Archaeal signal peptides—A comparative survey at the genome level (original) (raw)
Abstract
The correct delivery of noncytoplasmic proteins to locations both within and outside the cell depends on the appropriate targeting signals. Protein translocation across the bacterial plasma membrane and the eukaryal endoplasmic reticulum membrane relies on cleavable N-terminal signal peptides. Although the signal peptides of secreted proteins in Bacteria and Eukarya have been extensively studied at the sequence, structure, and functional levels, little is known of the nature of archaeal signal peptides. In this report, genome-based analysis was performed in an attempt to define the amino acid composition, length, and cleavage sites of various signal peptide classes in a wide range of archaeal species. The results serve to present a picture of the archaeal signal peptide, revealing the incorporation of bacterial, eukaryal, and archaeal traits.
Keywords: Archaea, protein translocation, secreted proteins, signal peptide
In each of the three domains of life, proteins destined for export from the cytoplasm are generally synthesized as preproteins, bearing a cleavable N-terminal signal peptide that serves to target the protein to the membrane-embedded export machinery (Rapoport et al. 1996; Eichler 2000; Manting and Driessen 2000). Preproteins destined for transfer across the eukaryal ER membrane via the Sec-based translocon are synthesized with the characteristic Sec signal peptide, composed of a basic n-region, a hydrophobic core, and a cleavage region (von Heijne 1990). In Bacteria, similarly composed signal peptides direct the posttranslational targeting of preproteins to SecYEG sites (Fekkes and Driessen 1999). Upon translocation across the membrane, the signal peptide is cleaved from the precursor via a membrane-bound signal peptidase. In Bacteria and eukaryal organelles of bacterial origin, preprotein export may also rely on the twin-arginine targeting (Tat) translocation pathway, so termed because of the presence of twin arginine or arginine-lysine residues in the signal peptides of Tat pathway substrates (Berks et al. 2000; Robinson and Bolhuis 2001). The export of bacterial type IV pilins is directed by yet a third class of signal peptides (Strom et al. 1994).
To date, little is known about protein export in Archaea, including the nature of signal peptides or range of secreted proteins. Indeed, because archaeal signal peptides have been identified and experimentally characterized in only a limited number of cases, it is unclear what archaeal signal peptides look like. The limited analysis performed thus far suggests that the mosaic character exhibited in other areas of archaeal biology also extends to archaeal signal peptides. On the one hand, features of archaeal signal peptidase I, the enzyme responsible for signal peptide cleavage, are eukaryal-like (Albers and Driessen 2002; Eichler 2002), suggesting that Archaea may rely on a eukaryal-like Sec signal peptide. In contrast, others have proposed that archaeal exported proteins may possess Gram-positive bacteria-like Sec signal peptides (Saleh et al. 2001). A bioinformatic study of signal peptides in the Euryarchaeote Methanococcus jannaschii suggested that Sec signal peptides in this organism may possess a hybrid character, combining a eukaryal-like cleavage site with a bacterial-like charge distribution, together with a hydrophobic region displaying a unique, archaea-specific composition. Traits of signal peptides unique to Archaea may, however, also exist (Nielsen et al. 1999). Indeed, most of these observations also hold true in the case of the Crenarchaeote Sulfolobus solfataricus (Albers and Driessen 2002). Moreover, although Sec-type signal peptides are predicted to predominate in archaeal genomes examined thus far (Rose et al. 2002; Dilks et al. 2003), studies addressing putative secreted proteins in Halobacterium sp. NRC-1 propose that this species makes predominant use of the Tat export system (Bolhuis 2002; Rose et al. 2002).
Since publication of the first studies addressing archaeal signal peptides, a significant number of additional archaeal genome sequences have become available. Hence, with the goal of a better description of archaeal protein export, the present report addresses, on a genome-wide level using Web-based programs, the predicted number of secreted proteins in a variety of Archaea, the nature of signal peptides employed, and finally, the types of proteins secreted.
Results and Discussion
Organisms studied
In this study, Web-based signal peptide and subcellular localization prediction programs were employed to screen 10 completed archaeal genomes in an attempt to identify putative archaeal secreted proteins. The organisms used in this study, along with some pertinent information regarding each species, are listed in Table 1. Two of the organisms examined belong to the Crenarchaeota (Aeropyrum pernix and S. solfataricus), while the other eight addressed are members of the Euryarchaeota, kingdoms within the domain Archaea (Woese et al. 1990). The genome size of the organisms under study ranges from 1.56 Mb for Thermoplasma acidophilum to 2.9 Mb for S. solfataricus, with the total predicted number of ORFs ranging from 1509 (T. acidophilum) to 2977 (S. sulfolobus). The studied organisms include seven heterotrophs, two autotrophs (the two methanogens), and one facultative autotroph (Archaeoglobus fulgidus). Three of the organisms listed are aerobes, five are anaerobes, while the two Thermoplasma species are facultative. All are thermophiles or hyperthermophiles except for the mesophilic Halobacterium sp. NRC-1. Furthermore, all of the organisms addressed in this study, with the exception of Methanothermobacter thermoautotrophicus, are flagellated and all are covered with surface (S)-layers except for the two Thermoplasma species and M. thermoautotrophicus, although the latter contains four predicted S-layer protein-encoding genes. Hence, given the broad range of physiological and structural characteristics borne by the 10 organisms considered, a wide spectrum of secreted proteins is likely to be expressed.
Table 1.
