A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing (original) (raw)
Abstract
Multiplexed high-throughput pyrosequencing is currently limited in complexity (number of samples sequenced in parallel), and in capacity (number of sequences obtained per sample). Physical-space segregation of the sequencing platform into a fixed number of channels allows limited multiplexing, but obscures available sequencing space. To overcome these limitations, we have devised a novel barcoding approach to allow for pooling and sequencing of DNA from independent samples, and to facilitate subsequent segregation of sequencing capacity. Forty-eight forward–reverse barcode pairs are described: each forward and each reverse barcode unique with respect to at least 4 nt positions. With improved read lengths of pyrosequencers, combinations of forward and reverse barcodes may be used to sequence from as many as _n_2 independent libraries for each set of ‘_n_’ forward and ‘_n_’ reverse barcodes, for each defined set of cloning-linkers. In two pilot series of barcoded sequencing using the GS20 Sequencer (454/Roche), we found that over 99.8% of obtained sequences could be assigned to 25 independent, uniquely barcoded libraries based on the presence of either a perfect forward or a perfect reverse barcode. The false-discovery rate, as measured by the percentage of sequences with unexpected perfect pairings of unmatched forward and reverse barcodes, was estimated to be **<**0.005%.
INTRODUCTION
With the advent of high-throughput sequencing, characterization of the nucleic acid world is proceeding at an accelerated pace. Three major high-throughput sequencing platforms are in use today: the Genome Sequencers from Roche/454 Life Sciences [GS-20 or GS-FLX; (1)], the 1G Analyzer from Illumina/Solexa (2) and the SOLiD System from Applied Biosystems (http://solid.appliedbiosystems.com). Comparison across the three platforms reveals a trade-off between average sequence read length and the number of DNA molecules that are sequenced. At present, the Solexa and SOLiD systems provide many more sequence reads, but render much shorter read lengths than the 454/Roche Genome Sequencers. This makes the 454/Roche platform appealing for use with barcoding technology, as the enhanced read length facilitates the unambiguous identification of both complex barcodes and sequences of interest.
The 454/Roche Genome Sequencers are called pyrosequencers because their sequencing technology is based on the detection of pyrophosphates released during DNA synthesis (3). A few sequencing runs using 454/Roche's pyrosequencing platform can generate sufficient coverage for assembling entire microbial genomes (4,5), for the discovery, identification and quantitation of small RNAs (6,7), and for the detection of rare variations in cancers (8), among many other applications. For analysis of multiple libraries, the currently available 454/Roche pyrosequencers can accommodate a maximum of only 16 independent samples, which have to be physically separated using manifolds on the sequencing medium. These separation manifolds occlude wells on the sequencing plate from accommodating bead-bound DNA template molecules, and thus restrict the number of output sequences. To overcome these limitations, we describe a high-information-content barcoding approach in which each sample is associated with two uniquely designed, 10-nucleotide barcodes. The presence of these assigned barcodes allow for independent samples to be pooled together for sequencing, with subsequent bioinformatic segregation of the pyrosequencer output. By not relying on physical separators, this procedure maximizes sequence space and multiplexing capabilities.
‘Barcodes’, or unique DNA sequence identifiers, have historically been used in several experimental contexts. In sequence-tagged mutagenesis (STM) screens, a sequence barcode acts as an identifier or type specifier in a heterogenous cell-pool or organism-pool. STM barcodes are usually 20–60 nt long, are pre-selected or follow ambiguity codes, and are present as one unit or split into pairs (9). Such long barcodes are not ideally suited for use with available pyrosequencing platforms because of restrictions on read length. Several recent papers have reported the use of very short (2- or 4-nt) barcodes with pyrosequencing platforms (6,10,11). Although such approaches are valuable, there are applications that call for a more definitive assignment of samples and/or for enhanced multiplexing capabilities. We describe here the design and testing of a series of 10-base barcodes that differ from each other at multiple positions (by at least 4 nt), and have a common specification that facilitates resolution of ambiguities. Using the GS20 platform (454/Roche), these barcodes allowed us to simultaneously pyrosequence small RNA libraries from 25 diverse samples for each pilot run, and unambiguously assign 99.8% of obtained barcoded sequences by bioinformatically probing for an error-free barcode at either end of the sequence of interest. The false-discovery rate, defined as rate of occurrence of mispaired forward and reverse barcodes, was 0.00042% (∼1 in 237 000 sequences) in run I, and 0.0044% (∼12 in 271 000 sequences) in run II. Discussed here are the modifications made for cloning and amplification protocols to handle highly complex nucleic acid pools, the considerations used in designing the barcodes, and the efficacy of the barcoding technology. We also speculate on the use of these pyrosequencer-tailored barcodes with other sequencing platforms.
