Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis (original) (raw)
Abstract
Expression and function of the transcription factor Sox10 is predominant in neural crest cells, its derivatives and in oligodendrocytes. To understand how Sox10 expression is regulated during development, we analysed the potential of evolutionary conserved non-coding sequences in the Sox10 genomic region to function as enhancers. By linking these sequences to a β-galactosidase marker gene under the control of a minimal promoter, five regulatory regions were identified that direct marker gene expression in transgenic mice to Sox10 expressing cell types and tissues in a defined temporal pattern. These possible enhancers of the Sox10 gene mediate Sox10 expression in the otic vesicle, in oligodendrocytes and in several neural crest derivatives including the developing peripheral nervous system and the adrenal gland. They furthermore exhibit overlapping activities and share binding sites for Sox, Lef/Tcf, Pax and AP2 transcription factors. This may explain high level and robustness of Sox10 expression during embryonic development.
INTRODUCTION
The transcription factor Sox10 is an important, pleiotropic regulator of neural crest and oligodendrocyte development in all vertebrate species so far analysed (1–7). It functions in stem cell maintenance (8), in several specification events (3,9) and during later phases of cell lineage progression including terminal differentiation (10–12).
Much sparser is the information about the factors that determine the spatiotemporal expression pattern of Sox10. In the neural crest, Sox10 expression is under the control of several extrinsic signals, such as BMPs, Wnts and FGFs (1,5,5). With regards to intrinsic signals, there is evidence that the related transcription factor Sox9 as well as FoxD3 are genetically upstream of Sox10 in the neural crest (13), while Sox9 and Olig2 appear to be involved in Sox10 gene induction in cells of the oligodendrocyte lineage (14–16). Whether these transcription factors influence Sox10 expression directly, is not clear at the moment and requires the identification of _cis_-acting sequences in the Sox10 genomic region.
BAC transgenic studies have shown that a genomic region encompassing ∼65 kb upstream as well as 55 kb downstream of the Sox10 gene are sufficient to reproduce the main features of Sox10 expression during embryonic development (17,18). Most of the important _cis_-acting elements furthermore appear to be localized in enhancers rather than the promoter region as the promoter failed to direct marker gene expression to most of the typical Sox10 expression sites (17).
The fact that coding exons and intervening introns could be removed from the Sox10 locus without significant alterations of the embryonic Sox10 expression pattern (9,19,19) argues that the major enhancers are localized to extragenic rather than intragenic regions. The existence of at least two such enhancers was recently proven in transgenic studies (17,21). Fortuitous deletion of genomic sequences from a _Sox10_-containing BAC transgene showed that important enhancer activity was localized in a 20–30 kb genomic interval upstream of the Sox10 promoter sequences and that this enhancer activity was strengthened by a second activity located in a 25–35 kb region further upstream (17). The latter region overlaps with a 16 kb deletion 47 kb upstream of the Sox10 gene which was generated by random insertion of a transgene into the Sox10 genomic region and had been found to cause partial depigmentation and colonic aganglionosis in the corresponding mouse mutant (21). Using bioinformatics, both studies have also detected several highly conserved sequence elements within the respective genomic intervals and postulated that these elements may possess enhancer activity (17,21). None of these elements has, however, been tested for its activity in the developing mouse embryo, and only one (alternatively named DCHCS-1 and MCS6) has been experimentally studied in cell culture and found to have enhancer activity in cultured melanocytes (21).
Here, we use mouse transgenesis to identify enhancer activity of evolutionary conserved non-coding sequences in the Sox10 genomic region in vivo. We have focused on those non-coding sequences that are conserved among amniotes and at the same time located in the genomic interval previously shown to be sufficient for the major aspects of Sox10 expression. Some of these sequences indeed functioned as cell type-specific enhancers and are good candidates for _bona fide cis_-acting gene regulatory elements of the Sox10 gene.
MATERIALS AND METHODS
Construction of transgene and generation of transgenic animals
Evolutionary conserved sequences U1, U2, U3, U4, U5, D6 and D7 (Figure 1A) were amplified with ∼0.15 kb flanking sequences on each side (Figure 1C) from 129 Sv mouse genomic DNA and inserted between SpeI and NotI sites of pBKS-hsp68-lacZ such that the evolutionary conserved sequences were placed immediately upstream of the hsp68-lacZ cassette (22). Each evolutionary conserved sequence was inserted separately. U3 and U4 were additionally cloned in combination upstream of the hsp68-lacZ cassette. The hsp68-lacZ cassette was located between HindIII and KpnI sites of pBKS (Figure 1B). The transgenes were separated from the vector backbone by a digest with SpeI and KpnI or NotI and KpnI, purified, and microinjected into the male pronucleus of fertilized FVB oocytes. Transgenic mice were generated from injected oocytes according to standard techniques. Founder mice and transgenic offspring were identified and genotyped by PCR analysis of DNA prepared from tail biopsies using a combination of the common reverse primer 5′-AGTAGCTGTCAGCGTCTGGT-3′ with one of the following transgene specific forward primers: 5′-GTAGGCCACAGGACATCAAC-3′ for U1, 5′-GGCACAGAAAGGTCTCTTTG-3′ for U2, 5′-GAGCCCTGCTCATAAACAAG-3′ for U3, 5′-ACTGCGACTCTGCTGTCTCT-3′ for U4, 5′-GCACTCCTCTATGCCT CAAG-3′ for U5, 5′-ATTCAGAGGAAGGCCAGAGG-3′ for D6 and 5′-AGAAGGCAGCTGAGGTGTCT-3′ for D7.
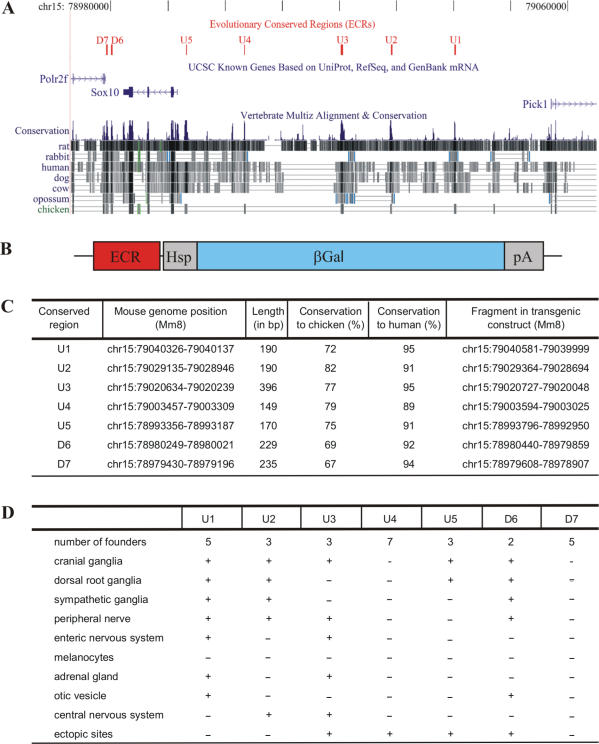
Figure 1.
