Alteration at a Single Amino Acid Residue in the T Cell Receptor α Chain Complementarity Determining Region 2 Changes the Differentiation of Naive Cd4 T Cells in Response to Antigen from T Helper Cell Type 1 (Th1) to Th2 (original) (raw)
Abstract
To study whether changes in the structure of a T cell receptor (TCR) at a single peptide-contacting residue could affect T cell priming with antigenic peptide, we made transgenic mice with a point mutation in the TCR α chain of the D10.G4.1 (D10) TCR and bred them to D10 β chain transgenic mice. The mutation consisted of a leucine to serine substitution at position 51 (L51S), which we had already established contacted the second amino acid of the peptide such that the response to the reference peptide was reduced by ∼100-fold. A mutation in the reference peptide CA134–146 (CA-WT) from the arginine at peptide position 2 to glycine (R2G) restored full response to this altered TCR. When we examined in vitro priming of naive CD4 T cells, we observed that the response to doses of CA-WT that induced T helper cell type 1 (Th1) responses in naive CD4 T cells from mice transgenic for the D10 TCR gave only Th2 responses in naive CD4 T cells derived from the L51S. However, when we primed the same T cells with the R2G peptide, we observed Th1 priming in both D10 and L51S naive CD4 T cells. We conclude from these data that a mutation in the TCR at a key position that contacts major histocompatibility complex–bound peptide is associated with a shift in T cell differentiation from Th1 to Th2.
Keywords: mice, transgenic, interferon type II, interleukin 4, peptides
Introduction
The TCR of CD4 T lymphocytes recognizes immunogenic peptide sequences bound within the groove of MHC class II molecules 1. Such recognition results in a signal that is transmitted through the TCR, activates intracellular signal transduction pathways, and culminates in the proliferation and differentiation of CD4 T cells into effector cells that execute many functions 2. Naive CD4 T cells may differentiate into either Th1 or Th2 cells 3. Th1 cells mediate inflammatory responses, activate macrophages, and stimulate cell-mediated immunity. Th2 cells activate B cells, stimulate humoral immunity, and are prominent mediators of allergic responses 4. The choice of differentiation into either Th1 or Th2 cells is a crucial step that determines the direction of the subsequent adaptive immune response. This choice is influenced by many factors including the interaction between the TCR and the peptide–MHC complex 5. The cytokine environment where the CD4 T cell meets its ligand is as important as the details of the direct engagement of its TCR by that ligand. In vitro studies have shown that the predominance of IL-4 or IL-6 drives Th2 differentiation, whereas the predominance of IFN-γ or IL-12 drives Th1 differentiation. The source of the initial IL-4 in vivo can be either NK1.1+ CD4 T cells 6, or naive CD4 T cells that have encountered IL-6–producing APCs 7. The source of the initial IFN-γ can be NK cells, CD8 T cells, or naive CD4 T cells that have encountered IL-12–producing APCs 8. Different pathogens trigger different innate immune mechanisms that favor one cytokine environment over the other. These mechanisms are probably related to the antigenic makeup of the pathogen, its route of entry, and the type and condition of the APC that initially responds to its presence 9 10.
We are beginning to understand the factors central to the TCR–peptide–MHC complex that influence CD4 T cell differentiation. In general, a longer duration of contact between the TCR and its ligand results in a full signal through the TCR and subsequent Th1 differentiation 5 11. Partial signals favor Th2 differentiation. The duration of contact is in turn influenced by several factors such as: (a) high density of peptide–MHC ligand on the surface of the APC, (b) the stability of the complex between the peptide and the MHC molecule, and (c) a longer half-life of TCR–peptide–MHC complexes.
Only three to five amino acids within a peptide bound in the groove of MHC molecules face outwards into the solvent for potentially direct contacts with TCR amino acids 12 13 14. Similarly, although many amino acids constitute the framework of the TCR α and β chains, only a few amino acids have been established to directly contact peptide amino acids 15 16 17 18 19 20 21. Functional studies that supported these findings came from experiments with altered peptide ligands (APLs). APLs are peptide variants that have been structurally designed to contain single amino acid substitutions at key TCR contact residues 22. APLs are still presented by the same MHC class II molecule and recognized by the same TCR. However, this recognition results in marked alterations in the nature of the signal received by the TCR. APLs were first shown to stimulate IL-4 production from Th2 clones that were capable of providing B cell help but failed to induce proliferation 23. APLs also stimulated Th1 clones to express IL-2 receptor and exhibit cytotoxic function without stimulating proliferation or cytokine production 24. The biochemical basis for the selective induction of these effector functions was shown to lie in incomplete phosphorylation of the CD3 chains, lack of recruitment and phosphorylation of Src homology 2 (SH2)-containing kinases, and distinct quantitative differences in calcium mobilization patterns 22. Whereas low doses of agonist peptides resulted in phosphorylation of tyrosine residues in the TCR-associated ζ chain and the tyrosine kinase ZAP-70, 100–1,000-fold higher doses of partial agonist peptides were required to observe the same effect.
It was further shown that stimulation of CD4 T cells by APLs versus agonist peptides influenced CD4 T cell differentiation. These observations supported previous studies where the MHC haplotype of the APC was found to influence CD4 T cell differentiation 25. I-As molecules that bound human collagen IV–derived immunogenic peptides 20,000 times more avidly than I-Ab favored differentiation of responding CD4 T cells along the Th1 path. Increasing the dose of antigen by immunizing I-Ab mice with higher doses of peptide was capable of generating Th1 responses. At a given dose, agonist peptides that formed high affinity interactions with the MHC molecule could engage the TCR longer, favoring Th1 differentiation, whereas APLs having altered affinities either to the presenting MHC molecule or to the TCR favored Th2 differentiation 26. Similarly, by varying the dose of a given peptide (agonist or APL), higher doses were shown to favor Th1 differentiation, whereas lower doses favored Th2 differentiation 27. To accomplish the same effect, the dose of the partial agonist peptide had to be 100–1,000-fold higher than the full agonist peptide. The differential outcome of CD4 T cell differentiation was also associated with biochemical alterations proximal to the TCR 28. Thus, the density and stability of interactions between the peptide and the MHC influence signals generated through the TCR.