MATERIALS AND METHODS
Cloning of small RNAs
We sought to identify novel small RNAs in 50 different RNA preparations derived from a diverse set of virus-infected and control cell and tissue samples. To pursue this analysis, small RNA fractions from each cell or tissue sample were independently isolated (using the miRvana small RNA isolation protocol (Ambion)), and processed to produce 50 individually barcoded cDNA libraries that could be sequenced using the Roche/454 platform. cDNA libraries were constructed (using slight modifications of standard procedures) as follows: RNA 3′ termini were coupled to adenylated DNA linkers as described (12,13), and subsequent ligation of the second linker was performed either directly to the 5′ termini of the RNAs [5′-P-dependent cloning; (12)], or to the 3′ termini of the cDNAs synthesized from the 3′-linkered RNAs [5′-P-independent cloning; (13)]. The following sets of linkers were used for cloning:
linkerA (RNA–DNA hybrid linker): ATCGTrArGrGrCrArCrCrUGrArArA
linkerB (IDT Linker 2): rAppCACTCGGGCACCAAGGA/3ddC/
linkerC (DNA linker): pCACTCGGGTGCCAAGGA/3ddC/
linkerD (IDT Linker 1): rAppCTGTAGGCACCATCAAT/3ddC/
Ribonucleotides are marked by the presence of ‘r’ before the nucleotide. ‘App’ is adenylated nucleotide; ‘ddC’ is dideoxycytidine and ‘p’ is phosphate group. Both linkerB and linkerD may be used with either cloning protocol. For our experiments, linkerA and linkerB were used for cloning 5′-monophosphated small RNAs, while linkerC and linkerD were used in constructing libraries using the 5′-P-independent cloning protocol. Published protocols for library construction (12,13) were modified to reduce the number of gel purifications. In particular, size fractionation on a 12% denaturing PAGE–urea gel was exclusively performed after the first ligation event. This substantially reduced the required amount of starting material (from several microgram to as little as 0.25 μg of the small RNA fraction), and minimized loss of material that would have otherwise occurred with the two additional gel elutions.
Overall design of barcoded primers used in amplifying cDNA libraries
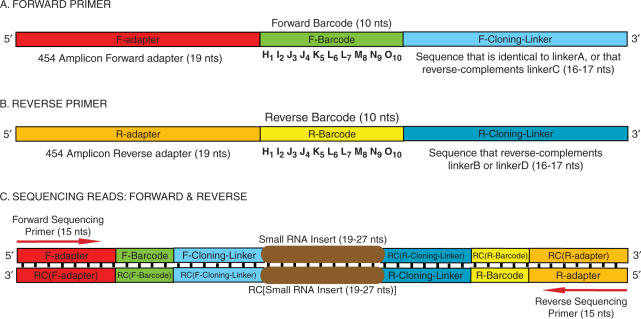
The barcoded primers were each 45–46 nt long and consisted of: (i) the 454 amplicon adapter (the amplicon sequencing primer annealing site; the ‘F-adapter or R-adapter’), (ii) the barcode (F-barcode or R-barcode) and (iii) the F-Cloning-Linker or R-Cloning-Linker (the sequences corresponding to the linkers used for construction of the initial nucleic acid library) (Figure 1).
Figure 1.
Design of forward and reverse primers. The synthesized primers are 45–46 nt long. (A and B) The template for the primers is: 454-Adapter:: Barcode:: Linker-Primer. Individual specifications for forward and reverse barcodes are also indicated. (C) Diagrammatic representation of the GS20 forward and reverse sequencing reads. ‘RC’ stands for Reverse Complement. Teeth denote base pairing.
Blueprint for barcodes
FORWARD barcode format: HIJJK LLMNO
REVERSE barcode format: HIJKK LMMNO
The quartets (H, I, J and K) and (L, M, N and O) each independently map to the four nucleotides (A, C, G or T). Guidelines for positional nucleotide restrictions in the barcodes are presented in Table 1.
Table 1.
