Characterization of the Expression, Intracellular Localization, and Replication Complex Association of the Putative Mouse Hepatitis Virus RNA-Dependent RNA Polymerase (original) (raw)
Abstract
Mouse hepatitis virus (MHV) RNA synthesis is mediated by a viral RNA-dependent RNA polymerase (RdRp) on membrane-bound replication complexes in the host cell cytoplasm. However, it is not known how the putative MHV RdRp (Pol) is targeted to and retained on cellular membranes. In this report, we show that a 100-kDa protein was stably detected by an anti-Pol antiserum as a mature product throughout the virus life cycle. Gradient fractionation and biochemical extraction experiments demonstrated that Pol was not an integral membrane protein but was tightly associated with membranes and coimmunoprecipitated with the replicase proteins 3CLpro, p22, and p12. By immunofluorescence confocal microscopy, Pol colocalized with viral proteins at replication complexes, distinct from sites of virion assembly, over the entire course of infection. To determine if Pol associated with cellular membranes in the absence of other viral factors, the pol domain of gene 1 was cloned and expressed in cells as a fusion with green fluorescent protein, termed Gpol. In Gpol-expressing cells that were infected with MHV, but not in mock-infected cells, Gpol relocalized from a diffuse distribution in the cytoplasm to punctate foci that colocalized with markers for replication complexes. Expression of Gpol deletion mutants established that the conserved enzymatic domains of Pol were dispensable for replication complex association, but a 38-amino-acid domain in the RdRp unique region of Pol was required. This study demonstrates that viral or virus-induced factors are necessary for Pol to associate with membranes of replication complexes, and it identifies a defined region of Pol that may mediate its interactions with those factors.
For all known positive-strand RNA viruses, RNA synthetic activity occurs on viral replication complexes that are derived from cellular membranes and is mediated by viral RNA-dependent RNA polymerases (RdRps). Recent evidence suggests that viruses in the order Nidovirales, including coronaviruses and arteriviruses, replicate their RNA at double-membrane vesicles in the host cell cytoplasm (19, 35). For the coronaviruses, a diverse family of positive-strand RNA viruses, it is not known how viral replication complexes are formed or how replicase proteins are recruited to and remain associated with cellular membranes. Although a core RdRp has been predicted based on sequence analysis, and membrane-associated RdRp activity has been detected in coronavirus-infected cells, the function of this putative enzyme has not been confirmed experimentally (1, 6, 16, 18, 27). As such, determination of the expression, intracellular localization, and membrane targeting of the coronavirus RdRp is central to understanding the functions of this protein during viral RNA synthesis.
Mouse hepatitis virus (MHV) is a well-established model for studies of the replication and pathogenesis of coronaviruses. MHV contains a positive-strand RNA genome of approximately 32,000 nucleotides (nt) that serves as a template for the synthesis of multiple RNA species during an infectious cycle. The first 22 kb of the MHV genome constitutes the replicase gene (gene 1), composed of two open reading frames, ORFs 1a and 1b, that are linked by a ribosomal frameshift (1, 7, 8, 34). Translation of ORF1a or the ORF1a-ORF1b fusion results in a possible 495- or 803-kDa polyprotein, respectively (27). These large polyproteins are not detected during an infectious cycle due to cotranslational proteolytic processing by viral proteinases to yield 15 mature protein products (3, 10-15, 17, 28-30, 40). While the viral proteinases are the only MHV replicase proteins with confirmed functions, sequence analysis of the gene 1 polyprotein reveals predicted helicase (Hel) and RdRp domains (18, 27). Specifically, the region immediately downstream of the ribosomal frameshift contains motifs that are conserved for all RdRps, including Mg2+ binding sites, an RNA-binding motif, and a catalytic Ser-Asp-Asp (SDD) core motif, as well as predicted RdRp unique, finger, palm, and thumb domains (18, 26, 33).
A 100-kDa protein expressed from the MHV RdRp domain (Pol) has been detected in MHV-infected cells by immunoprecipitation experiments (40). Immunofluorescence studies of Pol localization during MHV infection indicate that this protein colocalizes with newly synthesized viral RNA and gene 1 proteins in punctate cytoplasmic foci at 7 h postinfection (p.i.) (40, 44). However, the kinetics of Pol expression and intracellular localization during an entire MHV infectious cycle are not known. Previous data suggest that the MHV replication complex is composed of two distinct populations of proteins that separate at late times p.i. Specifically, immunofluorescence confocal microscopy experiments have established that the replicase proteins p22, p65, and Hel, as well as the viral nucleocapsid (N), colocalize at replication complexes distinct from sites of virion assembly at 5.5 to 6.0 h p.i. (4). However, at 9 h p.i., Hel and N are separate from p22 and p65, localizing to sites of virion assembly in the endoplasmic reticulum-Golgi intermediate complex (ERGIC), indicating that the two populations of proteins may serve different roles during the MHV life cycle (4). Moreover, after fractionation of membranes containing the MHV replication complex by density on an iodixanol gradient, newly synthesized viral RNA, Hel, N, and the amino-terminal replicase protein p28 cofractionate exclusively in the denser fractions of the gradient (41). This Hel-N-p28-RNA complex is distinct from p22, p65, and MP1, all of which fractionate in the less dense portions of the gradient (41). The association of the putative viral helicase, the viral nucleocapsid protein, and newly synthesized viral RNA together suggested that this dense protein population may be the enzymatically active portion of the viral polymerase which is tethered to cellular membranes via interactions with proteins in the less dense p65-p22-MP1 population (41).
Still, it remains unknown which population of proteins Pol is associated with during MHV infection. Biochemical extraction data reveal that Pol is tightly associated with cellular membranes but is not an integral membrane protein (19). Yet the mechanisms by which Pol is targeted to viral replication complexes and associates with membranes remain to be elucidated. Pol lacks conserved myristoylation and palmitoylation motifs and is not predicted to contain transmembrane domains, suggesting that protein-protein or protein-RNA interactions are required for the association of Pol with membranes of replication complexes. Nonetheless, Pol has not been shown to interact with any viral or cellular protein, nor has the ability of Pol to bind viral RNA been experimentally demonstrated.
In the present study, we sought to determine the kinetics of Pol expression and intracellular localization, as well as to characterize the cis and trans determinants of Pol association with the MHV replication complex. We defined the expression, processing, and stability of Pol by performing pulse-label and pulse-chase translation experiments. Using biochemical fractionation and immunofluorescence confocal microscopy, we have shown that Pol is associated with the population of proteins containing p65 and remains localized to replication complexes over the course of MHV infection. The results of biochemical extraction data further characterize the nature of Pol membrane association and elucidate protein interactions between Pol and several replicase proteins. Finally, using immunofluorescence confocal microscopy, we have established that targeting of a green fluorescent protein (GFP)-Pol fusion protein (Gpol) to replication complexes requires viral or virus-induced factors, as well as 38 amino acids (aa) (F411 to D448) of the Pol protein. Together, these results provide a foundation for biochemical and genetic studies of Pol interactions and functions during MHV replication.
MATERIALS AND METHODS
Virus, cells, and antisera.
Delayed brain tumor (DBT) cell monolayers (22) were infected with MHV A59 at a multiplicity of infection of 10 PFU in Dulbecco modified Eagle medium containing 10% fetal calf serum for all experiments.
