Inheritance of Polycomb-dependent chromosomal interactions in Drosophila (original) (raw)
Abstract
Maintenance of cell identity is a complex task that involves multiple layers of regulation, acting at all levels of chromatin packaging, from nucleosomes to folding of chromosomal domains in the cell nucleus. Polycomb-group (PcG) and trithorax-group (trxG) proteins maintain memory of chromatin states through binding at _cis_-regulatory elements named PcG response elements or cellular memory modules. Fab-7 is a well-defined cellular memory module involved in regulation of the homeotic gene Abdominal-B (Abd-B). In addition to its action in cis, we show here by three-dimensional FISH that the Fab-7 element leads to association of transgenes with each other or with the endogenous Fab-7, even when inserted in different chromosomes. These long-distance interactions enhance PcG-mediated silencing. They depend on PcG proteins, on DNA sequence homology, and on developmental progression. Once long-distance pairing is abolished by removal of the endogenous Fab-7, the derepressed chromatin state induced at the transgene locus can be transmitted through meiosis into a large fraction of the progeny, even after reintroduction of the endogenous Fab-7. Strikingly, meiotic inheritance of the derepressed state involves loss of pairing between endogenous and transgenic Fab-7. This suggests that transmission of nuclear architecture through cell division might contribute to inheritance of chromatin states in eukaryotes.
Keywords: Polycomb-group, nuclear architecture, chromosome pairing, chromatin/meiosis
Packaging of eukaryotic DNA into chromatin is achieved through its folding in a hierarchy of structures, such as nucleosomes, 30-nm chromatin fibers, higher order chromatin structures, and chromosomal domains. Chromatin folding organizes individual chromosomes in specific chromosomal territories in the cell nucleus (Cremer and Cremer 2001). Individual chromosomal territories are separated by an interchromatin compartment that contains diffusible components, protein complexes, and larger structures or nuclear bodies. All of these compartments are dynamic and continuously exchange components, although the motion of large bodies or chromosomal territories is constrained (Chubb and Bickmore 2003).
It has been suggested that nuclear architecture might play a regulatory role in gene expression and cellular differentiation. In this view, genes would be segregated in specific nuclear domains and regulatory changes might move these genes into different compartments. Evidence has accumulated for precise gene localization and for changes in their nuclear position upon regulatory transitions (Gasser 2001). In vertebrate cell types, transitions between active and silent transcriptional states were shown to be accompanied by shuttling between euchromatic and heterochromatic nuclear compartments (Brown et al. 1997; Francastel et al. 1999; Lundgren et al. 2000). Similarly, silencing of the Drosophila euchromatic brown gene by insertion of a 2-Mb block of heterochromatin (_bw_D allele) has been shown to correlate with its relocation into heterochromatin (Csink and Henikoff 1996; Dernburg et al. 1996).
Gene silencing can be induced by two largely nonoverlapping sets of proteins, namely components of heterochromatin and proteins of the PcG. Whereas heterochromatin is mainly found at centromeric and telomeric regions of the genome, PcG proteins are involved in silencing of a variety of euchromatic genes involved in developmental control, such as homeotic genes (Orlando 2003). In early Drosophila embryonic development, homeotic gene regulation depends on a regulatory cascade of spatially restricted transcription factors. When these factors disappear, the expression pattern of homeotic genes is maintained by a cellular memory system dependent on PcG and trxG genes. PcG proteins form large multimeric complexes conserved throughout evolution (Alkema et al. 1997; Saurin et al. 2001) that act on chromatin via associated histone deacetylase and methyltransferase activities (for review, see Orlando 2003) and via components of the transcriptional machinery (Breiling et al. 2001; Saurin et al. 2001). On the other hand, several trxG members encode for proteins involved in chromatin remodeling, histone acetylation, and methylation, which increase chromatin accessibility to transcription factors (Petruk et al. 2001; Beisel et al. 2002). PcG and trxG proteins maintain regulatory states of their target genes via _cis_-regulatory elements named PcG response elements (PRE) and trxG response elements (TRE). (Fauvarque and Dura 1993; Chan et al. 1994; Zink and Paro 1995). These sequences often overlap in the genome (Strutt et al. 1997; Orlando et al. 1998; Tillib et al. 1999), and it was shown that they are able to maintain both silent and active states, depending on early regulatory cues during development. For this reason, they have been named cellular memory modules (CMM; Cavalli and Paro 1998; Maurange and Paro 2002; Rank et al. 2002). Beside an action as PRE and TRE, DNA sequences contained within CMM have also been shown to behave as chromatin insulators, preventing enhancers from acting at downstream promoters when inserted between them (Hagstrom et al. 1996; Zhou et al. 1996; Mihaly et al. 1997). Thus, multiple functional elements are contained in regions defined as CMM, and the molecular and functional links between these elements are still poorly understood.
In addition to an action in cis on flanking genes along the chromosome, CMM-mediated regulation is enhanced by _trans_-interactions among chromosomes. A mini-white eye color reporter gene flanked by PRE/CMM sequences is more repressed when the transgene is in a homozygous state than in a heterozygous state (Fauvarque and Dura 1993; Chan et al. 1994; Zink and Paro 1995). This phenomenon is known as pairing-sensitive repression (for review, see Kassis 2002). It depends on somatic pairing of homologous chromosomes, a phenomenon that exists in many species and cell types, but is particularly pronounced in Diptera.
Silencing can also be enhanced by insertion of two CMM copies at different loci in the same chromosome and even in different chromosomes (Sigrist and Pirrotta 1997; Muller et al. 1999). Moreover, PcG proteins were shown to be involved in cosuppression of transgenes containing sequences from the white and Adh loci, even if these genes do not contain CMM elements per se (Pal-Bhadra et al. 1997). These phenomena of _trans_-sensing of homologous sequences (Henikoff and Comai 1998) are found in many species and they may involve (1) communication via diffusible components, such as post-transcriptional mechanisms of gene silencing mediated by RNA molecules (Hannon 2002), or (2) direct contacts between the region of DNA homology. In this work, we provide evidence that the Fab-7 CMM element from the Bithoraxcomplex (BXC) of homeotic genes can mediate long-distance interchromosomal interactions, and that these interactions may be heritable through mitotic and meiotic cell division.
