Priorities for nucleotide trace, sequence and annotation data capture at the Ensembl Trace Archive and the EMBL Nucleotide Sequence Database (original) (raw)
Abstract
The Ensembl Trace Archive (http://trace.ensembl.org/) and the EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/), known together as the European Nucleotide Archive, continue to see growth in data volume and diversity. Selected major developments of 2007 are presented briefly, along with data submission and retrieval information. In the face of increasing requirements for nucleotide trace, sequence and annotation data archiving, data capture priority decisions have been taken at the European Nucleotide Archive. Priorities are discussed in terms of how reliably information can be captured, the long-term benefits of its capture and the ease with which it can be captured.
INTRODUCTION
In Europe, the major public repositories for nucleotide trace, sequence and annotation are the EMBL Nucleotide Sequence Database (EMBL-Bank) and the Ensembl Trace Archive (ETA). Together, the two repositories are known as the European Nucleotide Archive (ENA). The remit of ENA is the capture and public presentation of nucleotide sequencing data, including traces, quality scores, assembly information, biological annotation and metadata. This data capture is achieved both by the provision of submission environments tailored to the needs of data generating communities and by close collaboration with the other major public repositories across the world, namely the DNA Databank of Japan (1) and GenBank (2), each of which are partners in the International Nucleotide Sequence Database Collaboration (INSDC), and the NCBI Trace Archive (3).
During 2007, the ENA has seen continued growth in both the volume and nature of data represented; at the time of going to press, the archive comprised over 1.7 billion records covering almost 1.7 trillion (1.7 × 1012) base pairs of sequence.
As the ENA continues to grow, new challenges are presented. Novel data types, including those from the ultra-high-throughput sequencing technologies and metagenomics, each demand dedicated data structures and procedures for their handling. The increased volumes of data demand rationalization of exactly which pieces of information are captured, which are validated manually or automatically and which are stored in the archives.
In this article, we present a summary of the major milestones and achievements of 2007, provide submission information and offer a summary of data presentation across ENA. In the remaining part of the document, we outline the data capture priorities that we have established to guide our activities in order to accommodate future growth in data volume and diversity.
MAJOR DEVELOPMENTS IN 2007
Both ETA and EMBL-Bank have continued to grow exponentially in 2007. At the time of going to press, ETA holds 1.6 billion traces from almost 1000 organisms, covering 1.5 trillion base pairs, while EMBL-Bank holds 103 million entries from ∼300 000 organisms, covering 182 billion base pairs. Strong areas of growth include metagenomic data and genomic data. The genomes webserver, http://www.ebi.ac.uk/genomes/, which offers a collection of genomes drawn from EMBL-Bank sequence entries, cites 4192 completely sequenced organisms, including 52 eukaryotes, 517 bacteria, 47 archaea and 1814 phage, virus and viroid genomes.
A number of key datasets have been newly presented in 2007 that are of importance to agriculture, medicine, industry, ancient DNA and ecology. Notable examples include a working draft WGS dataset from grape (Vitis vinifera cultivar PN40024, CAAP00000000), WGS data and first assembly of the horse genome (Equus caballus, CM000377), two further assembled and annotated genomes of the parasite Leishmania [_Leishmania infantum_, AM502219-AM502254 and Leishmania braziliensis, AM494938-AM494972; (4)], the industrially important fungus, Aspergillus niger CBS 513.88 [AM270980–AM270998, AM269948–AM270415; (5)], a key ancient DNA study, the mammoth fossil metagenome [CAAM00000000; (6)], the CAMERA global ocean survey metagenome trace and WGS data [AACY00000000; (7)] and human whole-genome re-sequencing project data (ABBA00000000).
Launched late in 2007, the EMBL submission portal (http://www.ebi.ac.uk/embl/Submission/index.html) provides a single point of entry for submitters of all ENA data types. A single user account is used across ENA submissions; users can register genome and metagenome projects, initiate submissions to ETA, launch and track Webin submission sessions for new EMBL-Bank data, report updates to existing records and launch and track Webin submission sessions for alignment data.
Finally, search tools and data downloads based around the ENA Project Database have been made available from the genomes webserver.