Organisms used in this study with pertinent traits
| Organism | Kingdom | Metabolism | Oxygen/temperature | S layer | Flagella | %G+C | Genome (MB) | ORFs | SignalP | Psort |
|---|---|---|---|---|---|---|---|---|---|---|
| Aeropyrum pernix | Crenarchaeote | Heterotroph | Aerobic hyperthermophile | Yes | Yes | 56.3 | 1.67 | 2694 | 672 (24.9%) | 121 (4.5%) |
| Archaeoglobus fulgidus | Euryarchaeote | Autotroph or heterotroph | Anaerobic hyperthermophile | Yes | Yes | 48.5 | 2.18 | 2436 | 382 (15.7%) | 79 (3.2%) |
| Halobacterium sp. NRC-1 | Euryarchaeote | Heterotroph | Aerobic mesophile | Yes | Yes | 67.8a | 2.57 | 2630 | 459 (17.5%) | 68 (2.6%) |
| Methanococcus jannaschii | Euryarchaeote | Autotroph | Anaerobic hyperthermophile | Yes | Yes | 31.4 | 1.67 | 1738 | 197 (11.3%) | 32 (1.8%) |
| Methanothermobacter thermoautotrophicus | Euryarchaeote | Autotroph | Anaerobic thermophile | Nob | No | 49.5 | 1.7 | 1855 | 268 (14.4%) | 68 (3.7%) |
| Pyrococcus abyssi | Euryarchaeote | Heterotroph | Anaerobic hyperthermophile | Yes | Yes | 45 | 1.76 | 1765 | 299 (16.9%) | 28 (1.6%) |
| Pyrococcus horikoshii | Euryarchaeote | Heterotroph | Anaerobic hyperthermophile | Yes | Yes | 41.9 | 1.74 | 2061 | 423 (20.5%) | 63 (3.1%) |
| Sulfolobus solfataricus | Crenarachaeote | Heterotroph | Aerobic thermophile | Yes | Yes | 36 | 2.9 | 2977 | 374 (12.6%) | 46 (1.5%) |
| Thermoplasma acidophilum | Euryarchaeote | Heterotroph | Facultative aerobe thermophile | No | Yes | 46 | 1.56 | 1509 | 176 (11.6%) | 26 (1.7%) |
| Thermoplasma volcanium | Euryarchaeote | Heterotroph | Facultative aerobic thermophile | No | Yes | 39.9 | 1.59 | 1522 | 169 (11.1%) | 24 (1.6%) |
Quantitation of the archaeal secretome
The archaeal genomic sequences were first addressed using SignalP v2.0 to detect the presence of predicted signal peptides and then to distinguish between signal peptides and noncleaved signal anchors (Nielsen et al. 1999). As only a limited number of archaeal secretory proteins have been characterized experimentally, automated computer programs such as SignalP can only be trained against reported or predicted eukaryal and bacterial (Gram-positive and Gram-negative) secreteomes. Accordingly, such genome-wide analysis of archaeal signal peptides would fail to detect the presence of uniquely archaeal putative signal peptides, that is, those that do not resemble either their eukaryal or bacterial counterparts. Despite this drawback, a significant number of signal peptide-bearing putative protein sequences were identified. Such analysis should, in theory, identify any protein targeted to the archaeal plasma membrane, be they either a secreted or membrane protein. In the next step of the analysis, those proteins predicted to contain signal peptides by at least one of the three SignalP data sets were then examined using Psort, a subcellular localization prediction program. Thus, in this study, proteins were considered secreted if they were predicted to bear a signal peptide by at least one of the three SignalP data sets and identified by Psort as either secreted in Gram-positive organisms, or localized to the periplasm or outer membrane of Gram-negative organisms. Proteins that were predicted to localize to the cytoplasmic membrane were excluded from this study, as were proteins listed by Psort as "unclear" using these criteria, although some of the latter may, in reality, correspond to true secreted proteins.
In the analysis performed using SignalP, from 169 (in Thermoplasma volcanium) to 672 (in A. pernix) proteins with potential signal sequences were identified (Table 1). From these values, it can be estimated that from 11.1% to 24.9% of the total number of predicted proteins are synthesized with signal peptides. The predictions made by SignalP were accepted regardless of the length of the signal peptide entailed, even though signal peptides greater than 50 amino acids in length, obtained in a few instances, are suspect. In Halobacterium sp. NRC-1, a number of examples were obtained, wherein, after removal of the predicted signal peptide, a secreted protein comprising only a very short peptide, from 10–31 amino acids in length, was predicted. These were all annotated as hypothetical proteins and seem unlikely to correspond to true proteins, being even smaller than known microhalocins, secreted protein antibiotics widespread in the haloarchaea (O’Connor and Shand 2002).
The lists of signal peptide-bearing proteins, which include secreted proteins, membrane proteins, and false positives, were then screened through Psort to theoretically include only secreted proteins. Taking into account that the caveats described above would lead to a small overestimation, a final tally ranging from 24 (T. volcanium) to 121 (A. pernix) secreted proteins was obtained. In terms of the percentage of total genome ORFs predicted by the screening approach employed, secreted proteins account for as low as 1.5% in S. solfataricus to as high as 4.5% in A. pernix. Thus, the two crenarchaeotes included in this study account for both the high and low end of secreted proteins, expressed as a percentage of the total genome ORFs.
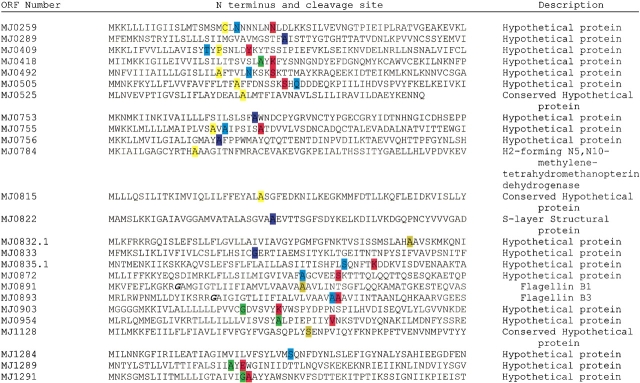
The lists of Psort-predicted secreted proteins from each genome are presented in Tables 2–11 (Table 2, showing the predicted secreted proteins from M. jannaschii, is presented whereas Tables 3–11, showing the predicted secreted proteins from the other strains are included in the Supplemental Material). Because of the uncertainty concerning the actual cleavage sites used in Archaea, the predicted signal peptide cleavage sites obtained using eukaryal and Gram-positive and Gram-negative bacteria trained systems are shown in a color-coded fashion in each case.
Table 2.
Signal peptide cleavage site prediction in the secreted proteins of Methanococcus jannaschii using HMM of SignalP
Using different computer programs (i.e., ExProt and LocateProtein), Saleh et al. (2001) predicted that A. pernix contained 712–857 exported/secreted proteins (26%–32% of the putative proteome), while the predicted number of exported/secreted proteins (expressed as percentage of the putative proteome) for other Archaea were: A. fulgidus 213–448 (9%–19%), M. thermoautotrophicus 208–250 (11%–13%), M. jannaschii 143–210 (8%–12%), Halobacterium sp. NRC-1 393–488 (19%–24%), and Pyrococcus horikoshii 331–461 (16%–22%), respectively. In the case of M. thermoautotrophicus, it was previously reported (Smith et al. 1997) that ∼12% of the proteins bore signal peptides. The values of both these studies are in good agreement with the SignalP results obtained in the present study, yet do not distinguish between proteins embedded in the plasma membrane, that is, exported, and those secreted, as differentiated here by the Psort program.
