Biology by design: reduction and synthesis of cellular components and behaviour (original) (raw)
Abstract
Biological research is experiencing an increasing focus on the application of knowledge rather than on its generation. Thanks to the increased understanding of cellular systems and technological advances, biologists are more frequently asking not only ‘how can I understand the structure and behaviour of this biological system?’, but also ‘how can I apply that knowledge to generate novel functions in different biological systems or in other contexts?’ Active pursuit of the latter has nurtured the emergence of synthetic biology. Here, we discuss the motivation behind, and foundational technologies enabling, the development of this nascent field. We examine some early successes and applications while highlighting the challenges involved. Finally, we consider future directions and mention non-scientific considerations that can influence the field's growth.
Keywords: synthetic biology, gene circuits, biological design, genetic engineering, computational biology, metabolic engineering
1. Introduction
Imagine you have been charged with building a robot capable of complex and autonomous operations in a dynamic environment. What are the most advantageous characteristics to build into such a machine? To perform work, energy will be needed—renewable energy extracted from the environment is ideal. To respond with meaningful behaviour, information gathering and possessing capabilities will be required. For coordinated operations, communication with other robots is essential. To maintain a long-term function, a self-contained repair or reproduction system will be necessary. To attempt some goals, the robot will need to be a minuscule. To achieve economic feasibility, production costs will have to be low. While all these requirements are significant hurdles to the robotics engineer on a budget, they are feats that life has accomplished time and time again.
Consider one of the simplest forms of life, bacteria. Bacteria, often only a few micrometres in length, are capable of many of the above requirements, including, entering minuscule environments, surviving on local nutrients and responding to fluctuations in their environment with adaptive behaviour (such as chemotaxis (Falke et al. 1997), altered nutrient utilization (Jacob & Monod 1961) and temperature-dependent gene expression (Yura & Nakahigashi 1999)). Many bacterial species communicate in order to produce coordinated behaviour (Bassler & Losick 2006) and with doubling times as fast as 20 min, their reproduction capacity is remarkable.
In fact, an engineer building a device on a bacterial ‘chassis’ would only need to build one functioning prototype, culture overnight in low-cost media and return the next morning to obtain trillions of virtually identical copies. In a sense, this is like programming a minuscule but complex computer that can also reproduce. As appealing as this concept may seem, several fundamental questions arise: what functions are we capable of programming into a living organism? To what extent will these functions be performed predictably and robustly? What is the best way to implement a pre-defined design goal and what challenges and opportunities may arise? These are some of the questions that the burgeoning field of synthetic biology is beginning to address.
Over the past few years, synthetic biologists have generated remarkable systems including: an expanded genetic code in Escherichia coli (Wang et al. 2001); various logic gates (Dueber et al. 2003; Rackham & Chin 2005_a_); rewired yeast mating and osmolarity response circuitry (Park et al. 2003); bistable switches in bacteria (Gardner et al. 2000; Isaacs et al. 2003); yeast (Becskei et al. 2001) and mammalian cells (Kramer & Fussenegger 2005); photographic bacteria (Levskaya et al. 2005); genetic and metabolic oscillators (Elowitz & Leibler 2000; Atkinson et al. 2003; Fung et al. 2005); artificial communication in bacteria (Bulter et al. 2004) and yeast (Chen & Weiss 2005); and many other interesting and useful systems.
Although there is a debate about the scope and boundaries of the field, some advocates supply that ‘synthetic biology’ is:
(A) the design and construction of new biological parts, devices and systems and (B) the re-design of existing, natural biological systems for useful purposes. (www.syntheticbiology.org, syntheticbiology.org:FAQ).
It is worth examining this definition more closely. Inherent in part (A) are engineering principles—the notions of abstraction and hierarchy. One level of abstraction consists of biological components with simple albeit well-defined functions, operating under defined conditions, i.e. parts. At a higher level of abstraction, parts can be combined to form devices. Similarly, devices come together to form systems on a third level of abstraction. The basic premise is that an individual researcher can work at one of these levels without necessarily requiring details about the precise mechanics of operation at another level (Endy 2005).
Part (B) states that biology is being redesigned for ‘useful purposes’. What purposes you might wonder? The first purpose may be obvious, and it is the practical application of biologically modified organisms in human life. Although our ancestors did not possess the advanced genetic tools available today, the litany of domesticated species including fermentation yeasts, crop grains and silkworms is a testament to the vast utility of modified living organisms to humans. However, modification of living organisms by traditional means, i.e. artificial selection, is an incremental and slow process with limited pay-offs during an individual's lifetime. For example, it has taken approximately 15 000 years of domestication by selective breeding to turn wolves into present-day dogs (Leonard et al. 2002), a process which grouped desirable genes in particular breeds. Improvements in DNA synthesis and genomic engineering methods have enabled the introduction of genetic changes in relatively short time frames. Such technologies will engender the practical application of modified biological systems to new areas, such as therapeutics, renewable energies and others. The practical applications of modified biological systems represent the first useful purpose behind a redesign.
Of course, even possessing large-scale DNA technology capable of making the changes needed to produce a guide dog from a wolf is not enough. The necessary DNA changes have to be known in advance in order to be made. This is far from the case—especially for a complex organism like the dog. Comparative genomics can elucidate the differences between the organisms, but does not yield the full understanding needed to prospectively say ‘If I want to program guide animal functions into organism X, here are the changes I will make and this is how those changes work.’ In the venerable words of physicist Richard Feynman, ‘what I cannot create, I do not understand’ (Hawking 2001). The laws of physics and chemistry apply to living systems just as they apply to non-living things, such as mechanical engines. Yet, designing and constructing even simple biological systems remains a major challenge, whereas mechanical engines can be predictably engineered. Feynman would conclude that there must exist fundamental gaps in our understanding of how biological systems operate. Synthetic biology is exploring these gaps in understanding by attempting to build and apply such systems.
Scientific experiments are run under specific conditions with the hope that the conclusions drawn will be applicable in a broader context. The creation of biological systems by using currently accepted (or debated) principles would test the limitations and applicability of those principles. Likewise, implementation of existing genes, proteins and pathways in non-native settings can help elucidate their functions and reveal unknown requirements for their operation. Synthetic biologists therefore aim not only to produce interesting and useful designs, but also to simultaneously develop a greater understanding of biological components and design principles in general (Sprinzak & Elowitz 2005). Therefore, the second, and equally important, purpose of synthetic biology is to gain the biological insight that arises from testing our knowledge during the design and implementation process.
2. Foundational technologies
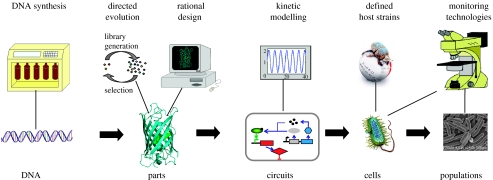
Just as the development of the microscope made the discovery of cells possible (Dunn & Jones 2004), new technologies are providing the critical foundation needed for synthetic biology. Here, we discuss the following four major advances that have produced enabling tools for experimentation and analysis in this regard: DNA synthesis; parts and devices design and optimization; systems modelling; and observational capabilities. Figure 1 illustrates an overview of where these technologies interact with synthetic biology.
Figure 1.
Interplay between engineering tools with a biological hierarchy. In order to simplify biological design, it is valuable to use an abstraction hierarchy. DNA codes for ‘parts’ that interact with each other to form circuits. The totality of all circuits and structures forms a single cell, which interacts with its neighbours and environment to form a population. At each level, technologies have been developed to assist and enable design. Shown here are the major advances that significantly reduce longstanding design, analysis and production barriers. Together, these technologies are helping to make integrated biological design a reality.
2.1 DNA synthesis
At the core of every living thing, dictating that organism's characteristics and behaviour is a string of nucleotide bases—its DNA. To reprogram an organism, that DNA needs to be altered or supplemented. Until recently, DNA manipulations were almost exclusively done in a ‘copy, cut and paste’ manner using polymerases, restriction endonucleases and ligases, respectively. While this enzymatic approach has produced a wealth of scientific advances, implementing a complicated biological design by these means is the literary equivalent of writing a paper using a photocopier, scissors and a stick of glue. Recently, however, biologists have received their metaphorical ‘typewriter’. Tian and colleagues developed a large-scale DNA synthesis method by using parallel oligonucleotide synthesis on a programmable microfluidics chip, followed by PCR amplification (Tian et al. 2004). In order to reduce the error rate to 1 in 1394 bases, the authors hybridized their ‘construction oligos’ against complimentary ‘selection oligos’ and washed away mismatches. Construction oligos were assembled into larger genes using polymerase assembly multiplexing (PAM), an overlap PCR-based method. Using their chip-based technology, the authors simultaneously synthesized and optimized all 21 genes encoding the 30S ribosomal subunit from E. coli.
In a different study, Jacobson and co-workers developed a method that can further improve the removal of error-containing DNA fragments (Carr et al. 2004). Using this method, which exploits the gel mobility shift apparent when MutS binds a mismatched double-stranded DNA, they were able to obtain an error rate as low as 1 in 10 000 (the average length of a prokaryotic gene is 924 bp, while that of a eukaryotic gene is 1346 bp; Xu et al. 2006). By applying these and other technologies, commercial companies are now able to offer large-scale (multi-kilobase and up) DNA synthesis for under $1 per base, with a two- to four-week turnaround time. Whereas these prices do not make a 10 kb construct inexpensive for most researchers, they imply that commercial synthesis has begun to rival the equipment, materials, labour and validation costs incurred by traditional cloning and construction means for select applications. It may be possible to further reduce these costs by 10–100-fold, i.e. 1–10 cents per base, within the next decade by fully automating and streamlining new high-throughput techniques (J. Tian 2006, personal communication).