DATA SUBMISSION
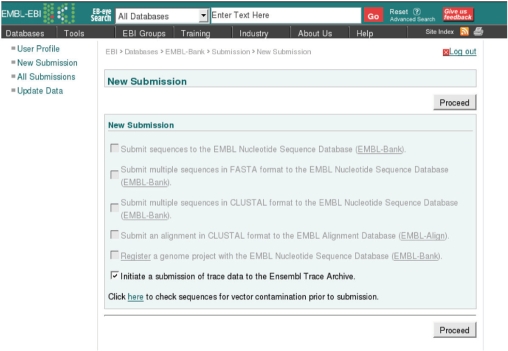
All registration, submission and update functionalities are available from the EMBL submission portal (Figure 1 and Table 2).
Figure 1.
EMBL submission portal. The screenshot shows the submission portal top page, where submission options are presented to the user.
Table 2.
Points of entry to the European Nucleotide Archive—submissions, retrieval and support
Project registration
For large-scale genome and metagenome sequencing projects, projects are registered by submitters as early as possible. At the time of registration, a unique project identifier is issued that can be used to refer collectively to data submitted as part of that project. The project identifier is of value for users of trace data presented as part of the ETA and sequence and assembly data presented as part of EMBL-Bank. At the time of registration, various project metadata are collected that cover the taxonomic details of the project, the purpose of the study and for single organism genome projects, details of replicons and expected sizes. As time goes on, metadata can be updated by submitters. When sequence data are generated, they are associated with the project identifier through the ‘Universal_project_ID’ field in ETA and in PR lines in EMBL flatfile format. Further details are available at: http://www.ebi.ac.uk/embl/Documentation/project_guidelines.html.
EMBL-Bank submission
The central submission tool for annotated sequence is Webin. While Webin has been in service for many years, it is undergoing redevelopment. Webin currently offers small-scale submission, ‘bulk’ submission, where the submitter describes a representative sample entry and submits variable field data subsequently using web forms or file upload and submission of novel sequence in multiple alignment formats. In the coming year, the underlying technology will be replaced to allow for new functionalities to be implemented. These new functionalities will be rolled out over time in due course, but will include the option to update existing records, an intuitive wizard to describe splicing and coding events and the increased use of standard and personalised templates for the rapid submission of multiple sequences from large-scale studies.
Trace submission
Files for trace data submission created by data submitters can be integrated into ETA. Ahead of submitting data, if the data are from a new sequencing project, then an ENA Project should be registered. Submitters should then e-mail trace-submission@ensembl.org, or follow links from the EMBL submission portal, to discuss the logistics of the submission with ETA staff.
DATA PRESENTATION
Data are presented in a host of ways in order to satisfy as many diverse user needs as possible. Traces, sequence entries and project records are available for retrieval by identifier; sequence similarity search tools are offered for both ETA and EMBL-Bank data; the term search tool, SRS, is offered for EMBL-Bank and project records. For large-scale users, datasets are available for FTP download or can be sent out in tape format. Where entries are organized into genomes, genome level services are presented at the EMBL genomes webserver. URLs to the appropriate resources are given in Table 2.
DATA CAPTURE PRIORITIES
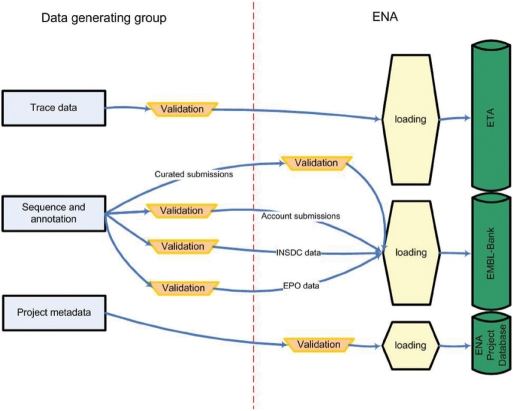
Data flow into the European repositories through a variety of pipelines (Figure 2). For ETA, data are submitted directly by e-mail or FTP and additional data are captured through regular exchange of new records with the NCBI Trace Archive. For EMBL-Bank, there are curated submissions, account submissions, daily exchange of new data with DDBJ and GenBank, European Patent Office data input and cross-reference mapping data captured from external bioinformatics resources.
Figure 2.
