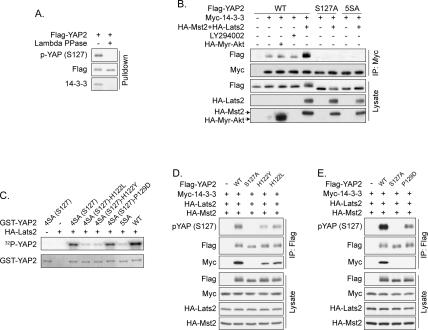
Figure 3.
Lats inhibits YAP by phosphorylating HXRXXS motifs. (A) YAP2 contains five HXRXXS motifs. The yeast Dbf2 optimal target sequence was aligned with the five HXRXXS motifs of human YAP2. (B) Ser127 is the major phosphorylation site in YAP2. Wild-type or mutant Flag-YAP2 was cotransfected with HA-Mst2 and HA-Lats2 as indicated. YAP2 mobility shift was determined by anti-Flag Western blot. (C) Lats2 directly phosphorylates YAP2 on HXRXXS motifs. In vitro phosphorylation of YAP2 mutants with immunoprecipitated HA-Lats2 was performed. Phosphorylation of GST-YAP2 was detected by either 32P incorporation or anti-phospho-YAP (S127) Western blot. (Bottom panel) GST-YAP2 input was shown by Coomassie Blue staining. (D) YAP2 phosphorylation-defective mutants S127A and 5SA are resistant to inhibition by Mst2 and Lats2. The reporter assay is similar to those in Figure 2C. The fold activity inhibition of each mutant by Mst2/Lats2 is indicated at the top of this panel. (E) Coexpression of Mst2 and Lats2 increases YAP2 S127 phosphorylation. Flag-YAP2 was cotransfected with HA-Lats2 and Flag-Mst2 into HEK293 cells as indicated. Flag-YAP2 was immunoprecipitated and phosphorylation of S127 was detected by pYAP (S127) antibody. (F) Knockdown of Lats decreases endogenous YAP S127 phosphorylation. HeLa cells were transfected twice with small interfering RNA for Lats1 and Lats2 as indicated. Phosphorylation and protein levels of endogenous YAP were determined by Western blot. Knockdown of Lats was verified by the anti-Lats antibody, which recognizes both Lats1 and Lats2. (G) YAP S127 phosphorylation increases with cell density. NIH-3T3 and MEF cells were harvested at different densities, and YAP phosphorylation was assayed. (H) Lats2 kinase activity increases with cell density. NIH-3T3 cells were harvested at different densities. Endogenous Lats2 was immunoprecipitated and used in an in vitro kinase assay. Phosphorylation of GST-YAP2 was detected by anti-phospho-YAP (S127) Western blot. Rheb IP was included as a negative control.
We mutated YAP2 by replacing individual serine residues in the HXRXXS motifs with alanine. Among the single mutants tested, S127A, which is conserved in Drosophila Yki, was most resistant to Mst2/Lats2-induced mobility shift (Fig. 3B). Mutation of all five serine residues (YAP2-5SA) produced a YAP2 downshift more dramatic than that of any single mutant. Furthermore, λ phosphatase treatment abolished the Mst2/Lats2-induced mobility shift of YAP2, therefore verifying the role of phosphorylation in this mobility shift (Fig. 3B). These results indicate that Ser127 is the primary phosphorylation site in YAP2, while serines in other HXRXXS motifs may also be phosphorylated.
To further confirm the phosphorylation of YAP2 HXRXXS motifs by Lats2, an in vitro kinase assay was performed. Mutation of S127 reduced and mutation of all five serine residues abolished YAP2 phosphorylation by Lats2 as determined by 32P incorporation (Fig. 3C). Phosphorylation of S127 was also verified by immunoblotting with a phospho-YAP (S127)-specific antibody (Fig. 3C). The specificity of this antibody was confirmed by phosphatase treatment (Supplementary Fig. S2A). These data demonstrate that Lats2 directly phosphorylates YAP2 on S127 and other serine residues in the HXRXXS motifs.
The functional significance of YAP2 phosphorylation was evaluated by the TEAD4 reporter assay. As shown before, wild-type YAP2 was potently inhibited by coexpression of Mst2/Lats2; however, the S127A mutant showed resistance to this inhibition (Fig. 3D). Furthermore, the YAP2-5SA mutant was not only resistant to inhibition by Mst2 and Lats2, but also displayed an elevated basal activity (Fig. 3D). Together, our data demonstrate that YAP2 activity is inhibited by phosphorylation of the HXRXXS motifs, especially S127. Since YAP2-5SA was also partially inhibited by Mst/Lats, additional levels of regulation may exist.
To confirm the phosphorylation of YAP2 S127 in vivo by Lats2, we did coexpression in cultured cells. Cotransfection of Lats2 alone or together with Mst2 increased YAP2 S127 phosphorylation (Fig. 3E). Interestingly, expression of the inactive Lats2-KR mutant decreased YAP2 basal phosphorylation, perhaps through a dominant-negative effect. This supports a role of endogenous Lats in YAP2 phosphorylation. To further confirm this, Lats1 and Lats2 were down-regulated by RNA interference. Knockdown of Lats2 caused a significant reduction in S127 phosphorylation of transfected Flag-YAP2, while knockdown of both Lats1 and Lats2 abolished its phosphorylation (Supplementary Fig. S2B). Similarly, knockdown of both Lats1 and Lats2 decreased endogenous YAP phosphorylation (Fig. 3F), thus establishing an important role of Lats in YAP phosphorylation in vivo.
After the determination of a Lats target phosphorylation site on YAP, we re-examined the cell density-induced phosphorylation of YAP. Along with the reduced electrophoretic migration of YAP as shown in Figure 1C, we also observed that YAP S127 phosphorylation was increased by cell density in both NIH-3T3 and mouse embryonic fibroblast (MEF) cells (Fig. 3G). To determine whether cell density regulates Lats kinase activity, we immunoprecipitated endogenous Lats2 from NIH-3T3 cells and measured its kinase activity toward YAP in vitro. Interestingly, Lats2 from high-density culture displayed an elevated activity (Fig. 3H). This result directly suggests the activation of Lats and possibly the Hippo pathway under high cell density, which nicely explains the increased phosphorylation of YAP.
It has been previously reported that YAP2 S127 is phosphorylated by Akt/PKB (Basu et al. 2003). However, the reported YAP inhibition by Akt-dependent phosphorylation is inconsistent with recent genetic data that demonstrate YAP as an oncogene. We tested the function of Akt in YAP phosphorylation. Surprisingly, neither LY294002 nor wortmannin (two PI3K inhibitors) decreased YAP2 S127 phosphorylation, although they potently blocked the phosphorylation of Akt and GSK3, a physiological Akt substrate (Supplementary Fig. S3A). In addition, neither EGF nor insulin stimulated YAP2 phosphorylation, while both strongly stimulated the phosphorylation of Akt and GSK3 (Supplementary Fig. S3A,B). Phosphorylation of Akt T308 by PDK1 is essential for Akt activity (Williams et al. 2000). However, YAP phosphorylation was not affected by PDK1 knockout (Supplementary Fig. S3C). In addition, coexpression of wild-type or constitutively active myristoylated Akt did not increase YAP2 phosphorylation (Supplementary Fig. S3D). We also observed that Akt did inhibit YAP2 activity, but in a kinase activity-independent manner, suggesting that Akt overexpression could inhibit YAP2 indirectly (Supplementary Fig. S3E). Together, our results demonstrate that Akt is unlikely to be responsible for YAP2 S127 phosphorylation.