Results
Pairing dependent silencing of the sd locus by Fab-7
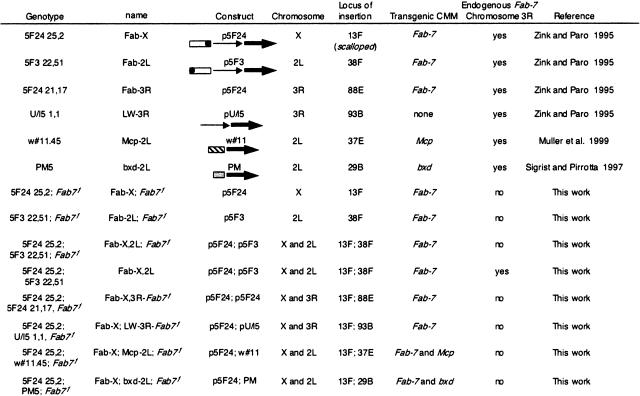
In most experiments throughout this work, two constructs were used carrying a 3.6-kb Fab-7 fragment cloned in two different orientations (p5F24 and p5F3 transgenes; Zink and Paro 1995; Table 1) upstream to lacZ and mini-white reporter genes. These transgenes were inserted at different genomic locations and combined with deletions or with mutations in PcG or trxG genes. For simplicity, transgenic lines will be named after the CMM element present in the transgene and the chromosomal arm of insertion of the transgenes. For instance, one previously published line, 5F24 25,2 (Zink and Paro 1995) will be renamed here as Fab-X, to indicate insertion of Fab-7 in the X chromosome (Table 1). As expected for CMM-mediated silencing, pairing-sensitive repression is observed and the eye color of homozygous females is strongly variegated, whereas in heterozygous females or in males (hemizygous) repression is weaker (Fig. 1A). Precise mapping of the transgene insertion indicated that the Fab-X line harbors two copies of the p5F24 transgene inserted in tandem 9.6 kb upstream of the scalloped (sd) gene (see Materials and Methods for details). The sd gene product is required for wingblade development in Drosophila (Campbell et al. 1992). Reduced expression of this gene leads to a characteristic wing phenotype (sd phenotype) that can have a different degree of severity, from small lesions in the wing margin to complete destruction of wing morphology. The insertion of the Fab-7 CMM at a distance of 18.4 kb from sd (8.8 kb DNA spanning the lacZ and mini-white regions plus 9.6 kb from the transgene insertion site to the sd promoter) induces a mutant phenotype, resulting in destruction of the wingblade. This phenotype is temperature sensitive and pairing dependent, as it is observed with a strong penetrance of up to 95% in homozygous females raised at 29°C, whereas it is almost absent in heterozygous females or hemizygous males (Fig. 1B). Both features are typical of PcG-mediated silencing and parallel effects on mini-white (Fauvarque and Dura 1993; Chan et al. 1994; Zink and Paro 1995). The incomplete penetrance of the sd phenotype does not depend on genetic heterogeneity of the flies, preventing silencing in a fraction of the population. When flies were raised at 29°C and Fab-X females with wild-type wings were selected and remated with Fab-X males, the next generation females showed a sd phenotype with similar penetrance as nonselected Fab-X females. Finally, as observed previously for the white eye phenotype in the Fab-X line (Cavalli and Paro 1999), the sd phenotype is strongly attenuated by mutations in PcG genes, whereas it is enhanced by a mutation in the trx gene (Fig. 1C).
Table 1.
Figure 1.
Pairing and PcG/trxG-dependent silencing mediated by the Fab-7 CMM in the Fab-X line. (A) Eye color phenotype of Fab-X flies grown at 25°C shows pairing-sensitive repression at the transgene. Genotypes are indicated above each panel. (B) Wing phenotype of Fab-X flies shows pairing-sensitive repression of the sd gene-flanking transgene insertion. The percentage of wings showing a sd phenotype at 29°C, and the number of wings analyzed are indicated. (C) Dependence of sd silencing on mutations in the PcG genes Pc, ph, Psc, and Pcl, and enhancement of silencing upon mutation of the trx gene.
Fab-7_-mediated repression at the X-linked_ sd locus is dependent on the endogenous Fab-7 located on the third chromosome
Surprisingly, repression of sd depends also on the presence of an intact copy of the endogenous Fab-7 element in the Abd-B locus, located in the right arm of the third chromosome (chromosome 3R). A genomic deletion of 4 kb encompassing the 3.6-kb Fab-7 transgenic element (Gyurkovics et al. 1990) was introduced in the homozygous state into the Fab-X line to give the Fab-X; Fab-7 1 line (Table 1; Fig. 2A). When raised at 29°C, Fab-X; Fab-7 1 females showed derepressed eye color and only 6%-12% of sd wing phenotype compared with 90%-95% of Fab-X females (Fig. 2A), suggesting that silencing of mini-white and sd is reduced. As a control, the presence of the Fab-7 deletion had no effect on the wing phenotype of a mutant line for the sd gene, indicating that endogenous Fab-7 does not play any role in sd regulation in the absence of the X-linked transgene (Fig. 2B). Moreover, a derepression both of mini-white and of sd was observed by introducing a homozygous Fab-7 12 deletion into Fab-X (13% sd phenotype, similar to the frequency observed in Fab-X; Fab-7 1). This mutation (Mihaly et al. 1997), deletes 1.5 Kb of DNA from the same region and has a similar effect on regulation of its endogenous target gene Abd-B, but it has an independent origin and genetic background from Fab-7 1. Therefore, derepression depends specifically on removal of Fab-7.
Figure 2.
Silencing of sd in the Fab-X line depends on the presence of the endogenous Fab-7 element. (A) Schematic drawing illustrating the genotype of the lines used in the experiments. The two copies of chromosome X and chromosome 3 are shown. The endogenous Fab-7 at chromosome 3 is shown in green. The Fab-7 1 deletion is represented as an empty triangle. The transgenic copy inserted in chromosome X is shown in red. (B) sd phenotype in a loss-of function sd mutant in the presence or absence of endogenous Fab-7. (C) In situ hybridization of sd in embryos labeled with an anti-MSL-1 antibody to distinguish female (in blue) from male (in brown) embryos. (D) Repression of sd transcript levels in the Fab-X line (middle) compared with control w 1118 (left) and with Fab-X, Fab-7 1 (right) embryos. (E) Repression of sd transcript levels in third instar imaginal wing discs from the Fab-X line (middle) compared with control w 1118 (left) and with Fab-X, Fab-7 1 (right) larvae. sd is normally expressed in the wing pouch of the imaginal disc and it is repressed in Fab-X, but not in Fab-X, Fab-7 1 larvae.
To further analyze the effect of the transgene and of the Fab-7 deletion on sd expression, we performed in situ hybridization using a sd mRNA probe. The results indicate that transgenic Fab-7 represses sd with increasing strength during developmental progression. In Fab-X female embryos of stage 9-11, sd expression is weakly reduced compared with control w 1118 and to Fab-X; Fab-7 1 embryos (Fig. 2D). At later developmental stages, silencing is more robust, as shown by a strong reduction in sd expression in ∼60% of the female wing imaginal discs from Fab-X third instar larvae (Fig. 2E). Both in embryos and in larvae, sd repression was lost upon deletion of endogenous Fab-7 in the Fab-X; Fab-7 1 line (Fig. 2D,E), although a weak residual silencing was visible in larval wing discs. Thus, sd silencing by the Fab-7 transgene depends on the presence of the endogenous Fab-7 of the BX-C at all stages of development. As expected, expression of sd was also restored in the context of a PcG gene mutation_, Polycomb-like_ (Pcl; data not shown).
Long-range pairing between transgenic and endogenous Fab-7
These data suggest that the exogenous Fab-7 at the sd locus may pair with the endogenous copy at the BX-C locus, and this pairing may be responsible for the strong silencing of the sd gene. To test this hypothesis directly, we analyzed the relative nuclear position of the two loci by two-color fluorescent in situ hybridization (FISH) of whole-mount embryos, followed by three-dimensional image analysis (Gemkow et al. 1996, 1998).
We used embryos at stage 9-11, a time when PcG-mediated sd gene silencing has already initiated. Image acquisition along the z-axis were done in ectodermal nuclei of parasegments (PS) 4 and 5 (this region of the embryo will give rise to the T2 segment and contains wing precursor cells). Using two probes specific for the Abd-B and the sd loci, we found that sd colocalizes only rarely with the BX-C in the wild-type situation. In wild-type female embryos, 7% pairing was observed (Fig. 3A-C). This frequency may depend on chromosomal movements occasionally bringing these two loci in the same nuclear neighborhood. On the other hand, Fab-7 inserted at the sd locus strongly promotes association to the BXC. The two loci colocalized in 23% of female embryonic nuclei (Fig. 3A-C; see also Supplemental Material). In contrast, colocalization is reduced to control levels in male embryos, correlating to the wing phenotype. To study whether association of the two loci depends on DNA sequence homology conferred by the presence of Fab-7, we analyzed long-distance pairing upon removal of the endogenous Fab-7. In Fab-X; Fab-7 1 female embryos, pairing was reduced to control wild-type level (Fig. 3A-C). These data show that long-distance interactions between Fab-7 CMM elements can occur in the nucleus and they stabilize the efficiency of PcG-mediated silencing.
