Alternative Mechanisms by Which Mediator Subunit MED1/TRAP220 Regulates Peroxisome Proliferator-Activated Receptor γ-Stimulated Adipogenesis and Target Gene Expression (original) (raw)
Abstract
Mediator is a general coactivator complex connecting transcription activators and RNA polymerase II. Recent work has shown that the nuclear receptor-interacting MED1/TRAP220 subunit of Mediator is required for peroxisome proliferator-activated receptor γ (PPARγ)-stimulated adipogenesis of mouse embryonic fibroblasts (MEFs). However, the molecular mechanisms remain undefined. Here, we show an intracellular PPARγ-Mediator interaction that requires the two LXXLL nuclear receptor recognition motifs on MED1/TRAP220 and, furthermore, we show that the intact LXXLL motifs are essential for optimal PPARγ function in a reconstituted cell-free transcription system. Surprisingly, a conserved N-terminal region of MED1/TRAP220 that lacks the LXXLL motifs but gets incorporated into Mediator fully supports PPARγ-stimulated adipogenesis. Moreover, in undifferentiated MEFs, MED1/TRAP220 is dispensable both for PPARγ-mediated target gene activation and for recruitment of Mediator to a PPAR response element on the aP2 target gene promoter. However, PPARγ shows significantly reduced transcriptional activity in cells deficient for a subunit (MED24/TRAP100) important for the integrity of the Mediator complex, indicating a general Mediator requirement for PPARγ function. These results indicate that there is a conditional requirement for MED1/TRAP220 and that a direct interaction between PPARγ and Mediator through MED1/TRAP220 is not essential either for PPARγ-stimulated adipogenesis or for PPARγ target gene expression in cultured fibroblasts. As Mediator is apparently essential for PPARγ transcriptional activity, our data indicate the presence of alternative mechanisms for Mediator recruitment, possibly through intermediate cofactors or other cofactors that are functionally redundant with MED1/TRAP220.
Peroxisome proliferator-activated receptor γ (PPARγ) is a key regulator of transcriptional pathways important for adipogenesis (34). _PPAR_γ−/− mice show a total absence of both brown and white adipose tissue. Furthermore, retrovirus vector-mediated ectopic expression of PPARγ alone can stimulate mouse embryonic fibroblasts (MEFs) to undergo adipogenesis. In such cells, the expression of CCAAT/enhancer-binding protein α (C/EBPα), another key transcriptional regulator of adipogenesis, and adipogenesis markers such as aP2, fatty acid synthase (FAS), and adipsin are induced in a PPARγ-dependent manner.
PPARγ and other nuclear hormone receptors comprise a superfamily of DNA binding transcription factors. However, they also require various transcriptional coactivators to activate, in a ligand-dependent manner, transcription of the specific target genes important for cell growth, homeostasis, and differentiation (36). These transcription coactivators often exist as multiprotein complexes. They may act either through chromatin remodeling and histone modification, after recruitment by promoter-bound nuclear receptors, or at steps involving subsequent preinitiation complex formation or function (transcription initiation and elongation). Transcription coactivators that act at the level of chromatin include ATP-dependent chromatin remodeling complexes and factors that contain (or interact with) histone acetyl transferases and methyltransferases (40).
The Mediator coactivator complex, in contrast to chromatin-modifying factors, acts more directly to facilitate promoter recruitment and function of RNA polymerase II and cognate general transcription factors. First identified as a defined complex in yeast, Mediator is evolutionarily conserved and contains approximately 30 subunits (7, 26). It is believed to connect transcriptional activators with the RNA polymerase II transcription machinery and appears to be essential for most (17), but not necessarily all (9), RNA polymerase II transcription. The mammalian Mediator/thyroid hormone receptor-associated protein (TRAP) complex was first isolated through affinity purification of an epitope-tagged thyroid hormone receptor α (TRα) from HeLa cells grown in the presence of a TRα ligand (10), and the complex is similar to or identical with other more recently described complexes that include the SRB/MED-containing cofactor complex (SMCC) and the PC2, NAT, mouse mediator, ARC, CRSP, DRIP, and human mediator complexes (26). These closely related mammalian Mediator complexes have been shown to interact, through distinct subunits, with diverse transcription activators that include nuclear receptors, Sp1, SREBP, NF-κB, p53, VP16, and E1A (reviewed in reference 4).
The MED1/TRAP220 subunit of the Mediator complex shows ligand-dependent interactions, through a region containing two nuclear receptor recognition (LXXLL) motifs, with multiple nuclear hormone receptors that include TRα, vitamin D receptor, PPARγ and PPARα, retinoic acid receptor α (RARα), retinoid X receptor (RXR), farnesoid X receptor, and estrogen receptor α and β (ERα and ERβ) (10, 20, 30, 32, 43, 44). A MED1/TRAP220 LXXLL-dependent interaction between the intact Mediator complex and TRα has also been demonstrated (25). These results have suggested a broad role for the Mediator complex in nuclear receptor function. The mouse MED1/TRAP220 subunit was independently isolated as a PPARγ-interacting protein in yeast two-hybrid screens and was shown to interact, in a ligand-dependent manner, with PPARγ. MED1/TRAP220 modestly increased the transcriptional activity with a PPARγ-responsive reporter, and a fragment of MED1/TRAP220 spanning the two LXXLL motifs acted as a dominant-negative repressor, suggesting that MED1/TRAP220 is a coactivator for PPARγ (44).
We recently showed that the MED1/TRAP220 subunit of the Mediator complex is essential for PPARγ-stimulated adipogenesis and expression of adipogenesis markers in MEFs, but not for MyoD-stimulated myogenesis (11). This finding provided an example of the regulation of cell-specific transcription and differentiation events through a distinct Mediator subunit. Further biochemical analyses showed (i) that PPARγ interacts directly with the purified Mediator complex in a ligand-dependent manner, (ii) that Mediator functions directly as a transcriptional coactivator for PPARγ on a DNA template containing three copies of the DR1 PPARγ recognition site in an in vitro transcription system reconstituted with highly purified factors, and (iii) that MED1/TRAP220 serves as an essential bridge for the interaction between Mediator complex and PPARγ in vitro (11). These data suggested a potential mechanism that may account for the inability of _MED1/TRAP220_−/− MEFs to undergo PPARγ-stimulated adipogenesis. However, the precise molecular mechanisms underlying the roles of MED1/TRAP220 and the associated Mediator complex in PPARγ-stimulated adipogenesis in vivo and the mechanism by which MED1/TRAP220 and Mediator regulate PPARγ transcriptional activity remain unclear.
Here, structural and functional analyses of MED1/TRAP220 indicate, surprisingly, that a strong, direct interaction of PPARγ with Mediator through the LXXLL motifs of MED1/TRAP220 is not required for PPARγ-stimulated adipogenesis of cultured MEFs and, furthermore, that PPARγ target gene expression and recruitment of Mediator to a PPARγ response element on the aP2 promoter in undifferentiated MEFs do not require MED1/TRAP220. The minimal region required for MED1/TRAP220 function in adipogenesis is mapped to an evolutionarily conserved 530-amino-acid N-terminal region that mediates the incorporation of MED1/TRAP220 into the Mediator complex. Our data thus suggest the existence of an alternative mechanism, involving other potentially redundant cofactors or intermediate cofactors, by which MED1/TRAP220 and the associated Mediator complex regulate the expression of known PPARγ target genes, as well as the possibility of as-yet-unidentified genes that require MED1/TRAP220 for expression in adipogenesis.
MATERIALS AND METHODS
Plasmids.
A PPARγ cDNA encoding the mouse PPARγ2 isoform (505 amino acids) was used in this study. For recombinant retroviruses, PPARγ cDNA was inserted into pMSCVpuro and pMSCVhyg (Clontech). PPARγ cDNA with an N-terminal FLAG tag was cloned into pWZLneo to generate pWZLneo-F-PPARγ. FLAG-tagged and hemagglutinin (HA)-tagged full-length wild-type and mutant MED1/TRAP220 cDNAs were cloned into pWZLhygro, as described previously (11). pMSCVpuro was used to express FLAG- and HA-tagged MED1/TRAP220 truncations. For glutathione _S_-transferase (GST) pull-down assays, the use of GST-TRα and GST-PPARγ fusions has been described previously (43). For in vitro transcription/translation, MED1/TRAP220 and Myc-tagged MED7 were cloned into pGEM7. For mammalian two-hybrid assays, full-length PPARγ or TRα cDNA was cloned into pACT (Promega) to generate pVP16-PPARγ or pVP16-TRα. MED1/TRAP220 cDNA encoding amino acids 1 to 670 [MED1/TRAP220(1-670)] was cloned into pBIND (Promega) to generate pGal4-MED1/TRAP220. For immunofluorescence, cDNAs of FLAG- and HA-tagged MED1/TRAP220 truncations were placed under the control of the cytomegalovirus promoter in the pIN vector, modified from pIRESneo (Clontech). All plasmids were verified by DNA sequencing.