Guidelines for barcodes
| Restrictions | Cloning protocol | Guidelines for restrictions |
|---|---|---|
| H ≠ (I, J, K) I ≠ (J, K) J ≠ K K ≠ L L ≠ (M, N, O) M ≠ (N, O) N ≠ O | 5-P and DL | Adjoining nucleotides must be different (except at dinucleotide positions). For each barcode, (H, I, J, K) and (L, M, N, O) uniquely map to one of (A, C, G, T). Each barcode has two distinct dinucleotide pairs: at positions (3,4) and (6,7) for the forward barcodes, and at positions (4,5) and (7,8) for reverse barcodes. |
| I ≠ (J, K) | ||
| J ≠ K | ||
| K ≠ L | ||
| L ≠ (M, N, O) | ||
| M ≠ (N, O) | ||
| N ≠ O | ||
| H ≠ G | 5-P and DL | The terminal nucleotide of the F-adapter and R-adapter (Figure 1) is a guanosine (in both (5′-phosphate-dependent and 5′-phosphate-independent protocols); ‘H’ cannot be part of a dinucleotide, and hence cannot be a guanosine. |
| O ≠ A | 5-P | First nucleotide of F-cloning-linker and R-cloning-linker is an adenosine (in the 5′-phosphate-dependent protocol). ‘O’ cannot be part of a dinucleotide, and hence cannot be an adenosine. |
| O ≠ T | DL | First nucleotide of F-cloning-linker and R-cloning-linker is a thymidine (in the 5′-phosphate- independent protocol). ‘O’ cannot be part of a dinucleotide, and hence cannot be a thymidine. |
Primer sequence corresponding to adjusted linker sequence (linker-primer)
linker-primerA (corresponds to linkerA): ATCGTAGGCACCTGAAA
linker-primerB (corresponds to linkerB): ACCGTCCTTGGTGCCCG
linker-primerC (corresponds to linkerC): TCCTTGGCACCCGAGTG
linker-primerD (corresponds to linkerD): TTGATGGTGCCTACAG
DNA amplification and pooling of independent samples
The linkered cDNA pool from each individual sample was amplified for 20 cycles [as described in (12)] with an unique pair of barcoded primers. A second round of amplification with temperatures and cycle lengths identical to the initial round, but with variable number of total cycles (six, eight and ten), was performed using 1/25th of the first PCR product, dNTPs, barcoded primers, buffer and Taq DNA polymerase in a 40 μl PCR mix. An analytical, non-denaturing 4% Nusieve gel was used to visually determine the fewest number of cycles that yielded a visible signal for PCR products containing inserts corresponding to small RNAs. DNA was extracted from the gel using the Qiaquick gel extraction kit (Qiagen). Although the instructions for gel extraction called for dissolution of the gel slice in Buffer QG at 50°C, we performed all manipulations of the DNA-containing buffers at room temperature, to ensure that the complex PCR mixtures were not denatured.
To obtain a rough quantitation of individual libraries, 10% of each recovered PCR product was separated on an analytical 2% agarose gel. Based on the intensities of bands on this gel, PCR products from 25 samples were pooled together in roughly equimolar ratios. Quality of the mixture was tested by conventional Sanger sequencing of 46 PCR products that had been cloned into TOPO-TA vectors (Invitrogen). If the sequence quality was satisfactory, 300 ng of the mixture was run on a non-denaturing 2% low melting point (LMP)-agarose gel (run I) or on a denaturing 6% PAGE–urea gel (run II; recommended) for quantification, and for further size-restricted purification of barcoded species that contain small RNA inserts. DNA was eluted from the PAGE–urea gel overnight at 4°C in 0.3 M NaCl.
Processing of pooled sample for high-throughput sequencing
Amplicon libraries prepared as described above may be entered directly into the 454/Roche sequencing pipeline, with the only major variable being the amount of DNA from the pooled sample used to seed the emulsion PCR reaction. Approximately 50 ng of the purified pool was set aside for each pyrosequencing run. For run I, DNA was not heat-denatured, and concentrations of both single-stranded (using Agilent 2100 Bioanalyzer's RNA chip assay; Agilent Technologies) and double-stranded (using the Quant-iT PicoGreen dsDNA Assay; Invitrogen) species were measured, before the DNA was channeled into the Roche/454 Amplicon sequencing pipeline. As estimating concentrations of appropriately sized molecules in each library has been most reliable following denaturation of samples, for run II (and for subsequent uses of this technology), we denatured input DNA (by flash heating and cooling) prior to both measurement of concentration (using the 2100 BioAnalyzer) and emulsion PCR. Post-denaturation, different ratios of DNA molecules to DNA-capturing beads were tested in an initial attempt to find a concentration of DNA that would result in a 10–30% bead recovery after emulsion PCR (1). For run II of this analysis, the DNA to bead ratio used was 3: 1. All subsequent procedures were carried out essentially as described in the GS20 guide-book for amplicon sequencing (454/Roche).
Data handling
Program ‘Barsort’ was used to resolve sequences into separate data bins based on perfect match to the 10-nt barcode sequence + adjoining ‘_x_’ nucleotides of the linker sequence (0 ≤ x ≤ length of linker). Insert sequences were then mapped to RNA databases using standard pattern-matching algorithms. The false-discovery rate was estimated based on sequence segregation into various subcategories using the program ‘Barverify’. Barsort and Barverify are available as source code and executables on request.