Characterization of evolutionary conserved non-coding sequences from the Sox10 genomic region and generation of transgenic lines containing these sequences. (A) Localization of seven ECR (U1, U2, U3, U4, U5, D6, D7, shown in red) in the Sox10 genomic interval on mouse chromosome 15 relative to Sox10 and the adjacent Polr2f and Pick1 genes (in blue). Sequence conservation among various mammalian species and chicken is evident from multiple sequence alignments. (B) Design of the transgenic constructs consisting of one of the seven ECR, the hsp68 minimal promoter (Hsp), the β-galactosidase marker gene (βGal) and a SV40 polyA signal (pA). (C) Position of each of the seven ECR in the mouse genome (Mm8), their lengths, conservation to chicken and man as well as the exact coordinates of the fragment present in the transgenic constructs. (D) Summary of independent founders obtained for each transgenic construct, resulting transgenic lines and their overall properties of transgene expression.
Sox10lacZ/+ mice in which Sox10 coding sequences have been replaced on one allele by lacZ marker sequences have been previously generated (9). In these mice, β-galactosidase expression reproduces the full pattern of Sox10 expression.
Tissue preparation, histological staining, immunohistochemistry and documentation
Embryos from 9.5 days post coitum (d.p.c.) to 16.5 d.p.c. were obtained from staged pregnancies and underwent fixation in 1% or 4% paraformaldehyde depending on their further usage. After fixation, embryos were immediately stained for β-galactosidase activity or were cryoprotected in sucrose and frozen at −80°C in Tissue Tec in preparation for cryotome sectioning.
Detection of β-galactosidase activity was performed after fixation in 1% paraformaldehyde on whole mount embryos (9.5 and 11.5 d.p.c.), isolated embryonic organs (16.5 d.p.c.) or 20 µm transverse cryosections (16.5 d.p.c.) by incubation in 1% X-gal for several hours at 37°C.
For immunohistochemistry, 10 μm cryotome sections from the forelimb level of genotyped, age-matched mouse embryos were used with the following primary antibodies: anti-β-galactosidase goat antiserum (1:500 dilution, Biotrend), anti-BFABP rabbit antiserum, (1:10 000 dilution, gift of C. Birchmeier and T. Müller, MDC, Berlin), anti-Olig2 rabbit antiserum (1:50 000 dilution, gift of D. Rowitch, UCSF, San Francisco), anti-Brn3.0 rabbit antiserum (1:2000 dilution, gift of Dr E. Turner, UCSD, San Diego), anti-Oct-6 rabbit antiserum [1:2000 dilution, (23)], anti-Lmx1b guinea pig antiserum (1:15 000 dilution, gift of C. Birchmeier and T. Müller, MDC, Berlin), anti-Sox10 guinea pig antiserum [1:2000 dilution, (24)]. Secondary antibodies conjugated to Cy2 and Cy3 immunofluorescent dyes (Dianova) were used for detection.
Samples were analysed and documented either with a Leica (Bensheim, Germany) inverted microscope (DMIRB) equipped with a cooled MicroMax CCD camera (Princeton Instruments, Trenton, NJ) or with a Leica MZFLIII stereomicroscope equipped with an Axiocam (Zeiss, Oberkochem, Germany).
Cell culture, transient transfection, extract preparation and EMSA
HEK293 cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% fetal calf serum (FCS) and transfected by the calcium phosphate technique using 10 µg plasmid DNA per 100 mm plate. Transfected plasmids contained expression cassettes for HA-tagged Sox9 (amino acids 1–190), HA-tagged Sox10 (amino acids 1–189) (12), T7-tagged Pax3 (25), T7-tagged AP2α (gift of H. Schorle, Bonn) and HA-tagged Lef1 (gift of J. Behrens, Erlangen) under the control of the cytomegalovirus immediate early promoter.
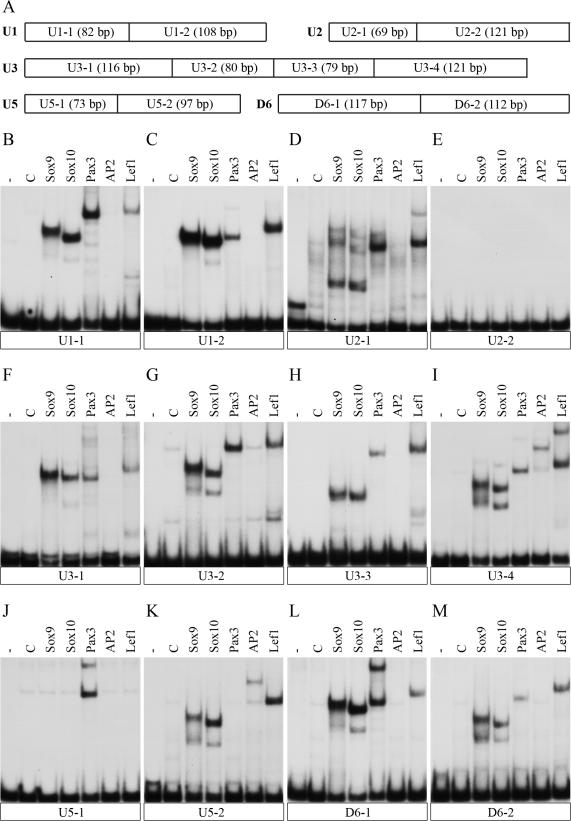
Forty-eight hours post transfection, cells were harvested for extract preparation (26) and ectopic expression of the respective transcription factor was verified by western blotting using anti-HA tag (1:200 dilution, hybridoma supernatant, clone 12CA5) or anti-T7 tag (1:10 000 dilution, Novagen) mouse monoclonals. With these extracts, electrophoretic mobility shift analyses (EMSA) were performed in the presence of poly(dGdC) as unspecific competitor in the case of Sox9, Sox10 and Lef1 or in the presence of poly(dIdC) in the case of Pax3 and AP2α using 32P-labelled fragments from the evolutionary conserved regions with a size between 69 and 121 bp (Figure 7A) (16).