Figure 3.
Long-range interactions between sd and the BX-C depend on sequence homology at Fab-7 and on the presence of PCL protein. (A) Two-color FISH in whole-mount female embryos. Examples of merged images of Dapi labeling (blue), the sd locus (green) and the BX-C (red) are shown. Projection of two _Z_-axis slices is shown. Embryo genotypes are indicated above each panel. Arrows indicate cases of colocalization between sd and the BX-C. (B) Single slices of individual nuclei show characteristic examples of data obtained in different lines. Dapi staining, the sd locus, the BX-C locus, and the merge of the three channels are shown. The genotypes and the presence of the endogenous or the transgenic Fab-7 copies are indicated. (C) Quantification of the percentage of colocalization of the two loci in stage 9-11 female embryos from different lines. The genotypes and the embryonic position of the analyzed nuclei are indicated below each bar. Error bars represent the standard deviation. At least 50 nuclei per embryo from four to sixembryos were analyzed in each experiment. (D) Same quantification as C, but from an independent experiment comparing female Fab-X embryos with mutant Fab-X; Pcl 10 embryos analyzed at stage 13-15 of development. (E) Quantification of long-distance Fab-7 pairing in third instar larval wing imaginal discs, measured as percentage of colocalization of the two loci in the different lines.
We then analyzed this phenomenon in the context of a Pcl null mutation (Pcl 10). Homozygous Pcl 10 mutant embryos were obtained by selecting non-GFP-labeled embryos from a line carrying the Fab-7 transgene and the Pcl mutation facing a balancer chromosome marked with a zygotically expressed GFP reporter gene. Association of the two loci was reduced to control levels in Fab-X; Pcl 10 embryos (Fig. 3B,D), showing that PcG proteins, in addition to sequence homology, are required for pairing.
It was reported recently that endogenous Fab-7 is transcribed in embryonic PS13 and PS14 (Rank et al. 2002). We confirmed this data by in situ hybridization using a Fab-7 probe and a comparison between wild-type and Fab-X embryos showed no difference in the staining pattern or intensity. The pairing frequency of sd with the BX-C was compared in PS4/PS5, in which endogenous Fab-7 is not transcribed, and in PS13/PS14, in which it is transcribed, but no difference was observed (Fig. 3C). We also tested whether genetic background has an influence on long-distance pairing. Both the transgenic lines and the Fab-7 1 deletion were originally obtained in a Oregon-R background. When this was changed to Canton-S (see Materials and Methods), both the sd phenotype and the strong increase in pairing between the sd and the BX-C loci were maintained in the presence of Fab-7 at the two loci (data not shown). Thus, transcription of the endogenous Fab-7 and strain-specific genetic variation have no major effects on pairing.
Long-distance Fab-7 pairing progression during development
Because in situ RNA hybridization showed that repression of sd in Fab-X is stronger in larvae compared with embryos, we analyzed whether the pairing efficiency of the BX-C with the sd locus is also increased at this stage. Pairing was analyzed by two-color FISH in imaginal wing discs from third instar female larvae (Fig. 3E). Control larvae showed pairing in only 5% of the cases, similar to what was seen previously in embryos (Fig. 3E). In contrast, in the Fab-X line association between the BX-C and the sd locus was found in 43% of the nuclei, a much higher frequency compared with embryos (Fig. 3, cf. E and C). This indicates that long-distance pairing is a dynamic process and its stability increases during later developmental stages. Long-distance pairing was severely reduced in larvae from the Fab-X; Fab-7 1 line, in which it reached 14%. However, this frequency is clearly higher than in the wild-type control. This suggests that, although sequence homology at Fab-7 is crucial for pairing, the transgenic Fab-7 might be able to establish interactions with other CMM in the BX-C to some extent.
Endogenous Fab-7 cosegregates in the cell nucleus with Fab-7 transgenes inserted at independent chromosomal loci
The _Fab-7_-dependent associations observed beetween the BX-C and the sd locus could be partly due to some specific features of one or the two loci. To test whether ectopic pairing depends on insertion of Fab-7 at the sd locus, we used the Fab-2L transgenic line (Zink and Paro 1995). Characterization of this line (see Materials and Methods) showed that the p5F3 Fab-7 construct is inserted in single copy at cytological position 38F, in the left arm of chromosome 2 (Table 1, for simplicity, this locus will be defined hereafter as 38F). Two-color FISH analysis with probes that recognize the 38F and BX-C loci showed a very strong increase in colocalization when Fab-7 is present at both loci (Fab-2L embryos) compared with wild-type embryos harboring no Fab-7 transgene at the 38F locus (11% vs. 0.7%; Fig. 4A,B). Thus, Fab-7 transgenes inserted at two independent loci can pair with the endogenous Fab-7 copy. To describe precisely the statistical distribution of the two loci, three-dimensional distances were measured in >200 nuclei of each sample. A global decrease in the three-dimensional distance was observed when Fab-7 is inserted at the 38F locus (Fig. 4B). This suggests that in addition to direct contacts, in a large fraction of the nuclei, the two loci are in close proximity. These nuclei may represent intermediate steps in the process of Fab-7 homology search.
Figure 4.
Long-range interactions between a Fab-7 transgene inserted at chromosome 2L and the BX-C. (A) Comparison of FISH signals from control w 1118 embryos and Fab-2L embryos at stage 9-11. (B) Quantitative analysis of the distribution of three-dimensional distances between the two loci in control w 1118 (brown bars) and Fab-2L (yellow bars) embryos. In the 0-0.5 μm distance range, two classes were distinguished, one class in which the two signals colocalized (11.0% for Fab-2L and 0.7% for w 1118, indicated by ticks within the bars), whereas in the other class, the two signals were close, but nonoverlapping.
Pairing of Fab-7 elements does not depend on features of the BX-C
We then investigated whether pairing depends on specific features of the BX-C, or instead represents pure affinity between homologous Fab-7 DNA sequences and chromatin structures. We reasoned that if pairing does not depend on specific features of the BX-C locus, strong sd silencing might be obtained by introduction of a second Fab-7 transgene in the absence of endogenous Fab-7. Therefore, we combined other Fab-7 transgene insertions with the Fab-7 transgene inserted at the sd locus. A Fab-7 1 deletion was first introduced into two transgenic lines. One was the Fab-2L line described above, whereas in the Fab-3R line, the p5F24 transgene is inserted at 88E in the right arm of the third chromosome (Table 1). These two lines were then crossed to the Fab-X line to obtain lines carrying two homozygous Fab-7 transgenes and the Fab-7 1 deletion; Fab-X,2L; Fab-7 1 and Fab-X,3R- Fab-7 1. As noted previously, deletion of endogenous Fab-7 leads to derepression of the sd gene in the Fab-X, Fab-7 1 line (Figs. 2A, 5A). However, the transgenic Fab-7 elements at chromosome 2L or 3R were able to complement the absence of endogenous Fab-7 and restore sd silencing (Fig. 5A). Fab-7 silencing was copy-number dependent. Indeed, we found that silencing of sd and of mini- white in the Fab-X,2L; Fab-7 1 line was weaker than in the related line Fab-X,2L, which carries the same transgenes in the presence of the endogenous copy of Fab-7.