RESULTS AND DISCUSSION
Design of barcoded primers
We have devised a barcoding approach to substantially enhance the scope and capacity of multiplexed high-throughput pyrosequencing. Specifications for the barcodes are based on details of pyrosequencing error characteristics. Sets of barcodes have been independently designed for the forward PCR primers (FORWARD barcodes) and the reverse PCR primers (REVERSE barcodes) used in library construction (Figure 1). Length of the barcode was chosen to allow for definitive assignment of barcodes, while minimizing the number of base reads used per barcode. The barcode design (Table 1) facilitated unambiguous assignment, with care taken to avoid homopolymeric runs that are particularly problematic in pyrosequencing technologies. All forward barcodes were of the type HIJJK LLMNO, while reverse barcodes were of the type HIJKK LMMNO. H, I, J, K and L, M, N, O are uniquely partnered with one of four nucleotides: A, C, G and T. The barcodes thus contained two instances of a 1–2 base run of each nucleotide (one from the H, I, J, K set, and the other from the L, M, N, O set), with no more than two successive occurrences of the same nucleotide (Table 1). This design avoids significant problems due to error rate at homopolymeric runs of nucleotides. The forward and reverse barcodes varied in dinucleotide positioning along the length of the barcode. Positions 3, 4 (nucleotide J) and 6, 7 (nucleotide L) were homopolymeric dinucleotides in the forward barcode, while positions 4, 5 (nucleotide K) and 7, 8 (nucleotide M) were homopolymeric dinucleotides in the reverse barcode.
There were several restrictions at specific positions in the barcode. The start position of the barcode (position H) was required to differ from the last nucleotide of the preceding adapter to avoid homodinucleotides at the Adapter::Barcode junctions (Figure 1). The terminal nucleotide of the barcode (position ‘O’) was constrained to prevent identity with the first nucleotide of both the F-Cloning-Linker and the R-Cloning-Linker. To ensure identical nucleotide possibilities at ‘O’ for all forward and reverse barcodes, priming sequences that correspond to the F-Cloning-Linker and to the R-Cloning-Linker were adjusted as necessary so that they started with the same base (refer to Methods section). These adjustments avoided any unnecessary restrictions on the last base of the barcode (‘O’), created independent sets of restrictions for positions H and O, eliminated the possibility for a dinucleotide at the Barcode::Cloning-Linker junctions and at the Adapter::Barcode junctions, and increased the total number of forward–reverse barcode pairs.
Sixty-four barcode pairs were thus designed, with any two forward or reverse barcodes differing in at least 4 nt: one doublet + two singlets (Table 2). These serve as strong unique identifiers for each of the forward and reverse barcode sets. Due to the terminal nucleotide restriction (on position ‘O’), only 48 barcodes may be used with a single 5′–3′ pair of linkers used in cloning. We make one additional note on the choice of barcode sequences: in applications where there is a need for fully optimized read length, an additional preference may be imposed for barcodes whose sequences best utilize the sequencing order (TACG for the GS20) so that a few more sequencing cycles may be available for the DNA of interest.
Table 2.
List of possible barcodes
| Forward barcodes | Reverse barcodes |
|---|---|
| AGCCTAAGCT | AGCTTAGGCT |
| AGTTCAAGTC | AGTCCAGGTC |
| ACTTGAACTG | ACTGGACCTG |
| ACGGTAACGT | ACGTTACCGT |
| ATCCGAATCG | ATCGGATTCG |
| ATGGCAATGC | ATGCCATTGC |
| CAGGTCCAGT | CAGTTCAAGT |
| CATTGCCATG | CATGGCAATG |
| CTAAGCCTAG | CTAGGCTTAG |
| CGAATCCGAT | CGATTCGGAT |
| TCAAGTTAGC | TAGCCTAAGC |
| TACCGTTACG | TACGGTAACG |
| TGAACTTGAC | TGACCTGGAC |
| TAGGCTTCAG | TCAGGTCCAG |
| AGCCTCCAGT | AGCTTCTTAG |
| AGCCTGGCAT | AGCTTGCCAT |
| AGTTCGGACT | AGTCCGAACT |
| AGTTCTTGAC | AGTCCTCCAG |
| ACGGTCCATG | ACGTTCAATG |
| ACTTGTTCAG | ACTGGTGGAC |
| ACTTGCCGAT | ACTGGCGGAT |
| ACGGTGGATC | ACGTTGAATC |
| ATCCGCCTAG | ATCGGCAAGT |
| ATCCGTTAGC | ATCGGTAAGC |
| ATGGCGGTAC | ATGCCGTTAC |
| ATGGCTTACG | ATGCCTAACG |
| CAGGTAAGTC | CAGTTAGGTC |
| CAGGTGGCAT | CAGTTGCCAT |
| CATTGAAGCT | CATGGAGGCT |
| CTAAGTTCAG | CTAGGTCCAG |
| CTAAGAACGT | CTAGGACCGT |
| CTGGATTGAC | CTGAATGGAC |
| CATTGTTAGC | CATGGTAAGC |
| CTGGAGGACT | CTGAAGAACT |
| CGAATAACTG | CGATTACCTG |
| CGAATGGATC | CGATTGAATC |
| CGTTAGGTAC | CGTAAGTTAC |
| CGTTATTACG | CGTAATAACG |