Total DNA synthesis can be used to alter or improve the sequences being built. In traditional cloning, targeted mutational changes are made only to small regions (approx. 20 bases) at once. Furthermore, each region altered imposes additional experimental steps. DNA synthesis methods, however, can synthesize an altered sequence with no more effort than that necessary to synthesize a wild-type sequence of the same length. For example, a protein-coding sequence can be matched with regard to codon usage in the host organism where it will be expressed. In this case, the sequence of amino acids in a protein is left unaltered by the modification, but translation efficiency can be improved by using codons whose cognate tRNAs are more abundant. Similarly, the sequence can be altered to remove or create mRNA secondary structures without changing the resulting amino acid sequence. Furthermore, a gene whose sequence is known, but whose DNA is hard to obtain, can be easily synthesized.
2.2 Design and optimization of parts
One level of abstraction from the DNA synthesis and manipulation is the parts production, which can be accomplished through either rational design or directed evolution. Recently, improved algorithms and processor power have allowed computational design efforts to achieve new milestones in reprogramming the function of many well-characterized natural proteins. In a series of studies integrating both computation and experiments, the Hellinga laboratory succeeded in introducing an allosteric control switch into the proton-ATP pump (in collaboration with the Montemango group; Liu et al. 2002); retooling sugar-sensing receptors to bind novel ligands, such as lactate, trinitrotoluene (TNT) and serotonin (Looger et al. 2003), and converting a receptor into a functional triose phosphate isomerase enzyme (TIM), catalysing a 105–106-fold rate improvement over the uncatalysed reaction (Dwyer et al. 2004). Significantly, they even demonstrate that designed parts are active in vivo and can be used to produce more complex systems. The TNT receptor and a designed Zn receptor were shown to induce gene expression in response to exogenous ligands when implemented in some of the earlier reported examples of synthetic signalling pathways (Dwyer et al. 2003; Looger et al. 2003). Likewise, the TIM enzyme was sufficiently active to complement its wild-type version and restore viability under gluconeogenic conditions.
Computational design has also found applications beyond altering the specificity or the enzymatic function. For example, the Baker laboratory has designed a new protein that folds to form a novel structure—matching their modelling predictions (Kuhlman et al. 2003). They also apply their computational methods to increase the thermostability of an enzyme by identifying key mutations. When some mutations were applied in concert, the result was a 30-fold increase in half-life at 50°C (Korkegian et al. 2005). The examples here illustrate altered specificity, novel functions and structures, improved stability and introduction of allosteric control. They highlight some of the contributions that computational protein design has made for parts generation and improvement. While we have only drawn examples from two research groups, computational protein design is a vast and growing field, with important contributions made by numerous other laboratories (Park et al. 2004).
Applying rational design to parts alteration or creation is advantageous, in that it cannot only generate products with defined function, but it can also produce biological insights into how the designed function comes about. However, it requires prior structural knowledge of the part, which is frequently unavailable. Directed evolution is an alternative method that can effectively address this limitation by allowing parts engineering without design. In essence, directed evolution begins with the generation of a library containing many different DNA molecules, often by error-prone DNA replication, DNA shuffling or combinatorial synthesis. The library is then subjected to high-throughput screening or selection methods that maintain a link between genotype and phenotype in order to enrich the molecules that produce the desired function. The process is then iterated to approach a desired endpoint (Arnold 2001; Kolkman & Stemmer 2001; Joyce 2004). A recent example of parts creation by directed evolution is the expansion and alteration of LuxR specificity for acyl-homoserine lactone ligands (Collins et al. 2005, 2006). LuxR is a transcriptional activator from the marine bacteria Vibrio fischeri, and it is naturally responsive to the signalling molecule 3OC6HSL. Collins et al. first employed a screening scheme to identify mutations that broadened the binding specificity of LuxR to other small molecules in the same class as 3OC6HSL (Collins et al. 2005). They then used a dual-selection method (Yokobayashi & Arnold 2005) to redirect LuxR specificity to one of those molecules, C10HSL (Collins et al. 2006). The result was a new protein that responds to the second chemical, but no longer to the first. These parts may be particularly beneficial to designers desiring multiple channels of simultaneous communication between cells. Directed evolution can also be applied at other levels of biological hierarchy, for example, to evolve entire gene circuits (Yokobayashi et al. 2002).
Rational design and directed evolution should not be viewed as opposing methods, but as alternate ways to produce and optimize parts, each with their own unique strengths and weaknesses. Directed evolution requires a high-throughput way to screen or select for a desired function and that functional mutants exist in the sequence space sampled. This second constraint becomes less likely as the desired function diverges further from the initial function. On the other hand, while rational design strategies can make multiple changes or large-scale alterations that incorporate scientific knowledge, these strategies are rarely precise enough to finely tune the system behaviour. Furthermore, it is difficult to know if additional optimization is possible when employing rational design. For these reasons, both methods can and should be used in conjunction and will hopefully continue to be applied in unison during the years to come.
Recent years have witnessed increasing interest in using parts based on RNA for intricate control of gene expression (Davidson & Ellington 2005; Isaacs et al. 2006). One particular line of research has been largely inspired from metabolite-controlled riboswitches prevalent in nature (Mandal & Breaker 2004; Nudler & Mironov 2004). RNA switches are advantageous in their fast response, broad applicability and chemical nature. RNA switches contain a ligand-binding region, or aptamer domain, that controls the function of an effector domain through binding-induced conformational changes. Strategies for the evolution of RNA aptamers and functional RNAs were developed early on (Ellington & Szostak 1990; Robertson & Joyce 1990; Tuerk & Gold 1990) due to the fact that the same molecule plays both functional and information-encoding roles (i.e. the genotype–phenotype link required for directed evolution schemes is intrinsic to the molecule). This allows the generation of a library directly from the products of a competitive screen in the previous round. Furthermore, the entire selection, amplification and iteration procedure can be economically accomplished in vitro. The chemical nature of RNA, with four bases possible at each position, means that a higher percentage of available sample space can be covered while evolving an RNA molecule than a protein of similar length (20 amino acid possibilities per position). Additionally, the interactions within an RNA molecule are largely driven by complementary base pairing. As a result, relatively accurate methods for the secondary structure prediction of RNA have been developed and are widely used (Mathews et al. 1999; Zuker 2003). Secondary structure information is valuable, because it can allow a researcher to make rationally guided changes.
In a recent work from the Smolke group (Bayer & Smolke 2005), switches were developed that exposed an antisense stem sequence upon binding a ligand, producing a riboregulator. Ligands, such as theophylline, controlled switches that turned on gene expression, as well as switches that turned off gene expression. These switches were shown to be tunable by making simple changes to the RNA sequence guided by thermodynamic properties. Multiple switches functioned independently in yeast even when binding similar molecules. Switches such as these may be useful in sensing cellular conditions and could also act as feedback mechanisms for tuning metabolic pathways in response to the depletion or accumulation of reactants, intermediates or products. Gallivan and colleagues demonstrate a synthetic RNA switch that is functional in prokaryotes and can be applied in screening or selection schemes that tie in vivo levels of small molecules to a reporter gene or cell survival, respectively (Desai & Gallivan 2004). In this manner, one could screen enzyme libraries for a desired catalytic function. Inversely, if the small chemical is supplied, then a library of riboswitches could be screened for binders that alter gene expression. Suess and colleagues, who first described a rationally designed in vivo RNA switch, implement it in such a way that it functions as a logic gate with another ligand, xylose (Suess et al. 2004). Perhaps, the best-known form of gene regulation by RNA, however, is the role of interfering RNA (Hannon 2002). Yokoboyashi and colleagues show that it is possible to modulate shRNA activity through the action of a small chemical by fusing the shRNA to an aptamer that responds to the chemical (An et al. 2006).
Synthetic riboregulators need not be ligand controlled. Collins et al. demonstrate a general method to introduce RNA-mediated post-transcriptional regulation into prokaryotic genes (Isaacs et al. 2004). They introduced a short sequence between the promoter and ribosome-binding site that when translated into mRNA folds into a hairpin with the adjacent ribosome-binding site, sequestering the site and preventing translation. Translation can be restored by expressing a trans-acting RNA that binds the hairpin and forms a more stable structure, which frees the ribosome-binding site.
These examples demonstrate that the cellular engineer of the future will not be restricted to the catalogue of known biological parts, but will also have the tools needed to supplement natural parts with custom-made parts for specific applications.
2.3 Modelling-guided circuit engineering
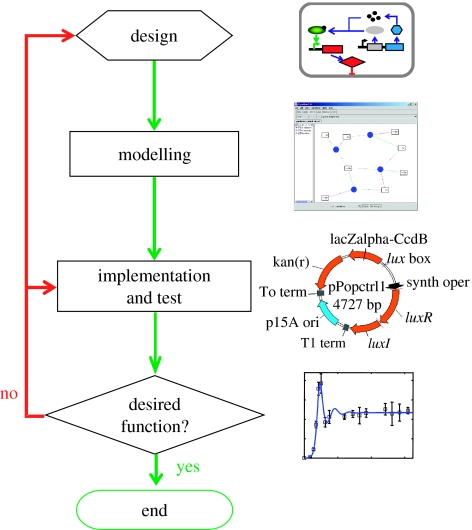
The engineering process usually involves multiple cycles of design, optimization and revision (box 1 and figure 2). This is particularly apparent in the process of constructing gene circuits. As the number of interacting parts and reactions increases, it becomes more difficult to intuitively predict circuit behaviour. Towards these ends, mathematical modelling is a useful design tool, in particular, for systems with complex dynamics, such as bistability and oscillations. The importance of mathematical modelling has been increasingly appreciated, as evidenced by its extensive application in systems biology as a way to decipher ‘design principles’ of natural biological systems (Asthagiri & Lauffenburger 2000; Tyson et al. 2001; Gilman & Arkin 2002; You 2004). In comparison, the utility of modelling in synthetic biology seems even more dominant (Hasty et al. 2002; Kaern et al. 2003).