Figure 5.
Association between transgenes carrying Fab-7. (A) In Fab-X; Fab-7 1 females, deletion of the endogenous Fab-7 derepresses sd (top, left). Strong repression is induced by introduction of a second Fab-7 transgene either in chromosome 2L (top, middle) or in chromosome 3R (top, right). Conversely, no repression is observed in the presence of the control pU/l5 transgene without CMM (bottom, left), or in the presence of a transgene carrying the bxd CMM (bottom, right). sd is slightly repressed upon introduction of a second transgene containing the Mcp CMM (bottom, middle). (B) Two-color FISH in female embryos carrying two Fab-7 transgenes, in the Fab-X,2L; Fab-7 1 line (top). Control female embryos are shown, carrying only one source of Fab-7, either at the sd locus (middle) or at the 38F locus in chromosome 2L (bottom). (C) Quantitative analysis of the distribution of three-dimensional distances between the two loci in control lines Fab-X; Fab-7 1 (blue bars) and Fab-2L; Fab-7 1 (brown bars) vs. the Fab-X,2L; Fab-7 1 line (yellow bars).
In contrast, Fab-X,LW-3R-Fab7 1 transgenic line (Table 1), carrying a second transgene without Fab-7 (Zink and Paro 1995), did not restore sd silencing (Fig. 5A), suggesting that silencing by multiple transgene copies requires the presence of Fab-7 in all transgenes. Two other CMM of the BX-C, such as Mcp and bxd, were also tested. bxd failed to complement the deletion of endogenous Fab-7, whereas a Mcp transgene had a weak effect, restoring sd silencing in 33% of the flies (Fig. 5A). Altogether, these results indicate that sequence homology at Fab-7 is the main determinant of long-distance interactions in our transgenic lines, although in the natural situation, interactions among heterologous CMM may occur.
To study whether two independent Fab-7 transgenes interact directly, we performed two-color FISH experiments in the Fab-X,2L; Fab-7 1 line, which carries two Fab-7 transgenes and no endogenous copy. As controls, we analyzed the single transgenic lines Fab-X; Fab-7 1 and Fab-2L; Fab-7 1 (Table 1), carrying Fab-7 at either of the two loci (Fig. 5B). For FISH, we used probes directed to the two loci of insertion, and we measured three-dimensional distances between them in the three lines. A dramatic difference was observed between control lines and the line containing Fab-7 at both loci (Fig. 5C). The two loci are much closer when they both carry Fab-7 than when they do not. Therefore, long-distance pairing can be mediated by Fab-7 alone, independent from its localization within the BX-C.
Pairing-dependent meiotic inheritance of Fab-7 chromatin states
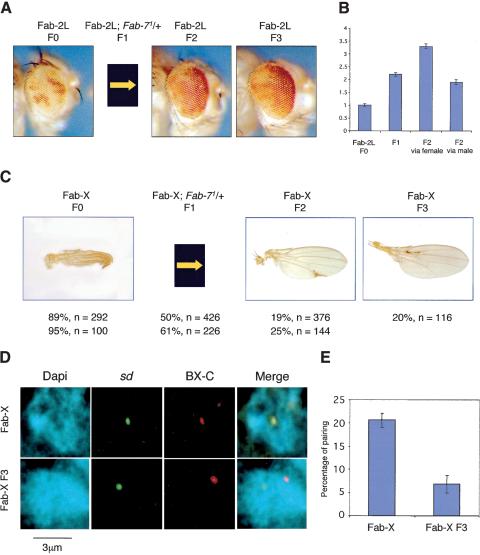
It was reported previously that the Fab-7 element is not only able to convey memory of silencing, but also of derepressed chromatin states. A strong derepression from silencing induced in Fab-7 transgenes by the yeast GAL4 factor could be inherited not only during development, but also through female meiosis into subsequent fly generations (Cavalli and Paro 1998). Because we observed loss of silencing upon unpairing of Fab-7 homologous elements, we tested whether derepressed states induced by unpairing could be inherited through meiosis. First, we generated Fab-2L flies containing a homozygous p5F3 transgene in the context of a heterozygous deletion of endogenous Fab-7 (F1 generation). The mini- white reporter gene was derepressed in these flies compared with a Fab-2L control. This was most evident at 18°C, a temperature at which PcG-mediated silencing is weaker (Fig. 6B). Then, the endogenous Fab-7 sequence was reintroduced by back-crossing the F1 flies in order to obtain F2 generation flies with a genotype equivalent to the Fab-2L F0 generation. Surprisingly, however, the derepressed mini-white state was maintained through meiosis in the F2 generation. Derepression was also maintained into the F3 generation upon recrossing derepressed F2 flies (Fig. 6A). Continued recrossing (at 18°C) showed maintenance of the derepressed state through at least 5 generations, showing that maintenance of derepressed chromatin is stable. Meiotic inheritance of the derepressed chromatin state was observed both from derepressed parental males and females, although maintenance was stronger through the female germ line (Fig. 6B). To quantify the fraction of flies that showed maintenance, we took advantage of a strong derepression that was observed in a subset of the flies. When these strongly derepressed (red eyed) flies were intercrossed, up to 60% of the progeny was still strongly derepressed, showing that inheritance can be robust. Maintenance of derepression is a reversible phenomenon, however. We reasoned that if one could boost PcG-dependent silencing in flies carrying a derepressed Fab-7 transgene, this may reset the chromatin state and restore the original level of silencing. Thus, strongly derepressed (fully red eyed) flies from the F4 generation were transferred to lay eggs from 18°C to 29°C. Because PcG-mediated silencing is stronger at higher temperatures, a decrease in maintenance of the derepressed state would be predicted in the following generation if maintenance of derepressed chromatin is reversible. Very strong silencing of mini-white was restored in 95% of the F5 progeny. Upon shift back to 18°C, subsequent generations showed strong mini-white silencing that was indistinguishable from the original line. This shows that inheritance of the derepressed state is a reversible process that depends on epigenetic determinants.
Figure 6.
Meiotic inheritance of chromatin states. (A) Inheritance of derepressed chromatin states in the Fab-2L line. Expression of the white gene is silenced in Fab-2L flies (F0). These flies were crossed with Fab-2L; Fab-7 1 in order to obtain a F1 progeny homozygous for the transgene and heterozygous for the endogenous Fab-7 copy. Females of this F1 generation were crossed with Fab-2L males. In the F2 generation, 50% of the progeny [identified by the absence of abdominal phenotypes (Gyurkovics et al. 1990) induced by the endogenous deletion of _Fab-7_] was genetically equivalent to the F0 (Fab-2L) generation, but was still derepressed. Derepression was maintained upon recrossing F2 Fab-2L flies (F3 generation). (B) Quantitation of eye pigment levels in control Fab-2L flies (F0 generation), in a F1 progeny homozygous for the transgene and heterozygous for the endogenous Fab-7 copy (Fab-2L; Fab-7 1/+), and in Fab-2L flies of the F2 generation obtained from a cross of Fab-2L; Fab-7 1/+ females with Fab-2L males (F2 via female) or from the reciprocal cross (F2 via male). (C) Inheritance of chromatin states in the Fab-X line. A similar crossing scheme was applied as in A and silencing of sd was analyzed. Genotypes, percentages of wings showing a sd phenotype, and numbers of wings analyzed are indicated. Two independent experiments are shown. sd derepression is inherited in F2 and F3 generation flies. (D) Two-color FISH in Fab-X F3 female embryos layed from F2 generation derepressed Fab-X flies (bottom), or from control Fab-X female embryos (top). (E) Quantification of the percentage of colocalization between the two loci in control Fab-X embryos (see also Fig. 3C for an independent experiment in the same condition), and Fab-X F3 embryos.