Figure 7.
Binding of neural crest derived transcription factors to evolutionary conserved non-coding sequences from the Sox10 genomic region. (A) Schematic representation of the fragments used for EMSA. The length of each fragment is given in bp. (B–M) Each of the radiolabeled fragments shown in (A) was incubated with control extracts (C) or extracts containing Sox9, Sox10, Pax3, AP2α or Lef1 protein before protein-DNA complexes were resolved from the unbound DNA by native gel electrophoresis. The unbound DNA is shown in the first lane of each panel (-) and runs at the bottom. Each panel represents the results from EMSA of a single fragment: (B) U1-1; (C) U1-2; (D) U2-1; (E) U2-2; (F) U3-1; (G) U3-2; (H) U3-3; (I) U3-4; (J) U5-1); (K) U5-2; (L) D6-1; (M) D6-2.
RESULTS
The Sox10 genomic region contains seven non-coding regions that are evolutionary conserved in mammals and birds
Sequence comparisons among orthologous genomic regions of mammalian and other vertebrate species have led to the identification of evolutionary conserved non-coding regions (ECR) that have an increased likelihood to function as transcriptional regulatory regions (27). Many of these regions are located in the vicinity of genes that code for important developmental regulators (28). Taking into account that Sox10 is such an important regulator and that its functions are highly conserved in vertebrate embryogenesis (29,30), comparative genomics should be suited to identify the gene's regulatory regions.
We concentrated on the genomic interval previously shown by BAC transgenesis to reproduce the main features of embryonic Sox10 expression (18). Using the ECR browser (http://ecrbrowser.dcode.org) and the human sequence as the base genome, we identified seven regions around the Sox10 gene that exhibited a greater than 70% sequence identity over at least 100 bp in comparison with the chicken genome (Figure 1A). Five, termed U1–U5, were localized in front of the Sox10 gene, two, called D6 and D7, were behind the gene. U1 corresponds to MCS6/DCHCS-1 in previous studies, U2 to MCS4, U3 to MCS3 and U4 to MCS2 (17,21). U5, D6 and D7 have not been described so far. Their length varied between 150 and 400 bp (Figure 1C). In comparisons between the mouse and chicken genome, these regions exhibited at least 67% identity over their whole length (Figure 1C and Supplementary Figures 1–6). Between mouse and human genome, the sequence identity was greater than 89% and there was additional strong sequence conservation in the flanking sequences. Both the lower sequence conservation and the shorter length in evolutionary distant species as compared to evolutionary close species has similarly been observed in a previous whole genome study (31). In contrast to this strong sequence conservation among mammals and between mammals and chicken, none of these regions exhibited a significant degree of conservation in Xenopus laevis, Danio rerio or Fugu rubripes. The conservation of the identified regions is thus restricted to amniotes.
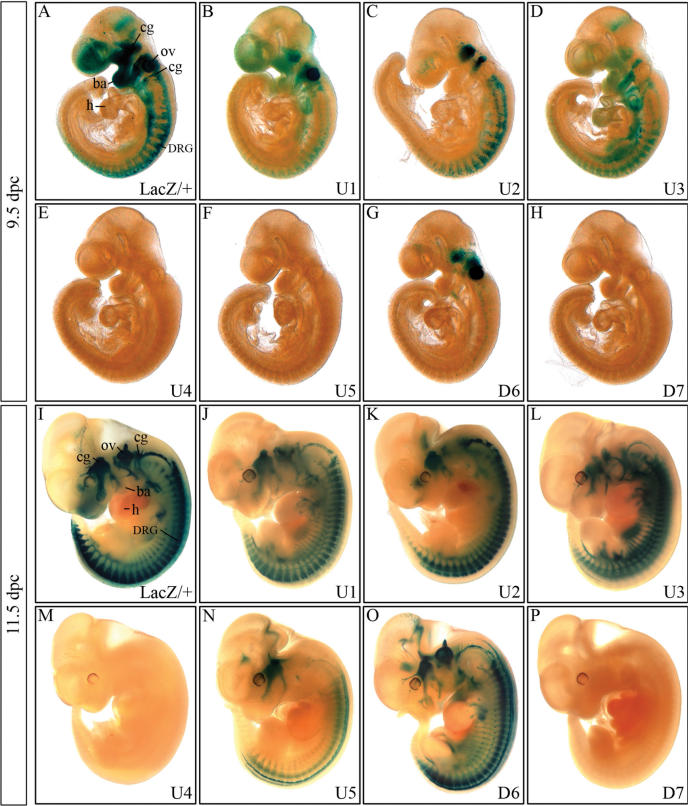
Figure 2.
Detection of β-galactosidase transgene expression in embryos at 9.5 and 11.5 d.p.c. β-Galactosidase activity was detected colorimetrically using X-gal substrate at 9.5 dpc (A–H) and at 11.5 d.p.c. (I–P) in Sox10lacZ/+ (A, I) and age-matched embryos carrying a hsp68-lacZ transgene driven by one of the following ECR from the Sox10 genomic region: U1 (B, J), U2 (C, K), U3 (D, L), U4 (E, M), U5 (F, N), D6 (G, O) and D7 (H, P). Transgenic embryos were stained in parallel for 9 h, Sox10lacZ/+ embryos for 3 h. No β-galactosidase staining was detected in wildtype littermates under the conditions applied. cg, cranial ganglia; DRG, dorsal root ganglia; ov, otic vesicle; ba, branchial arches; h, heart.
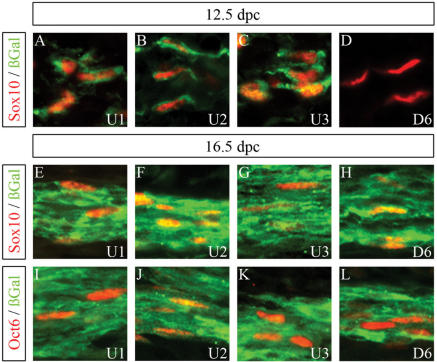
Figure 3.
Cellular expression pattern of β-galactosidase transgenes in peripheral nerves. Co-immunohistochemistry was performed on spinal nerves of embryos at 12.5 d.p.c. (A–D) and 16.5 d.p.c. (E–L) using antibodies directed against β-galactosidase (in green) in combination with antibodies directed against Sox10 (A–H) or Oct6 (I–L) (all in red). The embryos carried a hsp68-lacZ transgene driven by either the U1 (A, E, I), the U2 (B, F, J), the U3 (C, G, K) or the D6 (D, H, L) region.
