Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions (original) (raw)
Abstract
A set of oat–maize chromosome addition lines with individual maize (Zea mays L.) chromosomes present in plants with a complete oat (Avena sativa L.) chromosome complement provides a unique opportunity to analyze the organization of centromeric regions of each maize chromosome. A DNA sequence, MCS1a, described previously as a maize centromere-associated sequence, was used as a probe to isolate cosmid clones from a genomic library made of DNA purified from a maize chromosome 9 addition line. Analysis of six cosmid clones containing centromeric DNA segments revealed a complex organization. The MCS1a sequence was found to comprise a portion of the long terminal repeats of a retrotransposon-like repeated element, termed CentA. Two of the six cosmid clones contained regions composed of a newly identified family of tandem repeats, termed CentC. Copies of CentA and tandem arrays of CentC are interspersed with other repetitive elements, including the previously identified maize retroelements Huck and Prem2. Fluorescence in situ hybridization revealed that CentC and CentA elements are limited to the centromeric region of each maize chromosome. The retroelements Huck and Prem2 are dispersed along all maize chromosomes, although Huck elements are present in an increased concentration around centromeric regions. Significant variation in the size of the blocks of CentC and in the copy number of CentA elements, as well as restriction fragment length variations were detected within the centromeric region of each maize chromosome studied. The different proportions and arrangements of these elements and likely others provide each centromeric region with a unique overall structure.
The discovery of maize chromosome retention in oat “haploids” after oat × maize crosses and the subsequent recovery of stable maize chromosome-addition oat lines (1, 2) enabled us to develop a system of maize chromosome analysis similar to that available in mammalian hybrid cell systems (3, 4). We have already used it for the construction of maize chromosome-specific libraries of cloned DNA fragments (5), for characterization of chromosome-specific knob regions (6), and for introgressions of small segments of a maize chromosome into oat that provide tools for physical mapping of the maize chromosome (O. Riera-Lizarazu, M. I. Vales, E.V.A., R.L.P., and H.W.R., unpublished results). The maize chromosome-addition oat lines also provide an opportunity for analysis of individual centromeres of maize chromosomes.
Little is known about the organization of maize centromeric regions. The first maize centromere-associated sequences were only recently identified from B chromosomes (7) and from A chromosomes (8). The two A chromosome centromeric sequences, MCS1a and MCS1b, belong to families of repetitive elements that have a significant level of conservation across the cereals. However, the organization of these sequences within centromeric regions of maize chromosomes is not known.
In this research, a probe homologous to the MCS1a sequence (8) was used to identify and isolate several DNA segments from the centromeric region of maize chromosome 9. Six cosmid clones were isolated from a cosmid library made from DNA of a maize chromosome 9 addition line of oat. We found that the centromere-associated sequences MCS1a and MCS1b (8) are parts of long terminal repeats (LTRs) of a retrotransposon-like element, termed CentA. This element belongs to the medium-repetitive class of elements. The CentA element, or parts of it, were found in all six cosmid clones, and they are interspersed with other retrotransposable elements including the previously known retrotransposons Huck and Prem2 (9, 10).
A class of tandem repeats, which we named CentC (156 bp in length), was found in two of the six cosmid clones. The CentC elements are capable of forming long tandem arrays and are interspersed with blocks of other retrotransposable elements. Fluorescence in situ hybridization (FISH) indicates that the CentC and the CentA elements are localized at centromere regions of each maize chromosome, but in different relative proportions at the different centromeres. Southern blot hybridization of these two elements to a blot panel of maize chromosome addition lines revealed restriction fragment length variations of these elements in each of the maize chromosomes. These observations allow us to conclude that each maize centromeric region has a unique organization, even though they are all composed of the same or related building blocks.
MATERIALS AND METHODS
Maize and Oat Strains.
Plant materials used in this study included maize hybrid Seneca 60, maize inbred A188, oat cultivar Starter-1, and oat–maize chromosome addition lines for maize chromosomes 2, 3, 4, 7, 8, and 9. Oat–maize addition lines were derived from plants recovered following sexual crosses of oat by maize. Incomplete elimination of maize chromosomes in embryos produced from these crosses allowed for the recovery of stocks, each with an individual maize chromosome isolated in an oat genomic background (2).
DNA Purification and Analysis.