| TAGGCAAGCT | TAGCCAGGCT |
| TACCGCCATG | TACGGCAATG |
| TACCGAAGTC | TACGGAGGTC |
| TGAACGGCAT | TGACCGCCAT |
| TGAACAATCG | TGACCATTCG |
| TGCCACCGAT | TGCAACGGAT |
| TGCCAGGACT | TGCAAGAACT |
| TCGGACCTAG | TCGAACTTAG |
| TCAAGCCAGT | TCAGGCAAGT |
| TCAAGAATGC | TCAGGATTGC |
| CTGGACCTGA | CTGAACTTGA |
| CGTTACCGTA | CGTAACGGTA |
| TGCCATTGCA | TGCAATGGCA |
| TCGGATTCGA | TCGAATCCGA |
| AGCCTGGCTA | AGCTTGCCTA |
| AGCCTCCTGA | AGCTTCTTGA |
| ACTTGTTCGA | ACTGGTCCGA |
| ATCCGCCGTA | ATCGGCGGTA |
| ATCCGTTCGA | ATCGGTCCGA |
| ATGGCGGTCA | ATGCCGTTCA |
| CAGGTGGCTA | CAGTTGCCTA |
| CTAAGTTGCA | CTAGGTGGCA |
| CGTTAGGTCA | CGTAAGTTCA |
| TCGGACCGTA | TCGAACGGTA |
| TCAAGCCTGA | TCAGGCTTGA |
| TGCCAGGTCA | TGCAAGTTCA |
Testing barcoded primers
The barcoded primers were 45–46 nt in length (Supplementary Table 1), had long overhangs (29–30 nt), and short regions of complementarity to the template (16–17 nt; did not span the entire Cloning-Linker sequence). Despite the extended length of the barcoded primers, complex pools of cDNA containing 19–27 nt small RNA inserts were efficiently amplified to yield PCR products that were 115–123 nt long.
Deciphering the nature of products amplified from complex pools of cDNA
Construction and sequencing of libraries from complex mixtures of RNA templates involves numerous steps that may skew the relationship between ratios of molecules in the initial pool and obtained ratios of corresponding sequences. Differential efficiencies of RNA extraction, linker addition and reverse transcription may skew the initial cDNA pool. Differences in amplification efficiencies of individual sequences in the cDNA pool, however slight, may bias the library population over many rounds of PCR. Sequence-specific differences in efficiency or fidelity of sequencing may distort the final proportions of assigned DNA sequences. These biases have been extensively discussed in literature; although potentially confounding, their effects may be controlled for to some extent, so that sequence representations in the cDNA and subsequent PCR-amplified libraries at least provide for an initial assessment of sequence incidence in the original small RNA populations.
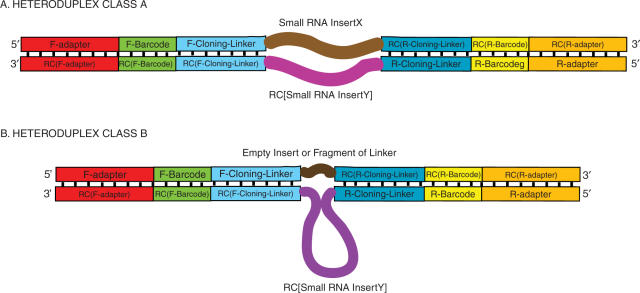
We note one additional concern that is highly relevant to preparation of libraries for high-throughput sequencing. This concern specifically relates to purification and handling of pooled DNA products following amplification of complex cDNA libraries. Amplified DNA pools generally consist of a mixture of single strands, perfect duplexes and partial duplexes. The overall structural composition of the amplified DNA pool is determined by the extent of amplification in the last round of PCR. DNA molecules that were substrates for polymerase extension during the last round of PCR will be double-stranded. Molecules that fail to extend in the last round of PCR may remain single-stranded, may form heteroduplexes (if they anneal to a distinct molecule during the last PCR cycle; Figure 2), or may form perfect duplexes (if they find and anneal to an exact complement during the last PCR cycle). This heterogeneity will be more prominent in situations where extension has become inefficient (e.g. PCR taken beyond a point of saturation), resulting in the majority of molecules being single stranded or heteroduplex during thermal cycling. Only abundant single-stranded species would be more likely to reanneal to form homoduplexes (Figure 1c). The low probability for rare single-stranded species to find perfect partner strands pushes them towards reannealing with non-identical PCR products (i.e. with library elements that have distinct inserts), resulting in the formation of heteroduplex structures (Figure 2).
Figure 2.
A diagrammatic representation of heteroduplexes that may be formed in an amplified pool. (A) Heteroduplexes formed between molecules from the same sample that have the same barcode, but different RNA inserts (X and Y). (B) Heteroduplexes formed between molecules with RNA inserts and molecules with no RNA inserts (or with fragments of linkers as inserts). Formation of these unusual duplexes may be facilitated by the 45–46 nt complementarity at either end of the insert. Thus, three types of molecules may be present during later stages of PCR: single-stranded, perfectly double stranded (Figure 1C) and heteroduplexes (shown here). The ratio of the three species is determined by the number of PCR cycles. As in Figure 1, ‘RC’ stands for Reverse Complement, and teeth denote base pairing.