Phosphorylation promotes YAP cytoplasmic localization and inhibits its transcription factor binding
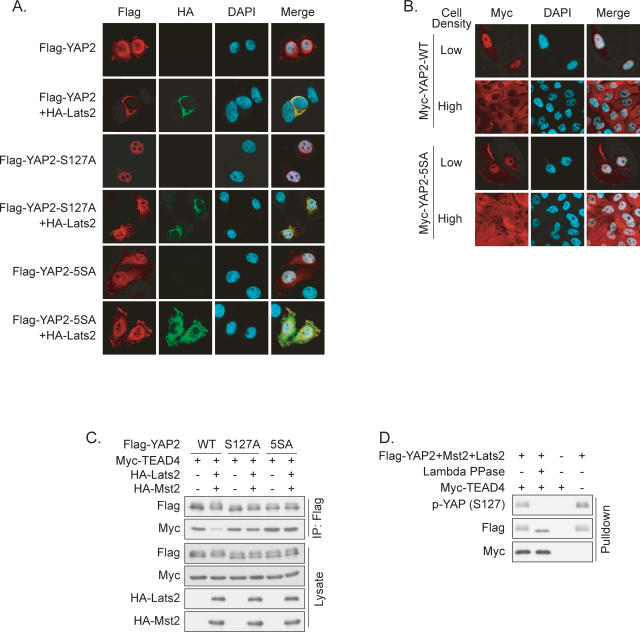
To directly determine the effect of phosphorylation on YAP localization, we performed immunofluorescence staining of transfected wild-type or phosphorylation-deficient YAP2. Flag-YAP2 showed prominent nuclear localization in transfected cells, while coexpression of Lats2 induced nearly complete cytoplasmic translocation (Fig. 4A). Interestingly, Lats2 had only minor effects on YAP2-S127A and 5SA localization.
Figure 4.
Phosphorylation promotes YAP cytoplasmic localization and inhibits transcription factor binding. (A) Ser127 is required for YAP2 cytoplasmic localization induced by Lats2. Flag-YAP2 wild type or mutants were transfected alone or together with HA-Lats2 into HeLa cells. Cells were stained with Flag and HA antibodies. (B) Phosphorylation is required for cell density-induced YAP2 cytoplasmic translocation. MCF10A cells stably expressing Myc-YAP2 or Myc-YAP2-5SA were cultured at low or high density. Myc-YAP2 was stained with anti-Myc antibody. (C) Lats and Mst decrease YAP2/TEAD4 interaction in vivo in a S127-dependent manner. Indicated plasmids were transfected into HEK293 cells. Flag-YAP2 was immunoprecipitated, and coprecipitated Myc-TEAD4 was detected by Western blot. (D) YAP2 dephosphorylation does not affect its interaction with TEAD4 in vitro. Flag-YAP2 (cotransfected with Mst2 and lats2) immunoprecipitated from HEK293 cells were treated with λ phosphatase as indicated and then used in an in vitro TEAD pull-down assay. Myc-TEAD4 was prepared from transfected HEK293 cells. The final products were analyzed by Western blot.
To determine whether Lats-dependent phosphorylation is indeed responsible for YAP translocation under high cell density, we examined MCF10A cells stably expressing Myc-YAP2 or Myc-YAP2-5SA. Similar to endogenous YAP, Myc-YAP2 showed density-dependent subcellular localization (Fig. 4B). In contrast, Myc-YAP2-5SA displayed both nuclear and cytoplasmic staining under high density. Together, our studies suggest that phosphorylation of HXRXXS motifs by Lats is at least in part responsible for the nuclear-to-cytoplasm translocation of YAP in response to cell contact signals.
YAP is a transcription coactivator; therefore, we hypothesized that the Lats-induced cytoplasmic translocation of YAP inhibits its function by attenuating its interaction with nuclear-localized transcription factors. Indeed, we observed that coexpression of Mst2 and Lats2 decreased the association between TEAD4 and YAP2 (Supplementary Fig. S4), but had no effect on the interaction between TEAD4 and YAP2-S127A or 5SA (Fig. 4C). To exclude the possibility that YAP2 phosphorylation directly affects YAP2/TEAD4 interaction affinity, we tested whether dephosphorylation affects YAP2/TEAD4 association in vitro. Immunoprecipitated YAP2 was treated with λ phosphatase and incubated with Myc-TEAD4 containing cell lysate. As shown in Figure 4D, dephosphorylation of YAP2 had little effect on its interaction with TEAD4 in vitro. Therefore, we conclude that YAP2 phosphorylation by Lats2 leads to decreased interaction with TEAD secondary to cytoplasmic retention.
S127 phosphorylation regulates YAP and 14–3–3 interaction
One commonly seen mechanism of cytoplasmic retention of nuclear proteins is 14–3–3 binding (Muslin and Xing 2000). Interestingly, YAP S127 phosphorylation has been reported to create a 14–3–3-binding site (Basu et al. 2003). We observed that YAP2 interacts with 14–3–3, and this interaction is completely abolished by phosphatase treatment (Fig. 5A). Furthermore, expression of Mst2 and Lats2, but not Akt, increased the interaction between YAP2 and 14–3–3 in a S127-dependent manner, as neither YAP2-S127A nor 5SA showed any binding to 14–3–3 (Fig. 5B). Our results suggest a model in which Lats2 promotes YAP2 cytoplasmic localization by increasing S127 phosphorylation and 14–3–3 binding.
Figure 5.
S127 phosphorylation regulates YAP and 14–3–3 interaction. (A) Dephosphorylation abolishes the interaction between YAP2 and 14–3–3 in vitro. Flag-YAP2 immunoprecipitated from transfected HEK293 cells was treated with λ phosphatase as indicated and then used to pull down endogenous 14–3–3 from HEK293 cell lysate. The products were analyzed by Western blot. (B) Lats2 but not Akt enhances YAP2 and 14–3–3 interaction. Flag-YAP2 plasmids were cotransfected with Myc-14–3–3 and other indicated plasmids into HEK293 cells. Myc-14–3–3 was immunoprecipitated and coimmunoprecipitated Flag-YAP2 was detected. (C) Mutation of H122 but not P129 decreases YAP2 S127 phosphorylation by Lats2. In vitro phosphorylation of YAP2 mutants by immunoprecipitated HA-Lats2 was performed. Phosphorylation of GST-YAP2 was detected by 32P incorporation. (Bottom panel) GST-YAP2 input was shown by Coomassie Blue staining. 4SA (S127) denotes that four of the five Lats phosphorylation sites were mutated to alanine except Ser127. (D) Mutation of His122 in YAP2 impairs Ser127 phosphorylation and 14–3–3 binding. Indicated plasmids were transfected into HEK293 cells. Flag-YAP2 was immunoprecipitated, and the immunoprecipitates were probed as indicated. (E) Pro129 of YAP2 is required for 14–3–3 binding. Experiments were similar to those in D.