Genomic DNA was purified as described (6). Cosmids containing maize centromeric DNA sequences were isolated from a cosmid library prepared from a maize chromosome 9 addition line in oat (6) that had been probed with a 32P-labeled DNA sample complementary to the MCS1a centromere repeat (8). The probe was amplified by PCR from maize genomic DNA by using two primers complementary to the MCS1a sequence (Cen-a-F: 5′-AGGTTCAGTGCTGTTCGTGG, and Cen-a-R: 5′-TGGAGGTGGATAACTTGTTGG). Gel-blot analysis of total genomic and cosmid DNA after gel electrophoresis was carried out as described by Sambrook et al. (11) with several modifications (12). A DNA blot panel of oat–maize chromosome addition lines for maize chromosomes 2, 3, 4, 7, 8, and 9 was created for chromosome assignment of cloned DNA sequences. DNA fragments and total genomic DNA were labeled by random primer extension. Some cosmid subfragments were cloned in pBS/KS (Stratagene) for subsequent sequencing. Sequencing was performed with the help of the KS and SK primers and by primer walking by using the Taq DyeDeoxy terminator cycle sequencing system (Applied Biosystems).
Restriction Mapping with Oligonucleotide Probes.
Restriction map construction was done according to the protocol adapted to the SuperCos1 vector following the manufacturer’s recommendations (Stratagene). Cosmid DNA was cut with _Not_I to excise the insert followed by partial digestion with an appropriate restriction enzyme. The digestion products were electrophoretically separated on an agarose gel, transferred to a nitrocellulose membrane, and probed with labeled oligonucleotide probe T3 (GGC CGC AAT TAA CCC TCA CTA AAG G) or oligonucleotide probe T7 (GGC CGC GAT ATA CGA CTC ACT ATA GG).
In Situ Hybridization.
DNA samples were labeled with fluorescein-12-dUTP or with tetramethylrhodamine-6-dUTP (Boehringer Mannheim), and in situ hybridization was performed according to the protocol provided by the manufacturer of the Prime-It Fluor Fluorescence Labeling Kit (Stratagene). Chromosomes were counterstained in antifade solution [25 mg/ml triethylenediamine in a 1:1 (vol/vol) glycerol and phosphate buffer] containing 20 ng/ml of 4′,6-diamino-2-phenylindole.
Metaphase chromosome spreads were prepared from maize Seneca 60 root tips pretreated with 0.05% colchicine for 2 hr. Root tips were fixed in 3:1 (100% ethanol/glacial acetic acid) overnight and stored in 70% ethanol. To prepare spreads of metaphase chromosomes, root tips were transferred into 45% acetic acid for 5 min and squashed in 45% acetic acid. The slides were frozen in liquid nitrogen and cover glasses were popped off with a razor blade. Slides were fixed in absolute ethanol overnight, dried, and stored until used for in situ hybridization.
RESULTS
Characterization of Cosmid Clones with Centromeric DNA Segments.
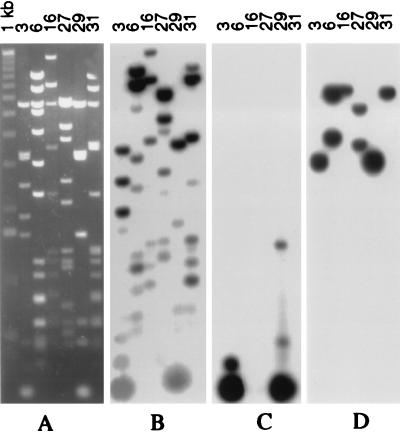
A cosmid library made from oat–maize chromosome 9 addition line DNA (6) was screened with a labeled probe that was complementary to the centromere-associated MCS1a sequence discovered earlier by Aragon-Alcaide et al. (8). Six positive cosmid clones were isolated and characterized. Digestion of these clones with different restriction enzymes revealed that each has a different restriction profile. However, some of the cosmid clones share similar or identical DNA subfragments; for example, there were several similar _Xmn_I DNA fragments in cosmids 3 and 29 (Fig. 1A). One or two restriction fragments in each clone cross-hybridized to the labeled MCS1a centromere-specific probe (Fig. 1D). Most of the fragments lacked the MCS1a sequence, but gave strong or medium hybridization signals after hybridization with labeled total maize genomic DNA (Fig. 1B). A cross-hybridization experiment to a collection of different repeated maize elements (6) identified Huck retroelements in cosmids 6 and 31 and Prem2 retroelements in cosmid clones 3 and 29 (data not shown). No cross-hybridization with any of the subfragments in these six cosmid clones was observed when knob-associated 180-bp (13) and TR1-repeated elements (14) were used as probes.
Figure 1.