In summary, all of the above studies have dissected the role of the trimolecular TCR–peptide–MHC complex on CD4 T cell differentiation by introducing changes in the peptide–MHC component. These changes are in turn interpreted by the TCR, as evidenced in qualitative and quantitative changes in the resultant intracellular biochemical signals. The direct change in the TCR itself described in this study is a novel approach to address the following question: Can two different TCRs recognize the same MHC–peptide ligand with different outcomes? Similar to the design of APLs, we have designed an altered TCR by substituting one amino acid in the TCR α chain at a key position that contacts MHC-bound peptide. We describe the behavior of this altered TCR compared with its parent TCR by generating transgenic mice bearing either TCR and studying the proliferation and differentiation of purified naive CD4 T cells from these two transgenic mice stimulated by the same peptide–MHC ligand. Whereas the parent TCR bearing CD4 T cells became Th1-like at high doses of peptide, the altered TCR bearing CD4 T cells became Th2-like at those same doses. Thus, subtle variations in the TCR can influence the outcome of CD4 T cell differentiation.
Materials and Methods
Mutagenesis of the D10.G4.1 TCR α Chain.
The mutagenesis of the D10.G4.1 (D10) TCR α chain has been described previously 29. In brief, mutagenesis was performed using the overlapping extension method. The D10 TCR α chain was used as a template DNA. The mutation was produced by standard PCR methods to produce a substitution at amino acid position 51 of leucine to serine (L51S) in the TCR α chain. The mutagenesis was verified by DNA sequencing.
Transgenic Mice.
The D10 transgenic mice were generated by microinjection of the rearranged genes encoding the D10 α and β chain TCR as described 15. Similarly, the L51S α chain carrying the transgene was microinjected into day 1 C3H/B6 embryos. The transgenic founder was backcrossed to B10.BR and further backcrossed to single D10 β chain transgenic mice (D10 β chain transgenic mice are described in reference 15). Both D10 and L51S TCR transgenic mice were further backcrossed to Cα−/− 129s mice. Homozygous Cα−/−, either carrying the transgenes or not, were selected for further breeding. The lines were maintained by breeding transgenic Cα−/− mice with nontransgenic Cα−/− mice. Transgenic mice were identified by screening tail DNA by PCR for the presence of the transgene using D10 TCR α and β chain–specific primers. For the α chain, the forward primer sequence was 5′ of the ATG, and the reverse primer sequence was 3′ of Jα within the intron between Jα and Cα. The sequences were as follows: D10 Vα forward primer, 5′tttctcccaaacttcagtcta3′, and D10 Vα reverse primer, 5′gctctggtcattggcacgat3′. For the β chain, the forward primer sequence was within the leader, and the reverse primer sequence was 3′ of Jβ within the intron between Jβ and Cβ. The sequences were as follows: D10 Vβ forward primer, 5′-gcattctagatggtcccaagatgggc-3′, and D10 Vβ reverse primer, 5′-ttaaggatccactctgctaaggttttctgc-3′. PCR conditions were 1 min at 94°C, 2 min at 55°C, and 2 min at 72°C for 40 amplification cycles.
Peptides.
Peptides were synthesized and purified by the W.M. Keck Biotechnology Resource Laboratory at the Yale School of Medicine using tBOC chemistry. Peptides were characterized by receptor protein (RP)-HPLC, amino acid analysis, and fast atom bombardment (FAB) mass spectroscopy. The sequence of the reference peptide CA134–146 (CA-WT) is HRGAIEWEGIESG, and of the mutated peptide with arginine to glycine at position 2 (R2G) is HGGAIEWEGIESG.
Isolation of Naive CD4 Transgenic T Cells.
12–14-wk-old D10.TCR or L51S.TCR transgenic mice were killed, and their spleens and LNs were removed. Single cell suspensions were prepared and enriched for CD4 T lymphocytes by magnetic bead depletion after staining with antibodies TIB-146 or TIB-164 (anti-B220), TIB-210 or TIB-105 (anti-CD8), 2.4-G2 (anti-Fc receptor), and 10.2.16 (anti–I-Ak), followed by anti–mouse IgG, anti–rat IgG, and anti–rat IgM coated magnetic beads (BioMag; PerSeptive Biosystems). Naive CD4 T cells were sorted by FACStarPLUS™ (Becton Dickinson) after staining with filter-sterilized biotinylated Mel-14 (anti–L-selectin), followed by PE-conjugated streptavidin (Caltag) and FITC–conjugated anti-Vα2 TCR (BD PharMingen). Vα2+Mel-14high cells were collected and cultured.
Flow Cytometry FACScan™ Analysis.
Single cell suspensions from the spleens and LNs of D10 or L51S TCR transgenic mice were stained either before or after enrichment of CD4 T cells with any of the following antibodies: biotinylated mAb 3D3 (clonotypic anti-TCR mAb specific to D10 and L51S TCRs) 30, FITC-conjugated anti-Vα2 TCR (BD PharMingen), biotinylated anti-Vβ8.1,8.2 TCR (BD PharMingen), Quantum red–conjugated anti-CD4 (Sigma-Aldrich), PE-conjugated anti-CD8α (GIBCO BRL and Life Technologies), or FITC-conjugated anti–TCR β chain (H57; BD PharMingen). Biotinylated mAb staining was followed by PE-conjugated streptavidin (Caltag). Stained cells were analyzed on a FACScan™ (Becton Dickinson), and collected events were analyzed using CELLQuest™ software (Becton Dickinson).
T Cell Proliferation Assays.
All T cell proliferation assays were performed in 96-well U-bottomed plates (Becton Dickinson) with 5 × 104 CD4 T cells per well. Total splenocytes from syngeneic nontransgenic Cα−/− littermates were used as a source of APCs after irradiation at 2,000 rads. 105 irradiated splenocytes were added per well followed by the addition of varying doses of CA-WT or R2G peptide ranging from 100 to 10−3 μM, in a final volume of 200 μl per well. The cultures were left undisturbed for 72 h, at which time each well was pulsed with 0.5 μCi of [3H]thymidine (NEN Life Science Products). The wells were harvested 18 h later onto printed filter mats (Wallac) using a 96-well cell harvester (TomTec). The filter mats were sealed in sample bags (Wallac), and the incorporated radioactivity was counted using a 1205 Betaplate liquid scintillation counter (Wallac).
Mini Double Cultures.