Polyclonal antisera used for biochemical experiments have been published previously. These include UP102 (anti-p28 [α-p28]-α-p65) (11), α-p65 (41), B1 (α-Hel) (13), α-p22, α-p12 (3), and α-3CLpro (29). Two monoclonal antibodies generated against the structural proteins nucleocapsid (α-N; J.3.3) and matrix (α-M; J.1.3) were generously provided by J. Fleming (University of Wisconsin, Madison). A rabbit polyclonal antiserum (VU145) was generated against the amino-terminal domain of Pol. All nucleotide and amino acid numbers correspond to the MHV A59 sequence modified by Bonilla et al. (1). Nucleotides 13696 to 14102 of gene 1 were amplified by reverse transcription-PCR (RT-PCR) from purified MHV A59 genomic RNA. The PCR product spanned 406 nt and comprised 134 aa of ORF1b (R4496 to K4630). Primer-generated restriction sites (5′ _Nco_I and 3′ _Hin_dIII) were used to subclone the PCR fragment into the pET23a bacterial expression vector (Novagen). A 16-kDa histidine-tagged protein was expressed in Escherichia coli BL21 cells, isolated by using nickel resin chromatography as described in the systems manual, and further purified by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) electroelution (Bio-Rad) as previously described (3). Rabbit antibodies were raised against this protein at Cocalico, Inc.
Radiolabeling of MHV proteins and immunoprecipitation.
Infection of DBT cells, radiolabeling, pulse-label and pulse-chase experiments, and immunoprecipitations were performed as previously described (13, 14, 30).
Cell fractionation and biochemical extraction.
Mock-infected or MHV A59-infected DBT cells were radiolabeled with 100 μCi of [35S]methionine-cysteine (TransLabel; ICN) per ml, lysed by ball-bearing homogenization, and subjected to differential centrifugation to obtain a nuclear fraction (NP), a large membrane pellet (P8), a small membrane pellet (P100), and a cytosolic fraction (S100) as previously described (41). To further separate components within P100, the small membrane pellet was subjected to fractionation by flotation on an iodixanol density gradient. Iodixanol (Optiprep; Nycomed Pharma) gradients were prepared as previously described (41) with 500 μl of the MHV-infected P100 pellet on the bottom of the gradient. The gradients were centrifuged at 35,000 rpm (116,000 × g) for 3 h at 4°C (in a Beckman SW55 rotor). Fractions were immunoprecipitated with antibodies against MHV proteins as described above.
Triton X-114 extractions were performed as described previously (2, 45). Briefly, the small membrane pellet (P100) from radiolabeled MHV-infected or mock-infected cells was treated with a 1% Triton X-114 solution (10 mM Tris [pH 7.2], 150 mM NaCl, 1% Triton X-114) on ice, incubated for 5 min at 37°C, centrifuged at 3,200 rpm (3,000 × g) for 5 min at room temperature, and then separated on a 6% sucrose cushion containing 10 mM Tris (pH 7.2), 150 mM NaCl, and 1% Triton X-114. The supernatant fraction was subjected to one additional round of Triton X-114 extraction. Detergent concentrations in the cytosolic fraction and membrane pellet fraction were equalized prior to immunoprecipitation. Sodium carbonate extractions were performed as described by Schmutz et al. (37) and van der Meer et al. (45). The P100 pellet from radiolabeled MHV-infected or mock-infected cells was resuspended in 1 M sucrose-Tris buffer containing 200 mM Na2CO3 at a pH of 11.0. Treated samples were then separated into phases on a 10% sucrose cushion at 100,000 rpm (320,000 × g) for 30 min at 4°C (in a Sorvall TLA-120 rotor). Fractions were subjected to immunoprecipitation as described above.
Immunoblot analysis.
Cell fractions from either mock-infected or MHV-infected cells were subjected to SDS-PAGE on a 5 to 18% acrylamide gradient gel. Proteins were transferred to a 0.45-μm-pore-size nitrocellulose membrane (Trans-Blot; Bio-Rad) at 100 V for 1.5 h in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) and immunoblotted with VU145 (α-Pol) antisera at a 1:1,000 dilution in 5% nonfat dry milk. A secondary anti-rabbit immunoglobulin conjugated to horseradish peroxidase (Amersham) at a 1:2,000 dilution allowed for the visualization of proteins after treatment with a chemiluminescence reagent (Perkin-Elmer). The blot was then stripped of α-Pol antibodies by using 100 mM β-mercaptoethanol, 2% SDS, and 62.5 mM Tris-Cl (pH 6.7) and was subsequently probed with the UP102 (α-p28-α-p65) antiserum at a 1:1,000 dilution.
Immunofluorescence assays.
DBT cells on glass coverslips were either mock infected or infected with MHV A59. At specific times indicated in the figures, cells were fixed with 100% methanol at −20°C. Indirect immunofluorescence assays were performed as described previously (4). Polyclonal antisera were used at a 1:100 dilution, whereas monoclonal antibodies were used at a 1:1,000 dilution. Immunofluorescence was detected at 488 and 543 nm by using a 40× oil immersion objective on a Zeiss LSM 510 confocal microscope. Image analysis and merging were performed using Adobe Photoshop, version 7.0.
Cloning and expression of Pol.
Nucleotides 13572 to 16338 were amplified from purified MHV A59 genomic RNA by using RT-PCR. The 5′ primer was designed to introduce mutations in the slippery sequence and complex RNA structures (pseudoknot) that would eliminate the ribosomal frameshift while maintaining the Pol coding sequence as predicted by Bredenbeek et al. (7). The sequence of the 5′ primer is 5′ GGA ATT CCA TGG CCT CAA AAG ACA CGA ACT TCT TGA ATC GGA TTC GCG GAA CAA GTG TAA ATG CCC GT 3′. Primer-generated restriction sites (5′ _Eco_RI and 3′ _Bam_HI) were also included for cloning of the PCR product into the pEGFP-C1 vector (Clontech). The designation of the resulting plasmid is pEGFP-C1-pol, and the protein is called Gpol.
Deletion mutagenesis of Gpol.
To delete portions of the carboxy terminus of Pol, pEGFP-C1-pol was digested with the following pairs of restriction enzymes: _Bam_HI and _Spe_I (GpolΔ1), _Bam_HI and _Apa_I (GpolΔ2), _Bam_HI and _Pst_I (GpolΔ3), _Bam_HI and _Ahd_I (GpolΔ4), and _Bam_HI and _Kpn_I (GpolΔ5). To delete amino acid residues D373 to R551 from Gpol, pEGFP-C1-pol was digested with _Apa_I and _Pst_I (GpolΔ373-551). The digested vectors were treated with T4 DNA polymerase (New England Biolabs) to create blunt ends while keeping the protein-coding region in-frame, gel purified, and subsequently ligated to circularize the plasmids. The 3′ ends of the pEGFP-C1-pol mutant cDNAs were sequenced by using primer 5′ GAC CGC TGC GCC TTA TCC 3′.
PCR was performed to fuse Pol amino acids (D373 to R551) to the carboxy-terminal end of GFP (GFP:373-551). Primers were designed to amplify nt 14686 to nt 15220, and pEGFP-C1-pol was used as the PCR template. Primer-generated restriction sites (5′ _Eco_RI and 3′ _Bam_HI) were included for cloning into pEGFP-C1 (Clontech). The sequence of the left primer was 5′ GAA TTC GAC CCT GCC CTT CA 3′, and the sequence of the right primer was 5′ GGA TCC CTT ATT CTT AGC ACT 3′. The PCR product was cloned into pGEM-T-Easy (Promega), sequenced by using primers T7 and SP6, then subcloned into pEGFP-C1-pol (Clontech) by using _Eco_RI and _Bam_HI sites, and sequenced across joins.