Composition of cloned centromeric DNA segments from maize chromosome 9. (A) Six cosmid clones (3, 6, 16, 27, 29, and 31) containing centromeric DNA sequence MCS1a have different restriction profiles after digestion with _Xmn_I restriction enzyme. (B) Hybridization with labeled genomic DNA indicates that almost all the subfragments contain medium or highly repetitive DNA sequences, some of which were identified as Huck and Prem2 retrotransposable elements. (C) Hybridization with the labeled CentC element shows that this repeated element is present in two cosmid clones, 3 and 29. Monomeric or dimeric units of this element are seen as a 156–312 bp group of fragments. (D) Hybridization with the labeled CentA element shows that it is present in all six cosmid clones, but in fragments of different lengths.
Subfragments 3–13 kb in length were found in cosmids 3 and 29 after digestion with _Sau_3A. These unusually long _Sau_3A subfragments were subcloned and partially sequenced from the ends. Sequence analysis revealed that they are composed of a class of tandem repeats with a unit length of 154–158 bp (GenBank accession nos. AF078918–AF078923). These repeats were termed CentC elements. Different copies of CentC elements have minor sequence differences in the form of point mutations. The CentC elements have no homology to any other maize DNA sequences in GenBank but possess partial identity to a tandem repeat and microsatellite of Oryza alta L. (GenBank accession no. X86001). The restriction enzyme _Xmn_I has one recognition site within this sequence and the corresponding monomeric unit of CentC may be seen in Fig. 1A as a thick band of approximately 150 bp in gel blots of restriction digests of cosmids 3 and 29. Blot hybridization indicated that, of the six cosmid clones containing MCS1a sequence, the CentC element was present only in these two cosmid clones (Fig. 1C). These two cosmids were characterized in detail.
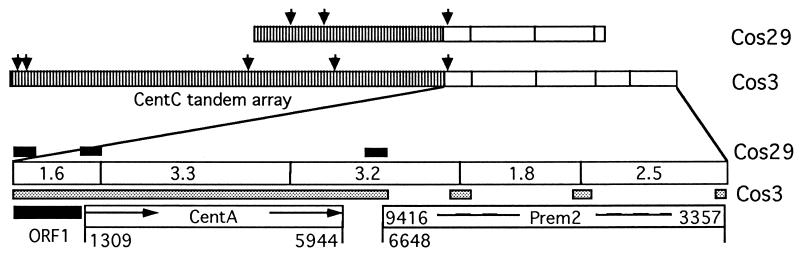
Restriction maps were constructed for cosmid clones 3 and 29 by using the _Xmn_I restriction enzyme (Fig. 2). The CentC element forms long tandem arrays of approximately 24 and 10 kb in cosmids 3 and 29, respectively. Although the _Xmn_I restriction maps for the right ends of the inserts are quite similar for cosmids 3 and 29, these two clones represent different regions of the chromosome 9 centromere because the tandem arrays of the CentC elements in these two cosmid clones have different _Sau_3A restriction sites (Fig. 2).
Figure 2.
Molecular maps of the centromeric DNA segments cloned in cosmids 3 and 29 isolated from maize chromosome 9. The _Xmn_I restriction sites are indicated by vertical lines within the bars. Positions for the _Sau_3A restriction sites are indicated by solid vertical arrows for the CentC tandem arrays only. The left vertically hatched parts of the cloned segments are composed of arrays of the CentC elements about 10 kb in length in cosmid 29 and 23 kb in length in cosmid 3. Some segments of the right portions of cosmids 3 and 29 were sequenced. Stippled bars represent sequenced segments of cosmid 3, and the upper, heavily shaded bars represent sequenced segments of cosmid 29 as they correspond to cosmid 3 sequences. A new retrotransposon-like element, CentA, was identified between nucleotides 1309 and 5944 of the large sequenced segment of cosmid 3; this element contains two LTRs, 1,304 and 1,305 bp in length, respectively (indicated by arrows). To the left of this element, ORF1 was identified. The right end of cosmid 3 contains a portion of the Prem2 retrotransposable (10) element that corresponds to the section of the published DNA sequence of this element included between nucleotides 3357 and 9416.
The 12 kb right portion of the centromeric DNA segment cloned in cosmid 3 (Fig. 2) was cut with _Xmn_I restriction enzyme into five subfragments, each of which was subcloned into pBS/KS in a _Sma_I site, and the ends of those fragments were sequenced (Fig. 2). Three _Xmn_I subfragments, 2.5, 1.8, and 3.2 kb in length, at the 3′ end of cosmid 3, revealed a high level of homology (90%) and colinearity to regions of the Prem2 retrotransposable element (10). This enabled us to predict that the end of the right LTR of the Prem2 element should reside within the 3.2-kb _Xmn_I subfragment. To sequence the junction point between the Prem2 right LTR and the adjacent DNA sequence, a primer (Prem2-R-LTR: 5′-ATTTTCAGTTTCGCCCTATTCACC) was designed that was complementary to a sequence close to the end of the right LTR of Prem2 and directed outwards from the element.