In quantifying the number of noncytoplasmic proteins in Archaea, Saleh et al. (2001) reported that, with the exception of A. pernix, Archaea contain fewer secreted or membrane inserted proteins than do Bacteria. In agreement with these results, G. Schneider (1999) predicted the fraction of noncytoplasmic proteins to be very low in Archaea. Despite these results, as well as those presented in the present study, one must be careful of generalizing that Archaea possess fewer secreted proteins than do Bacteria. A minimal number of secreted proteins may instead be a trait of (hyper)thermophilic organisms. Indeed, all the archaeal species examined in these earlier studies as well as in the present study (except Halobacterium sp. NRC-1) are thermophiles or hyperthermophiles, and show a low percentage of secreted proteins, as does the hyperthermophilic bacterium, Aquifex aeolicus (Schneider 1999). Moreover, as noted by G. Schneider (1999), hyperthermophiles tend to encode more charged amino acids than do mesophiles. The presence of charged residues could skew the predictions because the presence of such residues is an important factor in the LocateProtein prediction software used in earlier studies (Schneider 1999).
Classification of secreted archaeal proteins
Within each species, the vast majority (about 70%–90%) of the secreted proteins identified have thus far not been assigned a gene designation. The exception to this is in T. volcanium, where only 9 of 24 of the predicted secretory proteins have not been identified. Among those unidentified secreted proteins predicted in the various species by the present study, it was possible, in some instances, to assign putative functions by analyzing the hypothetical proteins through COGnitor (http://www.ncbi.nlm.nih.gov/COG/). For example, in the case of M. jannaschii, there are 28 of 32 secreted proteins that are annotated as hypothetical (Table 2). Upon analysis of the 28 hypothetical proteins through COGnitor, five were shown to be members of existing COGs, thereby hinting at their function. MJ0505 was assigned to COG 3233 and predicted to be adeacetylase; MJ0755 was assigned to COG 2247 and predicted to be a cell wall binding domain; MJ0815 was assigned to COG 1821 and predicted to be an ATP-utilizing enzyme of the ATP-grasp superfamily; MJ0872 was assigned to COG 0614 and predicted to be an ABC type cobalamin/Fe3+- siderophore transport system, periplasmic component; while MJ1502 was assigned to COG 1234 and predicted to be a metal-dependent hydrolase of the beta lactamase superfamily. In the case of T. volcanium, where only 9 of 24 secreted proteins were unidentified, one unidentified protein (TVN0969) was designated a member of COG 1136, proteins which serve as the ATPase component of an ABC-type transport system involved in lipoprotein release.
Of the 10 archaeal strains addressed in the current report, 7 are covered with S-layers. In addition, M. thermoautotrophicus contains genes annotated as encoding S-layer proteins. In contrast, the current annotated genome sequences of some of the S layer-possessing strains (i.e., A. pernix, Pyrococcus abyssi, P. horikoshii, and S. solfataricus) do not identify S-layer protein-encoding genes. Presumptive S-layer proteins have, however, been reported for P. abyssi (PAB1861) and P. horikoshii (PH1395) based on their significant homology to S-layer proteins in various methanococci (Claus et al. 2002). In Bacteria, S-layer proteins, located on the surface of the cell, would be recognized as secreted proteins by Psort. However, in the archaeal species studied here, only in M. jannaschii and M. thermoautotrophicus are the S-layer proteins designated as secreted by Psort. S-layer proteins in the other Archaea addressed here are predicted by Psort to be membrane proteins. Such assignment is likely a true reflection of the unusual wall structures of these Archaea, where the S-layer is in direct contact with the cytoplasmic membrane via a single transmembrane domain, without the intervening peptidoglycan of bacterial walls. Indeed, in the case of the Halobacterium sp. NRC-1 S-layer glycoprotein, the single transmembrane stretch, located between amino acid positions 817–833, has been directly shown to act as a membrane anchor (Lechner and Sumper 1987). Although translocated across the membrane, yet membrane-anchored via C-terminal membrane-spanning domains, these S-layer proteins cannot be considered as secreted by the criteria adopted for subcellular localization using Psort in this study. Still, archaeal S-layer proteins are among the few proteins whose signal proteins have been experimentally confirmed. As such, the finding that the signal peptides of these proteins were correctly predicted by SignalP in the present analysis is reassuring. Indeed, the N terminus of the mature M. jannaschii S-layer protein has recently been reported (Akca et al. 2002), leading to the prediction of a 28 amino acid residue-long signal peptide. An identical sequence was determined in the current study.
The cell envelope structure of the Thermoplasma species may also be implicated in the low number of secreted proteins predicted in these species. As mentioned, most of the archaeal strains addressed in the present study are surrounded by a protein-based S-layer without any intervening cell wall layer (Kandler and Konig 1993). The anchoring of S-layer proteins by C-terminal hydrophobic extensions has been speculated to lead to the presence of a pseudo-periplasmic space between the outermost canopy of the S-layer and the membrane (Kessel et al. 1988). Such a cellular compartment might house specific populations of secreted proteins, as does the periplasm of Gram-negative Bacteria (Sara and Sleytr 2000). In the case of the two Thermoplasma species, no cell wall of any type exists (Kandler and Konig 1993). Hence, the potential reservoir for secreted proteins, that is, the pseudo-periplasm, is absent, in turn perhaps contributing to the very low number of secreted proteins predicted for these two species, as reported in this study.
Although the lists of proteins identified as being secreted by the two-stage analysis employed in this study include many hypothetical proteins, numerous proteins of known or predicted identity, expected to be located beyond the cytoplasmic membrane, are also accounted for. These include various extracellular enzymes, binding proteins, adhesion molecules, etc. On the other hand, it is clear that some of the proteins listed in this report as secreted are unlikely to be so. Indeed, examples exist where all three trained data sets predicted the presence of a signal peptide on a protein not expected to be secreted. In P. abyssi there are three such proteins, PAB0512 (hydropantoate 2 reductase), PAB0936 (NADH oxidase), and PAB0763 (cytidylate kinase). In the cases of both PAB0763 and PAB0936, the amino acid sequences show excellent alignment over the entire length in a BLAST search to similar proteins in a wide variety of archaeal and bacterial species. However, the two P. abyssi proteins are extended by five or seven amino acids, respectively, at the N terminus compared to almost all homologs outside of other Pyrococcus species. In the case of PAB0936, translation is initiated at an alternate start codon, GTG. If five amino acids are removed from the N terminus of PAB0763, then it is considered to be a nonsecreted protein by all three trained data sets in SignalP. Under these circumstances, this protein would not be examined by Psort and, hence, would not have ended up on the secreted protein list amassed in this study. Similarly, if seven amino acids are removed from the N terminus of PAB0936 to align its start with the overwhelming majority of other homologs, then it is also considered nonsecretory by both bacterial-trained data sets, although not by the eukaryotic-trained data set. No alignment irregularity was noticed for the third protein PAB0512. Clearly, this small sampling indicates how small errors in the translation start sites could have significant effects on the SignalP determination.