To generate mutants of GpolΔ2, primers were designed to amplify the Gpol coding sequence with progressive truncations to the 3′ end of residues in the 178-aa region (D373 to R551). pEGFP-C1-pol was used as the PCR template. Primer-generated restriction sites (5′ _Eco_RI and 3′ _Bam_HI) were also included for cloning into pEGFP-C1 (Clontech). The sequence of the left primer used for all reactions was 5′ GAA TCC ATG GCC TCA AAA GAC 3′ at the start of the pol coding sequence. The sequences of the right primers were 5′ GGA TCC TGC CTC ATA ATA GAG 3′ (GpolΔ2-1), 5′ GGA TCC CAG TAA TAG CAG CAT 3′ (GpolΔ2-2), 5′ GGA TCC TTT CCA GGT TTA ACT 3′ (GpolΔ2-3), and 5′ GGA TCC CGC AAA TCA AGC AG 3′ (GpolΔ2-4). All PCR products were cloned into pGEM-T-Easy (Promega) and sequenced by using primers T7 and SP6. The polΔ2-1, polΔ2-2, polΔ2-3, and polΔ2-4 cDNAs were then subcloned into pEGFP-C1 (Clontech) by using _Eco_RI and _Bam_HI sites and were sequenced across joins.
Transfection and expression of GFP fusion proteins.
DBT cells were grown on glass coverslips for 24 h and then either transfected with pEGFP-C1, pEGFP-C1-pol, or cDNA encoding the Gpol deletion mutants or mock transfected according to the manufacturer's protocol (Effectene; Qiagen). Twenty-four hours posttransfection, cells either were infected with MHV A59 for 5.5 h or were mock infected. Cells were fixed with 100% methanol at −20°C, reconstituted with phosphate-buffered saline, and either imaged directly or probed with antibodies against MHV proteins by using indirect immunofluorescence as described above.
RESULTS
Pol detection, processing, and stability.
Pol has been detected in MHV-infected cells by immunoprecipitation, electron microscopy, and immunofluorescence using region-specific antibodies (19, 40, 44). However, there has been no description of a functional antibody able to detect Pol by multiple approaches. In the present study, we raised α-Pol rabbit polyclonal antibodies (VU145) against a recombinant six-histidine-tagged protein corresponding to amino acids R4496 to K4630 (nt 13696 to nt 14102) (1) of the gene 1 polyprotein within the Pol domain (Fig. 1A). This antiserum was shown to specifically detect Pol in MHV-infected cells by immunoprecipitation (Fig. 1B and C), immunoblotting (Fig. 2A), and immunofluorescence (Fig. 5A) assays.
FIG. 1.
MHV genome organization and Pol expression and stability. (A) MHV genome organization. Schematic shows the organization of the gene 1 polyprotein, as well as the location of genes 6 and 7, encoding structural proteins M and N, respectively. Confirmed or predicted mature gene 1 proteins are shown as boxes. Shaded boxes indicate replicase proteins of interest in the present study: the amino-terminal cleavage products (p28 and p65), the 3C-like proteinase (3CLpro), two carboxy-terminal ORF1a proteins (p22 and p12), the putative RdRp domain (Pol), and the putative helicase (Hel). The region of gene 1 used to generate the α-Pol rabbit polyclonal antiserum VU145 is indicated. (B and C) Pulse-label and pulse-chase translation of Pol. Cells were radiolabeled with [35S]Met-Cys at 5.5 h p.i., and either samples were withdrawn at the indicated times for pulse-labeling or, after 30 min of radiolabeling, cells were incubated in media containing excess unlabeled Met-Cys for 15 min to 16 h as described in Materials and Methods. Samples were immunoprecipitated with the α-Pol antiserum VU145, and proteins were analyzed by SDS-PAGE on a 5 to 18% acrylamide gradient gel followed by fluo-rography. For control lanes, cells were radiolabeled from 5.5 to 7.5 h after infection or mock infection. Lysates of mock-infected cells (M) were immunoprecipitated with VU145, and lysates of infected cells were also precipitated with preimmune sera (pre). Molecular mass standards (in kilodaltons) are shown to the left of the gel, and the locations of both Pol and a >150-kDa protein are shown on the right.
FIG. 2.
Detection of viral proteins after subcellular fractionation and differential centrifugation. MHV-infected or mock-infected cells were lysed by ball-bearing homogenization and subjected to differential centrifugation as described in Materials and Methods to obtain a nuclear pellet (NP), a large membrane pellet (P8), a small membrane pellet (P100), and a cytosolic fraction (S100) consisting of the supernatant after the 100,000-rpm spin. The fractions were subjected to SDS-PAGE on a 5 to 18% acrylamide gradient gel and then transferred to a nitrocellulose membrane for use in immunoblot assays with the α-Pol antiserum VU145 (A) or the α-p28-α-p65 antibody UP102 (B). Molecular mass standards (in kilodaltons) are indicated to the left of the blots, and the locations of Pol, p65, and p28 are indicated on the right.
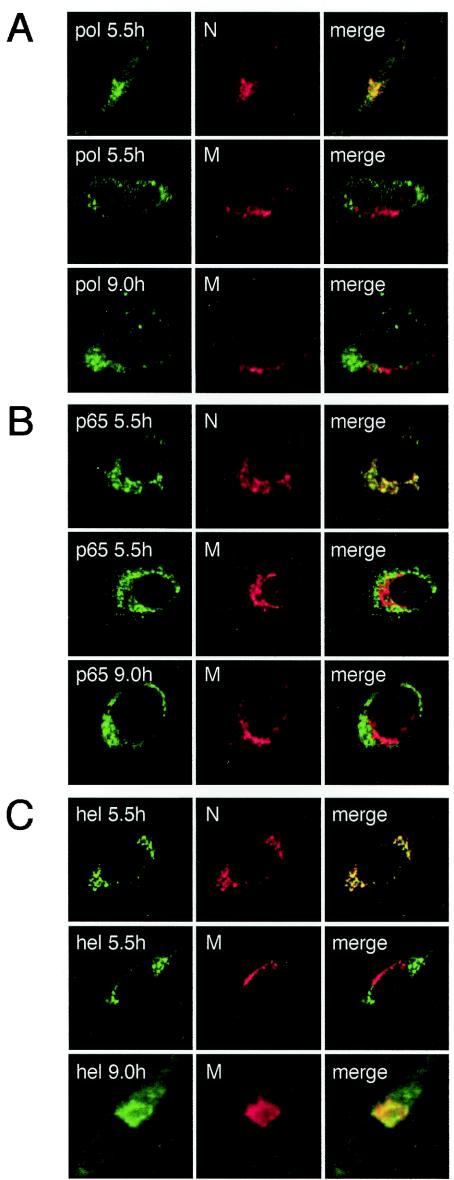
FIG. 5.
Time course of Pol localization during MHV infection. MHV-infected DBT cells were fixed at the indicated times p.i. prior to preparation for indirect immunofluorescence microscopy as described in Materials and Methods. Cells were imaged on a Zeiss LSM 510 confocal microscope. Images are single confocal slices taken by using a 40× objective and are representative of the cell population. (A) Dual-label imaging of Pol (green) and N (red) at 5.5 h p.i. or of Pol(green) and M (red) at 5.5 and 9 h p.i. (B) Dual-label imaging of p65 (green) and N (red) at 5.5 h p.i. or of p65 (green) and M (red) at 5.5 and 9 h p.i. (C) Dual-label imaging of Hel (green) and N (red) at 5.5 h p.i. or of Hel (green) and M (red) at 5.5 and 9 h p.i. Merged images are shown with areas of colocalization in yellow.