Input pipelines into ENA. Data input pipelines into ENA are shown; the left-hand side of the diagram represents the work of the data generating group and the right-hand side the work of ENA; validation refers to the semantic content of data transferred, particularly the biological content, and loading refers to syntactic checking of data and upload into the appropriate database.
At the start of data collection in the 1980s, extensive manual work was required to load data into the database. It soon became clear that the volume of data expected would require compromises with respect to the amount of time spent by staff validating data prior to loading. Clearly, the exponential growth of the database has not been reflected in a similar expansion of the team responsible for data loading. When automated sequencing became available, it was recognized that the newly established large sequencing centres would be able to provide ample bioinformatics expertise, given the scale of their operations and the nature of their funding, to produce validated database-ready files for loading with minimal intervention from database staff. This data input pipeline gave birth to the system of submission accounts, of which the database now has 104. Similar reasoning was applied when ETA was established in 2001 and responsibility for validation of submitted trace data lies with the submitting group.
While EMBL-Bank submission account holders and trace submitters have largely been able to satisfy the requirements of validation prior to database loading, changes in the nature of the sequencing community have begun to present new challenges. High-throughput sequencing became increasingly available to more and more research groups, through advancing technology and reduced cost. Currently, the sequencing of prokaryotic and simple eukaryotic genomes and the generation of large amounts of sequence data peripheral to genomes, such as those from EST and cDNA library sequencing and mapping projects, are not restricted to the large sequencing centres. Preferring to provide in-house validation rather than to rely on the now smaller and more constrained sequencing teams for these new datasets, EMBL-Bank chose to route this new breed of large-scale submissions through its curated submissions pipeline.
Although curated submissions to EMBL-Bank currently contribute only a limited share of data to ENA, we feel that the future growth of this pipeline and the impact it has on other data input pipelines (such as the transfer of practices and technology to other EMBL-Bank pipelines and the influence it has on practices and technology at collaborators’ sites), warrant its streamlining. Efforts have therefore been underway for some time in the development and implementation of validation technology and future developments in this area have now been prioritized. In the long term, we expect the curated submissions pipeline to draw on both robust high-capacity automation and manual curation in areas where biological decision-making and communications with submitters remain essential.
For each data element that might be captured by the database, three measures have been considered in the setting of priorities; the long-term value of capturing the data element, the ease with which it can be captured and the extent to which it can be validated semantically. High priority can easily be given to those elements with high long-term value, simple capture and high semantic validation potential. For those elements, however, that show limited long-term value, that are difficult to capture or have limited semantic validation potential, the decision to capture or not is less straightforward. Some of the priority decisions that ENA has made in this context are described in this section. For the purposes of this discussion, ‘validation’ refers to the checking of semantic integrity (both intrinsically and in relation to broader knowledge) and is mainly concerned with biological concepts, while ‘loading’ refers to the syntactic checking of content against database constraints and the subsequent population of database tables.
Trace and sequence data
ETA records present sequencing traces, quality scores and base calls (sequence). Next-generation sequencing technologies promise large volumes of very different data and it is likely that some representation of quality will be stored along with sequence, although the technology for dealing with these data types is still under development. While base quality scores can be stored, sequence alone is mandatory in EMBL-Bank records. Sequence is largely stored and presented as submitted to ENA, although for curated submissions to EMBL-Bank, sporadic vector contamination and source organism checks are run. Other than simple syntactic checking, traces, quality scores and sequence will not be further validated.
Trace data for experimental (non-assembly) applications
We expect that the decreasing cost of sequencing will herald extensive use of sequencing as a general assay for many types of experiment, whereas ‘traditional’ sequencing has generally been focused on generating assembled reference genome or mRNA sequence. Such experiments include sequencing for gene expression, the sequencing of chromatin immunoprecipitation preparations (so called ‘chIP-seq’) and sequencing of clonally barcoded populations (such as yeast knockout cell lines) in high-throughput experiments. Although the cost of sequencing for each application will be low (hence the feasibility of the experiment), the overall experiment is likely to remain comparatively expensive and the sequence will commonly be of interest to many researchers; this is particularly true of chIP-seq and gene expression datasets. We expect to store the trace information in an appropriate short-read format in ETA, and specifically not in EMBL-Bank, and associate the information with metadata relating to the parent experiment. In the case of gene expression, for example, metadata will conform to the ‘sample annotation’ components of the MIAMI standard (8). It is likely that the needs of the experimental community will drive the development of new standards, in particular through the interactions between reviewers and journals in publications covering standard classes of experiment. We expect from informal publisher contacts that the journals will have continued demand for the archiving of these experimental data for future use.