Box 1. A recipe for engineering gene circuits (also see figure 2).
- Design
- Determine the design goal
- For the purposes of this tutorial we will attempt to construct a population of cells that restricts its cell density below that imposed by nutrient limitations (You et al. 2004). The implementation discussed below is a revised version (Balagadde et al. 2005).
- Pick suitable host organisms/strains
- Key characteristics to consider here are: ease of genetic manipulation, growth rate, survivability under the desired conditions, and endogenous machinery you wish to exploit. E coli could be used for this application.
- Identify necessary ‘parts’
- Available places to draw from include the literature, genome sequences, colleagues, and the MIT registry (The_BioBricks_Foundation). Recall that: (i) Parts need not be from the host organism. While native parts are likely to function properly, they can lead to crosstalk with endogenous systems. (ii) Parts need not exist; they can be developed by rational design or directed evolution. (iii) The better characterized the parts, the easier your job will be. (iv) It is advantageous to include parts as reporters. In this tutorial we will pick the quorum sensing genes luxR and luxI, as well as the toxin gene (CcdB) from F plasmid segregation.
- Determine the design goal
- Modeling
- Build a mathematical model
- Start with the simplest model that can capture the circuit dynamics (for example a simplifying assumption might be to assume a protein's production rate depends on a transcription factor rather than explicitly modeling mRNA production, translation, and decay).
- Explore circuit dynamics in silico
- Address questions like: can the network architecture give you the function you want? What parameters are most critical for success? How do circuit dynamics change with parameters?
- Build a mathematical model
- Implementation, testing, and debugging
- Determine the DNA implementation of your circuit
- In our case we will implement our circuit on a plasmid and need decide on copy number, what promoters, RBSs, transcription terminators, and perhaps degron tags to use. Anther choice at this time is to decide if any components need to be expressed together on a polycistronic RNA. In this example, the circuit is implemented in a medium copy number plasmid (p15a origin) which the luxR and luxI gene are co-expressed by a P_lac/ara_ promoter. The CcdB gene is controlled by a PluxI promoter. Kanamycin resistance is used as a selection marker.
- If modeling indicates that a particular parameter is critical, build multiple versions
- It is rare for all parameters to be perfectly balanced on the first experimental implementation. Designing multiple circuits at once to sample a critical parameter space can increase the chance for initial success. It may also yield interesting information about whether that particular parameter is truly critical.
- Test your circuit and decide whether to retest, revise, or redesign
- If it works as predicted you can continue to fully characterize it. If not, can you fit your model to explain the behavior that is observed? What parameters may need altering to generate the desired function? At this point you can: (i) redesign the circuit to address critical parameter changes, and perhaps ‘fine tune’ the circuit function by directed evolution; or (ii) test the circuit in other strains or growth conditions.
- Determine the DNA implementation of your circuit
A working design usually requires multiple rounds of iteration of steps listed above, which is often the most time consuming portion of biological design.
Figure 2.
The typical process for engineering gene circuits (see box 1 for more details).
Various mathematical formulations can be used to model gene circuits. At the population level, gene circuits can be modelled using ordinary differential equations (ODEs). In an ODE formulation, the dynamics of the interactions within the circuit are deterministic. That is, given the same initial condition and numerical configurations, different rounds of simulations will lead to exactly the same results. In other words, the ODE formulation ignores the randomness intrinsic to cellular processes and is convenient for circuit designs that are thought to be less affected by noise or when the impact of noise is irrelevant. For instance, ODE models have been used to guide experimental efforts to program population dynamics in the temporal domain (You et al. 2004; Balagadde et al. 2005) or the spatial domain (Basu et al. 2004, 2005). Importantly, an ODE model facilitates further sophisticated analyses, such as sensitivity analysis and bifurcation analysis. Such analyses are useful to determine how quantitative or qualitative circuit behaviour will be impacted by changes in circuit parameters; this has been almost a standard practice in engineering of most gene circuits accomplished so far (box 1). For instance, in designing a bistable toggle switch, bifurcation analysis was used to explore how qualitative features of the circuit may depend on reaction parameters (Gardner et al. 2000). Results of the analysis were used to guide the choice of genetic components (genes, promoters and ribosome-binding sites) and growth conditions to favour a successful implementation of designed circuit function.
In a single cell, however, a gene circuit's dynamics often involve small numbers of interacting molecules. Such small numbers will result in highly noisy dynamics even for expression of a single gene (Elowitz et al. 2002; Ozbudak et al. 2002). For many gene circuits, the impact of such cellular noise may be critical and needs to be considered. This can be done using stochastic models (Rao et al. 2002). Different rounds of simulation using a stochastic model will lead to different results each time, which presumably reflect aspects of noisy dynamics inside a cell. For synthetic biological applications, the key of such analysis is not necessarily to accurately predict the exact noise level at each time point. This is not possible even for the simplest circuits due to the ‘extrinsic’ noise component for each circuit (Elowitz et al. 2002). Rather, it is a way to determine to what extent the designed function can be maintained and, given a certain level of uncertainty or randomness, to what extent additional layers of control can minimize or exploit such variations. For instance, a number of computational studies have been conducted to analyse the potential of cell–cell communication to synchronize intrinsically noisy and unreliable oscillators (Mcmillen et al. 2002; Garcia-Ojalvo et al. 2004).
Mathematical models, either stochastic or deterministic, can be digitally ‘evolved’ in silico to generate optimal circuit designs that satisfy a particular objective. Francois and Hakim used genetic algorithms to design gene regulatory networks that exhibited hysteresis or oscillations (Francois & Hakim 2004). Initially, a pool of gene circuits was constructed from basic reactions representing activation, repression and post-translational modification. These circuits were subsequently evolved using numerical simulations to obtain a desired output by repeated rounds of digital ‘mutations’ and functional ‘screening’. Several unique designs were generated that satisfy each design goal. These designs could serve as alternatives to consider, model or test during the circuit engineering cycle.
One of the most exciting aspects of synthetic biology is the multiple avenues being used to address questions. While some researchers may only apply a particular method for a given application, the domain as a whole will benefit from the use of these complementary approaches. For example, a simple linear cascade can be implemented using transcriptional regulation or reversible protein modification, both of which are prevalent in nature. Implementation by transcriptional control is appealing, because it is generally easier to stitch multiple DNA elements together. However, multi-component transcriptional cascades can introduce a significant time delay, as shown by Hooshangi et al. (2005). In this work, a one-stage cascade reached its half-maximal activation in minutes, whereas a three-stage cascade took several hours. Rosenfeld & Alon found that long transcriptional cascades are rare in the sensory systems of relatively short lived E. coli and Saccharomyces cerevisiae (Rosenfeld & Alon 2003). Protein modification-based circuits can offer much faster temporal response (Kholodenko 2006). As the field matures, it is probable that synthetic circuits, like nature, will integrate both DNA and protein regulatory logic in their design. The combination will exploit advantages of each method while mitigating their weaknesses. These choices will require mathematical modelling to ensure that the circuits can perform on the desired time-scale for a particular operation.
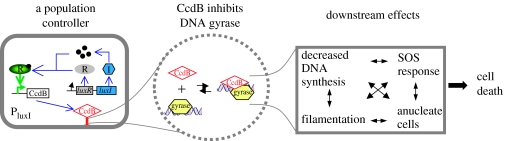
In most attempts to engineer gene circuits, mathematical models are often purposefully simplified to capture the qualitative behaviour of the underlying systems. Simplification is beneficial partially due to the limited quantitative characterization of circuit elements, one limitation that the BioBricks project aims to address (The BioBricks Foundation, Registry of Standard Biological Parts.), and partially because simpler models may better reveal key design constraints. The caveat, however, is that a simplified model may fail to capture richer dynamics intrinsic to a circuit. When engineering a population controller, we built a highly simplified kinetic model to capture the essence of the circuit dynamics, including cell growth, signal accumulation, killer protein accumulation and subsequent cell killing. The model predicts that the system will always lead to a stable regulated state, and this prediction was supported by the observations made in batch cultures (You et al. 2004). Yet, later, we observed sustained oscillations when cells expressing the circuit were grown in a microchemostat (Balagadde et al. 2005). One way to reconcile the experimental and modelling results was to introduce an extra step of regulation in our model, which indeed resulted in sustained oscillations for biologically feasible parameters. We note that still more layers of regulation are involved, further complicating the modelling analysis (figure 3).
Figure 3.
Complexity and uncertainty in a biological circuit design. Although we can build and model the circuit from box 1, it is remarkably difficult to capture even all the known interactions (let alone the unknown interactions). In our model, we have a single killing term that sets the rate of cell death proportional to the product of the killing rate constant, CcdB level and cell number. In reality, the situation is far more muddled. CcdB operates on DNA gyrase in a manner whose mechanistic details are still open to debate. The downstream effects of CcdB are plural and interrelated, and each of these involves many components. For example, the SOS response involves over a dozen players. Attempting to incorporate all the partially understood downstream effects would complicate the model with no guarantee of improving its accuracy. Nevertheless, by omitting them, we make the implicit assumption that they do not affect system dynamics.