To determine the kinetics of Pol expression, processing, and stability during MHV infection, the α-Pol antiserum VU145 was used to immunoprecipitate Pol from MHV-infected DBT cells after pulse-label and pulse-chase radiolabeling (Fig. 1). For these experiments, translation was synchronized to ensure that the proteins detected were de novo translation products. The timing of Pol expression was determined by pulse-labeling infected cells with [35S]Met-Cys followed by immunoprecipitation (Fig. 1B). In MHV-infected cells probed with VU145, a 100-kDa band consistent with the predicted mass of Pol was first detected after 60 min of labeling and accumulated without evidence of further processing, demonstrating that this was a mature protein product and that protein processing was occurring cotranslationally. No 100-kDa protein was detected following immunoprecipitation of mock-infected cell lysates with VU145 or following immunoprecipitation of MHV-infected cell lysates with preimmune sera from the same rabbit, indicating that VU145 was specific for Pol. To define the stability of Pol and to further characterize precursor-product relationships, MHV-infected cells were labeled with [35S]Met-Cys for 30 min at 5.5 h p.i., followed by a chase from 15 min to 16 h using media containing excess unlabeled methionine and cysteine (Fig. 1C). In agreement with the results of the 60-min pulse-label, Pol was detected after the 30-min chase. The Pol band was stable or increasing for as long as 5 h of chase (11 h p.i.) and was still detectable even after the 16th h of the chase (22 h p.i.), when the cell monolayer was completely involved in syncytia. These results suggested that previously processed molecules of Pol were resistant to degradation even following disruption of cell monolayers and cessation of viral replication. During both the pulse-label and the pulse-chase labeling experiments, a protein band with a calculated mass of >150 kDa was specifically detected in MHV-infected cells immunoprecipitated with VU145 (Fig. 1B and C). During the pulse-chase experiment, the >150-kDa protein decreased over time while Pol remained stable or increased. Although the >150-kDa protein was less abundant than Pol, the pattern suggested a possible precursor-product relationship with Pol. In contrast, it has been reported previously that a protein of >300 kDa was present as a Pol precursor (40). In our experiments, no protein of this size was detected in MHV-infected cells only; rather, a >300-kDa protein was detected in both mock-infected and infected cells probed with VU145 and was also detected by preimmune serum.
Pol is associated with the protein population containing p65.
Newly synthesized viral RNA and all tested replicase proteins, as well as N, have been shown to associate with a small membrane pellet following differential centrifugation of homogenized MHV-infected cells (41). To determine if Pol was associated with the same membrane population as other MHV replicase proteins, MHV-infected or mock-infected DBT cells were lysed by ball-bearing homogenization and subjected to three rounds of differential centrifugation to obtain a nuclear pellet (NP), a large membrane pellet (P8), a small membrane pellet (P100), and a cytosolic fraction (S100). The proteins in the membrane and cytosolic fractions were separated by SDS-PAGE and immunoblotted with the α-Pol antiserum VU145 and an antibody that recognizes both p28 and p65 (UP102) (Fig. 2). When the fractions were analyzed, all detectable Pol, p65, and p28 was associated with membrane fractions, with predominant localization to the P100 small membrane pellet. The pattern of Pol fractionation was identical to that of p65, with a majority in the P100 pellet and smaller amounts detected in the nuclear pellet and large membrane pellet. This result suggested that Pol was in the same membrane population as other replicase proteins and viral RNA.
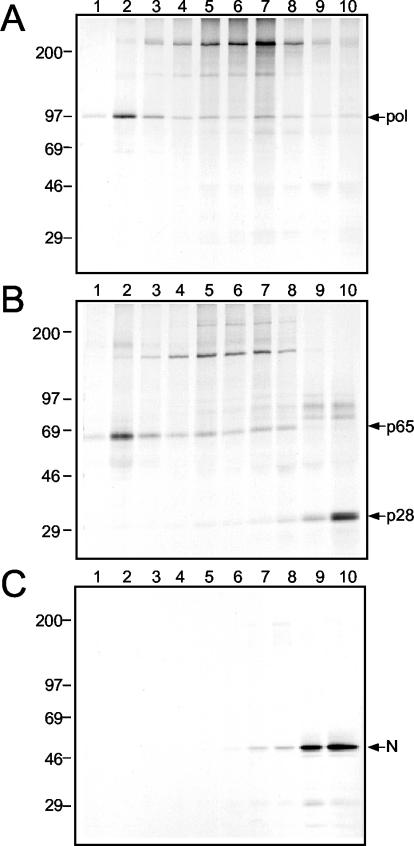
The replicase proteins in the P100 pellet segregate to two distinct populations of proteins (41). To determine the protein population with which Pol associated, the P100 pellet was resuspended and fractionated by flotation on an iodixanol gradient. Individual fractions were then immunoprecipitated by using either the α-Pol antiserum VU145, the α-p28-α-p65 antibody UP102, or a monoclonal antibody against N (J.3.3) (Fig. 3). Although Pol was detected in all gradient fractions, the majority of Pol was detected in the top, least dense fractions of the gradient (Fig. 3A). The pattern of gradient fractionation of Pol was almost identical to that of p65, indicating that these two proteins were associated with cellular membranes of the same density (Fig. 3B). In contrast, both p28 and N were detected in the bottom, densest fractions of the gradient (Fig. 3B and C). This result corroborated previous results of sedimentation analysis suggesting that p28 and N were readily dissociated from membranes containing p65 (41). Together, the results of differential centrifugation and gradient fractionation indicated that Pol was associated with membranes containing p65 and multiple other replicase proteins but not with the protein population containing p28 and N.
FIG. 3.
Gradient fractionation of membranes from MHV-infected DBT cells. The P100 pellet from MHV-infected cells was resuspended and fractionated on an iodixanol gradient as described in Materials and Methods. Gradient fractions were immunoprecipitated, separated by SDS-PAGE on a 5 to 18% acrylamide gradient gel, and subjected to fluorography. Lane 1 indicates the top of the gradient (less dense), and lane 10 indicates the bottom of the gradient (denser). Molecular mass markers (in kilodaltons) are given to the left of the gels, and proteins of interest are indicated on the right. (A) VU145 (α-Pol); (B) UP102 (α-p28-α-p65); (C) J3.3 (α-N).
Pol membrane association and protein interactions.
Since Pol remained associated with membranes during cell lysis, centrifugation, and fractionation, we next sought to define the mechanism of Pol membrane association. Previous studies have demonstrated that Pol lacks characteristics of an integral membrane protein (19). If so, then Pol association with membranes must occur by mechanisms such as protein-protein or protein-RNA interactions. To confirm the nature of Pol membrane association and to identify potential protein interactions, P100 pellets from radiolabeled MHV-infected cells were extracted with either Triton X-114 or sodium carbonate (Na2CO3) at pH 11.0 and were then immunoprecipitated with the α-Pol antiserum VU145 (Fig. 4). Following extraction with Triton X-114, Pol was detected exclusively in the aqueous phase (Fig. 4A). However, following extraction with Na2CO3, Pol was detected predominantly in the membrane pellet (Fig. 4B). Together, these results confirmed that Pol was tightly associated with membranes but was not an integral membrane protein. The results from this experiment were consistent with those reported by Gosert et al., suggesting that Pol retention on membranes may be mediated by _trans_-acting viral or cellular factors (19).
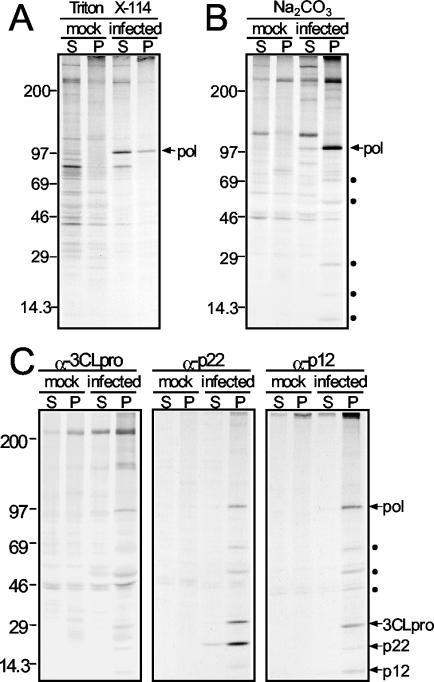
FIG. 4.