Figure 4.
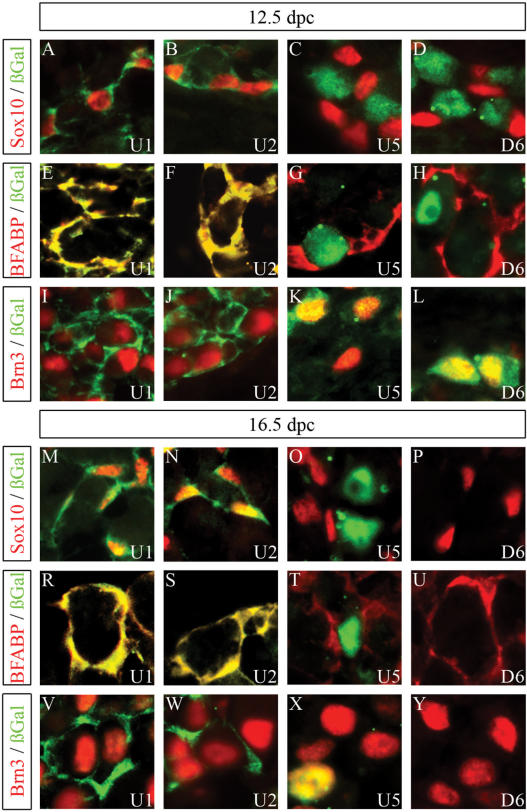
Cellular expression pattern of β-galactosidase transgenes in DRG. Co-immunohistochemistry was performed on DRG of embryos (forelimb level) at 12.5 d.p.c. (A–L) and 16.5 d.p.c. (M–Y) using antibodies directed against β-galactosidase (in green) in combination with antibodies directed against Sox10 (A–D, M–P), BFABP (E–H, R–U) or Brn3.0 (I–L, V–Y) (all in red). The embryos carried a hsp68-lacZ transgene driven by either the U1 (A, E, I, M, R, V), the U2 (B, F, J, N, S, W), the U5 (C, G, K, O, T, X) or the D6 (D, H, L, P, U, Y) region.
Figure 5.
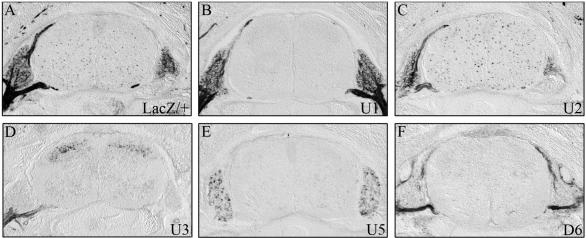
Detection of β-galactosidase transgene expression in embryos at 16.5 d.p.c. β-Galactosidase activity was detected colorimetrically using X-gal substrate on transverse sections from the forelimb level of Sox10lacZ/+ (A) and age-matched embryos carrying a hsp68-lacZ transgene driven by one of the following ECR from the Sox10 genomic region: U1 (B), U2 (C), U3 (D), U5 (E) and D6 (F). Embryo sections were stained in parallel for 2 h. No β-galactosidase staining was detected in wildtype littermates under the conditions applied.
Figure 6.
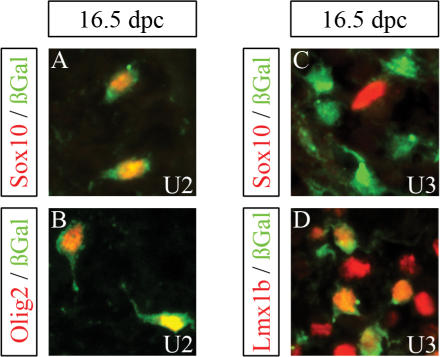
Cellular expression pattern of β-galactosidase transgenes in the embryonic spinal cord. Co-immunohistochemistry was performed on transverse sections of the embryonic spinal cord (forelimb level) at 16.5 d.p.c. using antibodies directed against β-galactosidase (in green) in combination with antibodies directed against the cell-type specific markers (in red) Sox10 (A, C), Olig2 (B) and Lmx1 (D). The embryos carried a hsp68-lacZ transgene driven by the U2 (A, B) or the U3 (C, D) region.
ECR of the Sox10 genomic region acquire their gene regulatory activity at different times of embryogenesis
To experimentally address the capacity of these seven evolutionary conserved regions to function as regulatory regions of the Sox10 gene, we cloned each element in front of a hsp68-lacZ cassette (Figure 1B) in which β-galactosidase expression is under the control of the hsp68 minimal promoter (22). We chose the hsp68 promoter because it has been extensively used in transgenic studies and has been shown to have no detectable basal activity in transgenic embryos (22).
We injected each construct in fertilized oocytes to generate transgenic lines. For each transgenic construct, at least two independent founders were obtained that stably transmitted the transgene to their offspring (Figure 1D). Transgenic progeny were collected at several stages of embryonic development followed by X-gal staining for the detection of the β-galactosidase transgene. Identical staining times were used for all transgenic embryos of a given age to obtain an approximation of the relative strength of transgene expression. Unless otherwise stated, staining patterns were highly reproducible between different lines of the same transgenic construct. Only staining intensity varied somewhat between the lines as a result of differences in copy number and integration site. Age-matched Sox10lacZ/+ embryos were stained in parallel to allow comparison with the regular Sox10 expression pattern (9).
At 9.5 d.p.c., Sox10 expression is visible in the frontonasal process, the otic vesicle, the cranial ganglia and branchial arches, in forming dorsal root ganglia (DRG) and sympathetic ganglia and further caudal in emerging neural crest cells (Figure 2A). A weak staining was also detected at the mid-hindbrain boundary.
Transgenic embryos fell into one of three categories at 9.5 d.p.c. Embryos carrying the U4, U5 or D7 transgene constituted one such category (Figure 2E, F and H). In this group, β-galactosidase staining and thus transgene expression was undetectable.
The second group consisted of embryos carrying the U2 or D6 transgene, and is characterized by a very restricted β-galactosidase expression pattern that corresponded to select parts of the Sox10 expression pattern (Figure 2C and G). The D6 transgene, for instance, was strongly expressed only in otic vesicle, trigeminal, facial and acoustic ganglia (Figure 2G). Very light β-galactosidase staining was additionally detected in the DRG area. DRG exhibited a much stronger β-galactosidase staining for the U2 transgene (Figure 2C). This staining decreased caudally in a manner similarly observed for the Sox10lacZ allele (Figure 2A). Other expression sites for the U2 transgene include the cranial ganglia, with particularly strong expression in the trigeminal, facial and acoustic ganglia (Figure 2C). Branchial arches and otic vesicle, in contrast, did not express the U2 transgene, and only very few cells were positive for U2 transgene expression in the frontonasal process.