The same experimental scheme was then repeated with the Fab-X line containing the Fab-7 element at the sd locus. Again, significant sd derepression was obtained upon removal of one endogenous Fab-7 in Fab-X F1 flies. The endogenous Fab-7 was reintroduced by back-crossing to obtain F2 flies of the same genotype as the Fab-X grandparents. Derepression of sd was observed in ∼80% of the F2 flies grown at 29°C, and it could be transmitted with high efficiency also to the F3 generation (Fig. 6C). In this case, no inheritance of the sd derepressed state was observed through the male germ line. However, one should note that Fab-7 does not significantly pair with the BX-C in males, and this is reflected in lack of silencing of the sd gene. Thus, males might be considered as neutral with respect to meiotic inheritance in this line.
Transmission of the derepressed state to a large majority of the progeny in the Fab-X line enabled us to compare the nuclear organization of the sd and BX-C loci in derepressed F3 female embryos with control Fab-X-repressed female embryos. Remarkably, long-range pairing of the two loci was reduced to background levels in the derepressed F3 progeny, although both the transgene, as well as the endogenous Fab-7 copies were now present in the genome (Fig. 6D,E). Therefore, meiotic inheritance of sd derepression is linked to loss of Fab-7 interchromosomal association. These data strongly suggest that long-distance pairing interactions mediated by the Polycomb pathway are a heritable feature of cell division.
Discussion
The results presented here show that a CMM element of 3.6 kb can mediate long-distance associations between distant chromosomal regions in embryonic nuclei. These interactions depend strongly on chromatin components of the PcG and on DNA sequence homology. Importantly, when disrupted in one generation, these pairing interactions are inefficiently re-established even upon reintroduction of sequence homology, and a large portion of the progeny maintains the loci unpaired in subsequent generations. This phenomenon is reversible, suggesting that PcG-mediated chromatin regulation is an equilibrium process that depends on the concentration of regulatory components and on the previous history of the cell. Perturbation of the balance between these regulatory cues might favor establishment as well as inheritance of active or repressed states at target genes. Several cases of inheritance of chromatin composition features have been reported in eukaryotes (Cavalli 2002). The data described here suggest that inheritance of Fab-7 regulatory states depends not only on chromatin components, but also on nuclear compartmentalization of chromosomal domains.
Nuclear compartmentalization of PcG target genes
PcG proteins have been implicated previously in phenomena involving long-distance interactions among independent loci. Phenotypic interactions were documented both at transgenes containing CMM elements (Sigrist and Pirrotta 1997; Muller et al. 1999) as well as in the phenomenon of cosuppression (Pal-Bhadra et al. 1997). In this last case, PcG proteins as well as mechanisms of RNA-dependent posttranscriptional gene silencing were shown to be involved (Pal-Bhadra et al. 2002). However, whether direct long-distance associations occur in these cases is presently unknown. On the other hand, long-distance pairing was observed previously in up to 30% of embryonic nuclei containing a euchromatic translocation of a region of ∼900 kb, spanning the BX-C and flanking genes from chromosome 3R to chromosome X (Gemkow et al. 1998). Although the DNA sequence determinants and the proteins responsible for pairing in this large chromosomal region were not identified, several CMM, including Fab-7, are present in the BX-C and could contribute to this interaction. On the basis of our results, we suggest that PcG proteins may mediate long-range pairing interactions in the case of Fab-7 as well as in transgenes containing other CMM and in some cases of cosuppression and silencing of repetitive DNA elements.
Long-distance associations may not only involve transgenes, but also natural genes regulated by the Polycomb pathway. PcG proteins are distributed in specific nuclear compartments that have been termed PcG bodies (Buchenau et al. 1998; Saurin et al. 1998). Although the significance of these bodies is presently unclear, we speculate that endogenous PcG target genes may undergo physical associations at nuclear PcG bodies dedicated to their regulation. Compartmentalization of PcG target genes may not be required for primary recruitment of PcG complexes, but it may rather stabilize PcG and trxG-mediated gene regulation. This phenomenon may not be unique in eukaryotic nuclei, as evidence for gene clustering at specific intranuclear organelles was found in vertebrates (for review, see Parada and Misteli 2002). These clustering phenomena were suggested to involve positioning of genes coregulated by the same set of proteins in the same nuclear compartments. PcG proteins may represent one class of factors acting in this manner.
A hint for involvement of the Fab-7 element in heterologous associations at PcG bodies comes from experiments showing that in Fab-X; Fab-7 1 larvae, the Fab-7 transgene pairs with the BX-C locus to some extent, even in the absence of the endogenous _Fab-_7. This suggests that in the absence of sequence homology, Fab-7 may interact with other CMM present in the BX-C, albeit more weakly. An increase in _Fab-7_-dependent sd silencing was detected in the presence of a transgene containing the Mcp sequence from the BX-C, suggesting that these two elements may be able to interact.
Molecular mechanisms underlying long-range pairing interactions
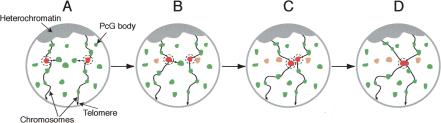
How is pairing achieved? First, homologous CMM must come in physical proximity. This may depend on constrained brownian motion of chromosomal territories (Marshall et al. 1997). However, other processes may help this long-distance search. In particular, it may be postulated that genes containing CMM localize to PcG bodies (Saurin et al. 1998). We speculate that these bodies might not be immobile, but, as in the case of splicing speckles (Misteli et al. 1997), they may rather undergo occasional movements, splitting, and mergers, although perhaps with different kinetics. Genes localized within these bodies may reside there for a certain time and then leave one PcG body to incorporate another one (Fig. 7). Such a dynamic behavior may allow PcG target genes to explore part of the nucleus, but would, at the same time, allow them to stay in the vicinity of other PcG target genes and prevent them from diffusing away randomly in the nucleoplasm. This may increase the probability for a CMM to explore contacts with other CMM.
Figure 7.
A PcG body-hopping model for establishment of long-range chromatin-pairing interactions. (A) Nuclei are represented in a Rabl configuration (Rabl 1885), in which centromeres are assembled near the apical pole of the nucleus, pointing toward the surface of the embryo, whereas telomeres point toward the opposite pole. Two chromosomal arms projecting from centromeric heterochromatin toward the basal pole of the nucleus are shown. PcG target genes are postulated to locate at PcG bodies (in green). Each PcG body may contain one or several of these genes. Two loci containing homologous CMM are depicted in red. CMM located at PcG bodies may thus be constrained, but they may still be capable of exploring the nuclear territory in the neighborhood of their PcG body (dashed circle around the CMM). (B) Chromosome movements may lead to occasional PcG body contacts. During these contacts, CMM may move from a PcG body into another one (the old CMM locations are shown in orange). (C) This step may be repeated several times and may thus allow a CMM to explore significant areas in the cell nucleus, containing preferentially other PcG target genes. (D) This PcG body-hopping process may favor establishment and maintenance of specific interactions between homologous CMM.