The minicultures were adapted from a protocol designed by Dr. Kim Bottomly's research group 25 as a method to provide naive CD4 T cells with both a primary and a secondary stimulation in vitro, and assess their subsequent differentiation into either Th1 or Th2 cells. All mini double cultures were performed in duplicate in 96-well U-bottomed plates (Becton Dickinson) and in a total volume of 200 μl/well. 5 × 104 FACS®-sorted naive D10 TCR or L51S TCR transgenic CD4 T cells were cultured with 105 irradiated syngeneic nontransgenic Cα−/− littermate splenocytes and 25 U/ml IL-2 in Click's modified EHAA medium supplemented with 5% FCS (Gemini Bioproducts, Inc.). CA-WT or R2G peptide was added to each well at varying doses ranging from 100 to 10−3 μM. Cells were rested on day 4 by removing the entire 200 μl of culture supernatants for cytokine analysis, and adding fresh 105 irradiated syngeneic Cα−/− splenocytes without IL-2 for 2 d. The cultures were then restimulated on day 6 by removing 100 μl of the culture supernatant and replacing it with 100 μl medium containing 105 irradiated syngeneic Cα−/− splenocytes to a final concentration of 25 U/ml IL-2 and 100 μM of R2G peptide. Culture supernatants excluding the cells were again removed for cytokine analysis on day 4 after the secondary stimulation, and cultures were thus terminated on day 10.
ELISA.
The levels of IL-4 or IFN-γ in the culture supernatants after primary or secondary stimulation were measured by ELISA. 96-well Nunc-Immuno plates (Nalge Nunc International) were coated with 100 μl/well of capturing anti–IL-4 mAb (11B11) and anti–IFN-γ mAb (HB-170) at 6 μg/ml in PBS at 4°C for 18 h. The plates were washed with 0.1% solution of Tween 20 (Sigma-Aldrich) in PBS, and blocked with 1% BSA (Sigma-Aldrich) in PBS for 1 h at 37°C. 50 μl of culture supernatants and IL-4 or IFN-γ standards was then added and incubated at 4°C for 18 h. Twofold dilutions of murine IL-4 or IFN-γ (BD PharMingen) of known concentrations were used as standards. Cytokine–antibody complexes were detected by the addition of biotinylated anti–murine IL-4 or IFN-γ (BD PharMingen), followed by peroxidase-conjugated streptavidin (Zymed Laboratories), and visualization after the addition of _o_-phenylenediamine dihydrochloride from tablets (Sigma-Aldrich). Color development was stopped with 3 M H2SO4. Absorbance at 492 nm was measured on an ELISA reader (ELx800; Biotek Instruments, Inc.). The supernatant concentrations of IL-4 or IFN-γ were calculated by extrapolating absorbance values from the standard curve plotting concentration versus absorbance.
Intracellular Cytokine Staining.
Double cultures were set up with naive sorted CD4 T cells and irradiated syngeneic Cα−/− splenocytes in the presence of 25 U/ml IL-2, in 24-well flat-bottomed plates (Becton Dickinson), and in a total volume of 2 ml EHAA medium/5% FCS (Gemini Bioproducts, Inc.). The cultures were primed with 10 μM CA-WT peptide and restimulated with 0.05 μg/ml PMA (Sigma-Aldrich) and 0.5 μg/ml Ionomycin (Calbiochem). 10 μg/ml brefeldin A (Epicenter Technologies) was added 4 h later, and the cells were stained after an additional 4 h using the FIX & PERM Cell Permeabilization kit (Caltag) and the following antibodies: Quantum red–conjugated anti-CD4 (Sigma-Aldrich), FITC-conjugated anti–IFN-γ (IgG1; BD PharMingen), PE-conjugated anti–IL-4 (IgG2b; BD PharMingen), and the isotype control immunoglobulin antibodies PE-conjugated rat IgG2b, and FITC-conjugated rat IgG1. Stained cells were immediately analyzed on a FACScan™ (Becton Dickinson). CD4+ T cells were gated and analyzed using CELLQuest™ software (Becton Dickinson).
Results
The Basis for Selecting the Amino Acid Substitution for the Generation of the Altered TCR.
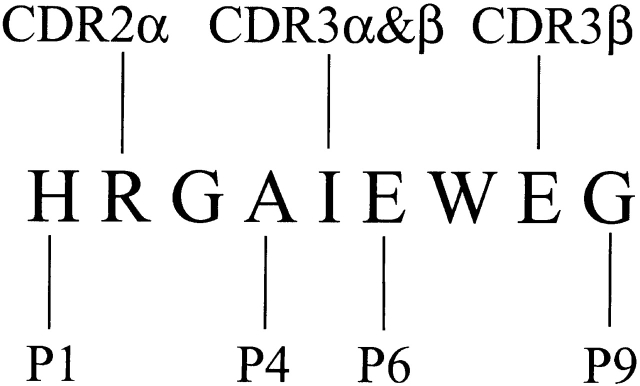
D10 is a cloned T cell line that recognizes the core nine–amino acid peptide sequence derived from a peptide within the chicken conalbumin protein (CA134–146) and presented by the MHC class II molecule I-Ak 29 30 31. The clone also exhibits alloreactivity towards other MHC class II molecules such as I-Ab, v, p, q, d. We have previously described the full characterization of the interaction of the D10 TCR with its peptide–MHC ligand by extensive mutagenesis of the TCR 15 29, its MHC ligands 16, and the peptide itself 29 32. The nine–amino acid core of the peptide derived from conalbumin has previously been referred to as CA-WT and has the sequence HRGAIEWEG. We have previously identified positions p1, p4, p6, and p9 of the CA-WT peptide as MHC binding residues, and p2, p5, and p8 of the peptide as TCR contact residues 15 (Fig. 1). The contact of peptide position 2 with the α chain CDR2 was later confirmed by site-directed mutagenesis of amino acid 51 of the TCR α chain 29.
Figure 1.
A schematic representation of the CA-WT 9-mer peptide amino acid sequence showing both the D10 TCR contact and I-Ak MHC class II contact residues. The figure is adapted from reference 15. The peptide amino acid residues that contact I-Ak are at positions 1, 4, 6, and 9, and labeled P1, P4, P6, and P9. Those that contact the TCR are at positions 2, 5, and 8. The L51S mutation affects the CDR2 region of the D10 TCR α chain, which contacts position 2 in the CA-WT peptide.
Another T cell clone, AK8, described previously by our laboratory, is related to D10, and both clones have the same TCR β chain but differ in their α chain by 7 amino acids within the first NH2-terminal 70 residues, including 1 that was in the α chain CDR2 at position 51 16. A separate mutation was thus introduced at this position from the leucine present in D10 to the serine present in AK8. This mutated α chain was transfected along with the parent β chain into 4G4 T cell hybrids to generate the T cell hybrid L51S 29. We showed that amino acid 51 in the CDR2 of the TCR α chain interacted with the peptide at p2 29. The proliferative response of L51S T cells was 150-fold lower to CA-WT than that of the parent D10 T cells. One particular peptide variant, R2G, was of interest since it restored the proliferative response of L51S T cells to levels and at doses similar to those obtained with D10 cells.