The final category consisted of U1 and U3 transgenic embryos in which transgene expression reproduced most of the Sox10 expression pattern (Figure 2B and D). In U1 transgenic embryos, all Sox10 expressing tissues were β-galactosidase positive with exception of the mid-hindbrain boundary (Figure 2B). For most tissues, expression levels were, however, significantly lower than in the Sox10lacZ/+ embryos. Only the otic vesicle appeared strongly stained in all U1 transgenic lines.
The otic vesicle was the sole site of Sox10 expression that did not express the U3 transgene at 9.5 d.p.c. (Figure 2D). For all other Sox10 expressing tissues, β-galactosidase staining was visible in U3 transgenic embryos. In the trunk region, it appeared more intense than the corresponding staining for U1 transgenic embryos (compare Figure 2D with B). The frontonasal process was only weakly stained by β-galactosidase activity in U3 transgenic embryos. Notably, U3 transgenic embryos also exhibited β-galactosidase staining at several sites where Sox10 expression does not normally occur, including cells in the heart. However, even this ectopic β-galactosidase staining was highly reproducible between the different U3 transgenic lines.
Gene regulatory activities of the ECR in the Sox10 genomic region converge during mid-embryogenesis
At 11.5 d.p.c., expression of the various transgenes had undergone a number of significant changes. β-Galactosidase staining in the branchial arches and the frontonasal process was no longer visible in transgenic embryos (Figure 2J–P). In age-matched Sox10lacZ/+ embryos, β-galactosidase staining had similarly disappeared from the branchial arches and was strongly decreased in the frontonasal process (Figure 2I) arguing that extinction of transgene expression recapitulates downregulation of Sox10 in these areas.
U1, U2, U3 and D6, the four transgenes that were already expressed at 9.5 d.p.c., still exhibited an intense β-galactosidase staining (Figure 2J–L and O). This was particularly strong throughout the developing peripheral nervous system including ganglia as well as nerves. Intriguingly, the staining pattern for the U1, U2, U3 and D6 transgenic embryos resembled each other much more than they did at earlier times arguing that these four evolutionary conserved non-coding sequences overlap significantly in their gene regulatory activity at this embryonic age.
Nevertheless, closer inspection revealed minor, but clear differences among U1, U2, U3 and D6 transgenic embryos. Compared to U1, U3 and D6 transgenic embryos, U2 transgenic embryos had consistently lower β-galactosidase staining in the cranial ganglia (compare Figure 2K with J, L and O). U1 and U2 transgenic embryos furthermore exhibited intense β-galactosidase staining in both DRG and nerves (Figure 2J and K), whereas staining in U3 transgenic embryos was more pronounced along the nerves (Figure 2L). D6 transgenic embryos in contrast showed preferential β-galactosidase staining in DRG (Figure 2O). Significant expression in cells of the enteric nervous system was only observed for the U1 and U3 transgenes.
Expression in the otic vesicle was consistent with observations on 9.5 d.p.c. in that U2 and U3 transgenic embryos did not exhibit β-galactosidase staining in this structure (Figure 2K and L). For U1 transgenic embryos, β-galactosidase staining in the otic vesicle was still detectable but much less pronounced than at 9.5 d.p.c. arguing that U1 activity is already downregulated at 11.5 d.p.c. (Figure 2J). D6 activity, in contrast, was still high according to the strong β-galactosidase staining in D6 transgenic embryos at 11.5 d.p.c. (Figure 2O).
Of the three ECR that had not exhibited gene regulatory activity in transgenic embryos at 9.5 d.p.c., U5 now started to become detectable in the trigeminal ganglion and in DRG arguing that U5 obtains gene regulatory activity later than U1, U2, U3 and D6 (Figure 2N). Additional U5 expression was observed in the dorsal spinal cord. Although also detected in D6 transgenic embryos (compare Figure 2N with O), the dorsal spinal cord is not a main expression site of Sox10.
U4 and D7 transgenic embryos, in contrast, failed to produce any β-galactosidase staining under standard staining procedures. Even under significantly prolonged staining times, no β-galactosidase activity was detectable in D7 transgenic embryos at 11.5 d.p.c. or any other earlier or later time during embryonic development including 18.5 d.p.c. This was also the case for embryos derived from five out of seven U4 transgenic lines. In contrast, embryos from one U4 transgenic line developed weak midline staining, whereas embryos from yet another U4 founder showed blood vessel staining upon extended staining times (data not shown). We believe that this weak staining is rather indicative of the site at which transgene integration occurred in the respective U4 line than of bona fide gene regulatory activity of U4. In embryos, in which the hsp68-lacZ cassette was under the simultaneous control of U3 and U4, transgene expression was both quantitatively as well as qualitatively identical to U3 transgenic embryos (data not shown). This corroborates the absence of any enhancer activity in U4 and at the same time argues against a general silencer activity of this evolutionary conserved sequence element. In summary, we failed to detect any consistent gene regulatory activity for the non-coding regions U4 and D7 despite their evolutionary conservation.
Gene regulatory activities of the ECR in the Sox10 genomic region diverge again during later stages of embryogenesis
β-Galactosidase expression in the various transgenic lines had not changed dramatically from 11.5 to 12.5 d.p.c. Within the developing peripheral nervous system, the U1, U2 and U3 transgenes were all still expressed in peripheral nerves (Figure 3A–C), whereas DRG expression was detected for U1, U2, U5 and D6 (Figure 4A–D). Those cells that expressed the β-galactosidase transgene under control of the U1, U2 or U3 element in peripheral nerves or ganglia also contained Sox10 (Figures 3A–C and 4A, B) and the early glial marker BFABP (Figure 4E, F and data not shown), but not the neuronal marker Brn3.0 (Figure 4I and J) indicating that transgene expression reflects expression of the endogenous Sox10 even at the cellular level. In contrast, β-galactosidase-positive cells in DRG of U5 or D6 transgenic lines did not colabel with Sox10 or glial markers (Figure 4C, D, G and H), but rather with neuronal markers (Figure 4K and L).