Once proximity is established, strong association might be established by regulatory components of CMM chromatin. The identification of a 3.6-kb DNA sequence as a sufficient region of homology to induce long-range physical associations will allow, for the first time, to dissect DNA sequences and chromatin factors responsible for pairing at high resolution. The 3.6-kb Fab-7 element contains a chromatin boundary that can attenuate enhancer-promoter communications (Hagstrom et al. 1996; Zhou et al. 1996; Mihaly et al. 1997), and a PRE (Hagstrom et al. 1997). GAGA factor binds to both the PRE and the boundary region of Fab-7 (Cavalli and Paro 1998). Moreover, this protein is able to bind cooperatively to DNA to form oligomers, bringing distant DNA sequences close together (Mahmoudi et al. 2002). Thus, GAGA factor-binding sites may be partly responsible for long-range interactions. Similarly, putative binding sites for Zeste, a protein that mediates _trans_-sensing phenomena (Pirrotta and Rastelli 1994), are also present in Fab-7, and they may contribute to pairing. Other chromatin components described previously to act at this element, such as proteins of the trxG (Orlando et al. 1998), chromatin condensation proteins, and DNA topoisomerase II (Lupo et al. 2001) may also be involved in pairing of Fab-7.
However, all of these proteins associate also with other CMM in the genome. How do they distinguish between DNA sequence homologous and nonhomologous CMM? One possibility is that chromatin regulation and the DNA sequence determine a specific array of proteins and of histone modifications associated with it. For a given locus, this may result in the formation of a unique order of chromatin tags that can only be found at loci sharing strong sequence homology. Some of these components may undergo dimerization or oligomerization, leading to specific contacts that may maintain homologous chromatin stably associated. Similar contacts may also be involved in chromosome pairing in somatic cells or during meiosis (Cook 1997).
Meiotic inheritance of chromosomal architecture
A remarkable finding involving long-distance pairing of the Fab-7 CMM is transmission through meiosis. What could be the role of this meiotic inheritance of chromatin states? This is particularly intriguing in the case of a CMM regulating a homeotic gene, as expression of homeotic genes must be reset at every generation in order to establish appropriate gene expression patterns along the anteroposterior embryonic axis. However, meiotic inheritance was reported previously to involve a phenotype associated with a chromosomal rearrangement at the BX-C locus, although no molecular determinant for this phenomenon could be found (Kuhn and Packert 1988). In Caenorhabditis elegans, PcG proteins establish a germ-line-specific gene silencing that is heritable through meiosis (for review, see Pirrotta 2002). One possible way in which meiotic inheritance could be important in Drosophila homeotic gene regulation is to maintain a default silenced state during early embryogenesis. At the onset of homeotic gene transcription, spatial-specific transcriptional repressors maintain homeotic genes repressed outside of the appropriate expression domains. Maintenance of repression is crucial, as failure could cause homeotic transformations. Inheritance of chromatin silencing may stabilize this repression and contribute to developmental homeostasis.
The fact that pairing interactions involving a chromosomal element regulated by PcG/trxG proteins are heritable raises the question of how transmission of chromatin architectural features is possible through cell division. Two different, but not mutually exclusive, mechanisms may contribute to explain this novel form of inheritance. Chromosomal contacts may depend on specific, heritable chromatin marks deposited in cis on the templates undergoing pairing. These marks may allow contacts to re-establish after they are broken during chromosome metabolism at mitosis and meiosis. Perturbation of these marks may change chromatin at Fab-7 and make it incapable of establishing pairing interactions with its homolog sequence in another chromosome. Chromatin marks, such as histone acetylation, histone methylation, and association of Swi6 protein to the mating type locus in Schizosaccharomyces pombe were shown previously to be heritable through meiosis (Ekwall et al. 1997; Nakayama et al. 2000). Our initial chromatin characterization in the presence or absence of _Fab-7_-pairing interactions showed recruitment of PcG proteins to the transgene and to the region surrounding its site of insertion in both cases, and did not reveal significant changes in PcG protein binding or in histone modifications.
A second mechanism for inheritance of long-range chromosomal interactions may depend on stable transmission of the relative chromosome positions and specific gene contacts through cell division. It was shown recently that global chromosome positioning can be transmitted in mammalian cells through the whole-cell cycle and mitosis (Gehrlich et al. 2003), although the fidelity of mitotic transmission may depend on cell type (Walter et al. 2003). Our data show that long-distance pairing is dynamic during development; it has a relatively weak frequency during embryonic stages, and it increases at larval stages. This dynamics may depend on the increased length of the cell cycle or on more robust PcG silencing in larvae, and it suggests that the actual physical contact between chromosomes may be lost, but regulation of nuclear compartmentalization may favor re-establishment of long-distance pairing at each cell generation.
In summary, the present study suggests that features of the nuclear architecture of PcG target genes can be transmitted through cell division. We propose that this may represent a novel form of epigenetic inheritance that may be used to convey cellular memory of chromatin states in eukaryotic organisms.
Materials and methods
Fly stocks and handling
Flies were raised in standard corn meal yeast extract medium. The Oregon-R w 1118 line was obtained from R. Paro (ZMBH, University of Heidelberg, Germany). Transgenic lines, obtained by injection of Oregon-R w 1118 embryos, are described in Table 1. Genetic homogenization of the Fab-X and w 1118; Fab-7 1 lines in Canton-S background was performed by seven rounds of crossing of Fab-X and w 1118; Fab-7 1 heterozygous females (5-6 in each cross) with w 1118 Canton-S males [kindly provided by J.M. Dura (Institute of Human Genetics, CNRS, Montpellier, France)]. By crossing these homogenized lines, a homogenized Fab-X; Fab-7 1 line was obtained. Mutant alleles used to test PcG and trxG effects are as follows: Pcl 10, Pc XL5, ph 410, Psc1, and trx E2. The sd ETX4 allele was described in Campbell et al. (1992). A stock carrying the KrGFP-CyO balancer chromosome (Casso et al. 2000) was used for selection of homozygous Pcl 10 mutants. Staged eggs were collected on agar plates with standard vinegar/fresh yeast medium. Eye pigment determination was done as described previously (Reuter and Wolff 1981). To map the insertion site of transgenes, we performed standard PCR rescue experiments, followed by DNA sequencing. The p5F24 construct in Fab-X is inserted in arm X at the sd locus, at position 98,830 of the GenBank Scaffold AE003500, in plus orientation, at 9,615 bp upstream of the sd transcription start site. DNA sequencing revealed that the p5F24 construct is inserted as an almost perfect tandem duplication in this line. The first p5F24 copy is complete, whereas the second copy (more proximal to sd) carries a deletion corresponding to nucleotides 100,229-101,165 of the AE003715 Scaffold. The p5F3 construct in Fab-2L is inserted in arm 2L at position 34,662 of the AE003661 Scaffold.
In situ mRNA hybridization
To produce a probe for sd, the 1.7-kb _Xho_I/_Eco_RI C-terminal fragment of the E21 sd cDNA (Campbell et al. 1992) was cloned into pBKS vector. Anti-sense RNA was produced by transcription from the T7 promoter. Sense RNA, used as a negative control for staining, was obtained by transcription from the T3 promoter. In situ hybridization on embryos and larvae were performed by a standard procedure (Tautz and Pfeifle 1989). To sexembryos after in situ hybridization, embryos were dehydrated in ethanol and rehydrated to proceed with immunostaining using the Male-Sex-Lethal-1 (MSL-1) antibody at a 1:500 dilution. MSL-1 is a member of the dosage compensation complex and is much more abundant in males than in females. The immunochemical reaction, performed with ABC elite kit (Vector laboratories) according to manufacturer's instructions, was stopped rapidly after 5 min, allowing brown staining in males to develop, whereas females were not stained. Embryos were mounted in a 1:1 mixture of glycerol and ethanol.