The entire segment between the end of the CentC tandem array and the right end of the Prem2 element was sequenced (GenBank accession no. AF078917). Two almost identical (99%) copies of a repeated element were identified within this segment in direct orientation (Fig. 2, indicated by arrows within the CentA box); these were separated by an unknown DNA sequence 2,026 bp in length. The region of DNA that extends from nucleotides 1309–5944 of this segment and includes two copies of the direct repeat and the internal 2,026-bp fragment was named the CentA element (Fig. 2). The structure of this element suggests that it could be a novel retrotransposon-like element with two LTRs, each about 1,300 bp in length. The internal 2,026-bp segment has no significant homology to any plant DNA sequence in GenBank. CentA-LTR-1304 and LTR-1305 have segments of homology (66–97% identity) to maize MCS1a and MCS1b, and rice, barley, and Brachypodium sylvaticum centromere-associated DNA sequences described by Aragon-Alcaide et al. (8) and have a short segment of homology (90%) to a Sorghum centromere-associated element (15). In general, the CentA element may be considered as a conserved centromere-associated repeated element. However, its partial and segmental homology to maize MCS1a and MCS1b centromere-associated sequences indicates that the related copies of this family of repeated elements are diverged.
The CentA elements of cosmid 3 and cosmid 29 have very similar structures based on their restriction profiles as well as on direct sequence comparison of certain regions within the clones (Fig. 2). Other centromeric clones contain diverged copies of CentA elements (Fig. 1D). A 638-bp _Eco_RI subfragment from cosmid 6 with homology to the CentA element was isolated and sequenced (GenBank accession no. AF082532). Sequence analysis indicates that this fragment has a high level of identity (96%) to the region included between nucleotides 2131 and 2764 of the large sequenced segment of cosmid 3; however, _Eco_RI restriction sites of the CentA copy on cosmid 6 are absent from both copies of the LTRs of the CentA element in cosmid 3.
Two segments of DNA in cosmid 3, one located between the end of the CentC tandem array and the beginning of the CentA element, and another located between the end of the CentA and the end of the Prem2 element, revealed significant homology to different segments of a Ty3/gypsy-type retrotransposable element (GenBank accession no. AF030633). The region between nucleotides 59 and 1309 of the large sequenced segment of cosmid 3 contains an open reading frame (ORF1), which may encode a polypeptide with high identity to a retrovirus-related DNA polymerase polyprotein found in Ananas comosus (GenBank accession no. Y12432) and many other organisms. These same two DNA segments also appear to be present in cosmid 29 (Fig. 2).
The nucleotide sequence analysis of cosmids 3 and 29 enabled us to identify at least two new types of repeating units: CentA, which is presumably a medium copy number retrotransposable element, and CentC elements, which can form tandem arrays. The cosmid clones 6, 16, 27, and 31 also contain CentA elements interspersed with copies of other medium and high-copy number repetitive elements of the maize genome (Fig. 1); some of these were identified as Huck or Prem2 retroelements, whereas others belong to unknown repetitive elements of the maize genome (Fig. 1B).
FISH of Different Repeated Elements Associated with Centromeric DNA.
In situ hybridization of different repeated elements found in cosmid clones isolated in this research was performed with metaphase chromosomes of maize root-tip cells. The 3.3-kb _Xmn_I subfragment of cosmid 3, which contains the CentA element, was labeled with fluorescein (green) and the CentC element was labeled with rhodamine (red). They were then hybridized simultaneously to the same cytological preparations (Fig. 3 A and B). Both probes hybridize to centromeric regions of all maize chromosomes. The CentC element (Fig. 3B) produced strong hybridization signals that are highly reproducible, whereas the CentA element (Fig. 3A) gave weaker hybridization signals. There is an obvious variation in the intensity of hybridization signals associated with different chromosomes, from small dots up to large blocks.
Figure 3.