Cell culture, retrovirus gene transfer, and induction of adipogenesis.
All cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (Mediatech). The HeLaS cell line stably expressing FLAG-tagged PPARγ (HeLaS/F-PPARγ) was established by using the retrovirus vector pWZLneo-F-PPARγ and following the protocol described previously (6). Retrovirus-mediated gene expression in self-immortalized wild-type and _MED1/TRAP220_−/− MEFs and the induction of adipogenesis were performed essentially as described previously (11). Briefly, 2 days after cells reached confluence, they were treated with a differentiation cocktail containing 0.5 mM 3-isobutyl-1-methyl-xanthine (Sigma), 1 μM dexamethasone (Sigma), 5 μg per ml insulin (Sigma), and 0.5 μM rosiglitazone or vehicle (dimethyl sulfoxide [DMSO]) alone. Two days later, cells were changed to medium containing 5 μg per ml insulin and 0.5 μM rosiglitazone or DMSO. The medium was replenished at 2-day intervals. At day 8 postinduction, cells were stained for lipid droplets with Oil Red O (Sigma) to check for morphological differentiation. Total RNA was purified with Trizol (Invitrogen) for Northern blot analysis.
Mammalian two-hybrid assay, luciferase reporter assay, and immunofluorescence.
For mammalian two-hybrid assays, 293T cells were seeded in 24-well plates at a density of 2 × 105 cells per well. Twenty-four hours later, cells were transfected by LipofectAmine 2000. For each well, a mixture of 50 ng of the reporter plasmid pG5luc (Promega), 200 ng of pBluescript as carrier, 250 ng of pVP16-PPARγ or pVP16-TRα, and 250 ng of pGal4-MED1/TRAP220 (wild type or mutant) was used. About 16 h after the start of transfection, cells were changed to phenol red-free DMEM (Invitrogen) supplemented with 1% charcoal-stripped fetal bovine serum (HyClone) and 1 μM T3 or rosiglitazone. Each type of transfection was done in triplicate. About 40 h after the start of transfection, cells were harvested, and cell extracts were analyzed for luciferase activity, using a dual-luciferase reporter assay system (Promega). Use of the luciferase reporter assay with _MED24/TRAP100_−/− MEF cells was described previously (19). Immunofluorescence was done as described previously (12).
Affinity purification of PPARγ-associated proteins and mass spectrometry.
HeLaS cells stably expressing FLAG-tagged PPARγ (HeLaS/F-PPARγ) or vector only (HeLaS/Vec) were grown to confluence on 400- by 15-cm dishes in DMEM containing 10% bovine serum (Invitrogen). Twelve hours before cells were harvested, 0.5 μM rosiglitazone was added to the medium. All solutions for nuclear extract preparation and affinity purification were supplemented with 0.5 μM rosiglitazone. Nuclear extracts were precleared with mouse immunoglobulin G (IgG) agarose and EZview Red protein A affinity gel (Sigma) in buffer A (20 mM HEPES [pH 7.9], 180 mM KCl, 0.2 mM EGTA, 1.5 mM MgCl2, 20% glycerol, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 0.1% NP-40) supplemented with the protease inhibitors aprotinin (1 μg/ml), leupeptin (2 μg/ml), and pepstatin (0.7 μg/ml) (Roche). Precleared nuclear extracts were incubated with the anti-FLAG M2 agarose (Sigma) in buffer A at 4°C overnight. After bound proteins were washed 12 times with buffer A, they were eluted twice with 0.25 mg/ml FLAG peptide (Sigma), concentrated with a Microcon column (Millipore), and resolved with 4 to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Bio-Rad). Protein identification by mass spectrometry was performed essentially as described previously (6).
Northern blotting and immunoblotting.
Northern and immunoblot analyses were performed as described previously (11). For Northern blotting, 10 μg of total RNA isolated with Trizol was loaded into each lane. Signal intensity was measured by PhosphorImager (Fuji) and normalized to the actin signal. For immunoblotting, rabbit anti-HA (catalog no. sc-805; Santa Cruz Biotechnology) and mouse anti-PPARγ (catalog no. sc-7273; Santa Cruz Biotechnology) antibodies were used at 1:400 and 1:200 dilutions, respectively. Mouse anti-FLAG (catalog no. F3165; Sigma) was used at a concentration of 1 μg ml−1. Rabbit anti-MED7, anti-MED12, anti-MED14, anti-MED17, and anti-MED21 were used at a 1:1,000 dilution as described previously (11).
GST pull-down and ChIP assays.
GST pull-down assays of in vitro-translated [35S]methionine-labeled proteins and Mediator from nuclear extracts were conducted essentially as described previously (11). For chromatin immunoprecipitation (ChIP) assays, mouse anti-MED23/Sur2 (catalog no. 550429) and anti-PPARγ (catalog no. sc-7273) were from BD Bioscience and Santa Cruz Biotechnology, respectively. ChIP was performed in triplicate (as described in reference 24), with minor modifications. In brief, MEFs were fixed for 10 min at 25°C with 1% formaldehyde in the culture medium. Cross-linking was stopped by the addition of glycine to a final concentration of 0.2 M. Cells were washed twice with phosphate-buffered saline, scraped down, and harvested by centrifugation. The cell pellet was washed once again with phosphate-buffered saline, resuspended in lysis buffer (50 mM HEPES, pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% Na deoxycholate, 1 μg/ml each of pepstatin, leupeptin and aprotinin, and 1 mM phenylmethylsulfonyl fluoride), and vortexed for 30 min at 4°C. Whole-cell lysates were sonicated on ice with 10 cycles of 20-s pulses on and 59-s pulses off, using a microtip probe on a sonication unit (Sonic dismembrator 500; Fisher) at a power setting of 22%. Sonicated lysates were clarified by centrifugation, and the supernatant was precleared with 200 μl of protein A-agarose (Upstate) for 2 h at 4°C. Each immunoprecipitation was performed with 3 to 5 μg of purified antibodies and 1 to 5 mg of precleared lysate. After the culture was incubated overnight at 4°C, 60 μl of protein A-agarose was added, and the incubation was continued for 1 h. Precipitates were sequentially washed twice in lysis buffer, twice in lysis buffer containing 500 mM NaCl, twice in wash buffer (10 mM Tris-HCl, pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Na deoxycholate, and 1 mM EDTA), and finally twice in Tris-EDTA. Bound complexes were eluted by incubating beads twice for 5 min at 65°C in 75 μl of elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS). Cross-linking of immunoprecipitates and input samples was reversed by an overnight incubation at 65°C, after which DNA was extracted with a PCR purification kit (Qiagen). Real-time PCR quantitation of the precipitated genomic DNA relative to that of the inputs was performed in triplicate using TaqMan probes on an ABI PRISM 7900HT model system. The sequences of TaqMan probes and primers are available upon request.
RESULTS
PPARγ associates with the Mediator complex in cells.
A HeLaS cell line that stably expresses full-length mouse PPARγ with an amino-terminal FLAG epitope tag (F-PPARγ) was established using retrovirus-mediated gene transduction. Nuclear extracts were prepared and incubated with anti-FLAG M2 agarose in the presence of 0.5 μM rosiglitazone, a synthetic PPARγ ligand. After bound proteins were washed extensively, they were eluted with FLAG peptide, resolved on SDS-PAGE, and identified by mass spectrometry. As shown in Fig. 1, anti-FLAG M2 agarose immunoprecipitated multiple proteins from F-PPARγ-expressing but not from vector-expressing cells. Mass spectrometry analyses of the readily visible bands indicated that the majority were subunits of the Mediator complex. These subunits included MED1/TRAP220, MED12/TRAP230, MED13/TRAP240, MED13L/THRAP2, MED14/TRAP170, MED15/PCQAP, MED23/TRAP150β, and MED24/TRAP100, with sequence coverage of 41.5%, 25.9%, 20.7%, 13.9%, 22.6%, 9.8%, 12.6%, and 17.0%, respectively (data not shown). An immunoblot analysis also confirmed the intracellular association of F-PPARγ with other Mediator subunits as well (data not shown). These results are consistent with our previous demonstration of an in vitro interaction between PPARγ and the Mediator complex (11).
FIG. 1.