Based on the information and results summarized above, we have generated L51S TCR transgenic mice in this study that bear the single mutation at position 51 of the TCR α chain and are restricted to I-Ak. These mice were bred onto a Cα2/2 background to ensure that no other endogenous α chains were used for pairing with the parent β chain. In addition, D10 TCR transgenic mice that have been described previously 15 were also bred onto a Cα−/− background for comparison. These mice serve as a good model for studying two TCRs that have the same ligand specificity and exploring different aspects of their thymic development as well as their behavior in secondary lymphoid tissues. In this study, we explore the influence of this single amino acid substitution at a peptide contact site on naive CD4 T cell differentiation.
An Amino Acid Substitution from L to S at Position 51 of the TCR α Chain Results in the Reduction of TCR Transgenic CD4 T Lymphocyte Numbers in Secondary Lymphoid Tissues.
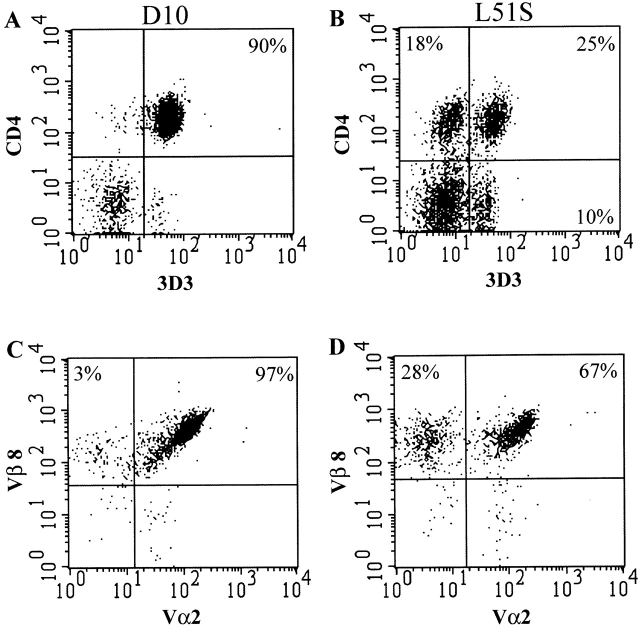
The percentage of transgenic T cells in the spleens and LNs was detected by staining with the clonotypic D10 TCR-specific mAb 3D3. Of the total LN cells, 15% were 3D3+ in L51S mice compared with 25% 3D3+ in D10 mice. Two-color FACS® analysis on CD4 T cell–enriched preparations from the LNs of L51S and D10 mice showed that after enrichment, 90% of the preparation was transgenic CD4+3D3+ in D10 (Fig. 2 A), and 25% was transgenic CD4+3D3+ in L51S mice (Fig. 2 B). The mean fluorescence intensities of 3D3 were similar: 56.36 for L51S, and 66.70 for D10. Furthermore, within the CD4 population of L51S LN cells, there were two subpopulations: CD4+3D3+ and CD4+3D3−. Therefore, in addition to the already reduced numbers of CD4 T cells in the peripheral lymphoid tissues of L51S mice, there was a further dilution in the percentage of transgenic CD4 T cells. The presence of a population of cells that is CD4−3D3+ is also of interest. We have performed three-color staining for 3D3, CD4, and CD8 to examine whether these cells were CD8+CD4−3D3+, and we found that they were CD8− CD4−3D3+ (data not shown). Although at present we do not know the significance of these cells, it is likely that these cells have downregulated CD4 expression to avoid deletion due to autoreactivity. The population of CD4 T cells that did not bear the transgenic TCR was 18% of the total CD4 enriched preparation, and it constituted 35–40% of the total CD4 population (Fig. 2 B).
Figure 2.
CD4 T cells in L51S TCR transgenic mice are markedly reduced in numbers. CD4 enriched cells from the LNs of D10 (A and C) or L51S (B and D) TCR transgenic mice were stained with Quantum red–conjugated anti-CD4 and biotinylated 3D3 followed by PE-conjugated streptavidin (A and B). LN cells were also stained simultaneously with Quantum red–conjugated anti-CD4, FITC-conjugated anti-Vα2, and biotinylated anti-Vβ8.1,8.2 followed by PE-conjugated streptavidin. CD4+ T cells were gated and their expression of the transgenic TCR Vα2+Vβ8+ is shown (C and D). Log fluorescence intensity is shown on both axes.
We further gated on CD4 T cells in the LNs of both L51S and D10 mice and examined their expression of the transgenic Vα2 and Vβ8 TCR chains. As expected, almost all (97%) CD4 T cells in D10 mice expressed both the transgenic Vα2 and Vβ8 TCR chains (Fig. 2 C). In sharp contrast, only 67% of the CD4 T cells in L51S mice expressed both Vα2 and Vβ8 TCR chains, and 28% expressed the Vβ8 TCR chain but not the Vα2 TCR chain (Fig. 2 D). The mean fluorescence intensities for Vα2 were similar: 113.26 for L51S, and 108.41 for D10. Further gating and simultaneous four-color staining showed that the CD4+3D3− cells were Vβ8+Vα2− (not shown). As these results were obtained in Cα2/2 mice, this suggests that such cells express an abnormal TCR made up of either a ββ homo- or heterodimer, or perhaps a βγ heterodimer.
The Amino Acid Substitution in the TCR α Chain at Position 51, One of Three Peptide Contact Sites, Alters the Proliferative Response of Transgenic CD4 T Cells.