At 16.5 d.p.c., β-galactosidase-positive cells had completely disappeared from DRG in D6 transgenic embryos (Figures 4P, U, Y and 5F). Although a fraction of sensory neurons within DRG still expressed the U5 transgene (Figure 4X), the overall β-galactosidase staining intensity had also dramatically decreased in DRG of U5 transgenic embryos (Figures 4O, T, X and 5E). Only DRG from U1 and U2 transgenic embryos still exhibited significant β-galactosidase staining at 16.5 d.p.c. (Figure 5B and C). Similar to DRG of age-matched Sox10lacZ/+ embryos, β-galactosidase was furthermore exclusively localized to Sox10- and BFABP-expressing satellite glia (Figure 4M, N, R and S) and not to Brn3.0-expressing sensory neurons (Figure 4V and W).
Strong β-galactosidase staining was also observed for the peripheral nerves in embryos from the U1, U2, U3 and D6 transgenic lines (Figure 5B–D and F). Whereas U1, U2 and U3 transgenes were already expressed in the peripheral nerve at 12.5 d.p.c., D6 transgene expression had been absent at these earlier times (compare Figure 4A–C with D). For all four transgenic lines, β-galactosidase was furthermore found in cells that express Sox10 as well as Oct6 (Figure 3E–L) arguing that the transgene in all four lines is properly expressed in Schwann cells.
By β-galactosidase staining, sympathetic ganglia were found to still express U1, U2 and D6 transgenes at 16.5 d.p.c., whereas the differentiating enteric nervous system and the medulla of the adrenal gland expressed β-galactosidase predominantly in the U1 and U3 transgenic lines (Figure 1D and data not shown).
Very little β-galactosidase staining was observed for most transgenic lines in the spinal cord at 16.5 d.p.c. In particular, the dorsal spinal cord expression observed for the U5 and D6 transgenic lines at 11.5 d.p.c. had mostly disappeared (Figure 5E and F). Only U2 and U3 transgenic embryos exhibited significant spinal cord expression (Figure 5C and D). In case of the U2 transgenic line, β-galactosidase-positive cells were scattered throughout the spinal cord parenchyma at 16.5 d.p.c. as expected for oligodendrocyte progenitors and observed in the age-matched Sox10lacZ/+ embryos (compare Figure 5C with A). Co-immunohistochemistry with Sox10 and Olig2 confirmed that the β-galactosidase expressing cells in the U2 transgenic line were oligodendroglial cells (Figure 6A and B). In contrast, β-galactosidase-positive cells in the U3 transgenic line were predominantly localized in the dorsal horn (Figure 5D) and expressed interneuron markers such as Lmx1b, but not Sox10 (Figures 5D and 6C, D). U2 is therefore the only identified evolutionary conserved non-coding sequence with regulatory activity in oligodendroglia.
Overlapping gene regulatory activities are reflected by shared patterns of transcription factor binding in the ECR
Despite the fact that several of the evolutionary conserved sequences exhibited overlapping enhancer activities in the early neural crest and several neural crest derivatives, we failed to detect any meaningful sequence similarities between these sequences by various bioinformatics approaches (data not shown). However, we found common binding motifs for transcription factors. Using the rVista2.0 program (http://rvista.dcode.org), numerous, well-conserved transcription factor binding sites were detected in all putative enhancers (Supplementary Figures 1–5). Those for U1, U2 and U3 have already been listed previously (17). Among them, binding sites for caudal-type homeodomain proteins, Sox proteins, Tcf/Lef proteins and Stat proteins were present in all enhancers with strong neural crest activity, i.e. U1, U2, U3 and D6. U1 and U3 as enhancers with particularly high expression in the early neural crest additionally carried potential binding sites for forkhead domain proteins, ikaros-type zinc finger proteins and engrailed-type homeodomain proteins. However, the relative arrangement of shared transcription factor binding sites was different in each enhancer.
We also tested several transcription factors with known roles in neural crest development directly for their ability to bind to the evolutionary conserved non-coding sequences (Figure 7). For EMSA, evolutionary conserved non-coding sequences were divided in subfragments that still allowed a good resolution of protein complexes after gel electrophoresis (Figure 7A). The tested transcription factors were Sox9, Sox10, Pax3, AP2α, Lef1, FoxD3 and Slug. With exception of the hind part of U2 (Figure 7E), all fragments bound at least one of the analysed transcription factors. None of the enhancers showed binding of FoxD3 and Slug (data not shown), only U3 and U5 bound AP2α (Figure 7I and K) and all had at least one binding site for Sox9, Sox10, Pax3 and Lef1 (Figure 7B–D and F–M). Binding was verified by supershift of the obtained complex with antibodies directed against the tag fused to each transcription factor (data not shown).
U1, U3 and D6 which are all enhancers with high neural crest activity possessed multiple binding sites for Sox proteins, Pax3 and Lef1 (Figure 7B, C, F–I, L and M). U2 which has also significant activity in the neural crest and its derivatives similarly has multiple Sox protein and Lef1 binding sites, but only a single Pax3 site (Figure 7D). In case of the Sox binding sites, some bound Sox protein dimers (Figure 7B–D, F, G, I, L and M), others bound monomeric Sox9 or Sox10 (Figure 7D and H) in accord with previous observations on other regulatory regions controlled by these proteins (30).
U5, in contrast, exhibits only weak neural crest expression. Correspondingly, it contained only a single dimeric Sox10-binding site, a single Pax3 and single Lef1 binding site in addition to its AP2α-binding site (Figure 7J and K). Thus, it appears that the evolutionary conserved non-coding sequences bind a similar set of neural crest-related transcription factors and that the number of binding sites within each element corresponds to the activity of each region within neural crest cells and their derivatives.
DISCUSSION
Over the last couple of years, a number of studies have shown that evolutionary conserved non-coding sequences in the vicinity of a gene are good candidates for the gene's regulatory regions (27,28). By applying this strategy to the Sox10 chromosomal region, we here identify five regions that have the capacity to direct transgene expression in tissues of mouse embryos in which Sox10 is normally expressed. Taking furthermore into account that Polr2f and Pick1, the two genes flanking the Sox10 genomic region, exhibit ubiquitous expression, these five evolutionary conserved non-coding sequences are good candidates for Sox10 gene enhancers. Similar to its close relative Sox9 (32,33), the Sox10 gene thus appears to be regulated by numerous enhancers that are distributed over large genomic regions.