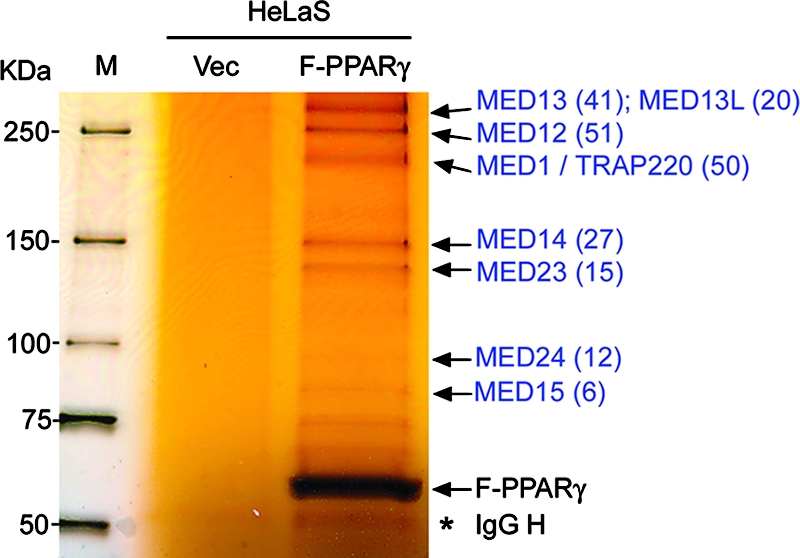
PPARγ associates with the Mediator complex in cells. Anti-FLAG M2 agarose was used to immunoprecipitate nuclear extracts of HeLaS cells stably expressing either FLAG-tagged PPARγ (F-PPARγ) or vector only (Vec). Immunoprecipitates were resolved on an SDS-PAGE gel and stained with silver. Mediator subunits identified by mass spectrometry are labeled in blue, with the number of matching tryptic peptides indicated in parentheses. M, protein molecular mass marker. The IgG heavy chain (IgG H) is marked with an asterisk.
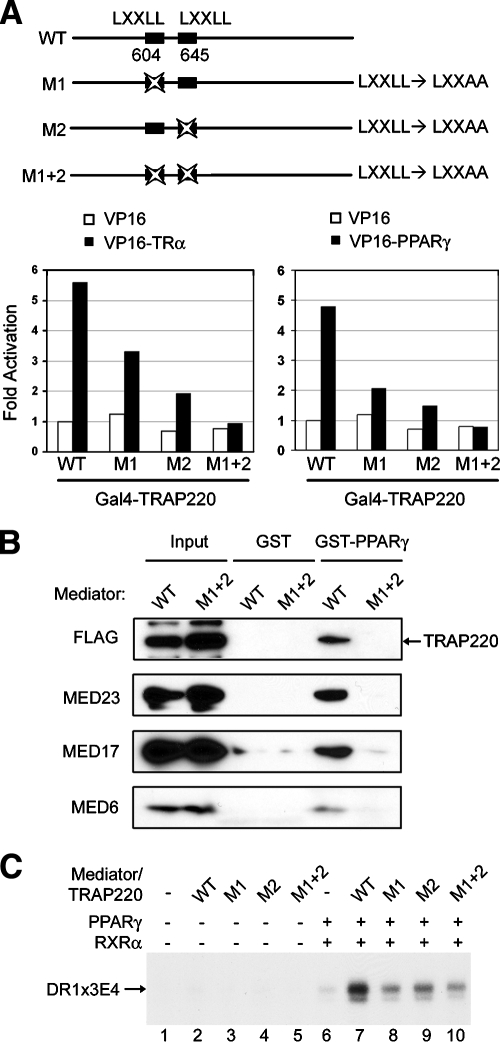
A demonstrable physical interaction between PPARγ and Mediator requires the two LXXLL nuclear receptor recognition motifs on MED1/TRAP220.
MED1/TRAP220 has two closely spaced LXXLL nuclear receptor recognition motifs (residue regions 604 to 608 and 645 to 649) in a central region that mediates ligand-dependent interactions between MED1/TRAP220 and the ligand binding domains of nuclear receptors. Ligand-dependent interactions of MED1/TRAP220 with PPARγ and (as a control) TRα in cells were confirmed by a mammalian two-hybrid assay (Fig. 2A). These interactions were significantly decreased by mutation of either of the MED1/TRAP220 LXXLL motifs to LXXAA and were essentially eliminated by the joint mutation of both motifs to LXXAA (Fig. 2A). Consistent with these results, a GST-PPARγ pull-down assay with in vitro-translated MED1/TRAP220 confirmed that the two LXXLL motifs are required for a demonstrable interaction between MED1/TRAP220 and PPARγ (data not shown).
FIG. 2.
Physical interaction between PPARγ and Mediator requires the two LXXLL motifs in MED1/TRAP220. (A) Physical interaction between PPARγ and MED1/TRAP220 requires both of the LXXLL motifs in MED1/TRAP220. Panel A top shows a schematic representation of the wild-type MED1/TRAP220 protein (WT) and the derived mutants with mutations in the two LXXLL motifs. The positions of the two LXXLL motifs are indicated. Panel A lower panels show mammalian two-hybrid assays. Plasmids expressing pVP16-PPARγ and MED1/TRAP220(1-670) fused to the Gal4 DNA binding domain (Gal4-TRAP220) were cotransfected with the reporter plasmid pG5luc into 293T cells. Interaction between TRα and the MED1/TRAP220 subunit is shown as a control. (B) PPARγ interaction with the Mediator complex requires the two LXXLL motifs in MED1/TRAP220. GST-PPARγ was incubated with nuclear extracts from _MED1/TRAP220_−/− cells expressing MED1/TRAP220(1-670), either with intact LXXLL motifs (WT) or with LXXAA mutations in both motifs (M1 + 2), in the presence of 1 μM rosiglitazone. Bound Mediator was monitored by immunoblotting with antibodies to indicated subunits. (C) The optimal PPARγ function in a purified transcription system requires both LXXLL motifs in MED1/TRAP220. Reactions with a PPRE-containing DNA template and with purified RNA polymerase II, general initiation factors, PC4, and Mediator were as described previously (11). Mediator complexes containing MED1/TRAP220(1-670) with mutations LXXLL to LXXAA in the first (M1), second (M2), or both (M1 + 2) LXXLL motifs were isolated as described previously (25).
MED1/TRAP220 serves as an essential bridge for a demonstrable physical interaction between PPARγ and the complete Mediator complex (11). To test whether the two LXXLL motifs carried on MED1/TRAP220 are also required for the interaction between the Mediator complex and PPARγ, GST-PPARγ was incubated with nuclear extracts from _MED1/TRAP220_−/− cells expressing MED1/TRAP220(1-670) with either the intact LXXLL motifs or the LXXAA mutations in both motifs. These derivatives are both efficiently incorporated into the Mediator complex (25). As shown in Fig. 2B, GST-PPARγ could bind only the endogenous Mediator complex from _MED1/TRAP220_−/− cells expressing the wild-type MED1/TRAP220(1-670). This result indicates that the PPARγ interaction with the Mediator complex requires the two LXXLL motifs on MED1/TRAP220. In an extension of previous studies showing that Mediator is required for optimal PPARγ function in a purified cell-free system (11), we analyzed Mediator complexes containing MED1/TRAP220(1-670) with the intact LXXLL motifs or with the mutated motifs (LXXAA). As shown in Fig. 2C, PPARγ (with RXRα) mediates only a minimal activation in the absence of Mediator (Fig. 2C, lane 6 versus lane 7). In contrast, Mediator with wild-type MED1/TRAP220(1-670) mediates a strong activation by PPARγ, as reported previously, whereas Mediator with a single mutation or joint LXXAA mutations in MED1/TRAP220(1-670) shows only a modest (severalfold reduced) activation by PPARγ (Fig. 2C, lanes 8 to 10 versus lane 7). Hence, while the LXXLL motifs are not absolutely required for significant Mediator-dependent activity in this purified system, which lacks many other nuclear cofactors, these motifs are required for optimal Mediator-dependent activity with PPARγ.
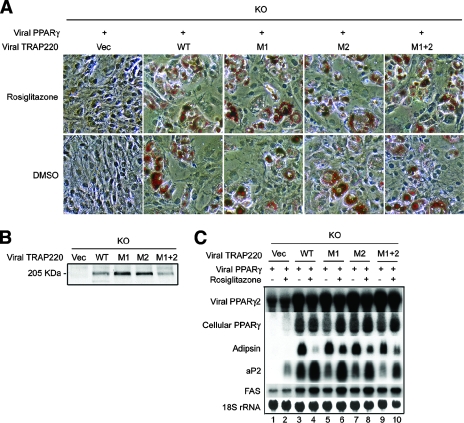
The two LXXLL motifs within MED1/TRAP220 are dispensable for PPARγ-stimulated adipogenesis.