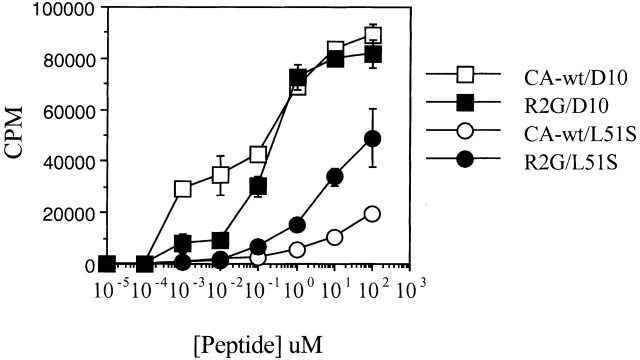
The proliferation of L51S CD4 T cells in response to CA-WT or R2G peptides was measured in the presence of irradiated syngeneic I-Ak APCs and compared with the proliferation of D10 CD4 T cells (Fig. 3). CD4 T cells from TCR transgenic mice had the same proliferative profile as L51S TCR–transfected cells 29. As expected, the proliferation of L51S CD4 T cells to the R2G mutant peptide was better than that to CA-WT peptide. In contrast, D10 CD4 T cells responded vigorously to both peptides. The substitution of R by G at p2 of the peptide did not influence recognition by D10 CD4 T cells, but enhanced the response of L51S CD4 T cells by 10-fold. This proliferative response to R2G was nonetheless 10-fold lower in L51S T cells than in D10, possibly reflecting the lower number of transgenic CD4 T cells in L51S CD4 enriched preparations. The proliferation of CD4 T cells from both D10 and L51S TCR transgenic mice in response to anti-CD3 mAb 2C11 was similar, indicating that the cells were equally responsive to maximal stimulation (data not shown, and Sant'Angelo, D.B., J.M. Blander, and C.A. Janeway, Jr., manuscript in preparation). The proliferation to allogeneic B6 splenocytes was similar (not shown), indicating that similar to the L51S TCR transfected cells 29, the L51S CD4 TCR transgenic cells retained the response to allogeneic MHC molecules (in this case to I-Ab).
Figure 3.
Proliferation of CD4 T cells from both D10 and L51S TCR transgenic mice in response to varying doses of CA-WT or R2G peptide in the presence of irradiated syngeneic splenocytes. The x-axis shows the concentration of peptide (in μM). The y-axis shows the counts per minute (CPM) of [3H]thymidine incorporated as a measure of proliferation.
CA-WT Peptide Primes Naive L51S CD4 T Cells to Become IL-4 Producers.
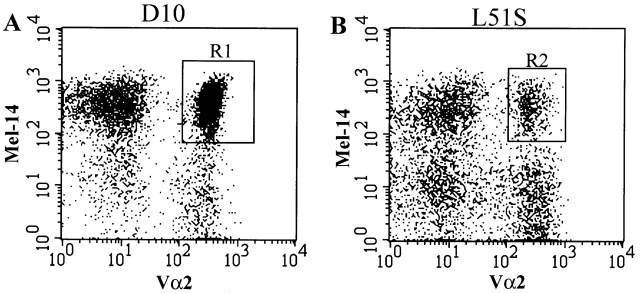
Naive transgenic CD4 T cells were isolated by sorting with FACS® based on their expression of Vα2 and L-selectin (Va2+Mel-14+) (Fig. 4A and Fig. B, gates R1 and R2; for D10 and L51S CD4 transgenic T cells, respectively). These markers were chosen for the following reasons. First, sorting based on Vα2 expression did not activate the T cells nonspecifically and excluded the population of L51S CD4 T cells that was Vβ8+Vα2−. Second, we have correlated the expression of L-selectin with naive CD4 T cell phenotype where the cells are CD44low CD45RBhighL-selectinhigh 33. For both L51S and D10 CD4 T cells, we established that L-selectin–expressing naive cells were also CD45RBhigh and CD44low (not shown).
Figure 4.
FACS® sorting of naive CD4 T cells from D10 or L51S TCR transgenic mice. Sterile sorting of naive transgenic CD4 T cells by FACS® based on the expression of the transgenic α chain (Vα2+) and the naive phenotype marker L-selectin (Mel-14+). CD4 T cell–enriched suspensions of spleen and LNs, D10 (A) and L51S (B), were stained with FITC-conjugated anti-Vα2 and biotinylated Mel-14 followed by PE-conjugated streptavidin. Naive transgenic Mel-14+Vα2+ cells were gated and collected. Log fluorescence intensity is shown on both axes.
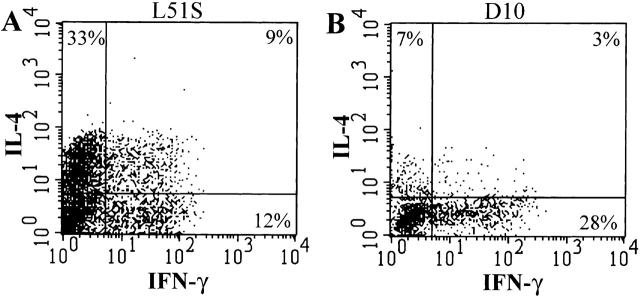
An in vitro system of priming naive CD4 T cells was established as described previously 27. Both L51S TCR transgenic and D10 TCR transgenic Vα2+Mel-14+ naive CD4 T cells were isolated and primed with 10 μM CA-WT peptide. After the second stimulation, CD4+ T cells were gated and examined for intracellular cytokine staining with antibodies to IL-4 and IFN-γ. Priming naive L51S CD4 T cells with 10 μM CA-WT peptide resulted in a major population (33%) of IL-4–producing CD4 T cells (Fig. 5 A). A discrete population (12%) produced IFN-γ, and 9% of the cells produced both IL-4 and IFN-γ. In contrast, a main population (28%) of D10 CD4 T cells produced IFN-γ when primed at the same dose of CA-WT peptide (Fig. 5 B).
Figure 5.
Intracellular cytokine staining of naive L51S and D10 CD4 T cells shows the predominant population of IL-4 producers in L51S cultures. Naive CD4 T cell cultures were primed with 10 μM CA-WT peptide and restimulated on day 6 with PMA and ionomycin, followed by treatment with brefeldin A. Cultured L51S naive CD4 T cells (A), or cultured D10 naive CD4 T cells (B), were stained 6 h later with Quantum red–conjugated anti-CD4, fixed with 1% paraformaldehyde, permeabilized, and stained with FITC-conjugated anti–IFN-γ, and PE-conjugated anti-IL-4. CD4+ T cells were gated, and their intracellular staining for IL-4 and IFN-γ is shown. The log fluorescence intensity is shown on both axes.