Clerical information
Clerical information includes all information that is added to archival records for the purposes of tracking, including submission information, entry and sequence versioning, EMBL-Bank taxonomic division and dataclass, trace identifiers and accession numbers. It is typically represented in the trace information section in ETA records and in the header section in EMBL flatfile format. Included in clerical information are also free text description fields and keywords (DE and KW lines in EMBL flatfile format), both of which serve to replicate information already associated with records, rearranged for the purposes of search and presentation. Clerical information can be generated in a highly automated way upon data loading and is of great user value, so ENA continues to build on existing automation in this area.
Source metadata
Biological source metadata provide details of the physical source of nucleic acid molecules represented in the nucleotide archives and are key to the interpretation of sequences in the analysis of expression, biodiversity, evolutionary biology and variation. Included are details of which molecule has been sequenced, library information, where the molecule was obtained, which isolate of the molecule's parent organism was used for extraction, taxonomy of the parent organism, details of the relationship to any host, ecology, community structure and environmental data and so on (Table 1). The information has great user value, both for highly specific searches and for the large-scale download of datasets to feed secondary resources. The extent to which data elements can be validated automatically is limited, though, and a diversity of validation procedures is required for breadth of coverage. Given the value, ENA has prioritized the development of procedures and supporting technology for the capture and validation of accurate source metadata, but expects that much of it will continue to require a degree of manual curation to achieve full value.
Table 1.
Description of source metadata in EMBL-Bank
| Qualifier type | Qualifiers |
|---|---|
| Molecule-related | /mol_type*, /chromosome, /segment, /clone, /clone_lib, /map, /PCR_primers***, /sub_clone, /tissue_library |
| ‘Parent’ setting | /virion, /proviral, /germline, /rearranged, /focus, /macronuclear, /organelle**, /plasmid |
| ‘Parent’ organism | /organism*, /strain, /variety**, /cultivar, /sub_species**, /sub_strain, /transgenic, /ecotype, /identified_by, /pop_variant, /serotype, /serovar, /sex, speciment_voucher***, /culture_collection***, /bio_material*** |
| ‘Parent’ resolution | /dev_stage, /tissue_type, /cell_line |
| Sampling | /environmental_sample, /lat_lon**, /collected_by, /isolate, /collection_date, /country***, /isolation_source, /lab_host |
| Integrative | /citation**, /db_xref, /label, /specific_host |
| Miscellaneous | /note |
One area of source annotation that has developed rapidly over the last few years is the description of environmental sampling associated both with anonymous sequencing from the environment and with ecological studies. Such ETA fields as Latitude and Longitude and EMBL-Bank qualifiers as /environmental_sample, /lat_lon and /collected_by have been introduced since 2001 to capture sampling information. Based on the information in these fields, such services as EMBL World have been developed (Table 2).
For access to source material for subsequent study, ENA values the use of those source fields that refer users to items in physical collections with long-term commitments to storage. Such centres include herbaria, culture collections, stock centres and museum collections. In EMBL-Bank, the qualifier/specimen_voucher has existed for some time, and in October 2007, a granular structure has been imposed, allowing users to cite source institute from a controlled list, collection and accession number. In December 2007, the new qualifiers /culture_collection and /bio_material will be introduced with similar structures.
Biological annotation
The understanding of the biological function of components of nucleotide sequence is the ultimate intention of many of the sequencing studies represented in ENA. Biological feature annotation is the representation of this understanding against coordinate-defined regions of submitted sequence. Within ENA, biological annotation is presented in EMBL-Bank, both in association with submitted sequence (primary records) and with Third Party Annotation [TPA, where researchers who have not generated sequence themselves present alternative annotation of existing sequence, derived from peer reviewed studies where direct, or inferred, experimental data are generated; (9)]. The EMBL feature table offers around 60 feature keys to describe biological features. Links to a feature definition browser and the Feature Table Definition document are given in Table 2.