Although inaccurately predicted translational start sites and other annotation errors might account for a few of these anomalies, the inclusion of such proteins in the list of Psort-selected secreted proteins is a likely reflection of the lenient criteria used here for selecting signal peptide-bearing proteins. In this study, any protein predicted to contain a signal peptide by only one of the three SignalP trained data sets was considered as secreted. In other words, there exist cases where a protein is predicted to bear a signal peptide by one data set yet not by the other two. As such, nonsecreted proteins would be erroneously included in the lists of secreted proteins. There is, however, an inherent failure rate with Psort as it was reported that the program successfully predicted the correct protein location 66% of the time for eukaryotic proteins and 83% for E. coli proteins (http://psort.nibb.ac.jp/helpwww.html; Horton and Nakai 1997). Thus, although the Psort program was successful in trimming the number of proteins bearing signal peptides from 11.1%–24.9% of the total number of proteins in the different genomes to a list of secreted proteins accounting for only 1.5%–4.5% of genomic ORFs, it is apparent that errors in designation in a number of cases have been made. However, although the selection strategy employed in this study implicitly leads to an overestimation of the actual number of secreted proteins, the predicted number of secreted archaeal proteins is still low (compared to Bacteria). Hence, it is possible that either the number of actual secreted proteins in Archaea is indeed extremely low, or that there exist significant numbers of secretory proteins synthesized with archaeal-specific signal peptides completely missed by currently employed trained data sets. On the other hand, it is also possible that the list of secreted proteins identified include a large number of candidates that, in fact, do not encode for true proteins. This point will be clarified as more is learned about Archaea and their proteins.
Nature of archaeal signal peptides
The amino acid composition and lengths of the predicted signal peptides within each organism were next compared. In all cases, the shortest average signal peptides were predicted by the eukaryotic training system and the longest by the Gram-positive bacterial trained data set. The values for each individual archaeon are listed in Tables 2–11, as are the compositions of the various signal peptides.
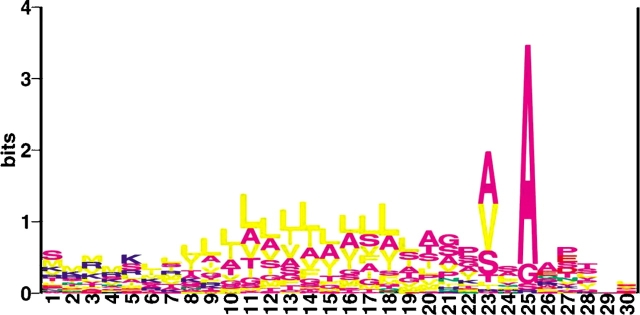
Of the total number of secreted proteins predicted from all 10 species, 78 contained a signal sequence cleavage site predicted by all three SignalP trained data sets, and as such, were more closely examined. The amino acid positions around these predicted cleavage sites are depicted in a LOGO (T.D. Schneider and Stephens 1990) in Figure 1 ▶. The −1 position is almost exclusively alanine (69 of 78), as is the case in Bacteria (Nielsen et al. 1997). The −3 position is dominated by approximately equal representation of alanine (29 of 78) and valine residues (26 of 78), although serine (13 of 78) is also common at this position. The +1 to +3 positions following the cleavage site have preferences again as observed in Bacteria but not in Eukarya. Alanine (19 of 78) is found most frequently at the +1 position, but is rarely, if ever, found at the +2 or +3 positions. Negatively charged amino acids (i.e., glutamic and aspartic acid) are also common at positions +1 (13 of 78) and +2 (20 of 78), but are rarely noted at position +3. The hydroxyl-bearing amino acids serine and threonine are commonly found at positions +2 (17 of 78) and +3 (20 of 78). The singularly most common amino acid at position +2 is, however, proline, found in 13 instances. It was also observed that in the predicted archaeal signal peptide h-region, leucine and alanine are the dominant amino acid residues, predicted to be present in approximately equal amounts, again as observed in Bacteria, although valine and isoleucine residues are also common. The predominance of leucine in the h-region, observed in Eukarya (Nielsen et al. 1997), is hence apparently not the case in Archaea. Thus, in those 78 proteins where all three SignalP data sets predicted the same cleavage site, the signal peptide resembles more closely the characteristic bacterial signal peptide than the typical eukaryal signal peptide. Examination of the sequence LOGO of 34 predicted signal peptides from M. jannaschii (Nielsen et al. 1999) revealed a dominance of alanine at the −1 position, a high isoleucine presence in the h-region, significant presence of valine, isoleucine, and leucine residues at the −3 position, a positively charged n-region with a lysine content and rare presence of arginine residues, as well as the occurrence of charged amino acids in the first positions following the cleavage site. In the 78 proteins studied in the current study, neither the high isoleucine presence in the h-region nor the significant presence of isoleucine or leucine residues at the −3 position were observed. Moreover, a positively charged n-region was observed that, as in Bacteria, included relatively equal arginine and lysine contents. However, because the earlier M. jannaschii study addressed all those proteins predicted to bear signal peptides by all three trained data sets, regardless of the final cellular destination of the protein, those results may not be directly comparable to the selected subset of secreted proteins addressed in the present report. In addition, some features observed by Nielsen et al. (1999) may be characteristic of M. jannaschii. For example, the high isoleucine content in the h-region of M. jannaschii signal peptides may be partly due to an overrepresentation of that amino acid in the genome as a whole (Nielsen et al. 1999).
Figure 1.
A sequence LOGO of 78 predicted archaeal signal peptides, aligned at their cleavage site (no gaps) using the default settings at the WebLogo site (http://www.bio.cam.ac.uk/cgi-bin/seqlogo/logo.cgi). The last amino acid in the signal peptide is position 25.