We previously showed that _MED1/TRAP220_−/− MEF cells are refractory to PPARγ-stimulated adipogenesis and that this defect can be rescued by the expression of exogenous full-length MED1/TRAP220 (11). To determine the role of the two MED1/TRAP220 LXXLL motifs in PPARγ-stimulated adipogenesis, we tested the ability of MED1/TRAP220 with the LXXAA mutations in either or both of the two LXXLL motifs to rescue the adipogenesis defect in _MED1/TRAP220_−/− cells. Consistent with our previous report (11), ectopic expression of the wild-type MED1/TRAP220 subunit restored PPARγ-stimulated adipogenesis in _MED1/TRAP220_−/− cells, while the vector alone did not. Surprisingly, ectopic expression of MED1/TRAP220 with the LXXAA mutations in either (M1 or M2) or both (M1 plus M2) of the two LXXLL motifs also rescued the defect in _MED1/TRAP220_−/− cells (Fig. 3A). Consistent with the morphological differentiation, adipogenesis marker genes, including endogenous _PPAR_γ, adipsin, aP2, and FAS, were induced at levels that were similar in both the mutant MED1/TRAP220-expressing cells and the wild-type MED1/TRAP220-expressing cells (Fig. 3C). Similarly, deletion of either one or both of the two LXXLL motifs or deletion of short adjacent regions had little effect on the ability of exogenous MED1/TRAP220 to rescue the adipogenesis defect in _MED1/TRAP220_−/− cells (data not shown), indicating that the two LXXLL motifs and the adjacent regions on MED1/TRAP220 are not essential for PPARγ-stimulated adipogenesis of MEFs. As the two LXXLL motifs mediate the observed physical interaction between PPARγ and MED1/TRAP220 (Fig. 2A and data not shown), these data indicate that any PPARγ-MED1/TRAP220 interaction due to these residues is dispensable for PPARγ-stimulated adipogenesis. As MED1/TRAP220 serves as an essential bridge for strong in vitro interactions between PPARγ and the Mediator complex (11) and as the demonstrated interaction between PPARγ and the Mediator complex requires the two LXXLL motifs in MED1/TRAP220 (Fig. 2B), these data also suggest that the physical interaction between PPARγ and Mediator through the LXXLL motifs is not required for PPARγ-stimulated adipogenesis of cultured MEFs.
FIG. 3.
The two LXXLL motifs in MED1/TRAP220 are dispensable for PPARγ-stimulated adipogenesis. _MED1/TRAP220_−/− (knockout [KO]) cells were infected with retroviruses expressing HA-tagged full-length wild-type (WT) or MED1/TRAP220 mutants, as shown in Fig. 2A. Cells were subsequently infected with MSCVpuro-PPARγ retroviruses and induced to differentiate for 8 days in the presence or absence of 0.5 μM rosiglitazone. (A) Morphological differentiation (Oil Red O staining). Vec, vector; M, motif. (B) Immunoblot analysis of ectopic MED1/TRAP220 expression by anti-HA antibody. (C) Northern blot analysis of adipogenesis markers.
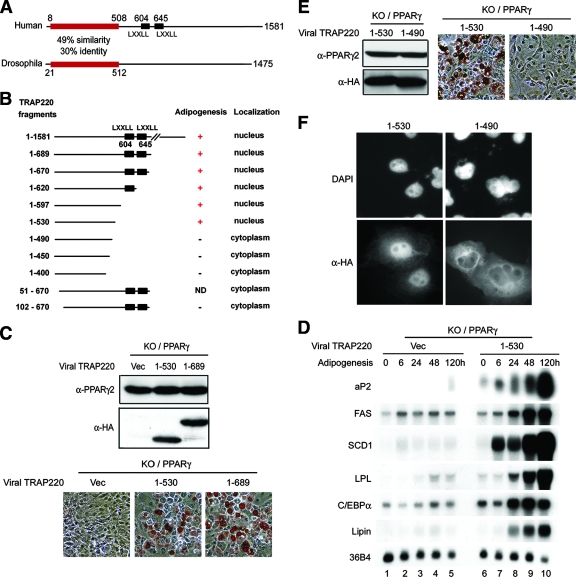
The conserved amino-terminal region (residues 1 to 530) of MED1/TRAP220 is sufficient to rescue the adipogenesis defect in _MED1/TRAP220_−/− cells.
Human and Drosophila melanogaster MED1/TRAP220 subunits share significant homology only at their N termini (Fig. 4A). To map the functionally critical region(s) within MED1/TRAP220 and to determine whether the demonstrated physical interaction between PPARγ and Mediator is required for PPARγ-stimulated adipogenesis, a series of HA-tagged MED1/TRAP220 truncations were introduced via retroviruses into _MED1/TRAP220_−/− cells that stably express ectopic PPARγ. An immunoblot confirmed the expression of MED1/TRAP220 truncations and similar levels of viral PPARγ expression in these cells (Fig. 4C and E and data not shown). These _MED1/TRAP220_−/−-derived PPARγ-expressing cells were subjected to the adipogenesis assay, and the results are shown in Fig. 4B. Interestingly, the expression of an N-terminal fragment, MED1/TRAP220(1-689), that contains the two LXXLL motifs was sufficient to rescue the adipogenesis defect in _MED1/TRAP220_−/− cells, suggesting that the C-terminal amino acids 690 to 1581 of MED1/TRAP220 are not required for PPARγ-stimulated adipogenesis (Fig. 4C, lower panel). Remarkably, an N-terminal fragment MED1/TRAP220(1-530) that contains only the evolutionarily conserved N-terminal region and is devoid of both LXXLL motifs was also sufficient to rescue the adipogenesis defect in _MED1/TRAP220_−/− cells (Fig. 4C, lower panel). Consistent with the morphological differentiation and with the LXXLL mutation analysis shown in Fig. 3C, the induction of adipogenesis marker genes was fully restored by expression of MED1/TRAP220(1-530) in _MED1/TRAP220_−/− cells (Fig. 4D). Similar results were obtained when such assays were repeated with MED1/TRAP220 fragments that contained residues 1 to 620, 1 to 597, and 1 to 560 and thus lacked either one or both of the two LXXLL motifs (Fig. 4B).
FIG. 4.
Residues 1 to 530 of MED1/TRAP220 are sufficient to rescue the adipogenesis defect in _MED1/TRAP220_−/− cells. (A) Schematic representation of homology between human and Drosophila MED1/TRAP220 proteins. The human MED1/TRAP220 protein sequence was used with BLASTp to query the NCBI protein database. (B) Summary of the structural and functional analyses of MED1/TRAP220 in the context of PPARγ-stimulated adipogenesis. _MED1/TRAP220_−/− cells expressing exogenous PPARγ were subsequently infected either with vector only or with retroviruses expressing HA-tagged MED1/TRAP220 fragments as indicated and induced to differentiate for 8 days in the absence of rosiglitazone. Similar results were obtained when the adipogenesis assay was done in the presence of 0.5 μM rosiglitazone (data not shown). (C) Residues 1 to 530 of MED1/TRAP220 are sufficient to rescue the adipogenesis defect in _MED1/TRAP220_−/− (knockout [KO]) cells. The experiments were done as described in the legend to panel B. (Top) Immunoblot before induction of adipogenesis. (Bottom) Morphological differentiation. Vec, vector. (D) _MED1/TRAP220_−/− cells expressing exogenous PPARγ were infected with retroviruses expressing MED1/TRAP220(1-530) and induced to differentiate in the absence of rosiglitazone. RNA samples collected at the indicated time points were subjected to Northern blot analysis. (E) Residues 1 to 490 of MED1/TRAP220 cannot rescue the adipogenesis defect in _MED1/TRAP220_−/− cells. The experiments were done as described in the legend to panel B. (Left) Immunoblot before induction of adipogenesis; (right) morphological differentiation. (F) Residues 1 to 490 of MED1/TRAP220 are localized to the cytoplasm. COS7 cells transfected with plasmids expressing HA-tagged MED1/TRAP220(1-530) or MED1/TRAP220(1-490) were stained with anti-HA antibody followed by Texas Red-conjugated secondary antibody. Nuclei were labeled with DAPI (4′,6′-diamidino-2-phenylindole).