2.4 Culturing and monitoring technologies
To determine if a synthetic circuit works as designed, one must be able to test it and observe its dynamics. These tasks have benefited from the rapid development of improved culturing and observational technologies. An ideal method for monitoring cellular dynamics over time should be easy to perform and should not significantly affect the properties being measured. One step towards this ideal has been the engineering of fluorescent protein variants (Giepmans et al. 2006). These proteins are genetically encoded and mature to functionality without requiring cofactors. Each variant fluoresces with a specific visible wavelength upon excitation, allowing multiple variants to be discerned in one cell.
Fluorescent proteins can directly report on protein levels when present in translational fusions or indirectly report when present in transcriptional fusions. A translational fusion is made by inserting a fluorescent protein into the reading frame of the target protein resulting in the translation of the fluorescent protein and target protein as one molecule. That is, one can tag a target protein with a fluorescent tail. In many cases, this does not significantly affect the function of either a target or a flourescent protein. A transcriptional fusion is made by co-expressing a fluorescent protein and a target protein by placing each behind the same promoter. While this strategy reports on promoter activity, a key determinant of intracellular levels, it fails to capture any post-transcriptional or post-translational regulation, such as the action of regulatory RNAs or proteases. With both transcriptional and translational fusions, fluorescence measurements are non-invasive to live cells, and the process can be automated for long-term measurements. Fluorescent proteins therefore represent an elegant solution for monitoring in vivo protein levels. Caution must be exercised with translational fusions, however, because even if the fluorescent tag does not alter the target protein's function per se, it may significantly impact its localization. Although many of such cases are unreported, the literature is spotted with examples of mis-localized or mis-transported fluorescent fusion proteins (Roucou et al. 2000; Hanson & Ziegler 2004). This is an important issue not only for studies that explore protein trafficking, but also for any system where altered localization will affect function.
A particularly appealing application of fluorescent proteins is to monitor single-cell dynamics in real time through optical microscopy. Single-cell measurements are critical for revealing heterogeneity in gene expression or differences in other phenotypic traits between the cells that are often masked in population-level measurements. In one of the earliest synthetic circuits published, Elowitz & Leibler built a circuit capable of producing oscillations in gene expression, but it was only through the microscopic tracking of individual lineages of bacteria that the oscillations became truly apparent (Elowitz & Leibler 2000). Similar techniques were used to characterize other oscillators implemented later (Atkinson et al. 2003; Fung et al. 2005). Recently, single-cell measurements have become the workhorse for a series of elegant experimental studies aimed at deciphering the origin and characteristics of cellular noise (Elowitz et al. 2002; Ozbudak et al. 2002; Blake et al. 2003; Raser & O'shea 2004; Hooshangi et al. 2005; Pedraza & Van Oudenaarden 2005; Rosenfeld et al. 2005; Austin et al. 2006; Guido et al. 2006; Volfson et al. 2006).
Remarkably, measurement capabilities are continuing to improve in resolution, as tools to track single molecules in vivo have also been developed. Building on previous mRNA visualization techniques (Bertrand et al. 1998), it is now possible to track individual mRNAs in vivo by using multiple fluorescent mRNA-binding proteins (Fusco et al. 2003; Golding & Cox 2004; Shav-Tal et al. 2004). Yu et al. show that it is even possible to detect a single fast maturing fluorescent protein by targeting it to the membrane (Yu et al. 2006). These detection methods improve researchers’ abilities to quantify the abundance and localization of cellular components. Researchers can then determine when and where the experimental system deviates from their expectations, improving their ability to test and troubleshoot designs.
It is a rare and joyous occasion when a synthetic genetic circuit actually works as expected for the first time. The laborious and time-consuming process of characterizing and debugging biological programs will become more significant as the circuits increase in complexity. This process is, by and large, the rate-limiting step for engineering gene circuits that program sophisticated dynamic behaviour (box 1). An important advance in this area is the miniaturization of characterization processes through microfluidics—the science and technology of systems that manipulate small amounts of fluids (10−9–10−18 l), using micro-sized channels (Quake & Scherer 2000; Hong & Quake 2003). Microfluidic metering enables ultra-low consumption of biological samples and reagents, allowing high-throughput research at low cost with short analysis time. Microfluidic miniaturization also facilitates automation and integration of complex chemical or biological procedures into a single process that is faster, more precise and more reproducible than its manual counterparts. Pioneered by the Quake laboratory, the development of actuatable pneumatic valves through multilayer soft lithography (MSL) has facilitated the design of complicated devices equipped with pumps, fluidic isolation and mixers (Unger et al. 2000).
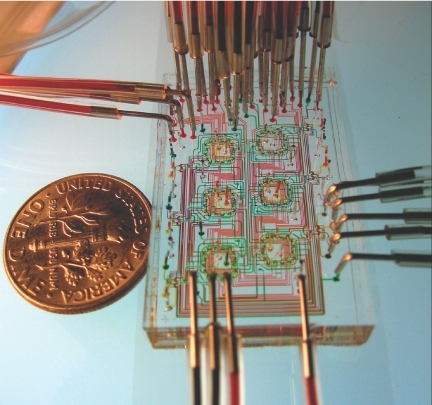
As a proof of concept for synthetic biological application, Balagadde and colleagues devised and implemented a miniaturized 16 nl bioreactor, called a microchemostat, that enables automated culturing and monitoring of small populations (102–104) of bacteria for hundreds of hours with single-cell resolution (Balagadde et al. 2005). By reducing the reactor volume by a factor of 105 when compared with traditional chemostats, microchemostat populations undergo proportionately fewer divisions per hour, which suppress the total mutation rate of the population. This, in turn, effectively insulates the micro-cultures from rapid evolution, prolonging monitoring of genetically homogeneous populations. The microchemostat system is automated by custom software which controls periodic media dilution, culture mixing, image acquisition and image analysis. Its unique design also allows multiple experiments to be run in parallel on the same chip (figure 4). In addition to the measurements of cell density and morphology, a recently improved chip design enables measurements of gene expression dynamics reported by fluorescence or luminescence (F. Balagadde 2006, unpublished data).
Figure 4.
A microfluidic chip with six parallel microchemostat reactors, used to study the growth of microbial populations. The coin is 18 mm in diameter.
In another microfluidics application, Thorsen and colleagues created a ‘comparator’ capable of screening individual cells for desired functionality in a high-throughput manner. In this device, two reagents can be separately loaded into 256 pairs of subnanolitre reaction chambers. Adjacent chambers are united allowing the reagents to mix and react. The products of each reaction can then be selectively recovered. This system was used to perform a high-throughput detection of single bacterial cells expressing recombinant cytochrome c peroxidase (Thorsen et al. 2002).
Fu and colleagues fabricated a microfluidic fluorescence-activated cell sorter (FACS) to sort live fluorescent E. coli cells. Compared with the conventional FACS machines, the microfluidic device allows for more sensitive optical detection of bacterial cells as well as DNA strands, and it is also capable of ‘reverse’ sorting. Reverse sorting is a procedure where cells are scanned at a high flow rate until a fluorescent cell is detected. Flow is then stopped and reversed, allowing the cell to be measured a second time and diverted into a collection tube. Reverse sorting is particularly useful for isolating rare cells or making multiple measurements on a single cell (Fu et al. 2002).
The aforementioned microfluidic devices can be used in stand-alone applications or as part of an integrated system. They are also disposable, which eliminates any cross-contamination in between the runs. These and many other microfluidic systems (Cookson et al. 2005; Groisman et al. 2005; Zhang et al. 2006) being actively developed will become important tools for synthetic biologists (El-Ali et al. 2006).
3. Applications
3.1 Green chemistry
Natural biological systems are astonishing production factories capable of synthesizing an impressive array of chemicals with relatively high yields. For example, the plant metabalome alone is estimated to contain over 1 million unique chemical species (Schwab 2003). Furthermore, all of these diverse chemical species are synthesized under ‘gentle’ conditions in the cells (i.e. in aqueous solutions and at mild temperatures). In contrast, current methods for organic synthesis often rely upon exotic solvents, reaction conditions and catalysts. Not only are such methods expensive, but they can also produce a variety of undesirable and toxic waste products. These problems can be alleviated through development of novel biological catalysts and synthetic metabolic pathways. Such advances could usher in a new era of environmentally friendly or ‘green’ chemistry by breaking our dependence on toxic solvents and catalysts while decreasing waste product formation.
It would be naive to think that custom metabolic synthesis will replace the majority of organic chemical synthesis in the near future. However, it can have an immediate impact on several areas. One such example is the production of artemisinic acid, a precursor to the antimalarial drug artemisinin. Originally discovered as a Chinese herbal therapy, artemisinin is currently isolated from the shrub Artemisia annua, but it is too expensive for most populations where malaria is a problem. Total chemical synthesis is difficult and costly, but researchers have recently reported the production of up to 100 mg l−1 artemisinic acid from an engineered laboratory yeast strain (Ro et al. 2006). To engineer the yeast strain, Keasling and colleagues first increased precursor production by manipulating the farnesyl pyrophosphate (FPP) pathway to augment FPP yield. They additionally downregulated a gene that diverts FPP to a sterol-producing pathway. They then added genes from A. annua to convert FPP to amorphadiene and subsequently convert amorphadiene to artemisinic acid. The authors report a simple purification scheme to recover the artemisinic acid, which can then be converted to artemisinin in a relatively straightforward chemical reaction. This would appear to be a vast improvement over their complementary work in E. coli that reported the introduction of a metabolic pathway capable of producing up to 24 mg l−1 of amorphadiene, an artemisinic acid precursor (Martin et al. 2003). It has recently been reported, however, that the engineered E. coli strain produces higher levels (500 mg ml−1) than previously measured (Newman et al. 2006). Measurement errors were due to the high volatility of amorphadiene in aqueous solutions. It therefore remains to be seen which organism will ultimately be the most useful as a bioreactor for this application. That virtually the same metabolic pathway can be built in two organisms from different taxonomic domains is perhaps an indication of the potential plasticity of cellular metabolism in the hands of a skilled practitioner.