In addition, it had been noted in earlier examinations of putative M. jannaschii and S. solfataricus secreted proteins (Nielsen et al. 1999; Albers and Driessen 2002) that an unusual feature of the predicted archaeal signal peptide cleavage site is the high occurrence of nearby tyrosine residues, often found at the +1 and −2 positions. This was not evident in the 78 secreted proteins examined in the current study, where only 1 protein contained a tyrosine at position +1, although 6 more contained tyrosine residues at position −2. If the search for tyrosine residues was extended to include any of positions −3 to +3, 15 of 78 proteins then included a tyrosine residue in the cleavage site region.
Twin arginine transport (Tat) signal peptides
In addition to the well-characterized prokaryotic Sec protein export system, a Sec-independent system has been recently described in Archaea, Bacteria, and chloroplasts (Berks et al. 2000; Robinson and Bolhuis 2001). Substrates of the so-called twin arginine transport (Tat) protein translocation system possess a signal sequence with n-, h-, and c-regions like those of Sec pathway proteins, yet possess several distinct Tat system-specific features. Foremost is the presence of a twin arginine motif, which is part of a larger conserved sequence (S/T)-R-R-x-φ-φ (where x is any amino acid and φ is a hydrophobic amino acid) found at the n-region/h-region boundary (Berks et al. 2000). In Gram-negative Bacteria, this sequence is usually (S/T-R-R-x-F-L-K). In addition, Tat signal peptides favor proline at position −6 and basic amino acids in the c-region proximal to the cleavage site. Tat signal proteins are also longer, on average, by 14 amino acid residues than Sec signal peptides. Archaeal reliance on the Tat pathway, however, appears to widely vary, as gauged by the predicted number of secreted archaeal proteins bearing a Tat signal peptide (Dilks et al. 2003). In Halobacterium sp. NRC-1, it has been proposed that the Tat system is the predominant protein export system, owing to the ability of the Tat pathway to translocate folded proteins. Such a property could complement the putative requirement for rapid folding of halophilic proteins so as to overcome dangers of biosynthesis in a highly saline cytoplasm as found in these cells (Bolhuis 2002; Rose et al. 2002). In other archaeal species, only limited use of the Tat system is predicted. For example, only three proteins with signal peptides that closely match the consensus for the twin arginine system have been reported in S. solfataricus (Albers and Driessen 2002). One of these is a Rieske iron sulfur protein (SSO2971), typical of the substrates for this system, that is, proteins that bind one or more cofactors in the cytoplasm and are folded before export (Berks et al. 2000). Interestingly, in accordance with the varied usage of the Tat system in Archaea, the presence of TatA and TatC, subunits of the Tat translocation machinery, was not detected in all archaeal species, specifically not in Pyrococcus species or the majority of methanogens (Yen et al. 2002; Dilks et al. 2003).
In their recent analysis, Rose et al. (2002) compiled a list of 64 putative Halobacterium sp. NRC-1 Tat substrates (compared to about 26 Tat substrates in Escherichia coli; Stanley et al. 2001; Robinson and Bolhuis 2001). Of these 64 candidates, recognizable homologs exist for 34 (13 binding proteins, 8 putative extracellular enzymes, 7 redox proteins, and 6 surface proteins). Of these 34 Halobacterium proteins, most of the putative extracellular enzymes were also identified as secreted in our analysis. These included Chi (chitinase), Vng0818c (chitinase A), Vng0819c (chitinase), Sub (subtilisin homolog), and Hly (halolysin). Moreover, these numbers reported by this earlier study are in line with a second recent study addressing the number of Tat pathway substrates predicted by the Halobacterium sp. NRC-1 genome (Bolhuis 2002). In that study, of the 103 proteins identified as bearing putative signal peptides, over 60% were predicted to target proteins to the Tat system. In the present analysis, numerous putative Tat system substrates were identified. These did not include many of the Tat substrates predicted in Halobacterium sp. NRC-1 and other archaeal species in earlier studies, as these were assigned an unclear designation by Psort in the present study, with either a cytoplasmic membrane or outer membrane location being predicted. As such, these were not included in the list of secreted proteins in the present report.
Flagellin-like signal peptides
It has been previously reported that the flagellins of Archaea are made as preproteins with short, atypical signal peptides which, in certain aspects, resemble signal peptides found in bacterial type IV prepilins (Faguy et al. 1994). It is a telling fact of the state of archaeal protein secretion that these atypical signal peptides are the best studied of all archaeal signal peptides. Key amino acids in and around the cleavage site have been identified by site directed mutagenesis (Thomas et al. 2001b). The enzyme involved in the processing, called FlaK or preflagellin peptidase, has also been identified in methanococci (Bardy and Jarell 2002) and homologs are present in several other flagellated archaea. FlaK is a prepilin peptidase equivalent (a member of the same COG 1989 "signal peptidases that cleave prepilin-like proteins"). As well as flagellins, this enzyme is also responsible for the processing of a small number of other proteins, at least some of which seem unrelated to flagellar assembly. Among such proteins with identified functions are a number of sugar binding proteins in S. solfataricus, containing highly similar signal peptides and experimentally shown to be processed at the correct site by N-terminal sequencing (Albers and Driessen 2002). Of the proteins determined in the current study to be secreted, several, such as sugar binding proteins in T. volcanium, can be readily detected by visual inspection as bearing the flagellin-like signal peptide/mature N-terminal sequence. However, in the autotrophic flagellated Archaea, such as M. jannaschii, the only proteins cleaved by FlaK may indeed be preflagellins.
Although flagellin sequences have a reported signal peptide of 11–12 amino acids in length (including M. jannaschii, Halobacterium sp. NRC-1, A. fulgidus, and various other Archaea not addressed in the present study; Thomas et al. 2001a), several others are annotated with much different, often extremely short, signal peptides of only 4–6 amino acids (see certain flagellins in P. abyssi, P. horikoshii, A. pernix, as well as other Archaea not included in this study). However, given that reducing the length of the signal peptide in an in vitro Methanococcus voltae preflagellin processing assay from 12 to 6 amino acids resulted in a loss of signal peptide cleavage (Thomas et al. 2001b), it may be that those very short predicted signal peptides, in fact, represent inaccurately predicted translation start sites. In contrast, some annotated flagellin sequences are predicted to have much longer signal peptides (e.g., Ta1407 in T. acidophilum and MJ0893 in M. jannaschii). Some of these sequences, however, contain in-frame methionines. Should these methionines correspond to true translation start sites, the signal peptides would be considerably shortened. This appears likely for MJ0893, where a well-positioned ribosome binding site is located upstream of an in-frame methionine residue, leaving an 11 amino acid residue signal peptide. At present, neither the biochemical nor genetic work needed to accurately identify the correct translation initiation site of most flagellins has been performed. Indeed, it is evident that translation start errors have been made in three of six flagellins in the annotated Halobacterium sp. NRC-1 sequence, as the three in question all start at methionine residues well into the N-terminal region of the mature processed proteins (compare, for example, the flagellin gene family reported in Halobacterium salinarum strain R1M1 by Gerl and Sumper 1988).