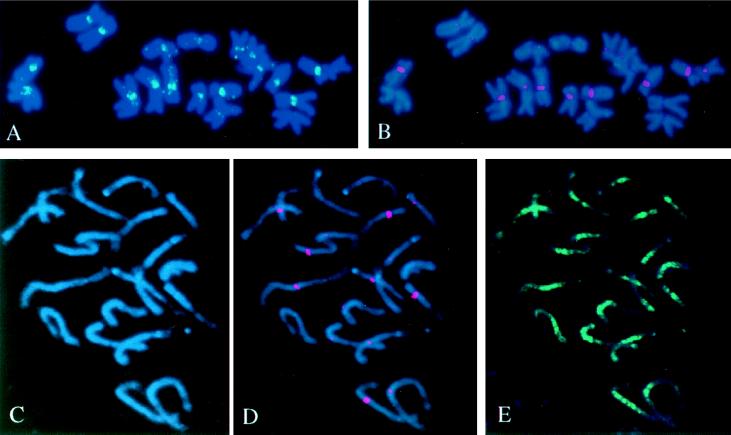
FISH of maize metaphase chromosomes with fluorescein-labeled CentA element (A) (the 3.3-kb _Xmn_I fragment on Fig. 2) and rhodamine-labeled CentC element (B). Both probes hybridize with centromeric regions. There is variation among different chromosomes in the intensity of hybridization signals (C_–_E). FISH of maize prophase chromosomes (C) (4′,6-diamino-2-phenylindole-counterstained after in situ hybridization) with rhodamine-labeled CentC element (D) and fluorescein-labeled Huck element (E). The Huck element is dispersed along all maize chromosomes with an increased concentration around centromeric regions.
The distribution along maize chromosomes was determined for two retrotransposable elements, Huck and Prem2, which are interspersed with the centromere-specific repeats, CentA and CentC. A 13-kb _Eco_RI DNA subfragment of cosmid 6 containing a portion of the Huck retroelement gave a diffuse pattern of hybridization (Fig. 3E), with a preferential concentration around centromeric regions as defined by CentC (Fig. 3D). DNA segments corresponding to the portion of the Prem2 element from cosmid 3 gave a diffuse pattern of hybridization along all chromosomes (not shown). Thus, the CentA and CentC elements are associated with centromeric regions, whereas the Huck retrotransposable elements are dispersed along all maize chromosomes with a preferential concentration around centromere regions.
Organization of the CentA and the CentC Elements in Centromeric Regions of Different Maize Chromosomes.
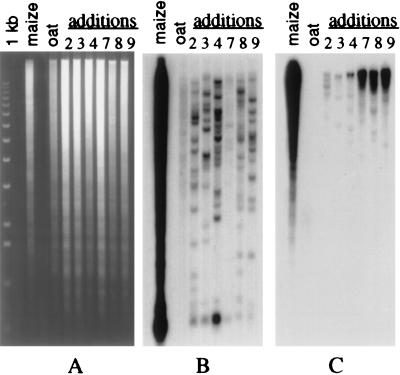
The availability of maize chromosome addition lines allowed comparative analysis of the organization of the CentA and the CentC elements in centromere regions of different maize chromosomes. The labeled CentA and the CentC elements were hybridized to gel blots of DNA samples from addition lines containing maize chromosomes 2, 3, 4, 7, 8, and 9 that were digested with _Eco_RI (Fig. 4). A relatively simple restriction profile composed of 10–20 bands for each maize chromosome indicates that there are only several dozen copies of this CentA element in each centromere region (Fig. 4B). Thus, this element may be considered as a medium repetitive element for the maize genome. The CentA element gave a weak diffuse hybridization pattern with oat DNA. This result indicates that CentA-related DNA sequences are present in the oat genome and is consistent with data of Aragon-Alcaide et al. (8) on the conservative nature of this element.
Figure 4.
Polymorphic organization of centromeric sequences of maize chromosomes. Southern blot hybridization of labeled CentA (B) and CentC (C) probes to a panel of chromosome addition lines of maize. (A) Ethidium bromide stained gel of _Eco_RI digested DNA samples from maize, oat, and six chromosome addition lines with maize chromosomes 2, 3, 4, 7, 8, and 9, respectively. The CentA element hybridizes to 10–20 polymorphic fragments in each maize chromosome. There is a faint hybridization with oat genomic DNA. The CentC element mostly hybridizes to high molecular weight DNA in each chromosome. Increased copy numbers exist in chromosomes 7, 8, and 9.
Hybridization of the labeled CentC element to the same blot panel as used with CentA revealed a significant variation in the intensity of hybridization signals among different maize chromosomes (Fig. 4C). This result correlates with the in situ hybridization data (Fig. 3 A and B). Most of the hybridizing bands correspond to high molecular weight DNA fragments in each chromosome, indicating that the CentC elements are capable of forming long arrays. Unlike CentA, no hybridization of CentC to oat genomic DNA was detected.