Triton X-114 and Na2CO3 extractions of DBT cells and detection of viral proteins. (A and B) The P100 pellet from radiolabeled MHV-infected or mock-infected cells was resuspended and treated with either a 1% Triton X-114 solution (A) or 200 mM Na2CO3 (B) at a pH of 11.0. The treated lysates were first separated into cytosolic, aqueous fractions (S) and membrane fractions (P) by centrifugation through sucrose cushions as described in Materials and Methods and then subjected to immunoprecipitation with the α-Pol antiserum VU145. Precipitated proteins were separated on a 5 to 18% acrylamide gel by electrophoresis and were visualized after fluorography. The location of Pol is shown to the right of the gels, and molecular size standards (in kilodaltons) are shown on the left. Coprecipitating proteins are indicated by solid circles on the right of the gel. (C) Na2CO3-treated lysates were immunoprecipitated with antisera against 3CLpro, p22, or p12 and analyzed as described above. The locations of Pol, 3CLpro, p22, and p12 are shown to the right of the gels. Coprecipitating proteins of unconfirmed identity are indicated by solid circles to the right of the gels. Molecular size standards (in kilodaltons) are shown on the left.
When the Na2CO3-extracted lysate was immunoprecipitated with the α-Pol antiserum VU145, several other proteins appeared to coimmunoprecipitate with Pol in the membrane pellet from MHV-infected cells (Fig. 4B). These proteins had calculated masses of 65, 29, 22, and 12 kDa. These sizes were consistent with those of known replicase proteins: p65 or Hel (65 kDa), p28 or 3CLpro (29 kDa), p22 (22 kDa), and p12 (12 kDa). This result suggested that the Pol interactions with other proteins were stringent, since the coprecipitation of the proteins occurred following Na2CO3 extraction and in the presence of SDS. To determine the identities of several of the proteins, the Na2CO3-extracted lysates were immunoprecipitated with antisera specific for 3CLpro, p22, and p12. Each of the antisera detected all of the other proteins as well as Pol (Fig. 4C). This result confirmed that the proteins were specific replicase proteins and that they were likely associated with Pol in replication complexes. Proteins with calculated masses of 65, 50, and 45 kDa also coprecipitated with antisera against 3CLpro, p22, and p12 specifically in the membrane pellet from infected cells. Although the identities of these proteins are unknown, the sizes were consistent with their being MHV proteins: p65 or Hel (65 kDa), N (50 kDa), and MP1 (45 kDa).
Pol is retained at MHV replication complexes throughout infection.
We next sought to determine if Pol was retained at replication complexes throughout infection or if Pol relocalized to sites of virion assembly in the ERGIC as has been previously shown for Hel and N (4). DBT cells on glass coverslips were either mock infected or infected with MHV for 5.5 or 9 h and subsequently probed with the α-Pol antiserum VU145 and antibodies against other replicase and structural proteins in indirect immunofluorescence assays (Fig. 5). In MHV-infected cells imaged by fluorescence confocal microscopy, Pol staining was detected as punctate, cytoplasmic foci, characteristic of viral replication complexes (Fig. 5A). At 5.5 h p.i., Pol, p65, and Hel colocalized with N at sites distinct from M accumulation (Fig. 5). At 9 h p.i., both Pol and p65 remained localized to punctate foci in the cytoplasm and showed no significant colocalization with M staining (Fig. 5A and B). In contrast, Hel made a transition from colocalization with proteins in replication complexes at 5.5 h p.i. to colocalization with M at 9 h p.i. (Fig. 5C). These data indicated that Pol remained associated with p65 in viral replication complexes and did not translocate to sites of virion assembly in the ERGIC.
Targeting of Gpol to MHV replication complexes requires viral infection.
Having confirmed that Pol is associated with cellular membranes and that Pol interacts with multiple replicase proteins, we next wanted to determine if these interactions were necessary for Pol association with the replication complex or if Pol was capable of independent association with membranes in the absence of other factors. To address this question, the pol domain (nt 13572 to nt 16354) (1) of gene 1 was cloned from purified MHV genomic RNA by RT-PCR. The pol domain is composed of 21 nt of ORF1a and 2,764 nt of ORF1b, requiring a −1 ribosomal frameshift 21 nt into the pol domain for the expression of the full-length protein (7, 27). The frameshift has been reported to be 35 to 40% efficient in vitro (7). While the ribosomal frameshift may be a critical regulatory feature of replicase protein expression, it is problematic for controlled expression of Pol from exogenous systems. To ensure expression of full-length Pol from a cytomegalovirus promoter-driven expression system, we mutagenized residues in the slippery sequence (UUUUUAAAC) and pseudoknot structure to eliminate the ribosomal frameshift and to fuse the coding sequence of pol as a single open reading frame (Fig. 6A) (8, 23). To track Pol localization in both mock-infected and MHV-infected cells, the pol coding sequence was cloned as a fusion, linking enhanced GFP to the amino terminus of Pol (Gpol).
FIG. 6.
Targeting of Gpol to replication complexes during MHV infection. (A) Cloning of MHV Pol. The pol region of gene 1 was cloned by RT-PCR. Primer-generated substitutions were made to nucleotides in both the slippery sequence (boldfaced) and the pseudoknot in order to eliminate the potential for a −1 ribosomal frameshift while maintaining the coding sequence of pol. Mutated residues are shown in red, and the resulting amino acid sequences for both wild-type Pol and cloned Pol are given above nucleotide codons. pol cDNA was subcloned into pEGFP-C1 and expressed as a fusion to the carboxy terminus of GFP (Gpol). (B and C) DBT cells on glass coverslips were transfected with cDNA encoding either GFP or Gpol. Twenty-four hours posttransfection, cells were either infected with MHV or mock infected for 5.5 h, fixed, and either imaged for GFP fluorescence (B) or subjected to indirect immunofluorescence as described in Materials and Methods by using antibodies against the MHV structural proteins M and N (C). Multinucleated cells are a cytopathic effect of viral infection. In all images, GFP is shown as green and antibody staining is shown as red. Merged images are shown with areas of colocalization in yellow.
DBT cells on glass coverslips were either mock transfected or transfected with plasmids encoding either GFP alone or Gpol. These cells were then either mock infected or infected with MHV and visualized by confocal microscopy (Fig. 6B). In both infected and mock-infected cells expressing GFP alone, the fluorescence was detected as a diffuse cytoplasmic and nuclear distribution without punctate foci, consistent with the known and previously observed pattern of GFP. When Gpol was expressed in mock-infected cells, a diffuse cytoplasmic distribution of fluorescence was observed. Notably, no punctate foci or reticular patterns of fluorescence were detected in mock-infected cells, indicating the lack of membrane association. When Gpol-expressing DBT cells were infected with MHV for 5.5 h, a significant change was detected in the pattern of Gpol localization. Distinct, punctate, cytoplasmic foci of fluorescence were detected, in addition to a diffuse fluorescence pattern likely due to high-level expression of Gpol in the cells.
To determine if the punctate foci in MHV-infected Gpol-expressing cells were viral replication complexes, the cells were probed with antibodies against the structural proteins N and M (Fig. 6C). Gpol colocalized with N in punctate cytoplasmic complexes but did not colocalize with M. These results demonstrated that MHV infection was required for targeting of Gpol to viral replication complexes, thus suggesting that viral or virus-induced cellular factors were required in order for Pol to associate with membranes.
Identification of a domain within Gpol required for replication complex association.