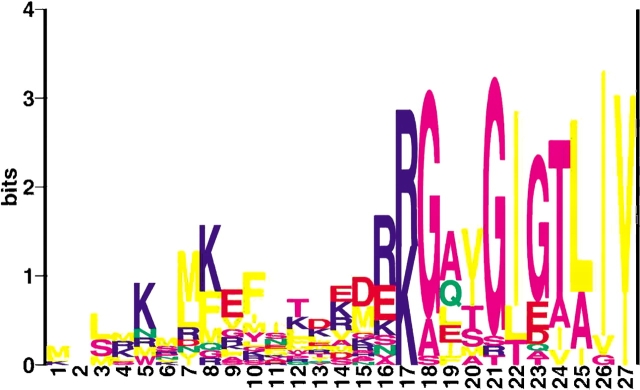
Available flagellin N-terminal sequences have been used to generate a LOGO diagram (Fig. 2 ▶). In generating the figure, translation start sites reported in the annotated genomes have been employed, with the exception of the three likely erroneous Halobacterium sp. NRC-1 sequences mentioned above. For these halobacterial flagellin sequences, we have instead used the identical signal peptide sequence reported for all five H. salinarum strain R1M1 flagellins, a sequence also reported for two of the flagellins annotated in the Halobacterium sp NRC-1 genome. The sixth predicted Halobacterium sp. NRC-1 flagellin has a signal peptide that is obviously different, yet which appears to have a correct initiation site, based on the conservation of sequence observed in all archaeal flagellins. This latter sequence has also been included in generating the LOGO diagram. Also included in the LOGO are the three S. solfataricus sugar-binding proteins (SSO0999, SSO2847, SSO3066) shown to be processed at the preflagellin-like cleavage site (Albers and Driessen 2002).
Figure 2.
A sequence LOGO of flagellin and sugar binding proteins signal peptides, aligned at their cleavage site (no gaps) using the default settings at the WebLogo site (http://www.bio.cam.ac.uk/cgi-bin/seqlogo/logo.cgi). The last amino acid in the signal peptide is position 18.
The LOGO diagram shows graphically the extreme conservation of the N terminus of the mature archaeal flagellin and sugar binding proteins and the conserved nature of the amino acids surrounding the cleavage site. In almost all cases, the signal peptide ends with two charged amino acid residues followed by a glycine at the −1 position. Usually the −2 and −3 positions are held by lysine or arginine, but rarely, as is the case in some of the halophilic and haloalkaliphilic flagellins, even positively charged amino acids can be found at these positions. The −1 position can be alanine in rare instances, and in one case (Ta1407 of T. acidophilum), a serine residue is detected.
The N-terminal 40–50 amino acids of the processed flagellins are highly conserved and hydrophobic. As with type IV pilins, the flagellins appear to have a typical signal peptide, which is, however, cleaved before the h-region rather than after it. Thus, many archaeal flagellins are predicted by SignalP to have a signal peptide (by one, two, or all three trained data sets), but in all cases, the cleavage site is incorrectly predicted. Considering the conservation of sequence at the N terminus, it was strange that, in many instances, the analysis identified only some of the multiple flagellins found within a given genome as secreted proteins. Although this may be considered a moot point because, as the signal peptides for all flagellins are incorrectly predicted by available programs, the fact that not even all flagellins are recognized by SignalP/Psort as secreted proteins does mean that other potential secreted proteins processed by FlaK and its homologs likely represent a missed subset of secreted proteins. Indeed, the sugar-binding proteins SSO2847 and SSO3066 shown in S. solfataricus as secreted and processed by the FlaK equivalent are not predicted to be secreted by the SignalP/Psort programs, as employed in the present study.
Lipoproteins
It seems increasingly clear that Archaea contain proteins that possess the LIPO box found in bacterial lipoproteins. This consensus sequence, [L/I/G/A]-[A/G/S]-C, contains an invariant cysteine at the +1 position, relative to the signal peptide cleavage site. In the mature lipoprotein, this cysteine residue is attached to a membrane-associated lipid anchor. Thus far, evidence for a lipoprotein-like lipid modification has been reported for halocyanin of Natranobacterium pharaonis (Mattar et al. 1994) and more recently in the H. salinarum binding proteins BasB and CosB (Kokoeva et al. 2002). Indeed, based on the lipoprotein signature, it has been suggested that all binding proteins annotated in the complete Halobacterium sp. NRC-1 genome are lipoproteins (Kokoeva et al. 2002). Earlier analysis of other complete genomes also predicted the existence of lipoproteins in various Archaea. A few potential lipoproteins have been suggested in S. solfataricus (Albers and Driessen 2002) and in both P. horikoshii (Kawarabayasi et al. 1998) and A. pernix (Kawarabayasi et al. 1999), many proteins have been reported to contain prokaryotic membrane lipoprotein lipid attachment site motifs.
Few, if any, lipoproteins would be expected to be found among the lists of secreted proteins generated in the present study since lipoproteins are generally embedded in either the cytoplasmic membrane (and, hence, not considered as secreted by the criteria used here) or in the outer membrane, which is not found in Archaea. Nonetheless, a screening of all the secreted proteins predicted in the present study at Prosite (PROSITE Release 17.42, of 06-Apr-2003; http://www.expasy.ch/prosite/), software designed to predict the presence of a prokaryotic lipoprotein signature motif, was performed. Only in the Pyrococcus species and A. fulgidus did Prosite predict the presence of lipoproteins among the secreted proteins. These predicted lipoproteins are P. abyssi PAB1651 (hypothetical protein), P. horikoshii PH0462 (hypothetical protein), PH1130 (hypothetical protein) and PH1190 (hypothetical protein), and A. fulgidus AF0397 (hypothetical protein). The three P. horikoshii proteins, as well as four others (PH1525, PH1690, PH1707, PH1929) included in the compilation of secreted proteins listed in this study, were among the many proteins listed previously as possessing the prokaryotic membrane lipoprotein attachment site in P. horikoshii (Kawarabayasi et al. 1998). However, the latter four proteins were not recognized as lipoproteins by Prosite. Similarly, eight proteins from A. pernix (APE0189, APE1303, APE1309, APE1348, APE1433, APE2040, APE2099, APE2434) also previously reported (Kawarabayasi et al. 1999) to contain the prokaryotic membrane lipoprotein attachment motif were also among those predicted secreted proteins for that organism in the present study. However, none were recognized as potential lipoproteins by Prosite. Interestingly, when all seven P. horikoshii potential lipoproteins as well as P. abyssi PAB1651, A. fulgidus AF0397, and the eight A. pernix putative lipoproteins were analyzed by a second program, namely the Lipop program, software found at the Psort site designed to detect lipoprotein signal sequences based on the consensus motif around the lipoprotein cleavage site as formulated by von Heijne (1989), none were identified as lipoproteins. It should, however, be pointed out that the reliability of both Web programs is questionable, because both BasB and the BasB equivalent in Halobacterium sp. NRC-1 (i.e., Vng1857C) are predicted by Lipop and Prosite to be lipoproteins, whereas both CosB and the CosB Halobacterium sp. NRC-1 equivalent (proX, a putative ABC transporter) are not recognized as lipoproteins by either program.