Two-color FISH
For the sd locus, we generated a probe of 25.5 kb made from seven genomic fragments, which were cloned into pBKS vector from λ clones of the sd region. For the BX-C locus, we generated a probe of 13.6 kb made from plasmids pBX-5989 and pBX-7652 (Strutt et al. 1997), which are located 23.5 and 37.6 kb from Fab-7, respectively. To enhance signals of the BX-C locus in larval discs, we generated a probe of 19.4 kb by adding fragments from plasmids pBX-1894 and pBX-3870 (Strutt et al. 1997), which are located 66.3 and 73.7 kb from Fab-7, respectively. For the 38F locus, five genomic PCR fragments of 1.2 kb and spaced by 2 kb from each other, were pooled for labeling. Probes were labeled by nick translation with biotin-14-dATP (Invitrogen life technologies) or digoxigenin-11-dUTP (Roche Diagnosis) according to manufacturer's instructions. Hybridization of each probe to the correct locus was checked by FISH in polytene chromosomes. Detailed coordinates of fragments used to produce probes are available upon request. FISH on whole-mount embryos was performed as described previously (Gemkow et al. 1996), with minor modifications as follows: for hybridization, the denatured probes were resuspended in 2× SSC, 10% dextran sulfate, 50% formamide, and 0.05% Salmon Sperm DNA hybridization buffer. Hybridization and post-hybridization washes were performed as in Gemkow et al. (1996), then embryos were blocked in PBS, 0.3% Triton, 1% BSA, 10% Donkey Serum for 2 h at room temperature. To detect the labeled probes, embryos were first incubated in 0.5 mL blocking buffer with the anti-digoxygenin-Rhodamine (Roche Diagnosis) at a 1:550 dilution, 1 h at room temperature, then with the anti-biotin-FITC (Vector laboratories) diluted 1:45 for 1 h. DNA was counterstained with DAPI, and embryos were mounted in Prolong Antifade medium (Molecular Probes). Larval imaginal discs were fixed in PBS, 0.1% Tween20, 4% formaldehyde for 20 min, and submitted to the same FISH protocol as embryos.
Microscopy and image analysis
Three-dimensional images were acquired with a cooled CCD camera (MicromaxYHS 1300, Roper Scientific) mounted on a DMRXA Leica microscope. Images were acquired with a 100× Plan/Apo Ojective (NA 1.4) mounted on a piezo electric (Roper Scientific). Optical sections were collected at 0.5-μm intervals along the z-axis. Distance measurements were performed in raw images and in images filtered using the nearest neighbor algorithm of Metamorph software (Universal Imaging Corporation). For each data set, statistical analysis was performed with stacks collected from 4-6 embryos or 3-4 wing imaginal larval discs, and FISH signals were analyzed in 50-80 nuclei per embryo or larval disc. The distance between the visually determined center of mass of dots corresponding to DNA hybridization signals were determined using the x,y,z-measured distance module of the Metamorph software. The average nuclear radius was 4.8 μm ± 0.3 μm, indicating that the embryonic nuclei analyzed were homogeneous in size. Distances between loci were therefore directly expressed in micrometers. A small fraction of nuclei, strongly deviating from overall circular shape, were not included in the analysis. Loci were considered as paired when the two signals overlapped. This is equivalent to a distance between the two centers of the signals lower than 0.3 μm. Due to incomplete somatic pairing in embryos, most, but not all nuclei contained one hybridization signal with each probe. Female embryos sometimes had nuclei with two unpaired sd spots, and could be distinguished easily from males that had one spot for the sd locus. This was confirmed by two-color FISH using the sd probe and a Y chromosome-specific probe from the Suppressor of Stellate locus. For figure display, single slices from z-stacks were deconvolved with Huygens MLE single tif procedure (Scientific Volume Imaging).
Acknowledgments
We thank R. Paro, F. Karch, J.M. Dura, and M. Muller for fly lines; A. Zider for the sd reagents; and M. Kuroda for MSL-1 antibody. We acknowledge R. Pepperkok (European Advanced Light Microscopy Facility), B. Geiger (Weizmann Institute of Science), P. Travo (CRBM), and the IFR N.3 Imaging Facility for help with imaging techniques. We thank P. Atger for artwork. F.B. was supported by the Fondation pour la Recherche Médicale (FRM), C.G. is supported by the Ministère de l'Enseignement Supérieur, and S.L. by the Association pour la Recherche sur le Cancer (ARC) and by CNRS. G.C. was supported by grants of the CNRS, the Human Frontier Science Program Organization, the FRM, and the ARC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.269503.
References
- Alkema M.J., Bronk, M., Verhoeven, E., Otte, A., van't Veer, L.J., Berns, A., and van Lohuizen, M. 1997. Identification of _Bmi1_-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes & Dev. 11**:** 226-240. [DOI] [PubMed] [Google Scholar]
- Beisel C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419**:** 857-862. [DOI] [PubMed] [Google Scholar]
- Breiling A., Turner, B.M., Bianchi, M.E., and Orlando, V. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412**:** 651-655. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Guest, S.S., Smale, S.T., Hahm, K., Merkenschlager, M., and Fisher, A.G. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91**:** 845-854. [DOI] [PubMed] [Google Scholar]
- Buchenau P., Hodgson, J., Strutt, H., and Arndt-Jovin, D.J. 1998. The distribution of Polycomb-group proteins during cell division and development in Drosophila embryos: Impact on models for silencing. J. Cell Biol. 141**:** 469-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S., Inamdar, M., Rodrigues, V., Raghavan, V., Palazzolo, M., and Chovnik, A. 1992. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes & Dev. 6**:** 367-379. [DOI] [PubMed] [Google Scholar]
- Casso D., Ramirez-Weber, F., and Kornberg, T.B. 2000. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech Dev 91**:** 451-454. [DOI] [PubMed] [Google Scholar]
- Cavalli G. 2002. Chromatin as a eukaryotic template of genetic information. Curr. Opin. Cell Biol. 14**:** 269-278. [DOI] [PubMed] [Google Scholar]
- Cavalli G. and Paro, R. 1998. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93**:** 505-518. [DOI] [PubMed] [Google Scholar]
- ____. 1999. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 286**:** 955-958. [DOI] [PubMed] [Google Scholar]
- Chan C.S., Rastelli, L., and Pirrotta, V. 1994. A Polycomb response element in the Ubxgene that determines an epigenetically inherited state of repression. EMBO J. 13**:** 2553-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb J.R. and Bickmore, W.A. 2003. Considering nuclear compartmentalization in the light of nuclear dynamics. Cell 112**:** 403-406. [DOI] [PubMed] [Google Scholar]
- Cook P.R. 1997. The transcriptional basis of chromosome pairing. J. Cell. Sci. 110**:** 1033-1040. [DOI] [PubMed] [Google Scholar]
- Cremer T. and Cremer, C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2**:** 292-301. [DOI] [PubMed] [Google Scholar]
- Csink A.K. and Henikoff, S. 1996. Genetic modification of heterochromatic association and nuclear organisation in Drosophila. Nature 381**:** 529-531. [DOI] [PubMed] [Google Scholar]
- Dernburg A.F., Broman, K.W., Fung, J.C., Marshall, W.F., Philips, J., Agard, D.A,, and Sedat, J.W. 1996. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell 85**:** 745-759. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Olsson, T., Turner, B.M., Cranston, G., and Allshire, R.C. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91**:** 1021-1032. [DOI] [PubMed] [Google Scholar]