Since Gpol was capable of being targeted to replication complexes in the presence of infection, we next sought to identify a specific domain or motif within Pol that was required for this association. Progressive carboxy-terminal truncations of Gpol were constructed (Fig. 7A) and used in experiments as described above for full-length Gpol. The proteins expressed from all truncated constructs showed similar patterns of diffuse cytoplasmic fluorescence in mock-infected cells. Several constructs showed evidence of aggregation of fluorescence when expressed at high levels in mock-infected and MHV-infected cells, suggesting exposure of a hydrophobic domain (Fig. 7B). However, only when cells expressing full-length Gpol or the truncation mutant GpolΔ1 or GpolΔ2 were infected with MHV were punctate perinuclear foci of fluorescence detected (Fig. 7B). In contrast, infected cells expressing GpolΔ3, GpolΔ4, or GpolΔ5 all were identical to mock-infected cells in that they showed no punctate perinuclear foci. Labeling of cells for N determined that the punctate foci in MHV-infected cells expressing Gpol, GpolΔ1, or GpolΔ2 were replication complexes (Fig. 7B). There was no colocalization of fluorescence from GpolΔ3, GpolΔ4, or GpolΔ5 with N (Fig. 7B), confirming their lack of replication complex association.
FIG.7.
Replication complex association of Gpol deletion mutants. (A) Gpol deletion mutagenesis. Progressive carboxy-terminal truncations were made to Gpol cDNA (pEGFP-C1-pol) by digestion of the plasmid with _Bam_HI and upstream restriction enzymes (listed above schematic) followed by blunt-end ligation after treatment with T4 DNA polymerase. Gene 1 nucleotide numbers of restriction cut sites are listed above the full-length Gpol diagram, and amino acid numbers within Pol are listed below. The predicted domains of Pol (RdRp unique, fingers, palm, and thumb) are indicated as differentially shaded regions on the Gpol schematic, and the location of the SDD core is shown above. GFP is shown as a green box fused to the amino terminus of the Pol protein and is not drawn to scale. The number of Pol amino acids remaining is given to the right of each mutant. The region between the dotted lines indicates a 178-aa domain within Pol (D373 to R551) that was required for replication complex association. (B) Immunofluorescence imaging of Gpol mutants. DBT cells on glass coverslips were transfected with cDNA encoding either Gpol or Gpol deletion mutants. Twenty-four hours posttransfection, cells were either infected with MHV or mock infected for 5.5 h and then subjected to indirect immunofluorescence with antibodies against N as described in Materials and Methods. In all images, GFP is shown as green and N staining is shown as red. Merged images are shown with areas of colocalization in yellow.
The difference between GpolΔ2 and GpolΔ3 was a 178-aa domain (D373 to R551) encoded by nt 14686 to nt 15220 of gene 1 (1), suggesting that a specific interaction domain within this region of the protein was necessary for association of Pol with the replication complex. To confirm that this 178-aa region was required for replication complex association, amino acids D373 to R551 were deleted from full-length Gpol (GpolΔ373-551) (Fig. 8A). In both mock-infected and MHV-infected cells, GpolΔ373-551 was detected as diffuse fluorescence in the cytoplasm with regions of punctate vesicular foci (Fig. 8B). However, the punctate foci observed in infected cells did not colocalize with N staining, demonstrating the lack of replication complex association of GpolΔ373-551 (Fig. 8B). This result supported the conclusion that residues D373 to R551 within Pol were required for targeting of Pol to replication complexes and further suggested that deletion of this region in the context of the full-length protein caused aberrant targeting or aggregation (Fig. 8B). To determine if the 178-aa region of Pol alone was sufficient for localizing GFP to the replication complex, this region was subcloned independently of the Pol protein as a fusion to the carboxy terminus of GFP (GFP:373-551) (Fig. 8A) and expressed in DBT cells that were subsequently either mock infected or infected with MHV. In both mock-infected and MHV-infected cells, GFP:373-551 was diffuse throughout the cytoplasm and did not colocalize with N (Fig. 8B). These results demonstrated that although the 178-aa domain (D373 to R551) was required, it was not sufficient for localizing Pol to replication complexes.
FIG.8.
Identification of a domain required for Gpol replication complex association. (A) Gpol mutagenesis. The 178-aa region (D373 to R551) either was deleted from full-length Gpol or was cloned as a fusion to the carboxy terminus of GFP. Truncations of this region (D373 to R551) within GpolΔ2 were generated by PCR as described in Materials and Methods. The predicted domains of Pol (RdRp unique, fingers, palm, and thumb) are indicated as differentially shaded regions on the full-length Gpol schematic, and the location of the SDD core is shown above. GFP is shown as a green box fused to the amino terminus of the Pol protein and is not drawn to scale. The number of Pol amino acids remaining is indicated to the right of each mutant. The region between the dotted lines represents a 38-aa domain within Pol (F411 to D448) that was required for replication complex association. (B) Immunofluorescence imaging of Gpol mutants. DBT cells on glass coverslips were transfected with cDNA encoding either full-length Gpol or Gpol deletion mutants. Twenty-four hours posttransfection, cells were either infected with MHV or mock infected for 5.5 h and then subjected to indirect immunofluorescence with antibodies against N as described in Materials and Methods. In all images, GFP is shown as green and N staining is shown as red. Merged images are shown with areas of colocalization in yellow.
To further define which residues of Pol were required for replication complex association, increasing deletions of the 178-aa region within GpolΔ2 were made from the carboxy terminus (Fig. 8A), and the GFP fusion proteins were used in experiments as described above. All constructs showed a diffuse fluorescence in transfected, mock-infected cells (Fig. 8B). In GpolΔ2-l- and GpolΔ2-2-expressing cells that were infected with MHV, punctate foci that colocalized with staining for N were detected in the cytoplasm (Fig. 8B). However, GpolΔ2-3 and GpolΔ2-4 remained diffuse in infected cells and did not colocalize with N (Fig. 8B). This finding demonstrated that a 38-aa domain (F411 to D448), encoded by nt 14911 to nt 14801 of gene 1, at the carboxy terminus of GpolΔ2-2 contained residues required for targeting Pol to membranes of replication complexes.
DISCUSSION
A characteristic feature of positive-strand RNA viruses is the formation of membrane-associated replication complexes in the cytoplasm of the host cell. The mechanisms of replication complex formation and function are as varied as the pathogenesis and diseases of different virus families. Yet there appear to be common themes of membrane association and membrane modification mediated by specific viral proteins. For the nidoviruses, there is an emerging view that replicase proteins translated and processed from the input genome form replication complexes on double-membrane vesicles (19, 35, 42). In addition, there is increasing evidence that replication and assembly are localized to different structures in the cell and are separated both temporally and physically (4, 41). However, the roles of individual replicase proteins in the formation and function of replication complexes have not been determined. One protein likely involved in replication complex function is Pol, the putative RdRp of MHV. The functional interactions of Pol with components of the replication complex during both genome replication and mRNA synthesis are predicted to be complex and highly organized. As such, it is expected that this 100-kDa protein is not merely an active site of a polymerase unit but also contains domains that mediate its efficient association with membranes, proteins, and RNA.
Pol remains detectable in MHV-infected cells as a mature product.