Study of the yeast Dbf2 kinase has shown that R at the −3 position of target S/T is critical for kinase recognition (Fig. 3A). However, the function of the H at the −5 position is unknown. We tested the importance of this histidine by examining the phosphorylation of YAP2-H122Y and H122L mutants. Mutation of H122 to either Y or L significantly decreased S127 phosphorylation in vitro and in vivo (Fig. 5C,D), indicating the importance of the histidine at the −5 position. These two mutations also attenuated interaction with 14–3–3 (Fig. 5D), which is likely due to decreased S127 phosphorylation. A proline at the +2 position of a phosphorylated serine is critical for 14–3–3 binding. To investigate the importance of this proline, we assayed 14–3–3 interaction with a YAP2-P129D mutant. As expected, mutation of P129 completely eliminated 14–3–3 binding and also decreased recognition by the pYAP antibody (Fig. 5E). However, in vitro phosphorylation assays showed that the P129D mutation did not affect phosphorylation by Lats2 (Fig. 5C). Based on the above data, we conclude that P129 is important for 14–3–3 binding, but is not directly involved in YAP phosphorylation by Lats. In contrast, H122 plays a critical role in YAP phosphorylation by Lats.
S127 phosphorylation regulates YAP and Yki biological function
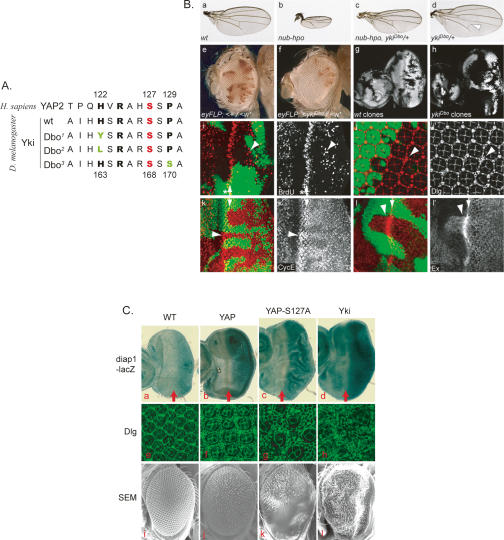
In a genetic screen for suppressors of phenotypes caused by Hippo overexpression, we recovered three alleles of yki. Remarkably, our yki alleles all affect the highly conserved region surrounding S168, which corresponds to S127 in YAP2. As shown in Figure 6A, two of the alleles harbor mutations of H163 to Y and L, while the other allele harbors a mutation of P170 to S. All three alleles strongly suppress the small wing phenotype caused by Hippo overexpression and exhibit weak semidominant phenotypes (Fig. 6B, panels a–d). The most noticeable dominant phenotypes were in the wings, which were slightly larger and often had defects in the posterior cross-vein. Due to the large wing phenotypes, we named these yki alleles after the large ears of the Dumbo cartoon character. The suppression of Hippo-hyperactivated phenotypes suggests that these mutant Yki proteins are constitutively active and evade suppression by Hippo signaling, which is consistent with our biochemical studies of YAP2. If this were the case, we would expect that cells homozygous mutant for these yki alleles phenocopy hippo loss-of-function mutations. Indeed, we found that ykiDbo mutant cells had a growth advantage over wild-type cells (Fig. 6B, panels e–h). This effect was apparent in adult eyes showing that ykiDbo mutant cells, marked by the lack of pigmentation, outcompeted red wild-type cells (Fig. 6B, panels e,f) as well as in developing eye tissues at larval stages, where _eyFLP_-induced ykiDbo mutant clones occupied nearly the entire disc tissues, in contrast to wild-type control clones, which occupied less than half of the discs (Fig. 6B, panels g,h). In addition, ykiDbo mutant eye tissue exhibited ectopic cell proliferation posterior to the morphogenetic furrow (Fig. 6B, panels i,i′), a region where wild-type cells exit the cell cycle and start to differentiate, and produced an excess number of interommatidial cells (Fig. 6B, panels j,j′). Characteristic for mutations in Hippo signaling components, cyclin E and ex, transcriptional targets of Hippo signaling, were up-regulated in ykiDbo clones (Fig. 6B, panels k,l). Interestingly, this up-regulation was also observed in heterozygous cells, perhaps due to the semidominant nature of the ykiDbo alleles.
Figure 6.
S127 phosphorylation regulates YAP and Yki biological function in vivo. (A) Alignment of the Homo sapiens YAP2 and the Drosophila melanogaster Yki wild-type and Dbo mutant proteins around the S127 (YAP2) residue. Mutated residues are shown in green. (B) Dominant active yorkie mutations around the phosphorylation site S168 mimic hippo loss-of-function phenotypes. (Panel a) Wild-type wing. (Panel b) Hpo overexpression driven by nubbin-Gal4. (Panel c) nubbin-Gal4 UAS-Hpo, ykiDbo/+. (Panel d) ykiDbo/+. (Panel e) A fly with an eye mosaic for a mutation in the white gene. Clones were induced using the eye-specific FLP driver (eyFLP), and a cell-lethal mutation on the homologous (w+) chromosome was used to eliminate twin spot clones, which increased the area of the w− cell clones. (Panel f) A fly with a mosaic eye induced by the same method as in e. However, this fly carries a ykiDbo mutation on the w− chromosome. (Panels g,h) Eye imaginal discs from third instar larvae containing wt and ykiDbo mutant clones that were marked by the absence of GFP (gray). (Panels i–l) ykiDbo mutant clones marked by the absence of GFP. (Panel i) Eye imaginal disc containing ykiDbo mutant clones and labeled for BrdU incorporation (red in panel i, and grayscale in panel i′). Asterisks indicate the morphogenetic furrow, arrows indicate the second mitotic wave, and arrowheads point to ectopic cell proliferation in ykiDbo mutant clones posterior to the second mitotic wave. (Panle j) Mid-pupal retina stained with Discs large (Dlg) antibodies to visualize cell outlines (red in panel j, and grayscale in panel j′). ykiDbo mutant clones showed extra interommatidial cells (arrowhead). (Panel k) ykiDbo mutant clones showed up-regulated expression of Cyclin E (arrowheads) (red in panel k, and grayscale in panel k′), most conspicuously behind the second mitotic wave (arrows). (Panel l) ykiDbo mutant clones showed increased Ex (red in panel l, and grayscale panel l′) levels in the eye imaginal disc. (C) The phosphorylation-defective YAP2-S127A is more active in promoting tissue growth in Drosophila. (Panels a–d) Third instar larval eye discs were analyzed for the transcriptional activities of diap1-lacZ reporter genes. Anterior is to the left. Red arrows indicate the morphogenetic furrow. (Panels e–h) Mid-pupal eye discs were stained with Discs large (Dlg) antibody to outline cells. SEM (scanning electron microscopy) images of fly adult eyes are presented in panels i–l. Genotypes of the fly tissues are GMR-Gal4/+; diap1-lacZ/+ (panel a), GMR-Gal4/UAS-Flag-YAP2; diap1-lacZ/+ (panel b), GMR-Gal4/UAS-Flag-YAP2S127A; diap1-lacZ/+ (panel c), GMR-Gal4/UAS-yki-V5; diap1-lacZ/+ (panel d), wild-type (Canton S) (panels e,i), GMR-Gal4/UAS-Flag-YAP2 (panels f,j), GMR-Gal4/UAS-Flag-YAP2S127A, (panels g,k), and GMR-Gal4/UAS-yki-V5 (panels h,l).