The heterogenous mixture of single strands, perfect duplexes and heteroduplexes skews the sequence incidence only if there is a step in the procedure that enriches for double-stranded molecules post-PCR. This is the case, however, with many gel isolation procedures that are employed to size-select for PCR products in a pre-defined size range. Biased selection in these protocols is attributed to a combination of factors: (i) differences in mobility between single-stranded, double-stranded and heteroduplex molecules on the gel, (ii) differential recovery from gel-isolation procedures, which may enrich for segments that have either formed perfect duplexes, or are capable of forming perfect duplexes.
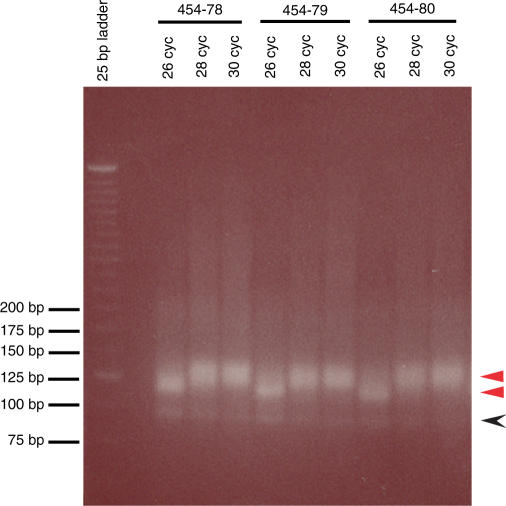
Our observations confirm that the overall nature of the DNA pool number was influenced by the extent of amplification, i.e. by the number of PCR cycles (Figure 3), since a migration shift on the agarose gel was evident upon the addition of two to four PCR cycles (with all other PCR conditions being constant). This shift is consistent with a transition between duplex structures (a relatively tight band), and a mixture of heteroduplex and single-stranded material (a more diffuse band starting above the presumed duplex band). In order to maximize the accuracy of sequence representation for each individual library, we took samples from PCR cycles prior to saturation. Products migrating with the expected double-stranded DNA population were then isolated from an agarose gel, recovered (by extraction without denaturation) and quantitated, before they were used for sequencing. Our samples were found to consist of approximately equal numbers of double-stranded and single-stranded DNA molecules by the Quant-iT PicoGreen dsDNA assay (Invitrogen), and the RNA BioChip assay (Agilent 2100 Bioanalyzer, Agilent Technologies), respectively (data not shown). Our efforts to avoid saturation of the PCR amplification, although not perfect, helped to maximize the proportion of homoduplexes formed in the last cycle of PCR, thus facilitating the recovery of both rare and common species in the library pool. All these observations stress the importance of close monitoring of complex DNA pools to ensure that overall quality is not compromised through excessive amplification of seed populations, and through subsequent manipulations of amplified populations.
Figure 3.
Visualizing the nature of amplified products from various cycles of PCR. With an increase in cycle number (twenty cycles of an initial round of PCR followed by six, eight or ten cycles of a second round of PCR), there is an evident shift in mobility of PCR products that contain a small RNA insert. Effect of PCR cycle number on three different samples is shown, stressing the importance of titrating the total number of DNA amplification cycles, to avoid saturation of the PCR amplification. Red arrows represent the two sizes of the insert-containing PCR products. PCR products without small RNA inserts migrate as faint bands between 75 and 100 bp (black arrow).
Analysis of data from two independent pyrosequencing runs
DNA from two independent sets of 25 barcoded libraries (prepared with 19–27 nts RNAs from different in vitro and in vivo virus-infected systems) were pooled for sequencing runs, and the data was used to evaluate the efficacy of the barcoding technology. Both 237 639 and 271 777 sequences from runs I and II, respectively, passed the GS20's quality control filters. Since six of the forward barcodes are exact reverse complements of six reverse barcodes, we added a number of adjoining nucleotides in the Cloning-Linker (0–17) to the barcode query to reduce ambiguity while segregating the sequences into independent datasets. These barcode + linker sequences are referred to herein as ‘search motifs’ or ‘barcode motifs’. Using 13-nt search motifs, Barsort and Barverify identified that 94.53% (run I) and 94.08% (run II) of the sequences had perfect forward and reverse barcode motifs, 2.90% (run I) and 3.24% (run II) of the sequences had perfect barcode motifs only at the 5′ end of the read, and 2.44% (run I) and 2.51% (run II) had perfect barcode motifs only at the 3′ end of the read (for data and terminology, refer Table 3).
Table 3.