In Bacteria, processing of lipoprotein signal peptides is performed by signal peptidase II (SPII). In SPII substrates, the cysteine residue is lipid-modified prior to the removal of the signal peptide by the enzyme. Despite the existence of SPII substrates in Archaea, to date, no archaeal SPII equivalent has been annotated in any completed genome sequences nor in the COG site database (http://www.ncbi.nlm.nih.gov/COG/). However, given that the lipid modification found on any archaeal lipoproteins is likely to be of the unusual C20 diphytanyl diether lipid type typical of Archaea (Kokoeva et al. 2002), it is not surprising that a SPII equivalent has not yet been reported upon examination of completed archaeal genomes.
Conclusions
In Archaea, numerous proteins, including a variety of enzymes and components of the protein-based S layer, must escape the confines of the cell. As in Bacteria and Eukarya, translocation of secreted proteins in Archaea requires that such proteins first be distinguished from the pool of cytoplasmic proteins, that they then be targeted to membranous translocation sites, and ultimately, that they be transported into and across the membrane. Signal peptides play an important role in each of these processes. To date, however, archaeal signal peptides have not been well characterized. Hence, towards reddressing this situation, signal peptides were selected from 10 completed archaeal genome sequences on the basis of their similarities to signal peptides of eukaryotic and Gram-positive and Gram-negative bacterial exported/secreted proteins. Such analysis revealed that archaeal signal peptides incorporate properties found in eukaryal and bacterial signal peptides, much like what is observed in other aspects of archaeal biology, including protein translocation. However, in the 78 proteins where the same cleavage site was predicted by all three trained data sets, the signal peptides more closely resembled the characteristic bacterial signal peptide rather than those found in eukaryal secreted proteins. This prediction, as well as the suggested utilization of a variety of different translocation systems, awaits experimental confirmation. Similarly, the existence of archaeal-specific signal peptides remains to be addressed. The selection criteria employed in this study, relying on similarities to identified eukaryal and bacterial signal peptides, would, by definition, fail to detect putative archaeal-specific signal peptides. Indeed, given the extremophilic nature of many Archaea, coupled with the unusual archaeal cell envelope (Kandler and Konig 1993) and unique membrane lipid structures (Kates 1993), it is conceivable that features of the different protein export systems, including their signal peptides, may be specific to this domain.
Materials and methods
Signal peptide determination
The 10 completed archaeal genomes (listed in Table 1) and deduced proteins used in this survey were obtained from the comprehensive microbial resource, found at the TIGR website (http://www.tigr.org). All predicted protein-encoding genes from each organism were analyzed using the automated program SignalP v2.0 (http://www.cbs.dtu.dk/services/SignalP-2.0), designed to detect the presence of signal peptides. In these analyses, protein sequences were examined using the hidden Markov model with truncation set to 70 amino acids, trained on the different data sets available (Eukarya, Gram-positive, or Gram-negative Bacteria). Proteins predicted to contain signal peptides by at least one of the three data sets were then examined using Psort (Nakai and Horton 1999; http://www.psort.nibb.ac.jp), a computer program designed to predict subcellular localization. Only those proteins considered to be secreted in Gram-positive organisms or localized to the periplasm or outer membrane of Gram-negative organisms were further examined.
Electronic supplemental material
Tables 3–11, listing the predicted secreted proteins, signal peptides, and putative cleavage sites of each of the archaeal genomes (except M. jannaschii, which is presented in Table 2 in the text) examined are included as supplementary material.
Acknowledgments
This work was supported by the Israel Science Foundation (Grant #291/99 to J.E.) and by the Natural Sciences and Engineering Research Council of Canada (NSERC; to K.F.J.). S.L.B. was supported by a postgraduate award from NSERC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03148703.
References
- Akca, E., Claus, H., Schultz, N., Karbach, G., Schlott, B., Debaerdemaeker, T., Declercq, J.P., and Konig, H. 2002. Genes and derived amino acid sequences of S-layer proteins from mesophilic, thermophilic, and extremely Thermophilic methanococci. Extremophiles 6 351–358. [DOI] [PubMed] [Google Scholar]
- Albers, S.-V. and Driessen, A.J.M. 2002. Signal peptides of secreted proteins of the archaeon Sulfolobus solfataricus: A genomic survey. Arch. Microbiol. 177 209–216. [DOI] [PubMed] [Google Scholar]
- Bardy, S.L. and Jarrell, K.F. 2002. FlaK of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiol. Lett. 208 53–59. [DOI] [PubMed] [Google Scholar]
- Berks, B.C., Sargent, F., and Palmer, T. 2000. The Tat protein export pathway. Mol. Microbiol. 35 260–274. [DOI] [PubMed] [Google Scholar]
- Bolhuis, A. 2002. Protein transport in the halophilic archaeon Halobacterium sp. NRC-1: A major role for the twin-arginine translocation pathway? Microbiology 148 3335–3346. [DOI] [PubMed] [Google Scholar]
- Claus, H., Akca, E., Debaerdemaeker, T., Evrard, C., Declercq, J.-P., and Konig, H. 2002. Primary structure of selected archaeal mesophilic and extremely thermophilic outer surface layer proteins. System. Appl. Microbiol. 25 3–12. [DOI] [PubMed] [Google Scholar]
- Dilks, K., Rose, R.W., Hartmann, E., and Pohlschroder, M. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: A genomic survey. J. Bacteriol. 185 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler, J. 2000. Archaeal protein translocation: Crossing membranes in the third domain of life. Eur. J. Biochem. 267 3402–3412. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Archaeal signal peptidases from the genus Thermoplasma: Structural and mechanistic hybrids of the bacterial and eukaryal enzymes. J. Mol. Evol. 54 411–415. [DOI] [PubMed] [Google Scholar]
- Faguy, D.M., Jarrell, K.F., Kuzio, J., and Kalmokoff, M.L. 1994. Molecular analysis of archaeal flagellins: Similarity to the type IV pilin-transport superfamily widespread in bacteria. Can. J. Microbiol. 40 67–71. [DOI] [PubMed] [Google Scholar]
- Fekkes, P. and Driessen, A.J. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerl, L. and Sumper, M. 1988. Halobacterial flagellins are encoded by a multigene family. Characterization of five flagellin genes. J. Biol. Chem. 263 13246–13251. [PubMed] [Google Scholar]
- Horton, P. and Nakai, K. 1997. Better prediction of protein cellular localization sites with the k nearest neighbor classifier. Intellig. Syst. Mol. Biol. 5 147–152. [PubMed] [Google Scholar]
- Kandler, O. and Konig, H. 1993. Cell envelopes of archaea: Structure and chemistry. In The biochemisty of archaea (archaebacteria) (eds. M. Kates et al.), pp. 223–260. Elsevier, New York.