A comparison of hybridization patterns of these two centromere-specific elements, CentA and CentC, to DNA of different maize chromosomes (Fig. 4 B and C) revealed a significant level of restriction pattern differences. This observation enabled us to conclude that each of the six maize chromosomes examined has a unique restriction fragment profile for the CentC as well as for CentA elements.
DISCUSSION
Only recently have the locations of maize centromeres been established on their respective molecular genetic maps (16) and the first A chromosome centromere-associated DNA sequences cloned and mapped by in situ hybridization (8). The reconstruction of the molecular organization of an individual centromere on a specific chromosome is a difficult task because centromeres of many eukaryotic organisms are composed of related repeated DNA sequences, some of which are capable of forming tandem arrays (17). Even in the case of Arabidopsis thaliana, the molecular organization of individual centromere regions (18) remains an unresolved problem in spite of the fact that physical maps have been constructed for all euchromatic parts of A. thaliana chromosomes (19, 20).
The oat–maize chromosome addition lines provide a unique opportunity to analyze an individual maize chromosome (2) and, consequently, an individual centromeric region. We have already demonstrated that the maize chromosome addition lines may be used for chromosome-specific library construction (5) and for analysis of knob regions from individual maize chromosomes (6, 14). In this research, our objective was to use similar techniques to isolate and characterize cloned DNA segments from a centromere region of maize chromosome 9. We have isolated and partially characterized six cosmid clones from maize chromosome 9 that overall contain approximately 180 kb of DNA sequences from the centromeric region. Most likely only a fraction of the entire centromere region of the maize chromosome 9 is present in these six cloned DNA segments because all cloned DNA segments have different restriction profiles, and we could not identify any obvious overlapping DNA segments.
Analysis of cloned centromeric DNA segments revealed that the centromere-associated repeated sequences may be subdivided into two classes: short repeating elements arranged in tandem arrays of different lengths (CentC) and centromere-associated retrotransposon-like elements (CentA). These two classes of centromere-associated repeats have interspersed among them large retrotransposable elements that are also dispersed throughout the entire maize genome. Some of those retrotransposable elements have been isolated and characterized previously, including Huck and Prem2 (9, 10).
The CentC element is a novel class of tandem repeats for the maize genome. It has partial homology to a tandem repeat isolated from the genome of upland rice, Oryza alta L. (GenBank accession no. X86001); however, there is no information available on the chromosome location of this repeat in O. alta. No related DNA elements were found in oat; thus, this family of elements may be considered as a maize-specific repeated element relative to oat, which evolved relatively recently in the evolution of cereals. According to Southern blot hybridization and in situ hybridization, these repeats form clusters of different sizes in different chromosomes and are limited to centromeric regions of maize chromosomes.
A second family of centromere-associated repeated elements, CentA, described here, has a structural similarity to retrotransposable elements because it is composed of a segment of DNA flanked by two LTRs. CentA has partial homology to sequences in rice, barley, B. sylvaticum, and maize centromere-specific DNA sequences MCS1a and MCS1b (8); the MCS1a sequence was used in this study as a probe to isolate the cosmid clones from the centromeric region of maize chromosome 9. CentA also has a short segment of homology to a Sorghum centromere-associated element described by Jiang et al. (15).
Other segments of DNA present in cosmid 3 included two segments that exhibited a significant level of homology to the Ty3/gypsy-type retroelement of the maize genome. The region between nucleotides 500 and 1130 of the segment of DNA flanked by CentC and CentA elements in the _Xmn_I 1.6-kb fragment in cosmid 3 (Fig. 2) revealed significant homology to Rle (GenBank accession no. AF057037) and Ty3/gypsy-type (GenBank accession no. AF030633) maize retrotransposable elements. In addition, significant homology was found between that segment and more than 40 other DNA sequences in GenBank. All of them have homology to retrotransposable elements found in many different species of plants, yeast, and animals. The region included between nucleotides 59 and 1309 contains an ORF1, which may encode a polypeptide with significant homology (52% identities, 73% positives) to a retrovirus-related DNA polymerase polyprotein (GenBank accession no. Y12432). Thus, this segment of maize centromeric DNA contains a section of a conserved retroelement that may be found in many different organisms and is most likely dispersed throughout the maize genome. The second segment, included between the right end of the CentA and the end of the Prem2 elements, has homology to a different segment of the same Ty3/gypsy-type retrotransposable element (GenBank accession no. AF030633) and to a repeated element of Sorghum centromeric DNA sequence (16). Four of the six cloned centromeric DNA fragments have no CentC elements and are composed of blocks of CentA elements mixed with other medium and highly repetitive dispersed elements of the maize genome, including Huck (9) and Prem2 (10) retrotransposable elements.