Artemisinic acid is not the first example of a therapeutic molecule produced in a cell culture. Many drugs currently on the market including insulin, erythropoietin and therapeutic antibodies are also made in cellular bioreactors. However, artemisinic acid is distinctly different from most biologically cultivated therapeutics, because it is not a protein and requires many more metabolic steps than simple transcription and translation. All these steps must be carefully regulated and balanced to control metabolic fluxes and maximize yield. This type of synthetic biology is deeply rooted in what many might call metabolic engineering (Bailey 1991; Stephanopoulos & Vallino 1991). Some may even argue that metabolic engineers have been doing synthetic biology far before the label became well established.
3.2 Therapeutics
Drug production is only one example of how synthetic biology can contribute to medicine. In this age of shots and pills, it is easy to forget that our bodies' defence system is predominantly composed of cells. Billions of immunological cells patrol our bodies at any given time on the lookout for antigens that indicate foreign cells or abnormal function. A key feature of our immune system is that it is predominantly targeted to the particular offending pathogens or region of infection through the use of cell surface receptors and signalling molecules (Goldsby 2003). Most drugs, however, are often taken systemically and can be damaging to unintended targets. For example, many chemotherapy treatments aim to control the fast proliferating cells in the cancer, but inadvertently destroy the rapidly dividing hair follicles and cells of gastric linings, resulting in hair loss and digestion problems.
Cells can be engineered to recognize specific targets or conditions in our bodies that are not naturally recognized by the immune system. Although some drugs can also be targeted to specific locations through aptamer (Mcnamara et al. 2006) or antibody conjugation (Schrama et al. 2006), a cell has the advantage of being able to interpret and respond to complex environmental signals. Anderson et al. (2005) engineered bacteria to invade tumours in response to specific extracellular conditions. By directing expression of the invasion gene from Yersinia pseudotuberculosis through promoters responsive to hypoxia, cell density or arabinose, they restricted bacterial invasion of mammalian tumour cells to these conditions. This is significant because the tumour environment is often hypoxic and allows for high bacterial cell densities due to depressed immune function in the tumour. Therefore, this work demonstrates, as a ‘proof of concept’, that one can potentially use engineered bacteria to target diseased cells without significantly impacting healthy cells.
A lot of synthetic biology research has been carried out in bacteria due to their ease of manipulation and simpler physiology when compared with mammalian cells. While engineered bacteria do have tremendous potential for therapeutic applications, as previously illustrated, the general public may feel more comfortable dealing with therapeutics derived from mammalian cells. Furthermore, mammalian cells are already closer to being optimized for functions in the human body. For these reasons, major advances in cell-based cancer therapeutics are being made through engineering of mammalian cells. Rosenberg and colleagues report the generation and application of tumour-specific T-cells in 15 metastatic melanoma patients (Morgan et al. 2006). To generate the cells, a T-cell receptor recognizing the tumour-associated antigen (TAA) MART-1 was transfected (using a retroviral vector) into the peripheral blood lymphocytes isolated from the patients. Patients received the engineered cells by adoptive cell transfer. Even though only 2 out of the 15 patients showed sustained regression, the work demonstrates the potential applicability of targeted therapy using engineered cells. The authors also indicate many possibilities for improvement in future trials, such as tighter binding TAA receptors or cytokine/tissue-homing mechanisms.
The aforementioned work used a retrovirus for integration of the transgene into patients' cells ex vivo. Viral vectors for gene therapy often insert DNA at a particular locus in the cells' chromosome. However, in many cases, it may be more desirable to actually replace a malfunctioning gene. Recent ‘parts design’ has produced a library of the so-called zinc-finger nucleases (ZFNs) that may enable in vivo human gene replacement. ZFNs join a type of DNA recognition element (zinc finger motifs) and a DNA-cleaving enzyme (nuclease) to target a specified sequence. Unlike many of the common bacterially isolated restriction enzymes, which recognize 4, 6 or 8 bp, ZFNs can recognize a sequence long enough for it to be unique in an organism's genome. Urnov et al. report the construction and application of a pair of ZFNs that recognize a 24 bp site in the human genome (Urnov et al. 2005). The ZFNs create a double-strand break in the chromosome. The break, in turn, induces cells' natural homologous recombination machinery to incorporate DNA from synthetic donor constructs. Twenty per cent of chromosomes successfully recombined, leading to 7% of cells homozygous for the correction in the absence of selection. ZFNs or other large and specific endonuclease design (Arnould et al. 2005; Ashworth et al. 2006) may hold the key to altering an organism's DNA post-development. Such in vivo genome alterations will enable therapeutic intervention ranging from simple replacement of mutant alleles with wild-type to controlled integration of novel multi-gene circuits. An intrinsic advantage of gene correction (over gene insertion) is that the replaced allele is present in its natural chromosomal locus, therefore increasing the chance it will be properly regulated.
3.3 Renewable energies
As the global supply of fossil fuels diminishes, alternative and renewable energy sources will become more critical than ever before. Production of bioethanol, ethanol derived from crops, has emerged as a potential way to convert abundant solar energy gathered by plants into easily stored fuel for combustion engines (Hahn-Hagerdal et al. 2006). Production of bioethanol relies upon micro-organisms, such as yeast, to ferment the plant materials. A current limitation, however, is that most naturally occurring or laboratory micro-organisms are incapable of converting all types of energy-storing compounds found in crops into ethanol. For this reason, sugarcane and corn are the major feedstocks for bioethanol conversion (both sucrose and starch can easily be converted to glucose). Consequently, bioethanol production is economically feasible only in the regions producing such crops, such as Brazil.
All human habitats have naturally thriving plants that contain other energy-storing compounds, including cellulose (40–50%), hemicellulose (25–35%) and lignin (15–20%; Grey et al. 2006). However, for this ‘cellulosic biomass’ to be used, improvements must be made in both the enzymatic degradation of these compounds into simpler sugars (including glucose) and the efficient conversion of non-glucose sugars to ethanol. Optimization of enzymes such as cellulose and hemicellulase can result in decreased costs and higher efficiency. Here, synthetic biology could play a role by either boosting expression through systems design or improving activity and stability through parts design.
Microbial strain engineering has already begun to tackle the issue of non-glucose sugar conversion (Jeffries 2006). The primary non-glucose sugar formed after enzymatic breakdown of cellulosic biomass is the pentose sugar xylose. As a result, early efforts have focused on the engineering of microbial strains capable of co-fermenting glucose and xylose simultaneously in order to increase yield and production rates. Ho and colleagues address the xylose utilization issue by introducing three xylose-metabolizing genes into the yeast chromosome at multiple copy numbers: xylose reductase (XR); xylitol dehydrogenase (XD) and xylulokinase (XK; Sedlak & Ho 2004). Together, these three enzymes convert xylose to xylulose-5-phosphate, a key metabolite in the yeast pentose metabolism pathway. Resulting strains produced ethanol levels in excess of 75% of the theoretical yield of sugars consumed. An alternate method, described by the Pronk laboratory, features the introduction of a fungal xylose isomerase from Piromyces and the overexpression of downstream pentose phosphate pathway genes: xylulokinase; ribose 5-phosphate isomerase; ribulose-5-phosphate epimerase; transketolase; and transaldolase (Kuyper et al. 2005). The GRE3 gene, which produces unwanted side product xylitol, was deleted from the strain. The resulting strain was capable of fast anaerobic growth with xylose as the sole carbon source, but still showed a strong preference for glucose in mixed carbon source cultures. In subsequent work, Pronk and colleagues employed long-term nutrient-limited chemostatic cultures to evolve strains with improved xylose uptake and usage kinetics, resulting in a strain that completely ferments both glucose and xylose in less than 25 h. While both sets of strains described here can benefit from further improvements, they demonstrate the progress being made towards expanding bioethanol production to more diverse crops. Future generations may be cultivating high yield and easy to grow species such as switchgrass or hybrid poplar trees to fuel the worlds growing energy needs.
3.4 Pattern formation
The human body is a complex system of specialized cells, tissues and organs. Remarkably, each highly specialized cell in our bodies arises from a single fertilized egg cell. This process of differentiation and morphogenesis is mediated by the delicate interplay of chemical gradients, cellular receptors, differential gene expression and cell migration (Gilbert 2000). The end result is that the 100 trillion cells of the adult human are neatly arranged and specialized in a way that allows for proper functioning of all bodily processes. Nature has produced incredibly complex systems, as well as a fantastic way of assembling them. This accomplishment is even more amazing considering that the overall robust system builds upon components that are often intrinsically ‘noisy’. With regard to this accomplishment, the synthetic biologist can ask ‘in what ways can we recreate or use the complex pattern formation systems found in nature, and to what ends?’