Despite the identification of a 100-kDa product in MHV-infected cells, the expression, processing, and stability of this putative core polymerase had not been characterized previously. Our results demonstrate that Pol is processed from the polyprotein, possibly by a cotranslational mechanism, and is detectable as a mature product at late times p.i. (up to 22 h p.i.). Moreover, Pol remains abundant in infected cells after a prolonged chase (5 h), indicating that the protein is extremely stable and resistant to degradation after processing. Even with this demonstrated stability of Pol, it has been shown that ongoing protein processing is required for MHV RNA synthesis to occur (25). These results may suggest that only newly processed molecules are able to function as the active form of the enzyme or that another protein interacts stoichiometrically with Pol and is the limiting factor in RNA synthesis. A report by van der Meer et al. showed evidence to suggest that Pol is secreted from the cell (44). Our data indicate that any Pol secretion that occurs does not qualitatively reduce the amount of Pol detectable within cells over time. Furthermore, a >150-kDa protein coprecipitates with Pol at various times during both the pulse-label and pulse-chase experiments. The amount of this large protein product decreased as Pol accumulated, suggesting a potential precursor-product relationship. The >150-kDa protein may be a Pol-Hel precursor that would serve as a depot for continuous Pol processing. Other precursors have been reported for Pol, including a >300-kDa protein extending from ORF1a into ORF1b (40). In our experiments, we did not detect a >300-kDa protein specifically in MHV-infected cells. Whether this reported difference is due to antibody specificity remains to be determined. A protein of this size was detected in both mock-infected cells and, to a greater extent, in infected cells, suggesting that the >300-kDa protein may be a cellular protein whose expression is induced during MHV infection. A protein of approximately 150 kDa has been reported as a precursor to the MP1-through-p15 region of the ORF1a polyprotein (36). It is possible that Pol coprecipitates with this precursor product. The demonstration that Pol coprecipitates with 3CLpro, p22, and p12, mature proteins from this region, favors this conclusion (see also below). Additional experiments will be necessary to unequivocally determine the relationship of this >150-kDa protein with Pol.
Pol remains associated with membranes of distributed cytoplasmic foci and does not translocate with Hel to sites of virion assembly.
The subcellular fractionation of MHV-infected cells confirms previous electron microscopic and biochemical data demonstrating that Pol is associated with membranes of the MHV replication complexes (19, 44). After differential centrifugation of MHV-infected cells, Pol cofractionates with viral RNA, all tested replicase proteins, including p28 and p65, and N in the P100 pellet; however, Pol is distinct from p28 and N after flotation on an iodixanol density gradient. The detection of Pol in the less dense iodixanol gradient fractions supports the conclusion that Pol is tightly associated with cellular membranes. Moreover, Pol cofractionates with p65, indicating that Pol is associated directly with a protein population containing p65, or that Pol and p65 are associated with membranes of homogeneous density. Due to the association of the viral helicase and newly synthesized viral RNA with the dense gradient fractions, it had previously been suggested that this soluble Hel-N-p28-RNA population may be the catalytic component of the MHV replication complex (41). However, the segregation of Pol to the distinct fractions containing p65 is evidence that Pol-containing membranes must be sites of viral RNA synthesis. Why then would newly synthesized viral RNA and the viral helicase be separate from Pol after fractionation of the replication complex on a density gradient? One possible explanation is that Hel and RNA are tethered to Pol-containing membranes by peripheral mechanisms, such as transient protein-protein and protein-RNA interactions, that are disrupted during the gradient fractionation procedure. Viral RNA serves as a template for both new RNA and protein synthesis, and genome-length RNA is also packaged into budding virus. As such, it is likely that viral RNA molecules disassociate from the membranes of the replication complex immediately after being synthesized. The time course of Pol, p65, and Hel localization supports this conclusion and suggests a dual function for the Hel-N-p28-RNA population in both RNA synthesis and delivery of genomic RNA to the ERGIC. Based on the demonstrated GTPase and RNA unwinding activity of the human coronavirus 229E Hel protein (38, 39), it is likely that MHV Hel has important functions in RNA synthesis. If Pol and Hel are required to associate with each other during RNA synthesis, the separation of Hel from the replication complex-bound RdRp at late times may function to temporally regulate RNA synthesis and virion assembly for MHV.
trans determinants of Pol-replication complex association.
Although Pol is tightly associated with cellular membranes, it lacks predicted transmembrane domains and consensus myristoylation or palmitoylation motifs. Thus, it is likely that the association of Pol with membranes of replication complexes requires interactions with membrane proteins. We have shown that the association of a GFP-Pol fusion protein (Gpol) with replication complexes requires viral infection, suggesting that viral or virus-induced factors are required for Pol membrane association. Although it is likely that virus-induced cellular factors may be involved, the timing of Gpol targeting to replication complexes and the results of coimmunoprecipitation experiments argue for a model in which viral factors, specifically viral replicase proteins, mediate this process. MHV replicase proteins identified during our coimmunoprecipitation experiments with Pol include 3CLpro, p22, and p12. The fact that the coimmunoprecipitations were reciprocal suggests that they are specific interactions within a complex of proteins and is interesting for several reasons. First, 3CLpro is flanked by two putative membrane-spanning proteins, MP1 and MP2, which are highly hydrophobic and have been proposed to be the anchoring proteins of the replication complex. Second, a 150-kDa protein has been identified as a precursor containing MP1, 3CLpro, MP2, p10, p22, p12, and p15, which is juxtaposed to Pol in the polyprotein and cleaved by 3CLpro (36). We detect a protein of approximately 150 kDa after immunoprecipitation with an α-Pol antiserum during pulse-label and pulse-chase translation experiments. In light of the fact that Pol coprecipitates with 3CLpro, p22, and p12, it is reasonable to propose that the ∼150-kDa protein is the MP1-through-p15 precursor.
A proposed model for the formation of the replication complex during virus infection would be the anchoring of the 150-kDa precursor to membranes via MP1 and MP2, with continued cotranslational and posttranslational cleavages by 3CLpro. In this setting, protein interactions would retain the mature protein components at the sites of translation and processing in the complex. The association of the poliovirus RdRp (3Dpol) with membranes occurs in this manner, whereby there is a direct interaction between 3Dpol and a membrane-bound viral protein (3AB) (31, 43). The demonstration that MHV Pol relocalizes from a diffuse distribution in mock-infected cells to viral replication complexes upon MHV infection, despite expression of the protein as a fusion with GFP (Gpol), indicates the specificity of the proposed interactions. Quantitative biochemical analysis of Gpol membrane association in infected cells has been problematic because studies are performed in a heterogeneous cell population in which only a fraction of the cells (less then 15%) both express Gpol and are infected with MHV. Therefore, in the present study, confocal immunofluorescence microscopy was the most suitable method for analyzing the determinants of Gpol targeting to the MHV replication complex. Although more experiments are required to determine the specific interactions responsible for Pol membrane association, this is the strongest evidence to date of interactions between MHV replicase proteins. Moreover, the data presented in this report demonstrate the ability to target exogenously expressed Pol to replication complexes, suggesting that _trans_-complementation of MHV mutants with defects in their Pol domains may be possible.
cis determinants of Pol replication complex association.
Deletion mutagenesis experiments have identified a 38-aa sequence within Pol (F411 to D448) that is required for targeting of Gpol to replication complexes. MHV Pol has been predicted to contain the same domains and structure as other RdRps such as poliovirus 3Dpol (20, 26, 33). Specifically, RdRps are predicted to have a “right-hand” structure with conserved finger, palm, and thumb catalytic domains that are homologous in DNA-dependent RNA polymerases and reverse transcriptases, as well as an upstream amino-terminal RdRp unique region that is evident only in RdRps (20, 26, 33). Direct comparison of the deduced MHV Pol amino acid sequence to that of 3Dpol would predict that residues F411 to D448 lie in the RdRp unique region of the protein (Fig. 8) (20, 26, 33). A 178-aa region (D373 to R551) that contains residues F411 to D448 is not sufficient for targeting GFP to the MHV replication complex (Fig. 8), indicating either that more than one domain within the RdRp unique region is required for efficient association of Pol with the replication complex or that the minimal targeting domain extends upstream of residue D373. Still, our deletion experiments demonstrate that the conserved putative enzymatic domains (fingers, palm, and thumb) are not involved in Pol replication complex association. The RdRp unique region of MHV Pol has extensive differences in amino acid sequence even from closely related viral species, such as the arteriviruses, yet is highly conserved among coronaviruses, including the newly identified human coronavirus associated with severe acute respiratory syndrome (32). Comparison of the 38-aa regions (F411 to D448) from several coronavirus species reveals a concentration of conserved hydrophobic amino acids (Fig. 9). Specifically, this region contains 16% phenylalanine residues, compared to 6% in the total protein. These residues may facilitate interactions of Pol with proteins or RNA. In fact, it has been shown for several viral RdRps that the amino-terminal RdRp unique region mediates important protein interactions. In particular, for brome mosaic virus, the helicase-like protein (1a) interacts with the amino terminus of the RdRp-like protein (2a) and recruits it to viral replication complexes on endoplasmic reticulum membranes (9, 24). Experiments are in progress to define the minimal residues within the RdRp unique region that are sufficient for Pol replication complex association, as well as to identify viral or cellular factors that interact with these residues. Nonetheless, the results of this report demonstrate that a critical region of Pol is required for replication complex association and may represent a potential target both for mutagenesis of the newly developed MHV infectious clone (47) and for pharmacological inhibitors of coronavirus RNA synthesis and viral spread.