Centromeric regions of maize chromosomes contain blocks of tandemly repeated CentC elements interspersed with blocks of retrotransposable elements. These blocks are unlikely to be random combinations of different retrotransposable elements. For example, we found that two cosmid clones, cosmids 3 and 29, have almost identical blocks of retrotransposable elements (Fig. 2), which are adjacent to different blocks of tandem arrays of CentC. This observation is compatible with the idea that a centromere region may have a hierarchical or subunit structure (21, 22); namely, certain segments composed of different elements may be repeated several times in a maize centromere. How many different genetic elements may be associated with maize centromeres is not clear. More extensive sequencing of other centromere DNA segments is necessary to answer this question.
This research indicates that the organization of centromeres of maize chromosomes resembles the organization of mammalian centromeres (for review see ref. 23) and the organization of centromeres in A. thaliana (18), where islands of middle-repetitive sequence elements have been found to be associated with tandem arrays of short repeats (19, 24–26).
Recently, we found that two other classes of tandemly arranged repeats in maize, the knob-associated 180-bp repeats (13) and TR1 elements (14), are also interspersed with retrotransposable elements (6). Most of the interspersed retrotransposable elements in the knob regions are full size, nonrearranged copies of retrotransposable elements such as Grande, Prem2, and Zeon, but no copies of Huck elements were observed in any of the 23 cosmid clones containing knob DNA segments. In contrast, we have found the Huck element in two cloned segments of centromere region DNA, and in situ hybridization indicates that Huck elements are concentrated around primary constrictions; thus, this is another example of region- or sequence-specific distribution of different retrotransposable elements along the maize chromosomes.
Knob regions of maize chromosomes may function under certain conditions as neocentromeres in meiotic cells in metaphase I (27, 28). One possibility is that the knob 180-bp tandem repeats may share a certain level of homology to centromere-associated tandem repeats (29). This suggestion was based on an indication that centromeric regions of B chromosomes may contain short DNA elements homologous to 180-bp knob-associated repeats (7). We could not detect the presence of 180-bp or TR1 knob-associated tandem repeats in cloned centromeric DNA segments in blot hybridizations done under standard washing conditions (0.015 M NaCl/0.0015 M sodium citrate at 60°C), nor could we find any obvious sequence homology between the CentC centromeric tandem repeat and the 180-bp and the TR1 knob-associated tandem repeats. Lack of obvious homology between the knob repeats and the centromeric repeats of A chromosomes suggests that unrelated DNA sequences are capable of performing the centromere function. In the case of oat–maize chromosome addition lines, it is obvious that maize and oat centromeres have a different composition. For example, the CentC element is present in all maize chromosomes and was not detected in oat chromosomes, whereas the CentA element was detected by Southern blot hybridization in oat DNA. In spite of these differences, maize centromeres function in oat cells with nearly the same efficiency as oat centromeres in established oat–maize addition lines. What kind of elements in the maize centromere region actually function as a centromere is not clear. It is possible that conservative, medium-repetitive elements found in centromeric regions of several different plant species play a key role in centromere function while arrays of tandem repeats play a supporting role.
There is significant variation among different maize chromosomes in the copy number of CentC elements associated with each centromere. This polymorphism is clearly seen after in situ hybridization (Fig. 3), as well as after gel-blot hybridization with the panel of DNA samples of oat–maize chromosome addition lines (Fig. 4C). The strongest signals were detected over chromosomes 7, 8, and 9 in the maize variety Seneca 60. Despite the fact that the majority of CentC elements have highly similar nucleotide sequences and form quite regular arrays of tandem repeats, it is possible to identify single base differences among the members of this family. Similarly, a significant level of variation in restriction fragment profiles was detected for CentA elements in different centromeres (Fig. 4B). Thus, despite the fact that maize centromeres are all composed of the same related elements, the differences in composition and mutual arrangements of those elements provide each centromere with a unique molecular organization.
Acknowledgments
This is a joint publication of the University of Minnesota and the U.S. Department of Agriculture–Agricultural Research Service and is paper 981130082, Scientific Journal Series, Minnesota Agricultural Experiment Station. This research was funded in part by Grant 96–35300-3775 of the U.S. Department of Agriculture National Research Initiative Competitive Grants Program.
ABBREVIATIONS
LTR
long terminal repeat
FISH
fluorescence in situ hybridization
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF078917–AF078923 and AF082532).