As a first step to address this question, Weiss and colleagues rewired cell signalling pathways to create a model system of chemical gradient-induced pattern formation in bacteria (Basu et al. 2005). ‘Sender cells’ produce a small membrane-diffusible chemical, acylhomoserine lactone (AHL), by expressing the luxI gene from V. fischeri. ‘Receiver cells’, in turn, respond to the signal through luxR activation upon AHL binding, which induces transcription from a lux promoter. By placing both a single repressor and a double repressor cascade behind lux promoters, Weiss and colleagues effectively created a band detector such that a downstream gene (gfp) is expressed only at intermediate concentrations of AHL. Furthermore, by creating variants of receiver plasmids through luxR mutagenesis and copy number reduction, receivers can be tuned to respond to different bands of AHL concentrations. Consequently, when a region of sender cells is placed within a lawn of receiver cells, fluorescence is observed only in a ring whose distance from the sender cells varies in accordance with the version of the receiver plasmid used. The visual result resembles a bullseye.
While the bullseye pattern is novel and interesting, one may be left wondering what use it can find. With a little imagination, however, one can envision using this pattern formation system to control a master regulatory gene capable of committing cells to a particular developmental fate. Higher-order functions in natural biological systems are associated with multi-cellularity and cellular specialization. To produce similarly complex functions, synthetic biologists will require mechanisms that produce and maintain differentiation patterns. These mechanisms may lead to highly sophisticated cellular systems for fabricating biomaterials with well-defined dimensions. This line of research may also synergize with research efforts focusing on regenerative medicine (Lagasse et al. 2001) and tissue engineering (Griffith & Naughton 2002), both of which hinge upon controlling differentiation and pattern formation. Biologists' continued efforts to implement synthetic multi-cellular systems will drive the production of new and better approaches to artificial cellular communication. Most communication systems employed by synthetic biologists thus far have made use of the small diffusible molecules from bacterial quorum sensing. Further developments may feature active and regulated transport of signalling molecules across the cell membrane and the use of cell surface receptors to recognize and send signals to adjacent cells.
4. Outlook
4.1 Standardization: promises and limitations
It has been suggested that many of the difficulties in the production and optimization of biological circuits are due to improper and incomplete description of parts (Endy 2005). These limitations are twofold: first, functional characteristics are often unknown for many parts; second, even if they are known, they are rarely described using standardized measures and are often buried in the literature. Towards addressing these limitations, the BioBricks Foundation has established a ‘registry of standard biological parts’ (http://parts.mit.edu/). The registry categorizes parts, devices and systems. Ultimately, the registry strives to provide information on not only sequence but also functional characteristics, and make information available through a central portal. Many of these parts have been cloned into plasmids that enable easy assembly. The plasmids are made available to students participating in the international Genetically Engineered Machine competition (iGEM). Members of the BioBricks Foundation hope that the registry will decrease the time and research costs needed to design and implement gene circuits. Such efforts are analogous in spirit to ongoing attempts to standardize mathematical models (Hucka et al. 2003) and formats for microarray data (Brazma et al. 2001). The limits in achieving parts standardization for E. coli and other organisms remain to be seen.
Even with a repository of information about standardized parts, a major challenge to applying this information will be developing strategies to deal with context dependence (Andrianantoandro et al. 2006; Arkin & Fletcher 2006). For example, synthetic gene circuits often exhibit varying behaviour in different cell strains. In some cases, this can be easy to rationalize by the presence or absence of a particular gene, or a documented difference in the growth rate. In other cases, causes of variability are much more difficult to ascribe due to many hidden interactions between the designed circuit and a far-from-elucidated host circuitry.
To address this issue, one may imagine selecting a standard cell strain, in which standard parts under standard conditions are to be quantified. A starting point for such a standard strain may be on its way. The Blattner group has recently engineered a series of multiple deletion strains (MDS) that have up to 15% of their parental MG1655 genome removed but maintain similar growth rates on minimal media (Posfai et al. 2006). Deletions were guided by comparative genomics with related strains. Removing ‘unnecessary’ portions of the genome can presumably reduce the number of hidden interactions. Notably, the deletions cleaned the cells of mobile DNA elements called insertion sequences (IS) that might reduce the genetic stability of a circuit by inserting themselves into and disrupting a DNA sequence unpredictably. Interestingly, the MDS strains produced some unanticipated benefits, including higher electroporation efficiencies than their parent strain and the ability to propagate some plasmids that the parent strain could not.
In an alternate approach, researchers at the Venter Institute have used Mycoplasma genitalium as a starting point in their attempts to determine a minimal gene set by systematically mutating every gene (Glass et al. 2006). Mycaplasma genilalium has the smallest known genome of organisms capable of growth in the absence of other species. They conclude that in a laboratory setting, only 382 of the strain's 482 genes are essential. Although a strain containing only this set of minimal genes has not yet been constructed, it could eventually serve as a bare bones platform upon which desired functionality can be added. Such a small number of genes might allow a greater percentage of the cell's molecular interactions and metabolic processes to be understood, making the strain more predictable and desirable as a starting point. However, of the 382 essential genes determined, 110 are annotated as hypothetical proteins or as proteins of unknown function, indicating that a truly complete cellular model, even for this simplest of cells, cannot yet be produced.
Despite characterizing parts in a standard strain under defined conditions, individual parts may impact the physiology of the host strain differently, for instance, by placing varying burdens on the host translation machinery. For this reason, one may wish to minimize such interactions by creating privileged sets of machinery. For instance, Rackham & Chin (2005_b_) describe the formation of orthogonal ribosome—mRNA pairs that could be used to keep a synthetic system and host more isolated. Using a dual positive–negative selection scheme, they isolated mRNAs with modified Shine–Dalgarno regions not recognized by endogenous ribosomes, but instead recognized by alternative ribosomes. Translation by orthogonal pairs should be unaffected by endogenous ribosomes and there should be no competition for ribosomes between orthogonal mRNAs and traditional mRNAs. In principle, multiple ribosome types can be implemented for a specified function, just as cells already possess multiple DNA or RNA polymerase types, which play specialized roles.
Previous and current progress promises an ever growing infrastructure that will no doubt tremendously benefit future synthetic biology research, fundamental and applied alike. Concerning standardization, however, two critical questions remain to be addressed by the community. First, given the amount of cell physiology (even for highly characterized organisms such as E. coli) that is still poorly understood, to what extent can we standardize parts or systems with confidence? Second, how much standardization can we afford and still hope to create useful systems that can work in complex environments such as in a cancer or a polluted environment?
There is little difficulty in unambiguously defining the DNA sequences that code for parts, be they proteins or RNAs. The true challenge lies at the functional levels. Parts will impact and be impacted by cell physiology, which also changes in response to the environmental conditions. In addition, parts tested in isolation may unpredictably impact each other's functions when combined. For example, connecting one part's DNA with another part's may introduce unintended regulation by introducing enough flexibility in DNA to allow DNA looping. For these reasons, one can rarely have complete confidence in the part's function even if he/she uses it in a standard strain characterized under a standard condition. Many such interactions are still poorly understood, complicating the use of standard parts. Yet, it is precisely this complexity that makes engineering biology challenging and interesting. Decoding this complexity is at least one important application of synthetic gene circuits. Without a much deeper understanding of cellular functions at all levels, it is difficult to even define standards meaningfully.
From a practical standpoint, too much standardization may remove flexibility in engineering useful systems. It would be illogical to rely only on standard strains that lack desirable properties for a particular application. Consider thermophilic bacteria, capable of life at temperatures as high as 113°C (Stetter 1999). The ability to thrive at elevated temperatures may be a useful property for synthetic organisms involved in chemical processing, because higher temperatures speed kinetic rates. Given the difficulty in thermostabilizing even a single protein, however, it is unlikely this quality can be engineered into a standard strain. For many applications, the researcher is left with no appealing options except to use non-standard strains. No single strain or growth condition can ever cover all potential synthetic biology applications.
If we remain dedicated to standardization, gathering standardized information for a set of potentially useful parts, in a set of useful strains, under a set of relevant conditions becomes a combinatorial nightmare. The inevitable result is that standards will only be available for a limited number of strains and conditions. Although some information is preferable to none, a rising danger is to place undue weight on the limited information available and assume that a part's behaviour will not vary significantly from the context in which it was described. In this situation, ‘significant’ is considered to be variation that exceeds the acceptable tolerance limits of a part in its new device. Accepting standardized information at face value, without acknowledging its limitations, will lead one to design many systems doomed to fail. However, being aware of the limitations allows one to use standard information without depending on it, to be guided by the information while simultaneously embracing strategies like combinatorial design (Guet et al. 2002) and directed evolution (Yokobayashi et al. 2002) of circuits—strategies that would be unnecessary in a fully standardized and predictable world.
4.2 De novo cells
Finally, synthetic biology may, in addition to redesigning cellular processes, contribute to producing artificial cells exhibiting all the qualities that we associate with life. For a good review of what characteristics such a cell would need and what progress has already been made, see Deamer (2005). It is probable that no matter what system is devised for artificial encapsulation of materials in membranes capable of self-reproduction, there will be argument as to whether life has truly been created. In fact, somatic cell nuclear transfer, best known for cloning Dolly, the sheep, has already accomplished a cellular ‘cold boot’. At the moment that the nucleus containing the DNA (software) is removed from the somatic cell, it is no longer living by standard definitions and could be considered a collection of nucleic acid and protein molecules. Similarly, an enucleated egg cell and cytoplasm (hardware) is not alive by consensus definitions. However, when the two are combined, life arises anew. The immediate retort might be that the system relies too heavily on cellular-derived components. Where does one draw the line however? Will it only officially be the ‘creation of life’ if each protein, nucleic acid or lipid in the new pseudo-cells is chemically synthesized from precursors? Will precursors themselves need to be produced from pure elements? In any case, de novo cell design can shed light on both the properties needed to produce life and how terrestrial life could have arisen initially.