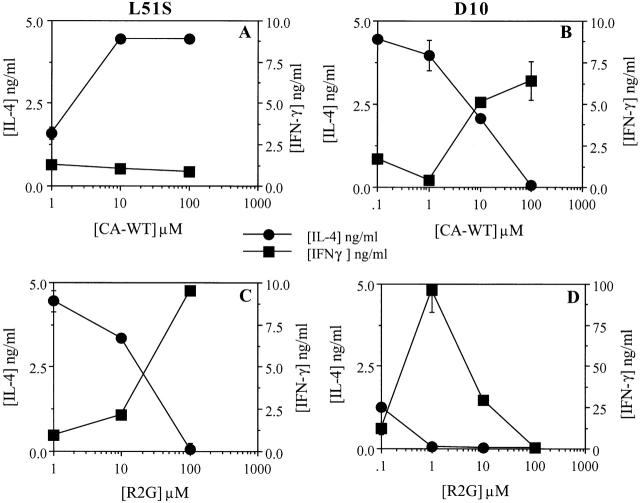
Naive CD4 T cells were also primed at different doses of CA-WT peptide. The dose of peptide varied 10-fold within a range of 100–0.1 μM. Culture supernatants were collected after the first peptide stimulation and tested for the presence of IL-4 and IFN-γ. As expected, neither D10 nor L51S naive CD4 T cells produced detectable levels of either cytokine after primary stimulation with CA-WT peptide (not shown). To promote maximal cytokine production and reveal the differentiated fate of these primed T cells, we chose to restimulate the cells with 10 μM R2G peptide, as the proliferative response to R2G was equal or better for D10 and L51S CD4 T cells, respectively. Priming L51S CD4 T cells with different doses of CA-WT peptide showed peak IL-4 production at higher doses of CA-WT peptide (100–10 μM) (Fig. 6 A, •). In contrast, IL-4 production by D10 CD4 T cells peaked at lower doses (1 and 0.1 μM) of CA-WT peptide (Fig. 6 B, •). When D10 and L51S CD4 T cells were primed with R2G peptide at different doses instead of CA-WT peptide, both L51S and D10 CD4 T cells preferentially produced IL-4 only at lower doses of peptide (Fig. 6C and Fig. D, respectively; •). Culture supernatants collected after the first stimulation with R2G contained no detectable levels of either IL-4 or IFN-γ cytokine (not shown).
Figure 6.
L51S TCR transgenic naive CD4 T cells differentiate into Th2-like IL-4–producing cells when primed in vitro with CA-WT peptide, and produce IFN-γ when stimulated with R2G. Naive CD4 T cells were cultured in the presence of varying doses of CA-WT (A and B) or R2G peptide (C and D), irradiated syngeneic splenocytes, and 25 U/ml IL-2. The primary supernatants were collected on day 4 and the cells were restimulated with 10 μM R2G peptide, after which the secondary supernatants were collected on day 4 and ELISAs were performed in duplicate to measure the concentrations of IL-4 and IFN-γ. (A and C) L51S CD4 T cell cytokine profiles; (B and D) D10 CD4 T cell cytokine profiles. IL-4 and IFN-γ concentrations are shown in ng/ml quantities. The concentrations of peptide used for priming are shown on the x-axes.
R2G Peptide Primes Naive L51S CD4 T Cells to Become IFN-γ Producers at High Doses and IL-4 Producers Only at Low Doses.
Priming L51S CD4 T cells with different doses of CA-WT peptide did not result in significant IFN-γ production (Fig. 6 A, ▪), whereas priming with increasing doses of R2G resulted in 9 ng/ml IFN-γ at the highest dose of 100 μM (Fig. 6 C, ▪). In contrast, D10 CD4 transgenic T cells primed with increasing doses of CA-WT or R2G peptide resulted in both cases in IFN-γ production at higher doses (Fig. 6B and Fig. D, respectively; ▪). Decreasing doses of both R2G and CA-WT peptide favored IL-4 production by D10 CD4 T cells (Fig. 6B and Fig. D, respectively; •).
Discussion
Recognition of immunogenic peptides by the TCR on CD4 T lymphocytes is a complex process. Engaging the TCR leads to the intracellular mobilization of protein kinases and phosphatases that initiate and modulate different signaling pathways. Activation through the TCR is not an “all or none” phenomenon, but rather a gradation of signaling that results in the selective stimulation of TCR-mediated effector functions 34. This has been clearly demonstrated by studies with APLs where the interaction between the peptide and the MHC is weakened, and by its influence on the signals generated by the TCR studied 22. Here we have shown that we can directly weaken the interaction between the TCR and the peptide–MHC and influence the type of differentiated effector CD4 T cell generated.
The mutation of the amino acid at position 51 of the TCR α chain seemed to lower the affinity of the D10 TCR to its CA-WT/I-Ak ligand, as suggested by the lower rate of proliferation of CD4 T cells carrying this mutated α chain. We had previously shown that the leucine at position 51 lies at the tip of the CDR2α region, and interacts with position 2 in the CA-WT peptide 29. Mice transgenic for the altered TCR and the mutated α chain and parent β chain were generated. These mice were used to examine whether a change within a region of the TCR actively involved in ligand recognition has any effect on the development, phenotype, and function of CD4 T cells bearing the altered TCR. Although we have introduced only a single change in the TCR, the influence on the development and behavior of CD4 T cells was profound. In TCR transgenic mice bearing the altered TCR, we observed an almost 60% reduction in the total numbers of CD4 T cells in the secondary lymphoid tissues. This is directly related to the selection events that these cells undergo in the thymus. Only a small percentage of the thymocytes in L51S TCR transgenic mice upregulate TCR expression and become CD4+CD8+. During positive selection, only 1% of these double positive cells proceed to become single positive CD4 thymocytes in contrast to 13% in D10 TCR transgenic mice (Sant'Angelo, D.B., J.M. Blander, and C.A. Janeway, manuscript in preparation). Thus, the peripheral T cell numbers in L51S TCR transgenic mice are low because of the inefficiency of positive selection in the thymus.
We had previously described the generation of transfected cells expressing either the D10 or the L51S TCR 29. In this study, we found that the pattern of proliferative response of CD4 T cells from L51S TCR transgenic mice was similar to that of L51S TCR–transfected cells. CD4 T cells from L51S TCR transgenic mice retained the reduced proliferative response to CA-WT. The cells did respond better to the peptide R2G, but this response was 100-fold lower than that of CD4 T cells from D10 TCR transgenic mice. In contrast, the proliferative response of L51S TCR–transfected cells to R2G was almost as good as that of D10 TCR–transfected cells 29. The proliferation data presented here suggested that the altered TCR expressed by CD4 T cells from L51S TCR transgenic mice possibly had a reduced affinity to the CA-WT by as much as 1,000-fold.
The novel approach employed in this study lies in comparing the responses of two TCRs, which recognize the same ligand: CA-WT peptide presented by I-Ak. The study shows the critical effect that a change as subtle as a single amino acid difference can have on the outcome of CD4 T cell differentiation. The position of the amino acid substitution at a peptide contact residue in the TCR highlights the importance of key amino acid residues in not only conferring T cell activation, but dictating the outcome of that activation. At present, we do not know of any naturally occurring in vivo correlates to our study. We may speculate that if there were such subtle variations in the T cell repertoire, a single immunodominant epitope presented by an APC could potentially be recognized by two CD4 T cell clones with very similar TCRs but which, at a given density of peptide–MHC complexes, differ in their potential to differentiate into either Th1 or Th2. The cytokine milieu created by the activated APC may then allow the further expansion and predominance of one clone over the other.