The combination of these phenotypes is very distinctive for loss of Hippo signaling, although they are not as severe as those observed for null mutants of hippo or wts. The ykiDbo alleles thus mimic hypomorphic hippo alleles. We conclude that the YkiDbo mutations produce dominant active proteins that are not as efficiently suppressed by Hippo signaling. This is likely due to reduced phosphorylation by Wts (Yki-Dbo1 and Dbo2) and reduced 14–3–3 binding (Yki-Dbo3), as observed for the respective YAP mutants.
In another line of evidence, we compared the activity of YAP/Yki and the phosphorylation-deficient S127A mutant in transgenic flies that overexpressed these proteins in developing eyes. As expected, overexpression of YAP2 or Yki increased the transcription of diap1-lacZ (Fig. 6C, panels a–d) and CycE-lacZ (Supplementary Fig. S5, panels a–d) reporter genes, transcriptional readouts for Yki activity (Huang et al. 2005). Overexpression of YAP2 had a moderate effect on eye size and slightly increased the size of larval eye discs and adult eyes (Fig. 6C, panels f,j). The phosphorylation-defective YAP2-S127A was more potent and caused a significant increase in the size of eye discs and in the number of interommatidial cells (Fig. 6C, panels g,k). The adult eyes of such animals were overgrown, but folded and had a severe morphological defect (Fig. 6C, panels a–c,i–k). All of these phenotypes are reminiscent of warts and mats mutants, and the YAP2-S127A was in fact as potent as the fly Yki protein in promoting tissue growth (Fig. 6C). These data, together with ones from the Dbo mutants, suggest a critical role of YAP/Yki phosphorylation by Lats/Wts in the negative regulation of YAP/Yki in vivo.
YAP regulates density-dependent gene expression and alteration of YAP activity affects cell contact inhibition
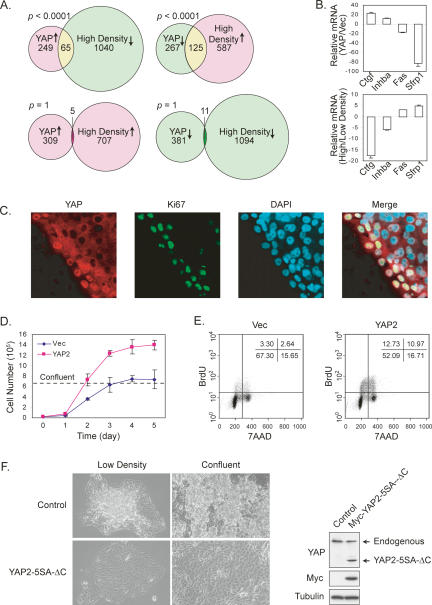
As a transcription coactivator, YAP functions by regulating gene expression. Gene expression microarray experiments were performed to compare genes that are regulated by YAP and by cell density. We found that the set of genes induced by YAP2 significantly overlaps with the set of genes that are repressed by high cell density (Fig. 7A). Similarly, the set of genes repressed by YAP2 (possibly by indirect means) significantly overlaps with the set of genes induced by high density. However, the set of genes induced (or repressed) by YAP2 does not significantly overlap with the set of genes induced (or repressed) by high density. The opposite regulation of gene expression by YAP and high cell density was confirmed by quantitative RT–PCR of selected genes (Fig. 7B). These observations indicate that YAP and cell density regulate many genes in opposite manners.
Figure 7.
YAP regulates density-dependent gene expression and alteration of YAP activity affects cell contact inhibition. (A) High cell density and YAP affect gene expression in opposite manners. YAP-regulated genes were revealed by microarray analyses of control and YAP-overexpressing NIH-3T3 cells. Density-regulated genes were also identified by microarray analysis of sparse and confluent cells. Genes that show more than twofold differences were used in the comparison. P values were calculated by Fisher exact test. (B) Quantitative RT–PCR confirmation of YAP and cell density-regulated genes. Total RNA isolated from NIH-3T3 cells stably expressing YAP2 or vector control (top chart) and from low- or high-density cultures (bottom chart) were analyzed by quantitative RT–PCR and normalized to HPRT (hypoxanthine phosphoribosyltransferase 1). (C) Correlation of cell proliferation and nuclear YAP localization. Confluent MCF10A culture was scratched. Six hours later, cells were fixed and stained for YAP and Ki67. (D) YAP promotes cell growth and elevates saturation density. Growth curves of NIH-3T3 cells stably expressing YAP2 or vector were determined. Confluent density is indicated. (E) YAP promotes proliferation of confluent cells. Vector and YAP-overexpressing NIH-3T3 cells were cultured to confluence. Cells at a similar density were pulse-labeled with BrdU followed by staining with anti-BrdU and 7-AAD (a fluorescent dye for total DNA) for flow cytometric analysis. (F) Dominant-negative YAP restores contact inhibition in ACHN cancer cells. ACHN cells stably expressing vector or Myc-YAP2-5SA-ΔC were cultured to low density or confluence. Cell morphologies are shown in the left panels. The loss of contact inhibition in ACHN cells is evidence that cells pile on top of each other. Myc-YAP2-5SA-ΔC expression level is shown by Western blot in the right panels.
Our data indicate that YAP may play a role in cell contact inhibition. To further investigate YAP regulation by cell contact, scratch wounds were generated in confluent cell cultures to relieve contact inhibition. As shown in Figure 7C, both YAP staining intensity and nuclear localization were significantly elevated in cells at the border of the wound, while cells further away showed cytoplasmic localization of YAP. Interestingly, the nuclear YAP-positive cells were also positive for Ki67, a marker of cell proliferation, indicating that these cells have re-entered the cell division cycle. The above data further demonstrate that YAP localization is regulated by cell density and that nuclear YAP may promote cell cycle entry.
To test the effect of increased YAP activity on contact inhibition, we stably expressed YAP2 in NIH-3T3 cells. YAP2-overexpressing NIH-3T3 cells kept proliferating even after reaching confluency and resulted in a higher saturation density than vector control cells (Fig. 7D). Confluent cells were also analyzed for cell cycle distribution by 5-bromo-2′-deoxyuridine (BrdU) incorporation assay. Many more YAP2-expressing cells (23%) were still in S phase compared with vector control cells (6%) (Fig. 7E). The above observations suggest that YAP overexpression may overcome contact inhibition, and therefore further supports an important role of YAP inactivation by the Hippo pathway in cell contact inhibition.
We tested whether interference of YAP activity was able to restore contact inhibition in cancer cells that are otherwise deficient of it. ACHN is a cancer cell line from a metastatic human renal adenocarcinoma. It has been reported to bear a deletion of Sav (Tapon et al. 2002), which suggests a deficient Hippo pathway. Consistently, the cells in this line clearly grow on top of each other and pile up, even under low cell density (Fig. 7F), suggesting loss of contact inhibition. We generated ACHN cells stably express YAP2-5SA-ΔC, which is the YAP2-5SA nucleus-localizing form with a deletion of the C-terminal transcription activation domain. This mutant YAP2 is insensitive to the Hippo pathway-induced cytoplasmic translocation and cannot activate gene expression, and therefore may act as a dominant-negative form. Although the expression of this mutant YAP2 was as low as endogenous YAP (Fig. 7F), its effect was dramatic. The YAP2-5SA-ΔC-expressing ACHN cells grow as a single layer and do not pile up even after confluent (Fig. 7F). This result indicates, at least in Hippo pathway-deficient cancer cells, that the loss of cell contact inhibition can be restored by blocking endogenous YAP function, and therefore further supports the function of YAP in contact inhibition.