4.3 Social impact
Synthetic biology will undoubtedly head in many unforeseen directions in the coming years and decades, but along the way, researchers in the field are paying particular attention to legal, ethical and political issues dealing with the redesign of life. At the second annual Synthetic Biology conference (SB2.0) in 2006, a full third of the time was devoted to issues of bio-safety, public perception, ownership and community organization. Even in the early stages of the field, the need for this discussion was apparent to many. Although screening and controls have been put in place by many DNA synthesis companies, DNA synthesis can allow for the creation of potentially dangerous genetic material. Nothing has illustrated this point more clearly than the 2002 production of poliovirus from synthetic DNA by the Wimmer laboratory (Cello et al. 2002). Using only synthetic oligos, cells expressing T7 polymerase and cell-free extracts, an ‘eradicated’ virus with the same pathogenic properties as the original was reproduced. Public perception issues were highlighted by the publication of an open letter from a group of NGOs, including Greenpeace and ETC that called for synthetic biologists to drop plans for self-governance and instead demand governmental supervision due to the ‘potential power and scope of this field’.
A consideration of ownership issues arises from the fact that if biological parts are owned and protected by various entities, it may be legally difficult to produce a complex system incorporating many of those parts—hindering innovation and potential societal good. Active analysis by legal scholars is needed to develop systems that ensure freedom to operate, but maintain incentives for invention and development. Some of this analysis is already underway (Rai & Boyle forthcoming). Finally, the need for community organization is evident in order to not only manage the issues of public perception, bio-safety and ownership, but also to guide the field in a way that reduces growing pains. Particularly important is the prevention of unrealistic expectations on the part of granting agencies and public. Synthetic biology holds a lot of promise, but none of the fields can address all problems and none have produced any answers overnight—despite popular hype. To achieve its vast potential, synthetic biology will need sustained support from governments and a public that understands progress is made in incidental steps.
Acknowledgments
We thank Jingdong Tian, Ron Weiss, Katarina Midelfort and Amelia Tauchen for their helpful discussions. Current research in the You laboratory is supported by the National Academies Keck Futures' Initiatives, the National Institute of Health, the National Science Foundation and the David and Lucile Packard Foundation.
References
- An C.I, Trinh V.B, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.C, Clarke E.J, Arkin A.P, Voigt C.A. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 2005;335:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E, Basu S, Karig D.K, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2:0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin A.P, Fletcher D.A. Fast, cheap and somewhat in control. Genome Biol. 2006;7:114. doi: 10.1186/gb-2006-7-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold F.H. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- Arnould S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- Ashworth J, Havranek J.J, Duarte C.M, Sussman D, Monnat R.J, Jr, Stoddard B.L, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthagiri A.R, Lauffenburger D.A. Bioengineering models of cell signaling. Annu. Rev. Biomed. Eng. 2000;2:31–53. doi: 10.1146/annurev.bioeng.2.1.31. [DOI] [PubMed] [Google Scholar]
- Atkinson M.R, Savageau M.A, Myers J.T, Ninfa A.J. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/S0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- Austin D.W, Allen M.S, Mccollum J.M, Dar R.D, Wilgus J.R, Sayler G.S, Samatova N.F, Cox C.D, Simpson M.L. Gene network shaping of inherent noise spectra. Nature. 2006;439:608–611. doi: 10.1038/nature04194. [DOI] [PubMed] [Google Scholar]
- Bailey J.E. Toward a science of metabolic engineering. Science. 1991;252:1668–1675. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- Balagadde F.K, You L, Hansen C.L, Arnold F.H, Quake S.R. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science. 2005;309:137–140. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- Bassler B.L, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Basu S, Mehreja R, Thiberge S, Chen M.T, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl Acad. Sci. USA. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Gerchman Y, Collins C.H, Arnold F.H, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- Bayer T.S, Smolke C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- Becskei A, Seraphin B, Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy S.M, Singer R.H, Long R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/S1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Blake W.J, Kaern M, Cantor C.R, Collins J.J. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Brazma A, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Bulter T, Lee S.G, Wong W.W, Fung E, Connor M.R, Liao J.C. Design of artificial cell–cell communication using gene and metabolic networks. Proc. Natl Acad. Sci. USA. 2004;101:2299–2304. doi: 10.1073/pnas.0306484101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr P.A, Park J.S, Lee Y.J, Yu T, Zhang S, Jacobson J.M. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cello J, Paul A.V, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- Chen M.T, Weiss R. Artificial cell–cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat. Biotechnol. 2005;23:1551–1555. doi: 10.1038/nbt1162. [DOI] [PubMed] [Google Scholar]
- Collins C.H, Arnold F.H, Leadbetter J.R. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol. Microbiol. 2005;55:712–723. doi: 10.1111/j.1365-2958.2004.04437.x. [DOI] [PubMed] [Google Scholar]
- Collins C.H, Leadbetter J.R, Arnold F.H. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat. Biotechnol. 2006;24:708–712. doi: 10.1038/nbt1209. [DOI] [PubMed] [Google Scholar]
- Cookson S, Ostroff N, Pang W.L, Volfson D, Hasty J. Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol. Syst. Biol. 2005;1:0024. doi: 10.1038/msb4100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.A, Ellington A.D. Engineering regulatory RNAs. Trends Biotechnol. 2005;23:109–112. doi: 10.1016/j.tibtech.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Deamer D. A giant step towards artificial life? Trends Biotechnol. 2005;23:336–338. doi: 10.1016/j.tibtech.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Desai S.K, Gallivan J.P. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13 247–13 254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- Dueber J.E, Yeh B.J, Chak K, Lim W.A. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301:1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- Dunn G.A, Jones G.E. Cell motility under the microscope: Vorsprung durch Technik. Nat. Rev. Mol. Cell Biol. 2004;5:667–672. doi: 10.1038/nrm1439. [DOI] [PubMed] [Google Scholar]
- Dwyer M.A, Looger L.L, Hellinga H.W. Computational design of a Zn2+ receptor that controls bacterial gene expression. Proc. Natl Acad. Sci. USA. 2003;100:11 255–11 260. doi: 10.1073/pnas.2032284100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer M.A, Looger L.L, Hellinga H.W. Computational design of a biologically active enzyme. Science. 2004;304:1967–1971. doi: 10.1126/science.1098432. [DOI] [PubMed] [Google Scholar]
- El-Ali J, Sorger P.K, Jensen K.F. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- Ellington A.D, Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Elowitz M.B, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Elowitz M.B, Levine A.J, Siggia E.D, Swain P.S. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- Falke J.J, Bass R.B, Butler S.L, Chervitz S.A, Danielson M.A. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois P, Hakim V. Design of genetic networks with specified functions by evolution in silico. Proc. Natl Acad. Sci. USA. 2004;101:580–585. doi: 10.1073/pnas.0304532101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A.Y, Chou H.P, Spence C, Arnold F.H, Quake S.R. An integrated microfabricated cell sorter. Anal. Chem. 2002;74:2451–2457. doi: 10.1021/ac0255330. [DOI] [PubMed] [Google Scholar]
- Fung E, Wong W.W, Suen J.K, Bulter T, Lee S.G, Liao J.C. A synthetic gene-metabolic oscillator. Nature. 2005;435:118–122. doi: 10.1038/nature03508. [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy S.M, Blanchard J.M, Singer R.H, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 2003;13:161–167. doi: 10.1016/S0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ojalvo J, Elowitz M.B, Strogatz S.H. Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proc. Natl Acad. Sci. USA. 2004;101:10 955–10 960. doi: 10.1073/pnas.0307095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner T.S, Cantor C.R, Collins J.J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Giepmans B.N, Adams S.R, Ellisman M.H, Tsien R.Y. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Gilbert S.F. Sinauer Associates; Sunderland, MA: 2000. Developmental biology. [Google Scholar]
- Gilman A, Arkin A.P. Genetic “code”: representations and dynamical models of genetic components and networks. Annu. Rev. Genomics Hum. Genet. 2002;3:341–369. doi: 10.1146/annurev.genom.3.030502.111004. [DOI] [PubMed] [Google Scholar]
- Glass J.I, Assad-Garcia N, Alperovich N, Yooseph S, Lewis M.R, Maruf M, Hutchison C.A, 3rd, Smith H.O, Venter J.C. Essential genes of a minimal bacterium. Proc. Natl Acad. Sci. USA. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I, Cox E.C. RNA dynamics in live Escherichia coli cells. Proc. Natl Acad. Sci. USA. 2004;101:11 310–11 315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsby R. W.H. Freeman; New York, NY: 2003. Immunology. [Google Scholar]
- Gray K.A, Zhao L, Emptage M. Bioethanol. Curr. Opin. Chem. Biol. 2006;10:141–146. doi: 10.1016/j.cbpa.2006.02.035. [DOI] [PubMed] [Google Scholar]
- Griffith L.G, Naughton G. Tissue engineering—current challenges and expanding opportunities. Science. 2002;295:1009–1014. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- Groisman A, Lobo C, Cho H, Campbell J.K, Dufour Y.S, Stevens A.M, Levchenko A. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods. 2005;2:685–689. doi: 10.1038/nmeth784. [DOI] [PubMed] [Google Scholar]