The stability and duration of the TCR interaction with its peptide–MHC ligand has been shown to play a crucial role not only in activating the T cell but also in affecting the type of effector CD4 T cell it can produce 11. Therefore, we predicted that the reduced proliferative response of the L51S TCR to CA-WT peptide, which may be due to a lower stability, duration, or affinity of the TCR interaction with its peptide–MHC ligand, would have an influence on the differentiation of these CD4 T cells. We examined this hypothesis in vitro under a set of well-defined conditions previously used to determine the influence of APLs on CD4 T cell differentiation. Based on the proliferative response of L51S CD4 T cells, we may consider R2G as the agonist peptide for the L51S TCR, and CA-WT as the partial agonist peptide. L51S CD4 T cells differentiated into IFN-γ producers when primed at higher doses of agonist, and IL-4 producers when primed at lower doses of agonist. This effect of peptide dose on CD4 T cell differentiation was similar to results obtained with naive CD4 T cells from TCR transgenic mice that recognize a peptide derived from tobacco horn-worm moth cytochrome c (pMCC) and presented by I-Eb 27. High doses of pMCC favored Th1 differentiation, whereas low doses favored Th2 differentiation. Furthermore, we showed that CD4 T cells from L51S TCR transgenic mice became IL-4 producers when primed with their APL CA-WT. This effect of APL recognition was similarly seen after in vivo priming of H-2S mice where immunization with a wild-type peptide derived from human collagen IV led to IFN-γ–producing effector cells, while immunization with variant APLs that bound I-AS less well led to IL-4–producing effector cells 26. Similarly, priming of naive CD4 T cells from TCR transgenic mice in vitro with pMCC peptide led to the same observations 35.
Our results in this study strengthen the notion that the nature of the signal transmitted by the TCR impacts downstream differentiation events. The results show that a mutation in the signal transmitting receptor itself, given the same exact ligand as the parent receptor, is sufficient to influence consequent cellular differentiation. We do not know at this time if such a mutation affects the structural conformation of the D10 TCR at its ligand binding sites. Recent findings using the A6 TCR of a CD8+ HLA-A2–restricted T cell clone showed that the different signals transmitted through this TCR upon encountering three different APLs could not be correlated with the three-dimensional structures of the TCR interacting with the three APL–HLA-A2 complexes 36. The similarity of the structures in all cases led to the conclusion that ligand-induced conformational changes in the TCR do not account for the generation of different signals. It remains to be seen whether the three-dimensional structure of the L51S TCR/CA-WT/I-Ak is similar to the recently reported crystal structure of the D10 TCR/CA-WT/I-Ak 37. The crystal structure of D10 TCR/CA-WT/I-Ak, however, indicates that the L at position 51 contacts the MHC, which is in direct contrast to our previous findings 15 16 29 32. Regardless, substituting an S for L at position 51 may have several effects, including loss of van der Waals bonds between the L of the TCR and the T at position 77 of the I-Ak β chain, a possible conformational change in the CDR2α loop, and resultant contacts with position 2 in the CA-WT peptide, and finally disruption of the interacting surfaces between the TCR and the MHC as a result of substituting an aliphatic residue with a small hydroxyl-bearing side chain (Reinherz, E., personal communication). It remains possible that a conformational change in the TCR chains may be sufficient to either generate a different signal or to induce changes in the subunits within the CD3 complex such that a larger and more stable signaling complex is not formed.
Acknowledgments
We thank Dr. Ellis Reinherz for valuable suggestions and comments, Tom Taylor for expert technical assistance with FACS® sorting, and Grigory Losyev for purifying and biotinylating mAbs 3D3 and Mel-14, and for providing antibodies 11B11 and HB-170 for ELISA, as well as the antibodies used for CD4+ T cell enrichment.
This work was supported in part by the Howard Hughes Medical Institute and by a National Institutes of Health grant (AI14579-22) to C.A. Janeway, Jr. J.M. Blander is a Fellow of the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper: APL, altered peptide ligand; D10, D10.G4.1; L51S, leucine to serine substitution at position 51; pMCC, moth cytochrome c peptide; R2G, arginine to glycine substitution at position 2.
References
- Germain R.N. Major histocompatibility complex-dependent antigen processing and peptide presentationproviding ligands for the clonal activation of T lymphocytes. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Jr., Bottomly K. Signals and signs for lymphocyte responses. Cell. 1994;76:275–285. doi: 10.1016/0092-8674(94)90335-2. [DOI] [PubMed] [Google Scholar]
- Mosmann T.R., Cherwinski H., Bond M.W., Giedlin M.A., Coffman R.L. Two types of murine helper T cell clones. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- Abbas A.K., Murphy K.M. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- Constant S.L., Bottomly K. Induction of Th1 and Th2 CD4+ T cell responsesthe alternative approaches. Annu. Rev. Immunol. 1997;15:297–322. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- Yoshimoto T., Bendelac A., Watson C., Hu-li J., Paul W.E. Role of NK1.1+ T cells in Th2 response and immunoglobulin E production. Science. 1995;270:1845–1847. doi: 10.1126/science.270.5243.1845. [DOI] [PubMed] [Google Scholar]
- Rincon M., Anguita J., Nakamura T., Fikring E., Flavell R.A. Interleukin (IL)-6 directs the differentiation of IL-4–producing CD4+ T cells. J. Exp. Med. 1997;185:461–469. doi: 10.1084/jem.185.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. Cytokines acting on or secreted by macrophages during intracellular infection (IL-10, IL-12, IFN-γ) Curr. Opin. Immunol. 1997;9:17–23. doi: 10.1016/s0952-7915(97)80154-9. [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Jr. Presidential Address to The American Association of Immunologists. The road less traveled bythe role of innate immunity in the adaptive immune response. J. Immunol. 1998;161:539–544. [PubMed] [Google Scholar]