Elevated YAP protein and nuclear localization in cancers
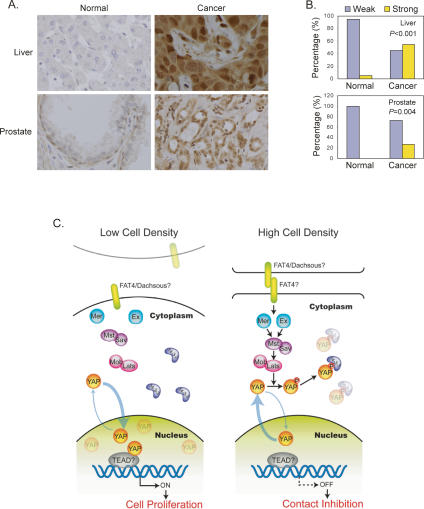
Although YAP has been implicated as a candidate oncogene, it has not been reported whether YAP is indeed activated in human cancers. We evaluated YAP expression in human cancers by immunohistochemical staining of tissue microarrays. Among the 115 cases of hepatocellular carcinoma (HCC) samples examined, 63 samples (54%) showed strong YAP staining, while 95% of normal liver tissue samples (40 out of 42 cases) showed very weak staining, indicating a significant difference in YAP protein levels between normal and cancerous tissues (P < 0.001, Fisher exact test) (Fig. 8A,B). Furthermore, the majority of HCC cells displayed stronger nuclear YAP staining. These observations show that dysregulation of YAP protein level and localization indeed occurs in human HCC. Similar observations were made in prostate cancer tissues (P = 0.004) (Fig. 8A,B). We speculate that YAP activation in cancer tissues is likely due to mutation or dysregulation of the Hippo pathway including YAP itself, and that uncontrolled YAP activation may contribute to cancer development.
Figure 8.
(A) Elevated YAP protein and nuclear localization in human cancers. Tissue microarrays of liver and prostate cancer were stained with anti-YAP antibody (brown). Cell nuclei were counterstained with Hematoxylin (blue). (B) Nuclear YAP protein is significantly elevated in human cancers. Samples were scored based on median nuclear staining intensity, ranging from 0 to 6 (0 for negative, and 6 for very strong staining). Strong staining was considered a score of 2 or higher for liver and 3 or higher for prostate. P values (Fisher exact test) indicate the differences in the proportions of strong YAP staining between cancer and normal specimens. (C) A model for YAP regulation by cell contact via the Hippo pathway.
Discussion
Recent studies have implicated YAP as an oncogene (Overholtzer et al. 2006; Zender et al. 2006). However, neither the precise biological function nor the physiological regulation of YAP is clear. Our study demonstrates that YAP functions downstream from and is inhibited by the Hippo pathway in cell contact inhibition (Fig. 8C).
It has been reported previously that YAP2 S127 is phosphorylated by Akt in response to growth factor stimulation (Basu et al. 2003). However, we found that phosphorylation of YAP2 S127 is not affected in PDK1 knockout cells, in which the Akt activity is abolished. Furthermore, EGF, insulin, PI3K inhibitors, and Akt overexpression did not affect YAP phosphorylation in our experiments. In contrast, we presented data to show that S127 in YAP2 is directly phosphorylated by Lats. Given the fact that YAP is an oncogene and its activity is inhibited by S127 phosphorylation, the previous model that Akt phosphorylates YAP S127 is inconsistent with the known function of Akt. We conclude that Lats plays a direct role in YAP phosphorylation and inhibition. However, it is still possible that YAP might be phosphorylated by Akt under some physiological or pathological conditions.
YAP in cell contact inhibition
An important observation described in this study is the dramatic translocation of YAP between the nucleus and the cytoplasm in response to cell density status. We propose a model that upon cell–cell contact, certain cell surface receptors (Fat is a possible candidate) are activated via interaction with other surface proteins (such as Dachsous) (Matakatsu and Blair 2004). The activated receptor then stimulates Merlin and Expanded, which in turn stimulate Mst/Sav protein kinase activity. Active Mst/Sav phosphorylates and activates the Lats/Mob complex, which directly phosphorylates YAP on HXRXXS motifs. Phosphorylated YAP then associates with 14–3–3 and is sequestered in the cytoplasm (Fig. 8C). Remarkably, genetic screens in Drosophila identified three Yki gain-of-function alleles, and mutation of the corresponding residues in YAP decreases phosphorylation or 14–3–3 binding. These results demonstrate the functional importance and the inhibitory nature of the phosphorylation of YAP-S127 (Yki-S168) by the Hippo pathway. However, additional mechanisms of YAP regulation may also exist, because YAP-5SA can be partially inhibited by Mst and Lats in the reporter assays, and the subcellular localization of this mutant still shows partial response to cell density.
Several lines of evidence support the function of YAP in contact inhibition. First, cell density regulates Lats kinase activity and YAP nuclear/cytoplasmic shuttling. Second, scratching of confluent cultured cells induces YAP nuclear localization in cells at the wound edge. Those cells with nuclear YAP also enter the cell cycle. Third, YAP-overexpressing cells fail to exit the cell cycle when confluent and grow to a much higher density. In fact, there is a strong correlation between nuclear YAP protein levels and staining with the proliferation marker Ki67 (data not shown). Fourth, expression of dominant-negative YAP restores contact inhibition in a human cancer cell line bearing deletion of Sav. Fifth, YAP regulates many genes in a manner opposite to high cell density. Furthermore, YAP is inhibited by Merlin, which has been implicated in mediating cell contact inhibition (Lallemand et al. 2003; Okada et al. 2005). Together, our study indicates that YAP plays a critical role in cell contact inhibition and that the Hippo pathway may relay cell contact signals to inactivate YAP, thereby inducing contact inhibition (Fig. 8C).
YAP in tumorigenesis
Our data show that YAP expression is frequently elevated in human cancers. More than 50% of HCC examined have increased nuclear YAP protein levels. Prostate cancers also have significant elevation of YAP protein levels and nuclear localization, although at a lower frequency. These data indicate that YAP may play an important role in human tumorigenesis. However, in spite of the high frequency of YAP overexpression we observed, a relatively low incidence (5%–15%) of amplification of the human chromosome 11q22 amplicon has been reported in human tumors (Baldwin et al. 2005; Snijders et al. 2005; Zender et al. 2006). Thus, we speculate that elevation of YAP protein levels in cancer is not entirely due to gene amplification, but may instead result from dysregulation of the Hippo pathway. For example, mutation of NF2 should result in inhibition of the Hippo pathway and subsequent activation of YAP by abrogation of inhibitory phosphorylation. We propose that Merlin functions as a tumor suppressor, at least in part by inactivating the YAP oncoprotein. It has also been reported that both Sav and Mob are mutated in tumor cell lines (Tapon et al. 2002; Lai et al. 2005). Therefore, mutation or dysregulation of Mst/Sav and Lats/Mob may contribute to uncontrolled YAP activation in human cancers.
Constitutive activation of YAP may cause evasion of contact inhibition, therefore providing a growth advantage for YAP-overexpressing cancer cells. YAP may also be activated in other cancer types. Future studies to elucidate the Hippo pathway and YAP regulation will not only provide new insights into cell growth regulation, especially cell contact inhibition, but will also be valuable in understanding tumorigenesis. Pharmacologic intervention in the Hippo pathway—for example, inhibition of YAP—may be an effective strategy to treat cancers exhibiting YAP activation and/or overexpression.