The capture of annotation of biological functions is important, although the extent to which many of the annotations can be validated computationally is limited. Transcription, splicing and coding, for example, are well understood and nature offers firm rules for such processes as translation that provide us with powerful validation tools. Other biological features, such as promoters and enhancers, on the other hand, offer limited known options for systematic validation. In general, there are two principles that can be followed to provide computational validation of annotation of biological features; ab initio rules and validation of annotation by inference from homologous sequences. Ultimately, it is experimental confirmation of function that gives credibility to functional annotation and therefore EMBL-Bank sees the capture of experimentally derived annotation as a high priority; the body of experimentally validated features in EMBL-Bank provides a clean dataset for the development of bioinformatic tools to establish function across the large volumes of unannotated sequence, for which the means are not available to run programmes of experimental interpretation.
To reflect the above, the recording of the nature of evidence for a particular annotated feature is considered to be very important to EMBL-Bank and is under continual review. The convention of annotation by inference, where the /inference qualifier is used to provide a controlled hierarchical description of the inference (such as details of an object in a remote database to which the sequence in question is related or the ab initio tool which has been used to interpret the sequence in question) has been further developed in 2007 to begin to restrict the second term of the controlled vocabulary. The description of experimental evidence for features, using the /experiment qualifier, does not as yet take advantage of a restricted vocabulary; while this would be desirable, ENA was not able to find a pre-existing vocabulary with the breadth required. Work continues within the submission environment development to ensure that generators of annotation are able easily to contribute /inference and /experiment information across their submissions.
Coding sequence annotation is among the most important areas in which ENA can serve users; not only for direct users of EMBL-Bank, but also for users of UniProtKB, for which EMBL-Bank coding regions are a primary source (10). Much is done to ensure that representations of coding sequence are biologically accurate. Coding sequence (CDS) features, for example, have for many years been validated in all entries for appropriate start and stop codons, taking into account translation table choice, any known translation exceptions, ribosomal slippage and RNA editing. More recently, technology has been implemented at EMBL-EBI that ensures that annotated post-translational cleavage events are validated for out-of-frame cleavage events. Where possible, gene symbols and product names from established nomenclatures are captured, along with systematic /locus_tag identifiers and EC numbers (see ‘Nomenclatures and Ontologies’, below).
In late 2007, EMBL-Bank introduced the new feature, ncRNA, for non-protein-coding RNA genes. This new feature consolidates a number of pre-existing non-coding gene features, scRNA, snRNA and snoRNA, and draws in gene types that previously did not have features, such as microRNA. Given their extensive existing use, rRNA and tRNA features will remain. While computational tools exist to validate some families of non-coding genes, their application in the validation of EMBL-Bank annotation has generally not yet taken place since specific technical implementation needs to be developed for each tool. For ribosomal RNA annotation, though, all curated 16S and 18S rRNA submissions are subject to homology comparison validation (for feature length and orientation) against reference sets of manually selected genes. We expect to be able to extend the reference set to cover additional rRNA genes and are currently working with DDBJ and GenBank to establish a minimal validation standard for all rRNA annotations.
Gene splicing patterns (shown as intron, exon and mRNA features in EMBL flatfile format) provide users with important information. While consensus splice sequences are known, the existence of consensus sites where splicing is not seen and of large numbers of splice sites that deviate significantly from the consensus make automated validation of annotated splice sites difficult. Clearly, for the coding regions of protein-coding genes, translation constraints provide some validation of splice patterns, but there is no systematic computational validation available outside these regions. Investigations into how evidence for splice sites can best be captured reliably (using /inference and /experiment qualifiers) from submitters will take place in the coming year, but it is likely that responsibility for validation of reported splice events will be delegated to such resources such as the Alternative Splicing and Transcript Diversity database (11) and Ensembl (12).
A number of structures exist for the representation of repeated sequence in EMBL-Bank, covering satellites, microsatellites, Alu repeats and so on. Work is currently underway at the INSDC to improve the classification of repeats. Although we will strive to provide the appropriate annotation structures for repeated sequences, because repeats are commonly imperfect, it is not currently considered practical to establish any further validation.