Summary of sequence distribution for both pyrosequencing runs as a function of barcode motif length
| Run number | Search motif | Perfect 5′ and 3′ Motifs | Mismatched 5′ and 3′ motifs | Perfect 3′ and imperfect 5′ motifs | Perfect 5′ and imperfect 3′ motifs | Duplicate perfect 5′ motifs | Duplicate perfect 3′ motifs | Total number of sequences |
|---|---|---|---|---|---|---|---|---|
| I | Barcode (10 nt) | 229 277 | 2349 | 2323 | 2923 | 1 | 0 | 23 6873 |
| I | Barcode (10 nt) + Cloning-Linker (1 nt) | 226 785 | 295 | 4339 | 5211 | 2 | 233 | 23 6865 |
| I | Barcode (10 nt) + Cloning-Linker (2 nt) | 226 108 | 4 | 5368 | 5858 | 0 | 0 | 23 7338 |
| I | Barcode (10 nt) + Cloning-Linker (3 nt) | 224 645 | 3 | 5800 | 6892 | 0 | 0 | 23 7340 |
| I | Barcode (10 nt) + Cloning-Linker (4 nt) | 223 744 | 3 | 6258 | 7335 | 0 | 0 | 23 7340 |
| I | Barcode (10 nt) + Cloning-Linker (COMPLETE) | 205 020 | 1 | 12 095 | 20 224 | 0 | 0 | 23 7340 |
| II | Barcode (10 nt) | 258 929 | 5061 | 3138 | 3736 | 2 | 179 | 27 1045 |
| II | Barcode (10 nt) + Cloning-Linker (1 nt) | 257 998 | 17 | 6128 | 7160 | 3 | 1 | 27 1307 |
| II | Barcode (10 nt) + Cloning-Linker (2 nt) | 257 150 | 12 | 6587 | 7557 | 2 | 1 | 27 1309 |
| II | Barcode (10 nt) + Cloning-Linker (3 nt) | 255 695 | 11 | 7175 | 8427 | 0 | 1 | 27 1309 |
| II | Barcode (10 nt) + Cloning-Linker (4 nt) | 254 491 | 11 | 7818 | 8988 | 0 | 1 | 27 1309 |
| II | Barcode (10 nt) + Cloning-Linker (COMPLETE) | 226 041 | 11 | 14 662 | 30 595 | 0 | 0 | 27 1309 |
Sequences that have a perfect match to only one of the expected motifs have been further subdivided into three classes based on whether the motif at the other end has: (i) perfect identity to an unexpected motif, resulting in an unassigned 5′–3′ motif combination (perfect, unexpected occurrence), (ii) a high-scoring partial match to the expected motif for that library (imperfect, expected occurrence) and (iii) no strong match to any barcode motif (imperfect, ambiguous occurrence). Perfect, ‘unexpected’ occurrences may be attributed to motifs with rare coincidental sequence matches (due to multiple fortuitous errors during barcode synthesis or sequencing), to contamination in PCR primer stocks, or to cross-contamination of template DNAs in the second round of PCR. The imperfect matches may arise due to errors in sequencing [particularly at the starts and ends of the sequencing reads; (1)], due to errors during conventional and/or emulsion PCR, or due to imperfections during the chemical synthesis of barcoded primers. With the barcode design used in this work (i.e. no homopolymer beyond 2 nt), we expected to minimize errors related to pyrosequencing. Indeed, the experimentally observed spectrum of errors did not match the types of homopolymer-related errors most frequently observed for pyrosequencing. Instead, the observed error spectrum appeared most closely to match that expected as a result of known imprecisions in the chemical synthesis of DNA primers used in standard PCR reactions (Supplementary Table 3). Adding to any potential protocol-related possibility of sample misassignment, we note that the highly sensitive nature of high-throughput sequencing needs to be considered in all analyses. In particular, any possible source for sample contamination upstream (or during) library construction needs to be carefully monitored.
For bioinformatic segregation of a barcoded dataset, a decision needs to be made as to how much of the (fixed) linker sequence will be required in addition to the (variable) barcode for definitive sample assignment. The false-discovery rate, defined by the percentage of ‘perfect and unexpected’ occurrences, was observed to decrease to <0.005% (Table 3), if three bases of the linker were used in addition to the 10-base barcode to assign sample identities. Thus a search motif of 13 nt was determined to be ideal for our studies in minimizing false-discovery rate, while maximizing the number of sequences with error-free hits identified by the program. Sequences segregated using motifs of this length were used for all further analyses. The low false-discovery rate for perfect matches strengthens the argument that any sequence with both 5′ and 3′ barcode motifs will be placed with reasonable accuracy into the correct bin. For our purposes, all sequences with a match to one of the two expected sequence motifs were considered as true members of the corresponding dataset. Run I (0.13%) and run II (0.17%) of sequences had no perfect matches to any of the sequence motifs (forward or reverse), and were discarded. Thus, data from two independent sequencing runs on pools of barcoded material from a total of 50 sample libraries (25 sample libraries per run) show similar patterns with respect to sequence recovery, binning and rate of errors that result in false discovery and misassignment.