Many important questions in the Hippo pathway remain to be addressed. For example, little is known about how Merlin is activated by cell contact. One possibility is CD44, a cell surface receptor implicated in cell contact inhibition, acts upstream of Merlin (Morrison et al. 2001). It is also possible that Fat4 may function upstream of Merlin or Ex1 to initiate the cell contact signaling pathway. Another key question is the mechanism of Mst activation by Merlin. Also elusive is the critical transcription factor(s) mediating the physiological function of YAP. We speculate that TEAD may have a role in YAP function. Interestingly, Scalloped, the Drosophila TEAD homolog, plays some roles in regulating cell proliferation and apoptosis (Delanoue et al. 2004), suggesting an intriguing possibility of Scalloped as a Yki target transcription factor. The bantam microRNA plays a critical role in Drosophila to mediate the Hippo pathway signaling, but there is no obvious bantam homolog in human genome (Nolo et al. 2006; Thompson and Cohen 2006). It will be interesting to see whether functionally similar microRNA exists in humans to mediate the physiological function of YAP.
In summary, our study demonstrates that inactivation of the YAP oncoprotein may play a critical role in cell contact inhibition. This is at least partially accomplished by the Hippo pathway-dependent phosphorylation that promotes YAP binding to 14–3–3 and cytoplasmic localization. Dysregulation of YAP evades contact inhibition and may contribute to tumorigenesis.
Materials and methods
Cell culture, transfection, and retroviral infection
HEK293 cells, 293T cells, HeLa cells, NIH-3T3 cells, MEF cells, ACHN cells, and the RT4-D6-P2T Schwannoma cells were cultured in DMEM (Invitrogen) containing 10% FBS (Invitrogen) and 50 μg/mL penicillin/streptomycin (P/S). MCF10A cells were cultured in DMEM/F12 (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, 10 μg/mL insulin, 100 ng/mL cholera toxin, and 50 μg/mL P/S. Transfection with lipofectamine was performed according to the manufacturer’s instructions.
To generate wild-type or mutant YAP2-expressing cells, retrovirus infection was performed by transfecting 293 Phoenix retrovirus packaging cells with empty vector or pQCXIH-YAP2 constructs. Forty-eight hours after transfection, retroviral supernatant was supplemented with 5 μg/mL polybrene, filtered through a 0.45-μm filter, and used to infect MCF10A, NIH-3T3, or ACHN cells. Thirty-six hours after infection, cells were selected with 200 μg/mL hygromycin (Roche) in culture medium. The RT4-D6-P2T Schwannoma cells with inducible Merlin expression have been described before (Morrison et al. 2001; Rong et al. 2004).
Immunofluorescence staining
For immunofluorescence staining, cells were cultured on coverslips to appropriate density. Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100. After blocking in 3% BSA for 30 min, slides were incubated with first antibody diluted in 1% BSA for 1.5 h. After washing with PBS, slides were incubated with Alexa Fluor 488- or 594-conjugated secondary antibodies (1:1000 dilution) for 1.5 h. The slides were then washed and mounted.
Immunoprecipitation and kinase assay
For the Lats2 and Mst2 kinase assays, HEK293 cells were transfected with HA-Lats2 or Flag-Mst2. Forty-eight hours post-transfection, cells were lysed with lysis buffer (50 mM HEPES at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10 mM pyrophosphoate, 10 mM glycerophosphate, 50 mM NaF, 1.5 mM Na3VO4, protease inhibitor cocktail [Roche], 1 mM DTT, 1 mM PMSF) and immunoprecipitated with anti-HA or anti-Flag antibodies. The immunoprecipitates were washed three times with lysis buffer, followed by once with wash buffer (40 mM HEPES, 200 mM NaCl) and once with kinase assay buffer (30 mM HEPES, 50 mM potassium acetate, 5 mM MgCl2). The immunoprecipitated Lats2 or Mst2 was subjected to a kinase assay in the presence of 500 μM cold ATP, 10 μCi [γ-32P]ATP, and 1 μg of GST-YAP2 expressed and purified from Escherichia coli as substrate. The reaction mixtures were incubated for 30 min at 30°C, terminated with SDS sample buffer, and subjected to SDS-PAGE and autoradiography. The same procedure was used for endogenous Lats2 kinase assay, except that endogenous Lats2 immunoprecipitated from NIH-3T3 cells was used.
Luciferase assay
For the luciferase reporter assay, 293T cells were seeded in 12-well plates. 5× UAS-luciferase reporter, CMV-β-gal, and indicated plasmids were cotransfected as described previously. Thirty-six hours after transfection, cells were lysed and luciferase activity was assayed using the enhanced luciferase assay kit obtained from BD Biosciences following the manufacturer’s instructions. All luciferase activities were normalized to β-galactosidase activity.
BrdU labeling and flow cytometric analysis
For cell cycle progression analysis, cells were cultured to desired confluence. Cells were then labeled with BrdU and analyzed by flow cytometry using the FITC BrdU Flow Kit obtained from BD Biosciences following the manufacturer’s instructions. Briefly, cells were pulse-labeled with 10 μM BrdU in culture medium for 30 min. After trypsinization and PBS wash, cells were fixed and permeabilized. Incorporated BrdU was exposed by DNase treatment and then stained by FITC-conjugated anti-BrdU antibody. Total DNA was stained by 7-AAD (7-amino-actinomycin D). Data were collected on a BD FACSCalibur and analyzed with CellQuest Pro software.
RNA isolation and real-time PCR
Total RNA was isolated from cultured cells using Trizol reagent (Invitrogen). cDNA was synthesized by reverse transcription using random hexamers and subjected to real-time PCR with gene-specific primers in the presence of Cybergreen (Applied Biosystems). Relative abundance of mRNA was calculated by normalization to hypoxanthine phosphoribosyltransferase 1 (HPRT) mRNA.
Gene expression microarray analysis
For analysis of gene expression in YAP overexpression cells, the cells were cultured to 90% confluency before harvest. For comparing gene expression in low- and high-density cultures, cells were seeded at different densities and harvested at 30% or complete confluency. Total RNA was extracted with Trizol (Invitrogen) followed by further purification using the RNeasy kit (Qiagen). Biotinylated cRNAs were then prepared according to the Affymetrix standard labeling protocol. The biotinylated cRNAs were then fragmented and hybridized to the Affymetrix GeneChip Mouse Genome 430 2.0 Array or the Human Genome U133 Plus 2.0 Array, respectively. Chips were washed and stained with Streptavidin R-phycoerythrin (Invitrogen). After scanning the chips, the data were analyzed using GCOS software. Scaling was performed with a target intensity of 500 to facilitate the comparison of multiple arrays. A cut-off value of 0.05 was applied to the detection P value to assign a present (P), marginal (M), or absent (A) call to each probe set. A signal value was calculated using the One-Step Tukey’s Biweight Estimate to represent the relative abundance of a transcript. Up- or down-regulation of a gene is determined by two criteria: first, at least one P call in the two samples being compared; second, at least a twofold change (or indicated) of the signal value. The microarray analysis was done at the Molecular Biology Core Laboratory (University of Michigan, School of Dentistry).