Signal and bind features, such as promoter, CAAT_signal, GC_signal, RBS, polyA_signal, attentuator, enhancer, primer_bind and protein_bind, have established structures within EMBL-Bank. For some, strong rules may be formulated for their prediction; these rules may require knowledge of adjacent features. For others, extensive experimental work along with the study of homologues will be required. These features, then, will be the focus of rationalization over the coming years; for those signals that are seen to be entirely predictable, we will either implement validation for stored annotation or will not store them, but rather generate them as part of data presentation; for those signals that cannot be predicted, we will investigate ways in which we can capture structured evidence for the submitted annotation.
Integration data
ENA, particularly EMBL-Bank, offers integration with broader bioinformatic data through a number of structures that are briefly reviewed in this section. The importance of coverage and synchrony will continue to guide the development of integration structures. EMBL-Bank approaches integration in a very different way from its INSDC partners; systematic cross-references are sought from external resources to which EMBL-Bank will cross-refer and are not accepted from sequence data submitters, internal cross-references to EMBL-Bank and ETA records are offered and integration with the suite of EBI tools is central to the integration service. These structures are presented in EMBL-Bank flatfile and XML formats and in SRS and DBfetch HTML views, are marked up with hyperlinks to the appropriate services.
References to the literature have been explicit in EMBL-Bank from the outset. With the increasing diversity of publication media and availability of traditional literature online, a number of developments at EMBL-Bank are underway to maximize the utility of literature references. Already in existence is technology that maps stored publications in EMBL-Bank to records from remote resources via the PubMed identifier system maintained by the US National Library of Medicine. This yields cross-references, seen as RX lines in EMBL flatfile format, from each publication cited in an EMBL-Bank sequence entry to online literature services at the EBI, namely SRS (Table 2) and CitExplore (http://www.ebi.ac.uk/citexplore/). These two services provide similar reciprocal cross-references from citations to EMBL-Bank entries. During 2007, work was undertaken to extend this mapping technology to capture links to publications indexed by resources other than PubMed. The first examples of such publications are indexed by the Agricola service from the US National Agriculture Library (NAL) of the US Department of Agriculture (USDA) and not by PubMed; at the time of going to press, there were 3769 literature items cited in EMBL-Bank from Agricola alongside 193 423 items from PubMed.
Internal cross-references are provided in EMBL-Bank sequence-annotation entries. These include links from segment entries to CON and ANN records, links from primary entries to TPA entries, links from TPA entries to component ETA traces, links to stored alignment data in EMBL Align and others.
External cross-references are under careful curation. In 2007, cross-references to a number of new resources were added to EMBL-Bank sequence entries. These resources are Genome Reviews, the IPD-Killer-cell Immunoglobulin-like Receptor Database, RFAM—the RNA families’ database (13), the Culture Collection of Algae and Protozoa (14) and the Alternative Splicing and Transcript Diversity database (11). The cross-referencing system allows entry-level and feature level cross-references, shown as DR lines and /db_xref qualifiers respectively in EMBL flatfile format. Feature-level cross-references are currently restricted to source and CDS features, although developments are underway to improve the technology to cover additional features. While explicit cross-references to specific functional resources, such as the wwPDB structural database, are attractive, links to these resources are largely the domain of more specialist domain databases, such as the UniProt Knowledgebase, and given an explicit link to the specific resource from the domain database, a link can be inferred from an EMBL-Bank entry that explicitly cross-refers to the domain database. Priority for the addition of new cross-references will be given to those resources that offer information on or physical links to collections of biological materials that serve as source for sequencing.
Whereas ETA records single unassembled reads, EMBL-Bank sequences may be assembled to various levels; anywhere from overlapping reads through scaffolds with sequencing gaps to complete chromosomes, such that contiguous sequence, regardless of length, can be represented in a single sequence-annotation entry. Explicit links between ETA and EMBL-Bank are limited; traces that have been assembled into reference sequence in EMBL-Bank can be linked through the optional Reference_Sequence field in ETA and EMBL-Bank TPA entries may refer to ETA traces that have been used in re-assemblies (in AS lines in EMBL flatfile format). However, links to reference sequence in ETA records are sparsely used and there are few EMBL-Bank TPA entries that cite ETA records. Above the level of assembly of ETA reads, EMBL-Bank offers explicit links between contigs and scaffold entries (CO lines in EMBL flatfile format) and between primary entries cited in TPA entries (AS lines in EMBL flatfile format). The value to users of being able to map from trace reads through contigs and upwards to annotated scaffolds is to be investigated across EMBL-Bank and ETA, with a view to the development of additional tools to extract and present this information.