- Kates, M. 1993. Membrane lipids of archaea. In The biochemisty of archaea (archaebacteria) (eds. M. Kates et al.), pp. 261–296. Elsevier, New York.
- Kawarabayasi, Y., Sawada, M., Horikawa, H., Haikawa, Y., Hino, Y., Yamamoto, S., Sekine, M., Baba, S., Kosugi, H., Hosoyama, A., et al. 1998. Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5 55–76. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi, Y., Hino, Y., Horikawa, H., Yamazaki, S., Haikawa, Y., Jin-no, K., Takahashi, M., Sekine, M., Baba, S., Ankai, A., et al. 1999. Complete genome sequence of an aerobic hyperthermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6 83–101. [DOI] [PubMed] [Google Scholar]
- Kessel, M., Wildehaber, I., Cohen, S., and Baumeister, W. 1988. Three-dimensional structure of the regular surface glycoprotein layer of Halobacterium volcanii from the Dead Sea. EMBO J. 7 1549–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoeva, M.V., Storch, K.-F., Klein, C., and Oesterhelt, D. 2002. A novel mode of sensory transduction in archaea: Binding protein-mediated chemotaxis towards osmoprotectants and amino acids. EMBO J. 21 2312–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, J. and Sumper, M. 1987. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J. Biol. Chem. 262 9724–9729. [PubMed] [Google Scholar]
- Manting, E.K. and Driessen, A.J.M. 2000. Escherichia coli translocase: The unravelling of a molecular machine. Mol. Microbiol. 37 226–238. [DOI] [PubMed] [Google Scholar]
- Mattar, S., Scharf, B., Kent, S.B.H., Rodewald, K., Oesterhelt, D., and Engelhard, M. 1994. The primary structure of halocyanin, an archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J. Biol. Chem. 269 14939–14945. [PubMed] [Google Scholar]
- Nakai, K. and Horton, P. 1999. PSORT: A program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 24 34–35. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10 1–6. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Brunak, S., and von Heijne, G. 1999. Machine learning approaches for the prediction of signal peptides and other learning sorting signals. Protein Eng. 12 3–9. [DOI] [PubMed] [Google Scholar]
- O’Connor, E.M. and Shand, R.F. 2002. Halocins and sulfolobicins: The emerging story of archaeal protein and peptide antibiotics. J. Indust. Microbiol. Biotechnol. 28 23–31. [DOI] [PubMed] [Google Scholar]
- Rapoport, T.A., Jungnickel, B., and Kutay, U. 1996. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membrane. Annu. Rev. Biochem. 65 271–303. [DOI] [PubMed] [Google Scholar]
- Robinson, C. and Bolhuis, A. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2 350–356. [DOI] [PubMed] [Google Scholar]
- Rose, R.W., Bruser, T., Kissinger, J.C., and Pohlschroder, M. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45 943–950. [DOI] [PubMed] [Google Scholar]
- Saleh, M.T., Fillon, M., Brennan, P.J., and Belisle, J.T. 2001. Identification of putative exported/secreted proteins in prokaryotic proteomes. Gene 269 195–204. [DOI] [PubMed] [Google Scholar]
- Sara, M. and Sleytr, U.B. 2000. S-Layer proteins. J. Bacteriol. 182 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, G. 1999. How many potentially secreted proteins are contained in a bacterial genome? Gene 237 113–121. [DOI] [PubMed] [Google Scholar]
- Schneider, T.D. and Stephens, R.M. 1990. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.R., Doucette-Stamm, L.A., Deloughery, C., Lee, H., Dubois, J., Aldredge, T., Bashirzadeh, R., Blakely, D., Cook, R., Gilbert, K., et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum delta H: Functional analysis and comparative genomics. J. Bacteriol. 179 7135–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley, N.R., Findlay, K., Berks, B.C., and Palmer, T. 2001. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotrophic defects in the cell envelope. J. Bacteriol. 183 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom, M.S., Nunn, D.N., and Lory, S. 1994. Postranslational processing of type IV prepilin and homologs by PilD of Pseudomonas aeruginosa. Methods Enzymol. 235 527–540. [DOI] [PubMed] [Google Scholar]
- Thomas, N.A., Bardy, S.L., and Jarrell, K.F. 2001a. The archaeal flagellum: A different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25 147–174. [DOI] [PubMed] [Google Scholar]
- Thomas, N.A., Chao, E.D., and Jarrell, K.F. 2001b. Identification of amino acids in the leader peptide of Methanococcus voltae preflagellin that are important in posttranslational processing. Arch. Microbiol. 175 263–269. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2 531–534. [DOI] [PubMed] [Google Scholar]
- ———. 1990. The signal peptide. J. Membr. Biol. 115 195–201. [DOI] [PubMed] [Google Scholar]
- Woese, C.R., Kandler, O., and Wheelis, M.L. 1990. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria and Eucarya. Proc. Natl. Acad. Sci. 87 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, M.-R., Tseng, Y.-H., Nguyen, E.H., Wu, L.-F., and Saier Jr., M.H. 2002. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 177 441–450. [DOI] [PubMed] [Google Scholar]