Acknowledgments
We thank Drs. Marius Sudol for the pCMV-Flag-YAP2 construct, Duojia Pan for a full-length yki cDNA, Tian Xu for Lats2, Brian Seed and Joseph Avruch for Mst2, Jiandie Lin for the 5× UAS-luciferase reporter and the Gal4-TEAD4 and Gal4-Foxo3 constructs, and Dario R. Alessi for the PDK1+/+ and PDK1−/− embryonic stem cell lysates. We thank the Developmental Studies Hybridoma Bank at the University of Iowa for Dlg antibody. We also thank Drs. Stephen J. Weiss, Patrick Hu, Ken Inoki, Chung-Han Lee, and Theresa A. Reno for critical reading of the manuscript. This work was supported by grants from NIH (to K.L.G), and the National Science Foundation (IOS-0641914) (to Z.-C.L.).
Footnotes
References
- Abercrombie M. Contact inhibition and malignancy. Nature. 1979;281:259–262. doi: 10.1038/281259a0. [DOI] [PubMed] [Google Scholar]
- Baldwin C., Garnis C., Zhang L., Rosin M.P., Lam W.L., Garnis C., Zhang L., Rosin M.P., Lam W.L., Zhang L., Rosin M.P., Lam W.L., Rosin M.P., Lam W.L., Lam W.L. Multiple microalterations detected at high frequency in oral cancer. Cancer Res. 2005;65:7561–7567. doi: 10.1158/0008-5472.CAN-05-1513. [DOI] [PubMed] [Google Scholar]
- Basu S., Totty N.F., Irwin M.S., Sudol M., Downward J., Totty N.F., Irwin M.S., Sudol M., Downward J., Irwin M.S., Sudol M., Downward J., Sudol M., Downward J., Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14–3–3 and attenuation of p73-mediated apoptosis. Mol. Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Bennett F.C., Harvey K.F., Harvey K.F. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Cho E., Feng Y., Rauskolb C., Maitra S., Fehon R., Irvine K.D., Feng Y., Rauskolb C., Maitra S., Fehon R., Irvine K.D., Rauskolb C., Maitra S., Fehon R., Irvine K.D., Maitra S., Fehon R., Irvine K.D., Fehon R., Irvine K.D., Irvine K.D. Delineation of a Fat tumor suppressor pathway. Nat. Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Conlon I., Raff M., Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Delanoue R., Legent K., Godefroy N., Flagiello D., Dutriaux A., Vaudin P., Becker J.L., Silber J., Legent K., Godefroy N., Flagiello D., Dutriaux A., Vaudin P., Becker J.L., Silber J., Godefroy N., Flagiello D., Dutriaux A., Vaudin P., Becker J.L., Silber J., Flagiello D., Dutriaux A., Vaudin P., Becker J.L., Silber J., Dutriaux A., Vaudin P., Becker J.L., Silber J., Vaudin P., Becker J.L., Silber J., Becker J.L., Silber J., Silber J. The Drosophila wing differentiation factor vestigial-scalloped is required for cell proliferation and cell survival at the dorso–ventral boundary of the wing imaginal disc. Cell Death Differ. 2004;11:110–122. doi: 10.1038/sj.cdd.4401321. [DOI] [PubMed] [Google Scholar]
- Eagle H., Levine E.M., Levine E.M. Growth regulatory effects of cellular interaction. Nature. 1967;213:1102–1106. doi: 10.1038/2131102a0. [DOI] [PubMed] [Google Scholar]
- Edgar B.A. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Halbleib J.M., Nelson W.J., Nelson W.J. Cadherins in development: Cell adhesion, sorting, and tissue morphogenesis. Genes & Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G., Hyun E., Tao C., Jafar-Nejad H., Halder G., Tao C., Jafar-Nejad H., Halder G., Jafar-Nejad H., Halder G., Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hariharan I.K.2006Growth regulation: A beginning for the hippo pathway Curr. Biol. 16R1037–R1039. . 10.106/j.cub.2006.11.012 [DOI] [PubMed] [Google Scholar]
- Harvey K., Tapon N., Tapon N. The Salvador–Warts–Hippo pathway—An emerging tumour-suppressor network. Nat. Rev. Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Harvey K.F., Pfleger C.M., Hariharan I.K., Pfleger C.M., Hariharan I.K., Hariharan I.K. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hergovich A., Stegert M.R., Schmitz D., Hemmings B.A., Stegert M.R., Schmitz D., Hemmings B.A., Schmitz D., Hemmings B.A., Hemmings B.A. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K., Pan D., Wu S., Barrera J., Matthews K., Pan D., Barrera J., Matthews K., Pan D., Matthews K., Pan D., Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M., Nolo R., Tao C., Verstreken P., Hiesinger P.R., Bellen H.J., Halder G., Nolo R., Tao C., Verstreken P., Hiesinger P.R., Bellen H.J., Halder G., Tao C., Verstreken P., Hiesinger P.R., Bellen H.J., Halder G., Verstreken P., Hiesinger P.R., Bellen H.J., Halder G., Hiesinger P.R., Bellen H.J., Halder G., Bellen H.J., Halder G., Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Lai Z.C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L.L., Li Y., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L.L., Li Y., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L.L., Li Y., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L.L., Li Y., Rohrbaugh M., Nikolaidis N., Ho L.L., Li Y., Nikolaidis N., Ho L.L., Li Y., Ho L.L., Li Y., Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lallemand D., Curto M., Saotome I., Giovannini M., McClatchey A.I., Curto M., Saotome I., Giovannini M., McClatchey A.I., Saotome I., Giovannini M., McClatchey A.I., Giovannini M., McClatchey A.I., McClatchey A.I. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes & Dev. 2003;17:1090–1100. doi: 10.1101/gad.1054603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A.S., Elia A.E., Devgan G., Ptacek J., Schutkowski M., Snyder M., Yaffe M.B., Deshaies R.J., Elia A.E., Devgan G., Ptacek J., Schutkowski M., Snyder M., Yaffe M.B., Deshaies R.J., Devgan G., Ptacek J., Schutkowski M., Snyder M., Yaffe M.B., Deshaies R.J., Ptacek J., Schutkowski M., Snyder M., Yaffe M.B., Deshaies R.J., Schutkowski M., Snyder M., Yaffe M.B., Deshaies R.J., Snyder M., Yaffe M.B., Deshaies R.J., Yaffe M.B., Deshaies R.J., Deshaies R.J. Substrate specificity analysis of protein kinase complex Dbf2–Mob1 by peptide library and proteome array screening. BMC Biochem. 2005;6:22. doi: 10.1186/1471-2091-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matakatsu H., Blair S.S., Blair S.S. Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development. 2004;131:3785–3794. doi: 10.1242/dev.01254. [DOI] [PubMed] [Google Scholar]