The regulatory activity of the identified Sox10 enhancers and their relative position to the Sox10 gene are also consistent with previous BAC transgene deletion studies (17). These studies found that deletion of most sequences upstream of the Sox10 promoter only leaves a weak residual Sox10 expression. On the basis of our study, most of this residual expression is likely mediated by D6 which was still present in the BAC deletion mutant, whereas U1, U2, U3 and possibly U5 were absent. The presence of 20–30 kb of additional upstream sequences in two further BAC deletion mutants led to the recovery of Sox10 expression, albeit at reduced levels (17). This interval contained U3 and possibly U5 so that the enhancer activities of these two elements are likely responsible for the observed recovery. Not contained, however, were U1 and possibly U2 arguing that the absence of these two enhancers in the BAC deletion mutants caused the quantitative difference to wildtype Sox10 expression levels.
In contrast to many other genes for key developmental regulators including Sox9 (28,32,34,35), non-coding sequences in the vicinity of the Sox10 gene are not conserved in all vertebrate species. The evolutionary most distant conservation is between mammals and chicken. No obvious sequence homologies in the Sox10 chromosomal region were obtained with Xenopus or zebrafish, although Sox10 similarly functions as a key regulator of neural crest development in all vertebrates (1–7). Interestingly, expression of the group B1 Sox genes Sox2 and Sox3 is also regulated by enhancer regions that are conserved in amniotes but not other vertebrates despite the crucial roles of these two genes in early embryogenesis and neurogenesis of all vertebrates (36,37).
The absence of evolutionary conserved Sox10 gene enhancers between mammals and fish agrees with previous experimental data. Whereas few kilobases of the zebrafish Sox10 upstream region allowed efficient expression in melanoblasts and oligodendroglial cells, this was not the case in mammals (17,38,38). Our results thus argue that there is not always a strict correlation between the conservation of a gene's function during development and the conservation of its regulatory elements. This has also been observed for a few other genes, most prominently for c-Ret (40).
In contrast to many other published cases where each regulatory element contributes a highly specific temporal or spatial aspect to the overall expression pattern of a gene (28,41,41), the regulatory elements of the Sox10 gene have overlapping activities. U1, U2, U3 and D6 are all simultaneously active at 11.5 d.p.c. in the developing peripheral nervous system or at 16.5 d.p.c. in Schwann cells of peripheral nerves. These overlapping activities are also reflected by the fact that similar combinations of transcription factors bind to U1, U2, U3 and D6. In particular, the presence of multiple binding sites for Sox proteins, Pax3 and Lef1 appears to be a hallmark of these enhancers. These sites may mediate effects of canonical Wnt signalling on Sox10 expression, Sox9-dependent induction and Sox10 autoregulation (1,5,5).
Sox10 expression in a particular tissue or cell type during embryonic development thus very often is the result of the composite activity of many enhancers. In this respect, regulation of Sox10 expression resembles closely the regulation of Sox2 expression in the developing neural tube (37).
Despite overlapping activity, there are also clear differences between the various enhancers. In Schwann cells, U1, U2 and U3 are, for instance, continuously active over a prolonged period, whereas D6 activity in peripheral nerves is interrupted. The fact that some regulatory regions show a continuous activity in a certain cell type, whereas others exhibit a more dynamic pattern may indicate that these regulatory regions are at least partly under control of different signals. Nevertheless, the simultaneous activity of many enhancers in the same cell type may be valuable as a fail-safe mechanism that guarantees continued Sox10 expression even upon inactivation or deletion of a particular regulatory region.
In case of Sox10, each regulatory region determines multiple aspects of the expression pattern. U2, for instance, drives Sox10 expression in satellite glia and oligodendroglial cells at the same time when it directs expression in Schwann cells. This contrast with other genes where the activity of a defined enhancer often correlates with expression in a certain cell type or tissue (28,41,41).
Several evolutionary conserved non-coding sequences also exhibited ectopic expression in our study. Some of this ectopic activity was clearly due to integration site effects as in the case of U4 where weak blood vessel or midline activity was detected selectively in the progeny of a single founder. In other cases, ectopic activity was reproducibly observed in all available founders. Thus U3 activity was present in the heart, and the dorsal spinal cord exhibited U3 and U5 activity at distinct phases of embryonic development. Additionally, some of the evolutionary conserved non-coding sequences were found to direct neuronal expression, although glia are the only differentiated cells in the nervous system that normally express Sox10 (9). These sites of ectopic activity very often correspond to cell types or tissues that originate from Sox10 expressing precursors (e.g. cardiac neural crest, progenitors to sensory neurons in DRG) or exhibit a very transient expression of low amounts of Sox10 (e.g. the dorsal spinal cord). It thus appears that the activity of some of the identified regulatory regions is not properly turned off when they are studied outside their normal genomic context.
The Sox10 genomic region therefore likely contains additional elements that help to restrict Sox10 expression during embryonic development in time and in space. Due to the design of our study, these regulatory activities would have escaped our detection. As a consequence, we should have found evolutionary conserved non-coding sequences in our study without apparent activity. This was indeed the case for U4 and D7 which may therefore function as negative regulatory elements. Whether these elements really function as silencers, has to be tested in future experiments. Our failure to detect U4-dependent silencing of U3 activity certainly argues that these elements could also have other less obvious functions.
The composite activity of the five identified evolutionary conserved non-coding sequences accounts for many aspects of the Sox10 expression pattern, although this does not exclude that there are more enhancers with similar and redundant activity yet to be discovered. Not reflected in the activity of the five evolutionary conserved non-coding sequences is the normally observed expression of Sox10 in the melanocyte lineage and in the early neural crest when cells change from a premigratory to a migratory state. Deal et al. reported that melanocyte expression is at least in part conveyed by the promoter region (17) which was not analysed in our study. The localization of the regulatory element for the early neural crest expression in contrast remains at large.