References
- Rines H W, Riera-Lizarazu O, Phillips R L. In: Modification of Gene Expression and Non-Mendelian Inheritance. Oono K, Takiwa F, editors. Tsukuba, Japan: NIAR; 1995. pp. 235–251. [Google Scholar]
- 2.Riera-Lizarazu O, Rines H W, Phillips R L. Theor Appl Genet. 1996;93:123–135. doi: 10.1007/BF00225737. [DOI] [PubMed] [Google Scholar]
- 3.Cox D R, Burmeister M, Price E R, Kim S, Myers R M. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 4.Olson M V. Proc Natl Acad Sci USA. 1993;90:4338–4344. doi: 10.1073/pnas.90.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ananiev E V, Riera-Lizarazu O, Rines H W, Phillips R L. Proc Natl Acad Sci USA. 1997;94:3524–3529. doi: 10.1073/pnas.94.8.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ananiev E V, Phillips R L, Rines H W. Genetics. 1998;149:2025–2037. doi: 10.1093/genetics/149.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfenito M R, Birchler J A. Genetics. 1993;135:589–597. doi: 10.1093/genetics/135.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. Chromosoma. 1996;105:261–268. doi: 10.1007/BF02524643. [DOI] [PubMed] [Google Scholar]
- 9.SanMiguel P J, Tikhonov A, Jin Y-K, Motchulskaia N, Zakharov D. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 10.Turcich M P, Bokhari-Riza A, Hamilton D A, He C, Messier W. Sex Plant Reprod. 1996;9:65–74. [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Helentjaris T University of Missouri-Columbia Maize RFLP Laboratory, editors. UMC Maize RFLP Procedures Manual. Columbia, MO: University of Missouri; 1995. p. 9. [Google Scholar]
- 13.Peacock W J, Dennis E S, Rhoades M M, Pryor A J. Proc Natl Acad Sci USA. 1981;78:4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ananiev E V, Phillips R L, Rines H W. Proc Natl Acad Sci USA. 1998;95:10785–10790. doi: 10.1073/pnas.95.18.10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J, Nasuda S, Dong F, Schrerer C, Woo S S, Wing R, Gill B, Ward D. Proc Natl Acad Sci USA. 1996;93:14210–14213. doi: 10.1073/pnas.93.24.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burr B, Burr F A, Matz F C. In: Genetic Maps. 6th Ed. O’Brien S J, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 6.190–6.204. [Google Scholar]
- 17.Moore G, Roberts M, Aragon-Alcaide L, Foote T. Chromosoma. 1997;105:321–323. doi: 10.1007/BF02529746. [DOI] [PubMed] [Google Scholar]
- 18.Round E K, Flowers S K, Richards E J. Genome Res. 1997;7:1045–1053. doi: 10.1101/gr.7.11.1045. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt R, West J, Love K, Lenehan Z, Lister C, Thompson H, Bouchez D, Dean C. Science. 1995;270:480–483. doi: 10.1126/science.270.5235.480. [DOI] [PubMed] [Google Scholar]
- 20.Zachgo E A, Wang M L, Dewdney J, Bouchez D, Camilleri C, Belmonte S, Huang L, Dolan M, Goodman H M. Genome Res. 1996;6:19–25. doi: 10.1101/gr.6.1.19. [DOI] [PubMed] [Google Scholar]
- 21.Zinkowski R P, Meyne J, Brinkley B R. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaszas E, Birchler J A. EMBO J. 1996;15:5246–5255. [PMC free article] [PubMed] [Google Scholar]
- 23.Pluta A F, Mackay A M, Ainsztein A M, Godberg I G, Earnshaw W C. Science. 1995;270:1591–1597. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 24.Richards E J, Goodman H M, Ausubel F M. Nucleic Acids Res. 1991;19:3351–3357. doi: 10.1093/nar/19.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelissier T, Tutois S, Tourmente S, Deragon J M, Picard G. Genetica. 1996;97:141–151. doi: 10.1007/BF00054621. [DOI] [PubMed] [Google Scholar]
- 26.Thompson H L, Schmidt R, Dean C. Nucleic Acids Res. 1996;24:3017–3022. doi: 10.1093/nar/24.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhoades M M, Dempsey E. J Hered. 1973;64:13–18. [Google Scholar]
- 28.Dawe R K, Cande W Z. Proc Natl Acad Sci USA. 1996;93:8512–8517. doi: 10.1073/pnas.93.16.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birchler J A. Genome Res. 1997;7:1035–1037. doi: 10.1101/gr.7.11.1035. [DOI] [PubMed] [Google Scholar]