Further deletion of 40 or more amino acids from the C terminus of MED1/TRAP220(1-530) resulted in a complete loss of the ability to rescue the adipogenesis defect in _MED1/TRAP220_−/− cells (Fig. 4B and E). To understand the underlying molecular mechanism, we examined the cellular localization of MED1/TRAP220 truncations. While residues MED1/TRAP220(1-530) and MED1/TRAP220(1-689) were localized almost exclusively within the nucleus, MED1/TRAP220(1-490) and shorter truncations resided predominantly in the cytoplasm (Fig. 4B and F). No classical nuclear localization signal was identified within MED1/TRAP220(1-530), and the addition of triplicate simian virus 40 (SV40) nuclear localization signals failed to relocate MED1/TRAP220(1-490) to the nucleus (data not shown), suggesting that the nuclear localization of MED1/TRAP220 may be facilitated by MED1/TRAP220-interacting subunits of the Mediator complex. As both the full-length MED1/TRAP220 subunit and the Mediator complex are known to be localized and functional within the nucleus, the cytoplasmic localization could explain, at least in part, the failure of residues 1 to 490 and shorter truncations of MED1/TRAP220 to rescue the adipogenesis defect in _MED1/TRAP220_−/− cells. Thus, MED1/TRAP220 requires only its evolutionarily conserved N terminus (residues 1 to 530) to support PPARγ-stimulated adipogenesis. As the demonstrated interaction of PPARγ with the Mediator complex is mediated by the two LXXLL motifs in MED1/TRAP220 (Fig. 2B), these data indicate that the strong ligand-dependent physical interaction between PPARγ and Mediator is not essential for PPARγ-stimulated adipogenesis in the cellular assays.
The conserved region (residues 1 to 530) of MED1/TRAP220 is incorporated into the Mediator complex but is unable to mediate a demonstrable interaction between Mediator and PPARγ.
Since residues 1 to 530 of MED1/TRAP220 comprise the evolutionarily conserved region and are essential for adipogenesis and in light of our observation that MED1/TRAP220 interacts with the Mediator complex through its N-terminal 670 amino acids (25), we speculated that residues 1 to 530 may suffice for the interaction between MED1/TRAP220 and the rest of the Mediator complex. Indeed, anti-FLAG antibody immunoprecipitated the endogenous Mediator complex from nuclear extracts prepared from _MED1/TRAP220_−/− cells expressing FLAG-tagged MED1/TRAP220(1-530) (Fig. 5A). To screen for the potential binding partner(s) of MED1/TRAP220 within the Mediator complex, in vitro-translated [35S]methionine-labeled MED1/TRAP220(1-530) and individual in vitro-translated Myc-tagged Mediator subunits were mixed and subjected to coimmunoprecipitation with anti-Myc antibody. Of the Mediator subunits tested, MED7 bound strongly to MED1/TRAP220(1-530) (Fig. 5B and data not shown). This result is consistent with the observation that yeast homologs of MED7 and MED1/TRAP220 interact directly and that both reside in the middle module of yeast Mediator (3). These data indicate that the conserved N terminus (amino acids 1 to 530) mediates MED1/TRAP220 interaction with the rest of the Mediator complex and furthermore suggest that the MED1/TRAP220 interaction with the Mediator complex is important for its function in adipogenesis. Consistent with our observation that the PPARγ interaction with the Mediator complex requires the two LXXLL motifs on MED1/TRAP220 (Fig. 2B), the conserved region containing residues 1 to 530 of MED1/TRAP220 was unable to mediate a demonstrable interaction between Mediator and PPARγ (Fig. 5C).
FIG. 5.
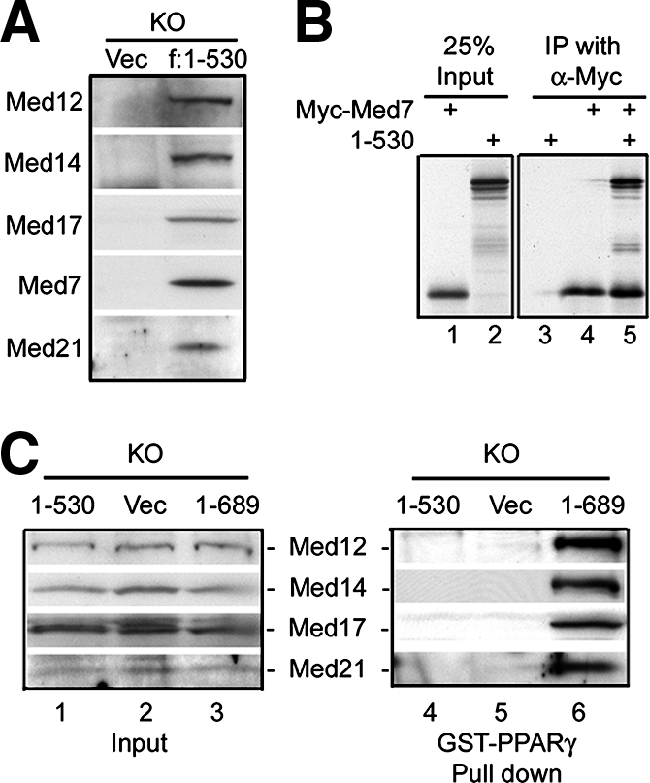
Residues 1 to 530 of MED1/TRAP220 are incorporated into the Mediator complex but are unable to mediate the interaction between Mediator and PPARγ. (A) Anti-FLAG M2-agarose was used to immunoprecipitate from nuclear extracts of _MED1/TRAP220_−/− (knockout [KO]) cells expressing FLAG-tagged MED1/TRAP220(1-530). Bound proteins were analyzed by immunoblotting with antibodies against the indicated Mediator subunits. Vec, vector. (B) The residue sequence 1 to 530 of MED1/TRAP220 interacts with the MED7 subunit of the Mediator complex. 35S-labeled in vitro-translated MED1/TRAP220(1-530) and Myc-tagged MED7 were mixed and immunoprecipitated with anti-Myc antibody. Bound proteins were washed three times with BC180/0.1% NP-40 and resolved with SDS-PAGE. (C) Mediator containing amino acids 1 to 530 of MED1/TRAP220 does not bind PPARγ. GST-PPARγ was incubated with nuclear extracts from _MED1/TRAP220_−/− (KO) cells expressing vector only, MED1/TRAP220(1-530), or MED1/TRAP220(1-689) in the presence of 1 μM rosiglitazone. Bound Mediator was monitored by immunoblotting with antibodies to the indicated subunits.
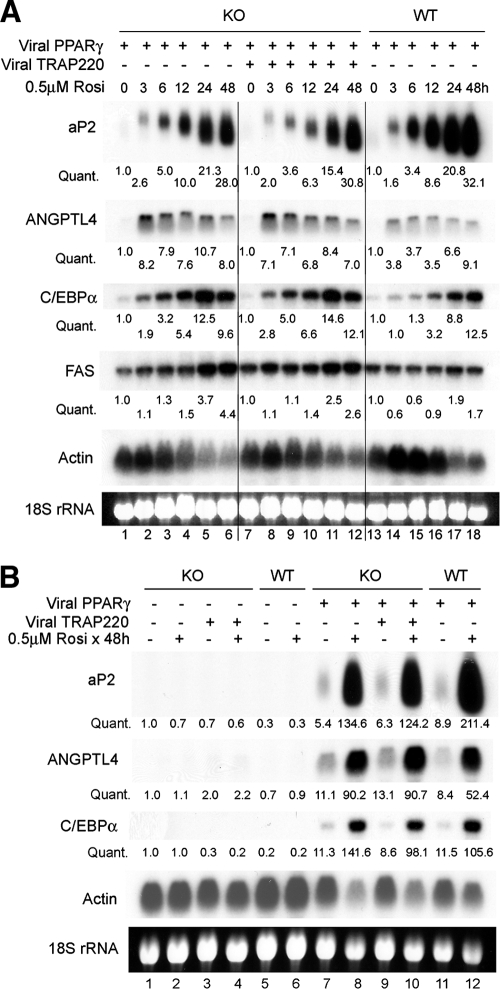
MED1/TRAP220 is not essential for PPARγ-dependent and ligand-dependent expression of PPARγ target genes in undifferentiated MEFs.
MED1/TRAP220 is important for the expression of adipogenesis markers and PPARγ target genes during adipogenesis (11). However, the dramatic induction of these genes during adipogenesis results from the combined action of multiple adipogenic transcription factors that include C/EBPα, C/EBPβ, and SREBP1 in addition to PPARγ (35). Also, there exists the possibility that the dramatic loss of the PPARγ target gene expression during adipogenesis may be an indirect consequence of the inability of _MED1/TRAP220_−/− cells to undergo adipogenesis and to express other factors important for optimal PPARγ target gene expression. Therefore, it is unclear from previous reports whether the MED1/TRAP220 requirement reflects a direct involvement in PPARγ-mediated activation of its target genes.