Interestingly, U1 had previously been implicated as DCHCS-1 in the regulation of Sox10 expression (21). Pigmentation defects and partial aganglionosis of the colon in a mouse model of Waardenburg/Hirschsprung disease had been attributed to random transgene insertion 47 kb upstream of the Sox10 gene and concomitant deletion of 16 kb. As U1 is present in the deleted region and exhibited enhancer activity in transfected melanocytic cell lines, U1 was postulated to be the melanocyte enhancer and possibly the enteric nervous system enhancer lost during transgene insertion. Our results support the role of U1 as an enteric nervous system enhancer. The link between the loss of U1 and the observed pigmentation defect in the mouse mutant is not as straightforward. Our results rather argue for a more complex scenario in which other regions deleted during transgene insertion, the rearrangement itself, or functional interactions between U1 and other regulatory regions contribute to the observed melanocyte defect. In future studies, it will be interesting to search for mutations or deletions in any of the identified enhancers in patients who do not carry mutations in the Sox10 coding region, but nevertheless exhibit the typical symptoms such as Waardenburg/Hirschsprung disease, peripheral neuropathies or central leukodystrophies (43,44).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
[Supplementary Data]
ACKNOWLEDGEMENTS
We thank C. Birchmeier, T. Müller, D. Rowitch and E. Turner for the gift of antibodies, J. Behrens, H. Schorle, D. Meijer and P. Labosky for plasmids. Supported by grants from the Deutsche Forschungsgemeinschaft (We1326/7 and We1326/8) to M.W. Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1.Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet JP. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev. Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- 2.Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 3.Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- 4.Herbarth B, Pingault V, Bondurand N, Kuhlbrodt K, Hermans-Borgmeyer I, Puliti A, Lemort N, Goossens M, Wegner M. Mutation of the Sry-related Sox10 gene in Dominant megacolon, a mouse model for human Hirschsprung disease. Proc. Natl Acad. Sci. USA. 1998;95:5161–5165. doi: 10.1073/pnas.95.9.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honore SM, Aybar MJ, Mayor R. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev. Biol. 2003;260:79–96. doi: 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 6.Kuhlbrodt K, Schmidt C, Sock E, Pingault V, Bondurand N, Goossens M, Wegner M. Functional analysis of Sox10 mutations found in human Waardenburg-Hirschsprung patients. J. Biol. Chem. 1998;273:23033–23038. doi: 10.1074/jbc.273.36.23033. [DOI] [PubMed] [Google Scholar]
- 7.Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 9.Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghislain J, Charnay P. Control of myelination in Schwann cells: a Krox20 cis-regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 2006;7:52–58. doi: 10.1038/sj.embor.7400573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peirano RI, Goerich DE, Riethmacher D, Wegner M. Protein zero expression is regulated by the glial transcription factor Sox10. Mol. Cell. Biol. 2000;20:3198–3209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung M, Chaboissier M-C, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
- 16.Stolt CC, Schlierf A, Lommes P, Hillgärtner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev. Cell. 2006;11:697–710. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Deal KK, Cantrell VA, Chandler RL, Saunders TL, Mortlock DP, Southard-Smith EM. Distant regulatory elements in a Sox10-betaGEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest-derived tissues. Dev. Dyn. 2006;235:1413–1432. doi: 10.1002/dvdy.20769. [DOI] [PubMed] [Google Scholar]
- 18.Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competition waves of oligodendrocytes in the forebrain and postnatal elimination of an early embryonic lineage. Nat. Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellerer S, Schreiner S, Stolt CC, Bösl MR, Wegner M. Functional equivalency of transcription factors Sox8 and Sox10 is tissue-specific. Development. 2006;133:2875–2886. doi: 10.1242/dev.02477. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig A, Schlierf B, Schardt A, Nave KA, Wegner M. A Sox10 rtTA mouse line for tetracycline-inducible expression of transgenes in neural crest cells and oligodendrocytes. Genesis. 2004;40:171–175. doi: 10.1002/gene.20083. [DOI] [PubMed] [Google Scholar]
- 21.Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin SQ, Green ED, Paisley D, Kelsh RN, Pavan WJ, et al. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum. Mol. Genet. 2006;15:259–271. doi: 10.1093/hmg/ddi442. [DOI] [PubMed] [Google Scholar]
- 22.Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 23.Friedrich R, Schlierf B, Tamm ER, Bösl MR, Wegner M. The class III POU domain protein Brn-1 can fully replace the related Oct-6 during Schwann cell development and myelination. Mol. Cell. Biol. 2005;25:1821–1829. doi: 10.1128/MCB.25.5.1821-1829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maka M, Stolt CC, Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 25.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlierf B, Ludwig A, Klenovsek K, Wegner M. Cooperative binding of Sox10 to DNA: requirements and consequences. Nucleic Acids Res. 2002;30:5509–5516. doi: 10.1093/nar/gkf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandelin A, Bailey P, Bruce S, Engstrom PG, Klos JM, Wasserman WW, Ericson J, Lenhard B. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelsh RN. Sorting out Sox10 functions in neural crest development. Bioessays. 2006;28:788–798. doi: 10.1002/bies.20445. [DOI] [PubMed] [Google Scholar]
- 30.Wegner M. Secrets to a healthy Sox life: lessons for melanocytes. Pigment Cell Res. 2005;18:74–85. doi: 10.1111/j.1600-0749.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- 31.Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, Pennacchio LA. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagheri-Fam S, Ferraz C, Demaille J, Scherer G, Pfeifer D. Comparative genomics of the SOX9 region in human and Fugu rubripes: conservation of short regulatory sequence elements within large intergenic regions. Genomics. 2001;78:73–82. doi: 10.1006/geno.2001.6648. [DOI] [PubMed] [Google Scholar]
- 33.Wunderle VM, Critcher R, Hastie N, Goodfellow PN, Schedl A. Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc. Natl Acad. Sci. USA. 1998;95:10646–10654. doi: 10.1073/pnas.95.18.10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blader P, Lam CS, Rastegar S, Scardigli R, Nicod J.-C, Simplicio N, Plessy C, Fischer N, Schuurmans C, et al. Conserved and acquired feautres of neurogenin1 regulation. Development. 2004;131:5627–5637. doi: 10.1242/dev.01455. [DOI] [PubMed] [Google Scholar]
- 35.de la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodriguez-Seguel E, Letizia A, Allende ML, Gomez-Skarmeta JL. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2006;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunelli S, Casey ES, Bell D, Harland R, Lovell-Badge R. Expression of Sox3 throughout the developing central nervous system is dependent on the combined action of discrete, evolutionary conserved elements. Genesis. 2003;36:12–24. doi: 10.1002/gene.10193. [DOI] [PubMed] [Google Scholar]
- 37.Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 38.Elworthy S, Lister JA, Carney TJ, Raible DW, Kelsh RN. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development. 2003;130:2809–2818. doi: 10.1242/dev.00461. [DOI] [PubMed] [Google Scholar]
- 39.Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat. Neurosci. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- 40.Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 41.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev. Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 42.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 43.Inoue K, Khajavi M, Ohyama T, Hirabayashi S-I, Wilson J, Reggin JD, Mancias P, Butler IJ, Wilkinson MF, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat. Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 44.Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Prehu M-O, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, et al. Sox10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]