Nomenclatures and ontologies
The adoption of appropriate community-accepted nomenclatures and ontologies is key to capturing useful source metadata and biological annotation. Systematic nomenclatures, such as gene symbol conventions and strain names, are crucial in the presentation of robust search tools. However, the breadth of coverage (of, for example, taxonomy, gene nomenclature and experimental techniques) at ENA requires the use of multiple, often overlapping, nomenclatures. Many of the source metadata structures in use have application across a broad taxonomic range and across a variety of research communities. In different taxa, standard ways of referring to concepts have developed, but they are typically not consistent between taxa. Similarly, in different communities of researchers, diverse systematic ways of referring to concepts have evolved.
Once database staff has established that there is an existing vocabulary that has community acceptance, we will aim to make mandatory the usage of the vocabulary in appropriate fields in affected records. Nomenclatures that are promoted during EMBL-Bank curated submissions processing are detailed at: http://www.ebi.ac.uk/embl/Documentation/useful.html.
Project metadata
As increasing numbers of genomes and metagenomes are sequenced, the presentation of metadata surrounding sequencing activities will become increasingly useful in searches and analyses. Improvement of the content of the ENA Project Database is possible, since although many metadata elements will be difficult to validate automatically, the database is currently comparatively small. ENA therefore plans to improve existing records over the coming years and follows keenly the standardization work of the Genomic Standards Consortium (15).
SELECTED DATA CAPTURE PRIORITIES
- Sequence.
- Sequencing-associated information (traces, quality scores, base calls).
- Coding sequence and related annotation.
- Experimentally confirmed annotation.
- The nature of evidence for annotation.
- Source metadata nomenclatures and ontologies.
- Cross-references to those resources that offer information on, or physical links to, collections of biological materials.
- Reference to the literature.
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by EMBL.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sugawara H, Ogasawara O, Okubo K, Gojobori T, Tateno Y. Nucleic Acids Res. 2008. DDBJ with new system and face. doi: 10.1093/nar/gkm889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler , et al. Nucleic Acids Res. 2008. GenBank paper. Database issue, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wheeler , et al. Nucleic Acids Res. 2008. NCBI Database resources paper. Database issue, in press. [Google Scholar]
- 4.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 6.Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, et al. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc. Natl Acad. Sci. USA. 2006;103:13578–13584. doi: 10.1073/pnas.0605327103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 9.Cochrane G, Bates K, Apweiler R, Tateno Y, Mashima J, Kosuge T, Karsch-Mizrachi I, Schafer S, Fetchko M. Evidence standards in experimental and inferential INSDC third party annotation data. OMICS. 2006;10:105–113. doi: 10.1089/omi.2006.10.105. [DOI] [PubMed] [Google Scholar]
- 10.The UniProt Consortium. Nucleic Acids Res. 2008. The Universal Protein Resource (UniProt) Database issue, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Texier V, Riethoven JJ, Kumanduri V, Gopalakrishnan C, Lopez F, Gautheret D, Thanaraj TA. AltTrans: transcript pattern variants annotated for both alternative splicing and alternative polyadenylation. BMC Bioinformatics. 2006;7:169. doi: 10.1186/1471-2105-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Cunningham F. 2008. Ensembl 2008. doi: 10.1093/nar/gkm998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gachon CM, Day JG, Campbell CN, Pröschold T, Saxon RJ, Küpper FC. The Culture Collection of Algae and Protozoa (CCAP): a biological resource for protistan genomics. Gene. 2007 doi: 10.1016/j.gene.2007.05.018. doi: 10.1016/j.gene.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Field D, Garrity G, Gray T, Selengut J, Sterk P, Thomson N, Tatusova T, Cochrane G, Glöckner FO, et al. eGenomics: cataloguing our complete genome collection III. Meeting report. Comparative and Functional Genomics. 2007 doi: 10.1002/cfg.494. doi: 10.1155/2007/47304, http://www.hindawi.com/GetArticle.aspx?doi=10.1155/2007/47304. [DOI] [PMC free article] [PubMed] [Google Scholar]