To begin to address this issue, we investigated the requirement of MED1/TRAP220 for PPARγ-induced activation of endogenous target genes in the absence of adipogenesis. Ectopic PPARγ was stably expressed at similar levels in wild-type cells, in _MED1/TRAP220_−/− cells, and in _MED1/TRAP220_−/− cells expressing exogenous full-length MED1/TRAP220, as described previously (11). Cells were treated with 0.5 μM rosiglitazone for 0, 3, 6, 12, 24, or 48 h, and the induction of the PPARγ target genes was monitored by Northern blotting. Importantly, to prevent the potential initiation of adipogenesis, cells were plated at low density, and samples were collected before cells reached confluence. As shown in Fig. 6A, rosiglitazone significantly induced expression of the endogenous PPARγ target genes aP2, ANGPTL4 (PGAR/FIAF), _C/EBP_α, and FAS. Strikingly, the ligand-dependent induction of endogenous PPARγ target genes was completely independent of MED1/TRAP220 status, with similar levels of induction and potency in the three cell lines. The induction of endogenous PPARγ target genes was strictly dependent on exogenous PPARγ and rosiglitazone, as no induction was detected in cells expressing the vector alone (Fig. 6B). Consistent with Northern blotting results, microarray analysis (using samples shown in Fig. 6B, lanes 2, 4, 8, and 10) failed to identify a MED1/TRAP220 requirement for the activation of any of the known PPARγ target genes by ectopic PPARγ and synthetic ligand (data not shown). Thus, MED1/TRAP220 is not essential for PPARγ to activate its target genes in MEF cells, at least under the cell culture conditions and ectopic PPARγ and synthetic ligand treatments employed. As MED1/TRAP220 serves as a bridge for a strong ligand-dependent interaction between PPARγ and Mediator, these results also indicate that a direct interaction between PPARγ and Mediator through MED1/TRAP220 is not essential for ectopic PPARγ-stimulated target gene expression in these cultured, undifferentiated MEFs.
FIG. 6.
MED1/TRAP220 is not essential for PPARγ-dependent and ligand-dependent expression of PPARγ target genes in undifferentiated MEFs. (A) MED1/TRAP220 is not essential for PPARγ-dependent and ligand-dependent expression of PPARγ target genes. Subconfluent undifferentiated MEFs were treated with 0.5 μM rosiglitazone (Rosi) for 0, 3, 6, 12, 24, or 48 h. Total RNA was extracted and subjected to Northern blotting. (B) The expression of PPARγ target genes in subconfluent MEF cells is strictly dependent on ectopic PPARγ and ligand. Cells were treated with 0.5 μM rosiglitazone for 0 or 48 h. Total RNA was extracted and subjected to Northern blotting. When total RNA was collected, cells were at ∼80 to 90% confluence (to prevent initiation of adipogenesis). KO, _MED1/TRAP220_−/− cells; WT, wild-type cells. Signal intensities after normalization to actin signal are labeled underneath.
The Mediator complex is thought to be generally required for transcription by RNA polymerase II (17). It has been shown previously that the MED24/TRAP100 component of the Mediator complex is essential in broad transcriptional events and is important for the integrity of the Mediator complex (19). Consistent with these observations, PPARγ showed significantly reduced transcriptional activity in embryonic cells deficient for MED24/TRAP100 (Fig. 7), suggesting that while the MED1/TRAP220 subunit is dispensable for PPARγ-mediated induction of target genes in undifferentiated MEFs, the intact Mediator complex is still required.
FIG. 7.
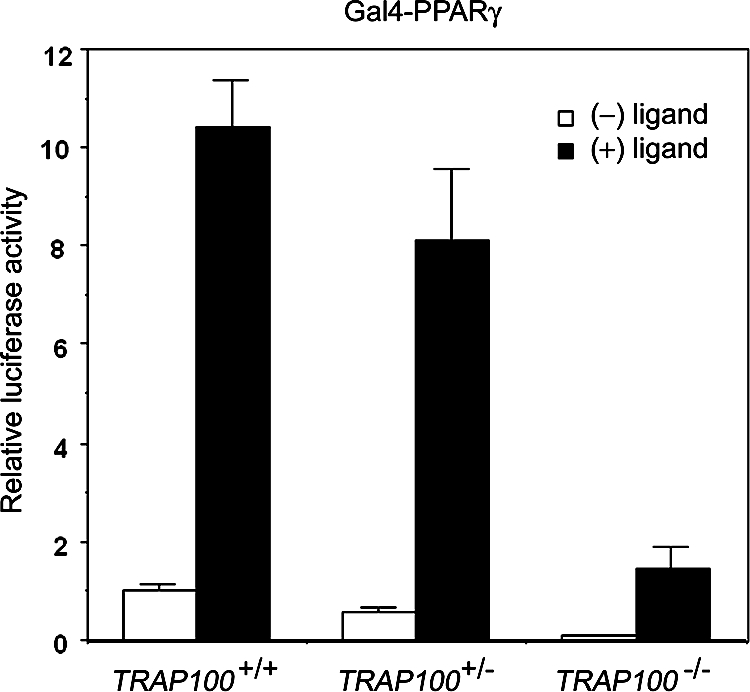
Defective PPARγ transcriptional activity in MED24/TRAP100-deficient embryonic cells. MED24/TRAP100+/+ or _MED24/TRAP100_−/− embryonic cells were cotransfected with a plasmid carrying a Gal4 DNA binding domain-PPARγ (Gal4-PPARγ) fusion protein and a luciferase reporter with Gal4-binding sites upstream of the SV40 promoter, as described previously (19). Reporter expression levels were normalized to the Renilla luciferase (control) expression levels, which were comparable for wild-type and Med24/TRAP100-deficient cells.
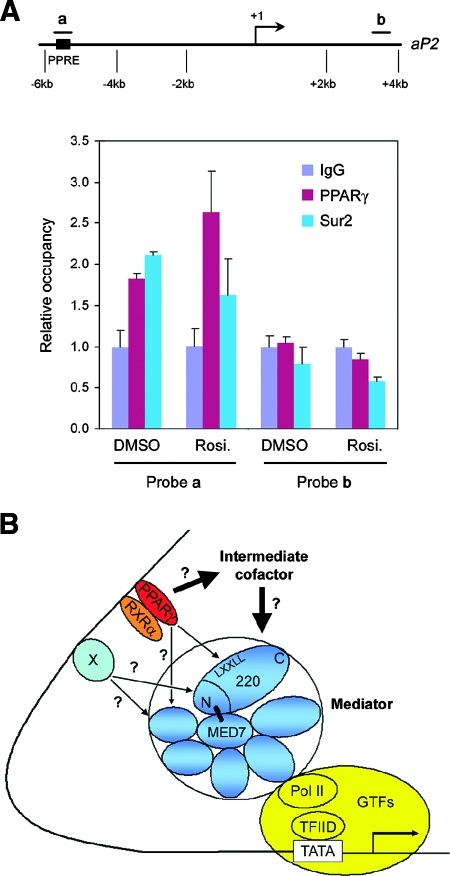
Mediator subunits, especially MED1/TRAP220, have been shown to be recruited to the PPARγ response elements (PPRE) of the PPARγ target gene aP2 (14, 31). To test whether MED1/TRAP220 is essential for the recruitment of Mediator to the aP2 promoter in undifferentiated MEFs, ChIP assays were performed with _MED1/TRAP220_−/− cells expressing exogenous PPARγ in the presence of 0.5 μM rosiglitazone. As shown in Fig. 8A, MED23/Sur2, which is an integral subunit of the Mediator complex, was recruited to the PPRE-containing region on the aP2 promoter, indicating that the Mediator complex can be recruited to the PPRE in the absence of a direct PPARγ interaction that involved MED1/TRAP220. Notably, however, PPARγ and Mediator are constitutively bound to the promoter in these cells. This result is consistent with the dispensability of MED1/TRAP220, which normally shows ligand-dependent interactions with PPARγ, and with Mediator recruitment through an alternative mechanism that could involve interactions of PPARγ or of other DNA-bound accessory factors with other Mediator subunits (see Discussion).
FIG. 8.
PPARγ and subunits of the Mediator complex are recruited to the PPARγ-response element of the aP2 enhancer in the absence of MED1/TRAP220. (A) ChIP assay. (Top) Schematic representation of the mouse aP2 promoter. The positions of TaqMan probes are indicated. (Bottom) Subconfluent undifferentiated _MED1/TRAP220_−/− MEFs expressing ectopic PPARγ were treated with 0.5 μM rosiglitazone (Rosi.) or vehicle (DMSO) alone for 24 h, followed by ChIP assays with the indicated antibodies. Immunoprecipitated DNA was quantified by real-time PCR with TaqMan probes a and b. (B) Model of mechanisms underlying MED1/TRAP220 and Mediator regulation of PPARγ-stimulated adipogenesis and gene expression.