- Guet C.C, Elowitz M.B, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- Guido N.J, Wang X, Adalsteinsson D, Mcmillen D, Hasty J, Cantor C.R, Elston T.C, Collins J.J. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- Hahn-Hagerdal B, Galbe M, Gorwa-Grauslund M.F, Liden G, Zacchi G. Bio-ethanol—the fuel of tomorrow from the residues of today. Trends Biotechnol. 2006;24:549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hanson D.A, Ziegler S.F. Fusion of green fluorescent protein to the C-terminus of granulysin alters its intracellular localization in comparison to the native molecule. J. Negat. Results Biomed. 2004;3:2. doi: 10.1186/1477-5751-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty J, Mcmillen D, Collins J.J. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- Hawking S.W. Bantam Books; New York, NY: 2001. The universe in a nutshell. [Google Scholar]
- Hong J.W, Quake S.R. Integrated nanoliter systems. Nat. Biotechnol. 2003;21:1179–1183. doi: 10.1038/nbt871. [DOI] [PubMed] [Google Scholar]
- Hooshangi S, Thiberge S, Weiss R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc. Natl Acad. Sci. USA. 2005;102:3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Isaacs F.J, Dwyer D.J, Collins J.J. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- Isaacs F.J, Dwyer D.J, Ding C, Pervouchine D.D, Cantor C.R, Collins J.J. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- Isaacs F.J, Hasty J, Cantor C.R, Collins J.J. Prediction and measurement of an autoregulatory genetic module. Proc. Natl Acad. Sci. USA. 2003;100:7714–7719. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jeffries T.W. Engineering yeasts for xylose metabolism. Curr. Opin. Biotechnol. 2006;17:320–326. doi: 10.1016/j.copbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Joyce G.F. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- Kaern M, Blake W.J, Collins J.J. The engineering of gene regulatory networks. Annu. Rev. Biomed. Eng. 2003;5:179–206. doi: 10.1146/annurev.bioeng.5.040202.121553. [DOI] [PubMed] [Google Scholar]
- Kholodenko B.N. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkman J.A, Stemmer W.P. Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 2001;19:423–428. doi: 10.1038/88084. [DOI] [PubMed] [Google Scholar]
- Korkegian A, Black M.E, Baker D, Stoddard B.L. Computational thermostabilization of an enzyme. Science. 2005;308:857–860. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B.P, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc. Natl Acad. Sci. USA. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton G.C, Varani G, Stoddard B.L, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Toirkens M.J, Diderich J.A, Winkler A.A, Van Dijken J.P, Pronk J.T. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 2005;5:925–934. doi: 10.1016/j.femsyr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Lagasse E, Shizuru J.A, Uchida N, Tsukamoto A, Weissman I.L. Toward regenerative medicine. Immunity. 2001;14:425–436. doi: 10.1016/S1074-7613(01)00123-6. [DOI] [PubMed] [Google Scholar]
- Leonard J.A, Wayne R.K, Wheeler J, Valadez R, Guillen S, Vila C. Ancient DNA evidence for Old World origin of New World dogs. Science. 2002;298:1613–1616. doi: 10.1126/science.1076980. [DOI] [PubMed] [Google Scholar]
- Levskaya A, Chevalier A.A, Tabor J.J, Simpson Z.B, Lavery L.A, Levy M, Davidson E.A, Scouras A, Ellington A.D, Marcotte E.M, Voigt C.A. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- Liu H, Schmidt J.J, Bachand G.D, Rizk S.S, Looger L.L, Hellinga H.W, Montemagno C.D. Control of a biomolecular motor-powered nanodevice with an engineered chemical switch. Nat. Mater. 2002;1:173–177. doi: 10.1038/nmat761. [DOI] [PubMed] [Google Scholar]
- Looger L.L, Dwyer M.A, Smith J.J, Hellinga H.W. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Martin V.J, Pitera D.J, Withers S.T, Newman J.D, Keasling J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Mathews D.H, Sabina J, Zuker M, Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mcmillen D, Kopell N, Hasty J, Collins J.J. Synchronizing genetic relaxation oscillators by intercell signaling. Proc. Natl Acad. Sci. USA. 2002;99:679–684. doi: 10.1073/pnas.022642299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcnamara J.O, 2nd, Andrechek E.R, Wang Y, Viles K.D, Rempel R.E, Gilboa E, Sullenger B.A, Giangrande P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Morgan R.A, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J.D, Marshall J, Chang M, Nowroozi F, Paradise E, Pitera D, Newman K.L, Keasling J.D. High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol. Bioeng. 2006;95:684–691. doi: 10.1002/bit.21017. [DOI] [PubMed] [Google Scholar]
- Nudler E, Mironov A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Ozbudak E.M, Thattai M, Kurtser I, Grossman A.D, Van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat. Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Park S.H, Zarrinpar A, Lim W.A. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- Park S, Yang X, Saven J.G. Advances in computational protein design. Curr. Opin. Struct. Biol. 2004;14:487–494. doi: 10.1016/j.sbi.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Pedraza J.M, Van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- Posfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- Quake S.R, Scherer A. From micro- to nanofabrication with soft materials. Science. 2000;290:1536–1540. doi: 10.1126/science.290.5496.1536. [DOI] [PubMed] [Google Scholar]
- Rackham O, Chin J.W. Cellular logic with orthogonal ribosomes. J. Am. Chem. Soc. 2005a;127:17 584–17 585. doi: 10.1021/ja055338d. [DOI] [PubMed] [Google Scholar]
- Rackham O, Chin J.W. A network of orthogonal ribosome x mRNA pairs. Nat. Chem. Biol. 2005b;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- Rai, A. K. & Boyle, J. Forthcoming. Synthetic biology: caught between property rights, the public domain, and the commons. PLoS Biol. [DOI] [PMC free article] [PubMed]
- Rao C.V, Wolf D.M, Arkin A.P. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- Raser J.M, O'shea E.K. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro D.K, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Robertson D.L, Joyce G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Alon U. Response delays and the structure of transcription networks. J. Mol. Biol. 2003;329:645–654. doi: 10.1016/S0022-2836(03)00506-0. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young J.W, Alon U, Swain P.S, Elowitz M.B. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Roucou X, Prescott M, Devenish R.J, Nagley P. A cytochrome c-GFP fusion is not released from mitochondria into the cytoplasm upon expression of Bax in yeast cells. FEBS Lett. 2000;471:235–239. doi: 10.1016/S0014-5793(00)01404-6. [DOI] [PubMed] [Google Scholar]
- Schrama D, Reisfeld R.A, Becker J.C. Antibody targeted drugs as cancer therapeutics. Nat. Rev. Drug Discov. 2006;5:147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- Schwab W. Metabolome diversity: too few genes, too many metabolites? Phytochemistry. 2003;62:837–849. doi: 10.1016/S0031-9422(02)00723-9. [DOI] [PubMed] [Google Scholar]
- Sedlak M, Ho N.W. Production of ethanol from cellulosic biomass hydrolysates using genetically engineered Saccharomyces yeast capable of cofermenting glucose and xylose. Appl. Biochem. Biotechnol. 2004;113-116:403–416. doi: 10.1385/ABAB:114:1-3:403. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy S.M, Fusco D, Janicki S.M, Spector D.L, Singer R.H. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D, Elowitz M.B. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G, Vallino J.J. Network rigidity and metabolic engineering in metabolite overproduction. Science. 1991;252:1675–1681. doi: 10.1126/science.1904627. [DOI] [PubMed] [Google Scholar]
- Stetter K.O. Extremophiles and their adaptation to hot environments. FEBS Lett. 1999;452:22–25. doi: 10.1016/S0014-5793(99)00663-8. [DOI] [PubMed] [Google Scholar]
- Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen T, Maerkl S.J, Quake S.R. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- Tian J, Gong H, Sheng N, Zhou X, Gulari E, Gao X, Church G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Tyson J.J, Chen K, Novak B. Network dynamics and cell physiology. Nat. Rev. Mol. Cell Biol. 2001;2:908–916. doi: 10.1038/35103078. [DOI] [PubMed] [Google Scholar]
- Unger M.A, Chou H.P, Thorsen T, Scherer A, Quake S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- Urnov F.D, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Volfson D, Marciniak J, Blake W.J, Ostroff N, Tsimring L.S, Hasty J. Origins of extrinsic variability in eukaryotic gene expression. Nature. 2006;439:861–864. doi: 10.1038/nature04281. [DOI] [PubMed] [Google Scholar]
- Wang L, Brock A, Herberich B, Schultz P.G. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- Xu L, Chen H, Hu X, Zhang R, Zhang Z, Luo Z.W. Average gene length is highly conserved in prokaryotes and eukaryotes and diverges only between the two kingdoms. Mol. Biol. Evol. 2006;23:1107–1108. doi: 10.1093/molbev/msk019. [DOI] [PubMed] [Google Scholar]
- Yokobayashi Y, Arnold F.H. A dual selection module for directed evolution of genetic circuits. Nat. Comput. 2005;V4:245–254. doi: 10.1007/s11047-004-7442-x. [DOI] [Google Scholar]
- Yokobayashi Y, Weiss R, Arnold F.H. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA. 2002;99:16 587–16 591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L. Toward computational systems biology. Cell Biochem. Biophys. 2004;40:167–184. doi: 10.1385/CBB:40:2:167. [DOI] [PubMed] [Google Scholar]
- You L, Cox R.S, 3rd, Weiss R, Arnold F.H. Programmed population control by cell–cell communication and regulated killing. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- Yu J, Xiao J, Ren X, Lao K, Xie X.S. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- Yura T, Nakahigashi K. Regulation of the heat-shock response. Curr. Opin. Microbiol. 1999;2:153–158. doi: 10.1016/S1369-5274(99)80027-7. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Boccazzi P, Choi H.G, Perozziello G, Sinskey A.J, Jensen K.F. Microchemostat-microbial continuous culture in a polymer-based, instrumented microbioreactor. Lab. Chip. 2006;6:906–913. doi: 10.1039/b518396k. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]