- Fauvarque M.O. and Dura, J.M. 1993. polyhomeotic regulatory sequences induce develpomental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes & Dev. 7**:** 1508-1520. [DOI] [PubMed] [Google Scholar]
- Francastel C., Walters, M.C., Groudine, M., and Martin, D.I. 1999. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell 99**:** 259-269. [DOI] [PubMed] [Google Scholar]
- Gasser S.M. 2001. Positions of potential: Nuclear organization and gene expression. Cell 104**:** 639-642. [DOI] [PubMed] [Google Scholar]
- Gehrlich D., Beaudouin, J., Kalbfuss, B., Daigle, N., Eils, R., and Ellenberg, J. 2003. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112**:** 751-764. [DOI] [PubMed] [Google Scholar]
- Gemkow M.J., Buchenau, P., and Arndt-Jovin, D.J. 1996. FISH in whole-mount Drosophila embryos. RNA: Activation of a transcriptional locus, DNA: Gene architecture and expression. Bioimaging 4**:** 107-120. [Google Scholar]
- Gemkow M.J., Verveer, P.J., and Arndt-Jovin, D.J. 1998. Homologous association of the Bithorax-Complex during embryogenesis: Consequences for transvection in Drosophila melanogaster. Development 125**:** 4541-4552. [DOI] [PubMed] [Google Scholar]
- Gyurkovics H., Gausz, J., Kummer, J., and Karch, F. 1990. A new homeotic mutation in the Drosophila bithoraxcomplex removes a boundary separating two domains of regulation. EMBO J. 9**:** 2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K., Müller, M., and Schedl, P. 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithoraxcomplex. Genes & Dev. 10**:** 3202-3215. [DOI] [PubMed] [Google Scholar]
- ____. 1997. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithoraxcomplex. Genetics 146**:** 1365-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G.J. 2002. RNA interference. Nature 418**:** 244-251. [DOI] [PubMed] [Google Scholar]
- Henikoff S. and Comai, L. 1998. Trans-sensing effects: The ups and downs of being together. Cell 93**:** 329-332. [DOI] [PubMed] [Google Scholar]
- Kassis J.A. 2002. Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 46**:** 421-438. [DOI] [PubMed] [Google Scholar]
- Kuhn D.T. and Packert, G. 1988. Paternal imprinting of inversion Uab1 causes homeotic transformations in Drosophila. Genetics 118**:** 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren M., Chow, C.M., Sabbattini, P., Georgiou, A., Minaee, S., and Dillon, N. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103**:** 733-743. [DOI] [PubMed] [Google Scholar]
- Lupo R., Breiling, A., Bianchi, M.E., and Orlando, V. 2001. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol. Cell 7**:** 127-136. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T., Katsani, K.R., and Verrijzer, C.P. 2002. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 21**:** 1775-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F., Straight, A., Marko, J.F., Swedlow, J., Dernburg, A., Belmont, A., Murray, A.W., Agard, D.A., and Sedat, J.W. 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7**:** 930-939. [DOI] [PubMed] [Google Scholar]
- Maurange C. and Paro, R. 2002. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes & Dev. 16**:** 2672-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J., Hogga, I., Gausz, J., Gyurkovics, H., and Karch, F. 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a _Polycomb_-response element. Development 124**:** 1809-1820. [DOI] [PubMed] [Google Scholar]
- Misteli T., Caceres, J.F., and Spector, D.L. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387**:** 523-527. [DOI] [PubMed] [Google Scholar]
- Muller M., Hagstrom, K., Gyurkovics, H., Pirrotta, V., and Schedl, P. 1999. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153**:** 1333-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Klar, A.J., and Grewal, S.I. 2000. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101**:** 307-317. [DOI] [PubMed] [Google Scholar]
- Orlando V. 2003. Polycomb, epigenomes, and control of cell identity. Cell 112**:** 599-606. [DOI] [PubMed] [Google Scholar]
- Orlando V., Jane, E.P., Chinwalla, V., Harte, P.J., and Paro, R. 1998. Binding of trithorax and Polycomb proteins to the bithoraxcomplex: Dynamic changes during early Drosophila embryogenesis. EMBO J. 17**:** 5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M., Bhadra, U., and Birchler, J.A. 1997. Cosuppression in Drosophila: Gene silencing of Alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent. Cell 90**:** 479-490. [DOI] [PubMed] [Google Scholar]
- ____. 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell 9**:** 315-327. [DOI] [PubMed] [Google Scholar]
- Parada L. and Misteli, T. 2002. Chromosome positioning in the interphase nucleus. Trends Cell. Biol. 12**:** 425-432. [DOI] [PubMed] [Google Scholar]
- Petruk S., Sedkov, Y., Smith, S., Tillib, S., Kraevski, V., Nakamura, T., Canaani, E., Croce, C.M., and Mazo, A. 2001. Trithoraxand dCBP acting in a complex to maintain expression of a homeotic gene. Science 294**:** 1331-1334. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. 2002. Silence in the germ. Cell 110**:** 661-664. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. and Rastelli, L. 1994. white gene expression, repressive chromatin domains and homeotic gene regulation in Drosophila. BioEssays 16**:** 549-556. [DOI] [PubMed] [Google Scholar]
- Rabl C. 1885. Über Zellteilung. In Morphologisches Jahrbuch (ed. C. Gegenbauer), pp. 214-330. Willhelm Engelmann, Leipzig, Germany.
- Rank G., Prestel, M., and Paro, R. 2002. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 22**:** 8026-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G. and Wolff, I. 1981. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol. Gen. Genet. 182**:** 516-519. [DOI] [PubMed] [Google Scholar]
- Saurin A.J., Shiels, C., Williamson, J., Satijn, D.P., Otte, A.P., Sheer, D., and Freemont, P.S. 1998. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142**:** 887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.J., Shao, Z., Erdjument-Bromage, H., Tempst, P., and Kingston, R.E. 2001. A Drosophila polycomb group complex includes Zeste and dTAFII proteins. Nature 412**:** 655-660. [DOI] [PubMed] [Google Scholar]
- Sigrist C.J. and Pirrotta, V. 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147**:** 209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H., Cavalli, G., and Paro, R. 1997. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 16**:** 3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. and Pfeifle, C. 1989. A non-radioactive method for the localisation of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98**:** 81-85. [DOI] [PubMed] [Google Scholar]
- Tillib S., Petruk, S., Sedkov, Y., Kuzin, A., Fujioka, M., Goto, T., and Mazo, A. 1999. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol. Cell. Biol. 19**:** 5189-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Schermelleh, L., Cremer, M., Tashiro, S., and Cremer, T. 2003. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell. Biol. 160**:** 685-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Barolo, S., Szymanski, P., and Levine, M. 1996. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes & Dev. 10**:** 3195-3201. [DOI] [PubMed] [Google Scholar]
- Zink D. and Paro, R. 1995. _Drosophila Polycomb_-group regulated chromatin inhibits the accessibility of a _trans_-activator to its target DNA. EMBO J. 14**:** 5660-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]