- Fearon D.T., Locksley R.M. The instructive role of innate immunity in the acquired immune response. Science. 1996;272:50–54. doi: 10.1126/science.272.5258.50. [DOI] [PubMed] [Google Scholar]
- Davis M.M., Boniface J.J., Reich Z., Lyons D., Hampl J., Arden B., Chien Y. Ligand recognition by alpha beta T cell receptors. Annu. Rev. Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- Bjorkman P.J., Saper M.A., Samraoui B., Bennett W.S., Strominger J.L., Wiley D.C. Structure of the human class I histocompatibility antigen HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Stern L.J., Brown J.H., Jardetsky T.S., Gorga J.C., Urban R.G., Strominger J.L., Wiley D.C. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- Stern L.J., Wiley D.C. Antigenic peptide binding by class I and class II histocompatibility proteins. Structure. 1994;2:245–251. doi: 10.1016/s0969-2126(00)00026-5. [DOI] [PubMed] [Google Scholar]
- Sant'Angelo D.B., Waterbury G., Preston-Hurlburt P., Yoon S.T., Medzhitov R., Hong S.-C., Janeway C.A., Jr. The specificity and orientation of a TCR to its peptide-MHC class II ligands. Immunity. 1996;4:367–376. doi: 10.1016/s1074-7613(00)80250-2. [DOI] [PubMed] [Google Scholar]
- Hong S.-C., Chelouche A., Lin R.-H., Shaywitz D., Braunstein N.S., Glimcher L., Janeway C.A., Jr. An MHC interaction site maps to the amino-terminal half of the T cell receptor α chain variable domain. Cell. 1992;69:1–20. doi: 10.1016/0092-8674(92)90618-m. [DOI] [PubMed] [Google Scholar]
- Jorgensen J.L., Esser U., Fazekas de St. Groth B., Reay P.A., Davis M.M. Mapping T cell receptor-peptide contacts by variant peptide immunization of single-chain transgenics. Nature. 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- Luescher I.F., Anguire F., Peitisch M.C., Jongeneel C.V., Cerottini J.C., Romero P. Structural analysis of TCR-ligand interactions studied on H-2Kd restricted cloned CTL specific for a photoreactive peptide derivative. Immunity. 1995;3:51–63. doi: 10.1016/1074-7613(95)90158-2. [DOI] [PubMed] [Google Scholar]
- Danska J.S., Livingstone A.M., Paragas V., Ishihara T., Fathman C.G. The presumptive CDR3 regions of both T cell receptor alpha and beta chains determine T cell specificity for myoglobin peptides. J. Exp. Med. 1990;172:27–33. doi: 10.1084/jem.172.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K.C., Degano M., Stanfield R.L., Brunmark A., Jackson M.R., Peterson P.A., Teyton L., Wilson I.A. An alphabeta T cell receptor structure at 2.5 Å and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- Garboczi D.N., Ghosh P., Utz U., Fan Q.R., Biddison W.E., Wiley D.C. Structure of the complex between human T-cell receptor, viral peptide, and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- Sloan-Lancaster J., Allen P.M. Altered peptide ligand-induced partial T cell activationmolecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 1996;14:1–27. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- Evavold B.D., Allen P.M. Separation of IL-4 production from Th cell proliferation by an altered T cell receptor ligand. Science. 1991;252:1308–1310. doi: 10.1126/science.1833816. [DOI] [PubMed] [Google Scholar]
- Evavold B.D., Sloan-Lancaster J., Hsu B.L., Allen P.M. Separation of T helper 1 clone cytolysis from proliferation and lymphokine production using analog peptides. J. Immunol. 1993;150:3131–3140. [PubMed] [Google Scholar]
- Pfeiffer C., Murray J., Madri J., Bottomly K. Selective activation of Th1- and Th2-like cells in vivo—response to human collagen IV. Immunol. Rev. 1991;123:65–84. doi: 10.1111/j.1600-065x.1991.tb00606.x. [DOI] [PubMed] [Google Scholar]
- Pfeiffer C., Stein J., Southwood S., Ketelaar H., Sette A., Bottomly K. Altered peptide ligands can control CD4 T lymphocyte differentiation in vivo. J. Exp. Med. 1995;181:1569–1574. doi: 10.1084/jem.181.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant S., Pfeiffer C., Woodard A., Pasqualini T., Bottomly K. Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J. Exp. Med. 1995;182:1591–1596. doi: 10.1084/jem.182.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin Y., Leitenberg D., Tao X., Bottomly K. Distinct biochemical signals characterize agonist- and altered peptide ligand-induced differentiation of naive CD4+ T cells into Th1 and Th2 subsets. J. Immunol. 1997;159:5802–5809. [PubMed] [Google Scholar]
- Hong S.-C., Sant'Angelo D.B., Dittel B.N., Medzhitov R., Yoon S.T., Waterbury G., Janeway C.A., Jr. The orientation of a T cell receptor to its MHC class II:peptide ligands. J. Immunol. 1997;159:4395–4402. [PubMed] [Google Scholar]
- Kaye J., Porcelli S., Tite J., Jones B., Janeway C.A., Jr. Both a monoclonal antibody and antisera specific for determinants unique to individual cloned helper T cell lines can substitute for antigen and antigen presenting cells in the activation of T cells. J. Exp. Med. 1983;158:836–856. doi: 10.1084/jem.158.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C.A., Jr., Bottomly K. Responses of T cells to ligands for the T-cell receptor. Semin. Immunol. 1996;8:109–115. doi: 10.1006/smim.1996.0013. [DOI] [PubMed] [Google Scholar]
- Dittel B.N., Sant'Angelo D.B., Janeway C.A., Jr. Peptide antagonists inhibit proliferation and the production of IL-4 and/or IFN-γ in T helper 1, T helper 2, and T helper 0 clones bearing the same TCR. J. Immunol. 1997;158:4065–4073. [PubMed] [Google Scholar]
- Swain S.L., Croft M., Dubey C., Haynes L., Rogers P., Zhang X., Bradley L.M. From naive to memory T cells. Immunol. Rev. 1996;150:143–167. doi: 10.1111/j.1600-065x.1996.tb00700.x. [DOI] [PubMed] [Google Scholar]
- Sloan-Lancaster J., Allen P.M. Significance of T-cell stimulation by altered peptide ligands in T cell biology. Curr. Biol. 1995;7:103–109. doi: 10.1016/0952-7915(95)80035-2. [DOI] [PubMed] [Google Scholar]
- Tao X., Grant C., Constant S., Bottomly K. Induction of IL-4-producing CD4+ T cells by antigenic peptides altered for TCR binding. J. Immunol. 1997;158:4327–4344. [PubMed] [Google Scholar]
- Ding Y.-H., Baker B.M., Garboczi D.N., Biddison W.E., Wiley D.C. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999;11:45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- Reinherz E.L., Tan K., Tang L., Kern P., Liu J., Xiong Y., Hussey R.E., Smolyar A., Hare B., Zhang R. The crystal structure of a T cell receptor in complex with a peptide and MHC class II. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]