- McClatchey A.I., Giovannini M., Giovannini M. Membrane organization and tumorigenesis—The NF2 tumor suppressor, Merlin. Genes & Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- Morrison H., Sherman L.S., Legg J., Banine F., Isacke C., Haipek C.A., Gutmann D.H., Ponta H., Herrlich P., Sherman L.S., Legg J., Banine F., Isacke C., Haipek C.A., Gutmann D.H., Ponta H., Herrlich P., Legg J., Banine F., Isacke C., Haipek C.A., Gutmann D.H., Ponta H., Herrlich P., Banine F., Isacke C., Haipek C.A., Gutmann D.H., Ponta H., Herrlich P., Isacke C., Haipek C.A., Gutmann D.H., Ponta H., Herrlich P., Haipek C.A., Gutmann D.H., Ponta H., Herrlich P., Gutmann D.H., Ponta H., Herrlich P., Ponta H., Herrlich P., Herrlich P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes & Dev. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin A.J., Xing H., Xing H. 14–3–3 proteins: Regulation of subcellular localization by molecular interference. Cell. Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- Nolo R., Morrison C.M., Tao C., Zhang X., Halder G., Morrison C.M., Tao C., Zhang X., Halder G., Tao C., Zhang X., Halder G., Zhang X., Halder G., Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Okada T., Lopez-Lago M., Giancotti F.G., Lopez-Lago M., Giancotti F.G., Giancotti F.G. Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. J. Cell Biol. 2005;171:361–371. doi: 10.1083/jcb.200503165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., You L., Giancotti F.G., You L., Giancotti F.G., Giancotti F.G. Shedding light on Merlin’s wizardry. Trends Cell Biol. 2007;17:222–229. doi: 10.1016/j.tcb.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Overholtzer M., Zhang J., Smolen G.A., Muir B., Li W., Sgroi D.C., Deng C.X., Brugge J.S., Haber D.A., Zhang J., Smolen G.A., Muir B., Li W., Sgroi D.C., Deng C.X., Brugge J.S., Haber D.A., Smolen G.A., Muir B., Li W., Sgroi D.C., Deng C.X., Brugge J.S., Haber D.A., Muir B., Li W., Sgroi D.C., Deng C.X., Brugge J.S., Haber D.A., Li W., Sgroi D.C., Deng C.X., Brugge J.S., Haber D.A., Sgroi D.C., Deng C.X., Brugge J.S., Haber D.A., Deng C.X., Brugge J.S., Haber D.A., Brugge J.S., Haber D.A., Haber D.A. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes & Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pantalacci S., Tapon N., Leopold P., Tapon N., Leopold P., Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Rong R., Surace E.I., Haipek C.A., Gutmann D.H., Ye K., Surace E.I., Haipek C.A., Gutmann D.H., Ye K., Haipek C.A., Gutmann D.H., Ye K., Gutmann D.H., Ye K., Ye K. Serine 518 phosphorylation modulates merlin intramolecular association and binding to critical effectors important for NF2 growth suppression. Oncogene. 2004;23:8447–8454. doi: 10.1038/sj.onc.1207794. [DOI] [PubMed] [Google Scholar]
- Silva E., Tsatskis Y., Gardano L., Tapon N., McNeill H., Tsatskis Y., Gardano L., Tapon N., McNeill H., Gardano L., Tapon N., McNeill H., Tapon N., McNeill H., McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Snijders A.M., Schmidt B.L., Fridlyand J., Dekker N., Pinkel D., Jordan R.C., Albertson D.G., Schmidt B.L., Fridlyand J., Dekker N., Pinkel D., Jordan R.C., Albertson D.G., Fridlyand J., Dekker N., Pinkel D., Jordan R.C., Albertson D.G., Dekker N., Pinkel D., Jordan R.C., Albertson D.G., Pinkel D., Jordan R.C., Albertson D.G., Jordan R.C., Albertson D.G., Albertson D.G. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- St John M.A., Tao W., Fei X., Fukumoto R., Carcangiu M.L., Brownstein D.G., Parlow A.F., McGrath J., Xu T., Tao W., Fei X., Fukumoto R., Carcangiu M.L., Brownstein D.G., Parlow A.F., McGrath J., Xu T., Fei X., Fukumoto R., Carcangiu M.L., Brownstein D.G., Parlow A.F., McGrath J., Xu T., Fukumoto R., Carcangiu M.L., Brownstein D.G., Parlow A.F., McGrath J., Xu T., Carcangiu M.L., Brownstein D.G., Parlow A.F., McGrath J., Xu T., Brownstein D.G., Parlow A.F., McGrath J., Xu T., Parlow A.F., McGrath J., Xu T., McGrath J., Xu T., Xu T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Miyoshi Y., Takahata C., Irahara N., Taguchi T., Tamaki Y., Noguchi S., Miyoshi Y., Takahata C., Irahara N., Taguchi T., Tamaki Y., Noguchi S., Takahata C., Irahara N., Taguchi T., Tamaki Y., Noguchi S., Irahara N., Taguchi T., Tamaki Y., Noguchi S., Taguchi T., Tamaki Y., Noguchi S., Tamaki Y., Noguchi S., Noguchi S. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin. Cancer Res. 2005;11:1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- Tapon N., Harvey K.F., Bell D.W., Wahrer D.C., Schiripo T.A., Haber D.A., Hariharan I.K., Harvey K.F., Bell D.W., Wahrer D.C., Schiripo T.A., Haber D.A., Hariharan I.K., Bell D.W., Wahrer D.C., Schiripo T.A., Haber D.A., Hariharan I.K., Wahrer D.C., Schiripo T.A., Haber D.A., Hariharan I.K., Schiripo T.A., Haber D.A., Hariharan I.K., Haber D.A., Hariharan I.K., Hariharan I.K. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Thompson B.J., Cohen S.M., Cohen S.M. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Tyler D.M., Baker N.E., Baker N.E. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev. Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan R.S., Kango-Singh M., Nolo R., Tao C., Halder G., Kango-Singh M., Nolo R., Tao C., Halder G., Nolo R., Tao C., Halder G., Tao C., Halder G., Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Vassilev A., Kaneko K.J., Shu H., Zhao Y., DePamphilis M.L., Kaneko K.J., Shu H., Zhao Y., DePamphilis M.L., Shu H., Zhao Y., DePamphilis M.L., Zhao Y., DePamphilis M.L., DePamphilis M.L. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes & Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Shimizu T., Lai Z.C., Shimizu T., Lai Z.C., Lai Z.C. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M., Hamaratoglu F., Kango-Singh M., Udan R., Chen C.L., Tao C., Zhang X., Halder G., Hamaratoglu F., Kango-Singh M., Udan R., Chen C.L., Tao C., Zhang X., Halder G., Kango-Singh M., Udan R., Chen C.L., Tao C., Zhang X., Halder G., Udan R., Chen C.L., Tao C., Zhang X., Halder G., Chen C.L., Tao C., Zhang X., Halder G., Tao C., Zhang X., Halder G., Zhang X., Halder G., Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Williams M.R., Arthur J.S., Balendran A., van der Kaay J., Poli V., Cohen P., Alessi D.R., Arthur J.S., Balendran A., van der Kaay J., Poli V., Cohen P., Alessi D.R., Balendran A., van der Kaay J., Poli V., Cohen P., Alessi D.R., van der Kaay J., Poli V., Cohen P., Alessi D.R., Poli V., Cohen P., Alessi D.R., Cohen P., Alessi D.R., Alessi D.R. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol. 2000;10:439–448. doi: 10.1016/s0960-9822(00)00441-3. [DOI] [PubMed] [Google Scholar]
- Wu S., Huang J., Dong J., Pan D., Huang J., Dong J., Pan D., Dong J., Pan D., Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Yagi R., Chen L.F., Shigesada K., Murakami Y., Ito Y., Chen L.F., Shigesada K., Murakami Y., Ito Y., Shigesada K., Murakami Y., Ito Y., Murakami Y., Ito Y., Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F., Pan D., Pan D. Fat flies expanded the hippo pathway: A matter of size control. Sci. STKE. 2007;2007:pe12. doi: 10.1126/stke.3802007pe12. [DOI] [PubMed] [Google Scholar]
- Zender L., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Luk J.M., Wigler M., Hannon G.J., Wigler M., Hannon G.J., Hannon G.J., et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]