Distribution of sequences across samples
The barcoding approach requires being able to establish a relationship between ratios of material from various libraries, and the ratios of obtained sequences that correspond to those libraries. Our pooling of equimolar ratios of DNA from independent libraries based on visual quantification was evidently less than perfect, as there was variance in the representation of individual libraries in the overall pool (Supplementary Tables 4D and 5D). If the pooling was indeed equimolar, the expected mean number of sequences per independent pool would be 9596 (run I) and 10 852 (run II), with expected binomial SD of 95.98 (run I) and 102.07 (run II). Instead, in run I, we observed a SD of 2909.93 from the mean, and the number of sequences per set varied from 2950 to 17 922. In run II, we observed a SD of 2801.64 from the mean, and the number of sequences per set varied from 5347 to 15 901. The observed fluctuations between samples are not statistical in nature and may primarily be due to inconsistencies in ethidium bromide (EtBr)-based quantitation of DNA. Much of the difference may result from subjectivity in visual quantification (2-fold differences or less are considered appropriate for this method). In addition, EtBr will fluoresce more effectively with double-stranded species than with heteroduplex or single-stranded species. Thus, ratio of single-stranded to duplex species may influence the intensity of the band during gel electrophoresis, and may contribute to the observed variation. Finally, the fraction of heteroduplexes and non-annealed single strands present in the pool has an impact on the compactness of the band, and this in turn may affect precise concentration approximation. Denaturing materials from individual samples, and measuring their concentrations using Agilent's 2100 BioAnalyzer (before pooling samples together) may overcome these drawbacks of visually estimating DNA concentrations.
CONCLUSION
We have described a pyrosequencing-tailored barcoding approach that allows for the unambiguous assignment of nucleic acid sequences from a mixture of libraries from up to 48 different samples. Lengthening the barcodes, or variations in the fixed forward and reverse linkers used to generate the initial cDNA libraries can easily expand the multiplexing capacity. A remarkably low misassignment rate (0.00042% and 0.0044% for two independent experiments) was obtained by requiring a match to the 10-base barcodes and additional three bases of the fixed linker. The low misassignment rate also indicates the potential utility of combinations of forward and reverse barcodes to generate up to 482 (= 2304) combination barcodes for this set of sequences.
Previously, multiplexed pyrosequencing using the 454 sequencing technology was implemented to analyze sequences from several datasets simultaneously (6,10,11). However, the barcodes used in these experiments are too short to allow for massively parallel runs involving large numbers of sample libraries. Shorter barcodes also have a steeper trade-off between number of possible barcodes and the minimum number of nucleotide differences between individual barcodes. Based on standard error rates for pyrosequencing and our observation that ∼6% of sequences have at least one error in either their forward or reverse 10-nt barcodes (Table 3, and previous paragraph), we can predict that if the minimum number of nucleotide differences is decreased, there would be an increase in the frequency of sequence misassignment.
Our barcode specification has been tailored for use with pyrosequencing technologies, particularly reflecting restrictions imposed on the generation of homopolymeric runs. These barcodes may also be compatible with other sequencing platforms. Optimally, however the precise structure of the barcodes might need to be revised to reflect sequencing error profiles of the various systems. If necessary, multiple sequencing primers may also be used to identify the barcode and the sequence of interest, to overcome limitations imposed by short read lengths. As the availability of high-throughput technology advances, massively multiplexed barcoded sequencing will be extremely useful in performing surveys of diverse systems for nucleic acid signatures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
[Supplementary Data]
ACKNOWLEDGEMENTS
This work was funded by Stanford University Department of Pathology, NIH ROI GM37706; SGTC: 1S10RR022982 and R01HG003571. We wish to thank Jonathan Gent, Scott Boyd, Julia Pak, Weng-Onn Lui, Sam Gu, Daniel Blanchard, Rosa Alcazar, Frederick Tan, Ky Sha, Steve Johnson, Jay Maniar, Cecilia Mello and Chaya Krishna for assistance with materials, technical aspects and discussion. P.P. is supported by a Stanford Graduate Fellowship. Funding to pay the Open Access publication charges for this article was provided by Stanford University Pathology Department.
Conflict of interest statement. None declared.
REFERENCES
- 1.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett ST, Barnes C, Cox A, Davies L, Brown C. Toward the 1,000 dollars human genome. Pharmacogenomics. 2005;6:373–382. doi: 10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- 3.Ahmadian A, Ehn M, Hober S. Pyrosequencing: history, biochemistry and future. Clin. Chim. Acta. 2006;363:83–94. doi: 10.1016/j.cccn.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 4.Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh JD, Kling-Backhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, Cordum HS, Wang C, Elliott G, et al. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl Acad. Sci. USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 8.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat. Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 9.Mazurkiewicz P, Tang CM, Boone C, Holden DW. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat. Rev. Genet. 2006;7:929–939. doi: 10.1038/nrg1984. [DOI] [PubMed] [Google Scholar]
- 10.Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P, Bushman FD. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm435. e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 13.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]