FIG. 9.
Amino acid alignment of the 38-aa Pol targeting domains from different coronavirus species. Shown is an alignment of amino acid residues 411 to 448 from MHV A59 (1) with the homologous sequences from the following coronaviruses: bovine coronavirus (BCoV) (46), severe acute respiratory syndrome coronavirus (SARS-CoV) (32), infectious bronchitis virus (IBV) (5), and human coronavirus 229E (HCoV229E) (21). Conserved residues are shaded, and the consensus sequence is given at the bottom.
Acknowledgments
This work was supported by Public Health Service grant RO1 AI-26603 (to M.R.D.). S.M.B. is also supported by a Training Grant in Mechanisms of Vascular Disease, Department of Pathology, Vanderbilt University School of Medicine (Richard L. Hoover, Program Director).
We acknowledge Kristen Bellenger for assistance and Erik Prentice, Rachel Graham, Amy Sims, and Paul McDonald for their support and critical reading of the manuscript. Confocal microscopy was performed in the Molecular Imaging Shared Resource of the Vanderbilt Ingram Cancer Center (IP30CA68485).
REFERENCES
- 1.Bonilla, P. J., A. E. Gorbalenya, and S. R. Weiss. 1994. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology 198**:**736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256**:**1604-1607. [PubMed] [Google Scholar]
- 3.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74**:**3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285**:**21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boursnell, M. E., T. D. Brown, I. J. Foulds, P. F. Green, F. M. Tomley, and M. M. Binns. 1987. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 68**:**57-77. [DOI] [PubMed] [Google Scholar]
- 6.Brayton, P. R., S. A. Stohlman, and M. M. Lai. 1984. Further characterization of mouse hepatitis virus RNA-dependent RNA polymerases. Virology 133**:**197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bredenbeek, P. J., C. J. Pachuk, A. F. Noten, J. Charite, W. Luytjes, S. R. Weiss, and W. J. Spaan. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV A59: a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 18**:**1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brierley, I., P. Digard, and S. C. Inglis. 1989. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57**:**537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74**:**4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denison, M., and S. Perlman. 1987. Identification of putative polymerase gene product in cells infected with murine coronavirus A59. Virology 157**:**565-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison, M. R., S. A. Hughes, and S. R. Weiss. 1995. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology 207**:**316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denison, M. R., A. C. Sims, C. A. Gibson, and X. T. Lu. 1998. Processing of the MHV-A59 gene 1 polyprotein by the 3C-like proteinase. Adv. Exp. Med. Biol. 440**:**121-127. [DOI] [PubMed] [Google Scholar]
- 13.Denison, M. R., W. J. Spaan, Y. van der Meer, C. A. Gibson, A. C. Sims, E. Prentice, and X. T. Lu. 1999. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73**:**6862-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denison, M. R., P. W. Zoltick, S. A. Hughes, B. Giangreco, A. L. Olson, S. Perlman, J. L. Leibowitz, and S. R. Weiss. 1992. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology 189**:**274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison, M. R., P. W. Zoltick, J. L. Leibowitz, C. J. Pachuk, and S. R. Weiss. 1991. Identification of polypeptides encoded in open reading frame 1b of the putative polymerase gene of the murine coronavirus mouse hepatitis virus A59. J. Virol. 65**:**3076-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennis, D. E., and D. A. Brian. 1982. RNA-dependent RNA polymerase activity in coronavirus-infected cells. J. Virol. 42**:**153-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong, S., and S. C. Baker. 1994. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology 204**:**541-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17**:**4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76**:**3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5**:**1109-1122. [DOI] [PubMed] [Google Scholar]
- 21.Herold, J., T. Raabe, B. Schelle-Prinz, and S. G. Siddell. 1993. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology 195**:**680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirano, N., K. Fujiwara, and M. Matumoto. 1976. Mouse hepatitis virus (MHV-2). Plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 20**:**219-225. [PubMed] [Google Scholar]
- 23.Jacks, T., H. D. Madhani, F. R. Masiarz, and H. E. Varmus. 1988. Signals for ribosomal frameshifting in the Rous sarcoma virus _gag_-pol region. Cell 55**:**447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao, C. C., and P. Ahlquist. 1992. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J. Virol. 66**:**7293-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J. C., R. A. Spence, P. F. Currier, X. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 208**:**1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72**:**2197-2206. [DOI] [PubMed] [Google Scholar]
- 27.Lee, H. J., C. K. Shieh, A. E. Gorbalenya, E. V. Koonin, N. La Monica, J. Tuler, A. Bagdzhadzhyan, and M. M. Lai. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180**:**567-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, D. X., H. Y. Xu, and T. D. Brown. 1997. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J. Virol. 71**:**1814-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, X., Y. Lu, and M. R. Denison. 1996. Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology 222**:**375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, X. T., A. C. Sims, and M. R. Denison. 1998. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J. Virol. 72**:**2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyle, J. M., A. Clewell, K. Richmond, O. C. Richards, D. A. Hope, S. C. Schultz, and K. Kirkegaard. 2002. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J. Biol. Chem. 277**:**16324-16331. [DOI] [PubMed] [Google Scholar]
- 32.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300**:**1399-1404. [DOI] [PubMed] [Google Scholar]
- 33.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252**:**287-303. [DOI] [PubMed] [Google Scholar]
- 34.Pachuk, C. J., P. J. Bredenbeek, P. W. Zoltick, W. J. Spaan, and S. R. Weiss. 1989. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology 171**:**141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73**:**2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242**:**288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmutz, C., L. Rindisbacher, M. C. Galmiche, and R. Wittek. 1995. Biochemical analysis of the major vaccinia virus envelope antigen. Virology 213**:**19-27. [DOI] [PubMed] [Google Scholar]
- 38.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6**:**1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seybert, A., and J. Ziebuhr. 2001. Guanosine triphosphatase activity of the human coronavirus helicase. Adv. Exp. Med. Biol. 494**:**255-260. [DOI] [PubMed] [Google Scholar]
- 40.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J. W. Oh, and M. M. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73**:**5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sims, A. C., J. Ostermann, and M. R. Denison. 2000. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J. Virol. 74**:**5647-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82**:**985-994. [DOI] [PubMed] [Google Scholar]
- 43.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271**:**26810-26818. [DOI] [PubMed] [Google Scholar]
- 44.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73**:**7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Meer, Y., H. van Tol, J. K. Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72**:**6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo, D., and Y. Pei. 2001. Full-length genomic sequence of bovine coronavirus (31 kb). Completion of the open reading frame 1a/1b sequences. Adv. Exp. Med. Biol. 494**:**73-76. [PubMed] [Google Scholar]
- 47.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76**:**11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]