DISCUSSION
In this report, structural and functional analyses of MED1/TRAP220 have been directed toward an understanding of the mechanisms by which the Mediator complex and the MED1/TRAP220 subunit regulate PPARγ-stimulated adipogenesis and target gene expression. We show (i) that direct, ligand-dependent physical interaction between PPARγ and Mediator (11) requires the two LXXLL motifs in MED1/TRAP220, (ii) that a direct physical interaction between PPARγ and Mediator through these motifs is not required for PPARγ-stimulated adipogenesis, (iii) that the minimal region of MED1/TRAP220 required for adipogenesis lies in the evolutionarily conserved amino-terminus (amino acids 1 to 530) that is incorporated into the Mediator complex but unable to mediate a demonstrable interaction between the PPARγ and the Mediator complex, and (iv) that in cultured, undifferentiated MEFs, MED1/TRAP220 is required neither for ectopic PPARγ-mediated activation of several known target genes nor for the recruitment of Mediator to the PPRE on the aP2 target gene. The implications of these findings for our current understanding of the Mediator function in the regulation of transcription, especially by nuclear receptors, are discussed.
Physical and functional interaction of PPARγ with mediator through the MED1/TRAP220 LXXLL motifs.
Given its critical role in activator-driven transcription of most (17), but not necessarily all (9), genes, a critical question concerns the mechanism by which Mediator is recruited to target promoters. Biochemical and genetic evidence strongly suggest a mechanism in which different (promoter-bound) activators recruit Mediator through interactions with distinct Mediator subunits (4, 21, 26). Relevant to the present study, many nuclear receptors have been shown to interact, in a ligand-dependent manner, with the Mediator MED1/TRAP220 subunit, through interactions between the liganded AF2 receptor domain and the MED1/TRAP220 LXXLL motifs (8, 20, 30, 32, 33, 38, 39, 43, 44), and studies of mutated Mediator complexes have established a key role for MED1/TRAP220 and resident LXXLL domains in strong interactions between TRα and the complete Mediator complex (25).
In the case of PPARγ, our previous studies have shown a direct ligand-dependent interaction between PPARγ and Mediator that is dependent upon MED1/TRAP220 (11). Similar to our earlier demonstration of an intracellular TRα-Mediator complex (10), we now have shown an intracellular PPARγ-Mediator interaction and, furthermore, that this interaction depends upon the LXXLL motifs in MED1/TRAP220. Beyond the physical interaction assays, transcription assays with mutant Mediator complexes in a cell-free system reconstituted with purified factors have shown that the intact MED1/TRAP220 LXXLL motifs are essential for optimal Mediator-dependent PPARγ function. These results clearly establish the potential for MED1/TRAP220 LXXLL-dependent Mediator function but do not eliminate the possibility that other cellular cofactors (not present in the purified system) might bypass the LXXLL motif requirement (see further discussion below).
In vitro differentiation of MEFs into adipocytes in the absence of the MED1/TRAP220 LXXLL motifs.
PPARγ plays a key role in adipogenesis, both in vivo and in vitro (cell culture) (reviewed in reference 34), and the in vitro MEF differentiation systems have provided powerful tools for analyzing PPARγ gene activation mechanisms through various cofactors (42). As a follow-up to our previous study demonstrating a key role for MED1/TRAP220 in PPARγ-dependent adipogenesis (11), we have analyzed the structure-function relationships of MED1/TRAP220. Remarkably, in the in vitro MEF differentiation assay, an N-terminal fragment (residues 1 to 530) that lacks both of the LXXLL motifs proved sufficient for normal levels of adipogenesis and for attendant PPARγ target gene expression. This fragment corresponds to the only phylogenetically conserved part of MED1 and, since it is incorporated normally into the Mediator complex, likely functions through the Mediator complex. MED1/TRAP220(1-530)-mediated adipogenesis is apparent in both the presence and the absence of an ectopic PPARγ ligand (rosiglitazone), such that this surprising result cannot be attributed to an abnormally (hyper-) activated PPARγ. From a mechanistic view, these results indicate that, at least in cultured cells, a strong PPARγ-Mediator interaction mediated by the MED1/TRAP220 LXXLL motifs is not required for the PPARγ functions important for adipogenesis. As discussed below, there may be alternate, possibly redundant, mechanisms for Mediator recruitment. Should this be the case and should these be completely independent of MED1/TRAP220, a key unanswered question concerns the role of the MED1/TRAP220(1-530) fragment that is essential for adipogenesis. Finally, while the MED1/TRAP220 LXXLL motifs are clearly dispensable for in vitro differentiation of MEFs to adipocytes, it remains to be determined if this is true in vivo.
PPARγ target gene activation and mediator recruitment in the absence of MED1/TRAP220.
Since the apparent MED1/TRAP220 subunit requirement for PPARγ target gene expression in adipogenesis might be indirect, a role for MED1/TRAP220 in the ligand-dependent function of ectopic PPARγ in subconfluent undifferentiated MEFs was explored. Remarkably, ligand- and ectopic PPARγ-dependent expression of a number of PPARγ target genes was found to be comparable in wild-type and _MED1/TRAP220_−/− cells. While these results do not rule out a direct role for MED1/TRAP220 in PPARγ target gene activation during adipogenesis, they clearly indicate the potential for MED1/TRAP220-independent PPARγ functions that might be relevant to endogenous PPARγ function in certain responses. Despite the lack of the MED1/TRAP220 requirement for ectopic PPARγ function in this assay, ChIP assays indicated Mediator recruitment, consistent with Mediator function, to the PPRE of the aP2 PPARγ target gene. The ligand-independent nature of this recruitment is consistent with a mechanism that does not involve MED1/TRAP220.
While the present results do not rule out Mediator recruitment and function through MED1/TRAP220 when this coactivator is present, they do suggest alternative, potentially redundant, pathways for Mediator recruitment and/or PPARγ function. Thus, as indicated in Fig. 8B, intermediate cofactors that are recruited directly by PPARγ may interact with and recruit Mediator. Such a model is also consistent with the recent report that both the SWI/SNF and the Mediator complexes can be targeted to a TR target gene through p300, which itself is recruited through interaction with SRC nuclear receptor coactivators (18). Similarly, others have reported interaction between Mediator and a group of factors that includes the nuclear receptor coactivator NCOA6/PRIP/ASC-2 (22, 37), which is a component of a histone methyltransferase complex (6, 13, 23). The demonstration of physical and functional interactions of Mediator with PPARγ coactivator α (PGC-1α) provides another example of a mechanism for indirect recruitment of Mediator (41). Significantly, however, and in agreement with the observed ligand-independent recruitment of Mediator to an RAR-activated promoter (29), Mediator recruitment to the rosiglitazone-activated aP2 promoter in undifferentiated MEFs is ligand-independent. This suggests recruitment via a PPARγ AF2 domain-independent mechanism. Potentially relevant, glucocorticoid receptor has been shown to interact in a ligand-independent manner with the MED14/TRAP170 component of Mediator (16), and recent studies indicate that MED1/TRAP220 and MED14/TRAP170 are differentially required for different GR target genes (5).
Hence, there exists the possibility of as-yet-undescribed interactions between PPARγ and other Mediator subunits. Such interactions might be substantially weaker than the ligand-dependent PPARγ-MED1/TRAP220 interaction and demonstrable only in the context of multicomponent receptor-cofactor complexes on the promoter. As indicated in the Fig. 8B model and given that most promoters employ multiple DNA-binding factors, the Mediator might be recruited by another constitutively bound factor but remain poised until other ligand-induced events occur (28, 29). Finally, recent demonstrations of Mediator-dependent basal (activator-independent) transcription and Mediator interactions with components of the general transcription machinery indicate the potential for activator-independent recruitment of the Mediator with target genes (1, 2, 27). MED1/TRAP220- and MED1/TRAP220 LXXLL motif-independent PPARγ target gene activation mechanisms, and the identification of relevant cofactors, must be the subject of future investigation.
Specialized roles for MED1/TRAP220 LXXLL domains?
The short LXXLL sequence has been identified as a signature motif that mediates the binding of a number of coactivators to the ligand binding domains of nuclear receptors (15), and there is detailed structural and functional information about the role of these motifs in nuclear receptor function in various (mostly cell-based) assays. In view of the nearly universal function of the Mediator complex in gene activation events in yeast (17), and presumably metazoans, it is notable that MED1/TRAP220 is the only Mediator subunit known to bind to the ligand-induced AF2 domains of nuclear receptors and that various in vitro assays have established the importance of the LXXLL domain for optimal receptor function. Nonetheless, as emphasized by the current study, the in vivo significance of the direct interactions between the PPARγ and the Mediator through the MED1/TRAP220 LXXLL motifs remains unclear. Such an interaction may be important for the expression of select (as-yet-unidentified) PPARγ target genes in a ligand- and-tissue-specific manner or for the expression of a broader set of target genes when the activities of alternate, potentially redundant cofactors are limiting. The resolution of this question will depend upon more complete information on all cofactors involved in PPARγ function and effects of combinatorial knockouts and (mutant) knockins of these factors.
Acknowledgments
This work was supported by NIH grant DK07190 to R.G.R. and by the Intramural Research Program of the NIDDK, NIH, to K.G.
Footnotes
▿
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Baek, H. J., Y. K. Kang, and R. G. Roeder. 2006. Human mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J. Biol. Chem. 28115172-15181. [DOI] [PubMed] [Google Scholar]
- 2.Baek, H. J., S. Malik, J. Qin, and R. G. Roeder. 2002. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol. Cell. Biol. 222842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumli, S., S. Hoeppner, and P. Cramer. 2005. A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J. Biol. Chem. 28018171-18178. [DOI] [PubMed] [Google Scholar]
- 4.Blazek, E., G. Mittler, and M. Meisterernst. 2005. The mediator of RNA polymerase II. Chromosoma 113399-408. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W., I. Rogatsky, and M. J. Garabedian. 2006. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol. Endocrinol. 20560-572. [DOI] [PubMed] [Google Scholar]
- 6.Cho, Y. W., T. Hong, S. Hong, H. Guo, H. Yu, D. Kim, T. Guszczynski, G. R. Dressler, T. D. Copeland, M. Kalkum, and K. Ge. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 28220395-20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conaway, R. C., S. Sato, C. Tomomori-Sato, T. Yao, and J. W. Conaway. 2005. The mammalian mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30250-255. [DOI] [PubMed] [Google Scholar]
- 8.Coulthard, V. H., S. Matsuda, and D. M. Heery. 2003. An extended LXXLL motif sequence determines the nuclear receptor binding specificity of TRAP220. J. Biol. Chem. 27810942-10951. [DOI] [PubMed] [Google Scholar]
- 9.Fan, X., D. M. Chou, and K. Struhl. 2006. Activator-specific recruitment of mediator in vivo. Nat. Struct. Mol. Biol. 13117-120. [DOI] [PubMed] [Google Scholar]
- 10.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 938329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge, K., M. Guermah, C. X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417563-567. [DOI] [PubMed] [Google Scholar]
- 12.Ge, K., and G. C. Prendergast. 2000. Bin2, a functionally nonredundant member of the BAR adaptor gene family. Genomics 67210-220. [DOI] [PubMed] [Google Scholar]
- 13.Goo, Y. H., Y. C. Sohn, D. H. Kim, S. W. Kim, M. J. Kang, D. J. Jung, E. Kwak, N. A. Barlev, S. L. Berger, V. T. Chow, R. G. Roeder, D. O. Azorsa, P. S. Meltzer, P. G. Suh, E. J. Song, K. J. Lee, Y. C. Lee, and J. W. Lee. 2003. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 23140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan, H. P., T. Ishizuka, P. C. Chui, M. Lehrke, and M. A. Lazar. 2005. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 19453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387733-736. [DOI] [PubMed] [Google Scholar]
- 16.Hittelman, A. B., D. Burakov, J. A. Iniguez-Lluhi, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 185380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95717-728. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Z. Q., J. Li, L. M. Sachs, P. A. Cole, and J. Wong. 2003. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and mediator for transcription. EMBO J. 222146-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, M., H. J. Okano, R. B. Darnell, and R. G. Roeder. 2002. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 213464-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, Y. K., M. Guermah, C. X. Yuan, and R. G. Roeder. 2002. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl. Acad. Sci. USA 992642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, Y. J., and J. T. Lis. 2005. Interactions between subunits of Drosophila Mediator and activator proteins. Trends Biochem. Sci. 30245-249. [DOI] [PubMed] [Google Scholar]
- 22.Ko, L., G. R. Cardona, and W. W. Chin. 2000. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. USA 976212-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S., D.-K. Lee, Y. Dou, J. Lee, B. Lee, E. Kwak, Y.-Y. Kong, S.-K. Lee, R. G. Roeder, and J. W. Lee. 2006. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc. Natl. Acad. Sci. USA 10315392-15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mai, B., S. Miles, and L. L. Breeden. 2002. Characterization of the ECB binding complex responsible for the M/G1-specific transcription of CLN3 and SWI4. Mol. Cell. Biol. 22430-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik, S., M. Guermah, C. X. Yuan, W. Wu, S. Yamamura, and R. G. Roeder. 2004. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol. Cell. Biol. 248244-8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30256-263. [DOI] [PubMed] [Google Scholar]
- 27.Mittler, G., E. Kremmer, H. T. Timmers, and M. Meisterernst. 2001. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, S. W., G. Li, Y.-P. Lin, M. J. Barrero, K. Ge, R. G. Roeder, and L.-N. Wei. 2005. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol. Cell 19643. [DOI] [PubMed] [Google Scholar]
- 29.Pavri, R., B. Lewis, T. K. Kim, F. J. Dilworth, H. Erdjument-Bromage, P. Tempst, G. de Murcia, R. Evans, P. Chambon, and D. Reinberg. 2005. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell 1883-96. [DOI] [PubMed] [Google Scholar]
- 30.Pineda Torra, I., L. P. Freedman, and M. J. Garabedian. 2004. Identification of DRIP205 as a coactivator for the Farnesoid X receptor. J. Biol. Chem. 27936184-36191. [DOI] [PubMed] [Google Scholar]
- 31.Qi, C., S. Surapureddi, Y. J. Zhu, S. Yu, P. Kashireddy, M. S. Rao, and J. K. Reddy. 2003. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor gamma (PPARgamma)-interacting protein, is required for PPARgamma-mediated adipogenesis. J. Biol. Chem. 27825281-25284. [DOI] [PubMed] [Google Scholar]
- 32.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Naar, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398824-828. [DOI] [PubMed] [Google Scholar]
- 33.Ren, Y., E. Behre, Z. Ren, J. Zhang, Q. Wang, and J. D. Fondell. 2000. Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol. Cell. Biol. 205433-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosen, E. D., C. H. Hsu, X. Wang, S. Sakai, M. W. Freeman, F. J. Gonzalez, and B. M. Spiegelman. 2002. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 1622-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosen, E. D., and B. M. Spiegelman. 2000. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 16145-171. [DOI] [PubMed] [Google Scholar]
- 36.Rosenfeld, M. G., and C. K. Glass. 2001. Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem. 27636865-36868. [DOI] [PubMed] [Google Scholar]
- 37.Surapureddi, S., S. Yu, H. Bu, T. Hashimoto, A. V. Yeldandi, P. Kashireddy, M. Cherkaoui-Malki, C. Qi, Y. J. Zhu, M. S. Rao, and J. K. Reddy. 2002. Identification of a transcriptionally active peroxisome proliferator-activated receptor alpha-interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proc. Natl. Acad. Sci. USA 9911836-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taatjes, D. J., T. Schneider-Poetsch, and R. Tjian. 2004. Distinct conformational states of nuclear receptor-bound CRSP-Med. complexes. Nat. Struct. Mol. Biol. 11664-671. [DOI] [PubMed] [Google Scholar]
- 39.Treuter, E., L. Johansson, J. S. Thomsen, A. Warnmark, J. Leers, M. Pelto-Huikko, M. Sjoberg, A. P. Wright, G. Spyrou, and J. A. Gustafsson. 1999. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J. Biol. Chem. 2746667-6677. [DOI] [PubMed] [Google Scholar]
- 40.Vaquero, A., A. Loyola, and D. Reinberg. 2003. The constantly changing face of chromatin. Sci. Aging Knowledge Environ 2003:RE4. [DOI] [PubMed]
- 41.Wallberg, A. E., S. Yamamura, S. Malik, B. M. Spiegelman, and R. G. Roeder. 2003. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell 121137-1149. [DOI] [PubMed] [Google Scholar]
- 42.Wu, Z., E. D. Rosen, R. Brun, S. Hauser, G. Adelmant, A. E. Troy, C. McKeon, G. J. Darlington, and B. M. Spiegelman. 1999. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 3151-158. [DOI] [PubMed] [Google Scholar]
- 43.Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 957939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu, Y., C. Qi, S. Jain, M. S. Rao, and J. K. Reddy. 1997. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 27225500-25506. [DOI] [PubMed] [Google Scholar]