High resolution protein structure prediction and the crystallographic phase problem (original) (raw)
. Author manuscript; available in PMC: 2008 Aug 11.
Published in final edited form as: Nature. 2007 Oct 14;450(7167):259–264. doi: 10.1038/nature06249
Summary
We describe a new approach to refining protein structure models that focuses sampling in regions most likely to contain errors while allowing the whole structure to relax in a physically realistic all-atom force field. In applications to models produced using NMR data and to comparative models based on distant structural homologues, the method can significantly improve the accuracy of the structures in terms of both the backbone conformations and the placement of core side chains. Further, the resulting models satisfy a particularly stringent test: they provide significantly better solutions to the X-ray crystallographic phase problem in molecular replacement trials. Finally, we show that all-atom refinement can produce de novo protein structure predictions that reach the high accuracy required for molecular replacement. Phases for diffraction data for a 112-residue protein have been determined without any experimental phase information and in the absence of any templates suitable for molecular replacement from the Protein Data Bank. These results suggest that the combination of high resolution structure prediction with state-of-the-art phasing tools may be unexpectedly powerful in phasing crystallographic data for which molecular replacement is hindered by the absence of sufficiently accurate prior models.
Introduction
High resolution prediction of protein structures from their amino acid sequences and the refinement of low resolution protein structure models to produce more accurate structures are long-standing challenges in computational structural biology.1 The refinement problem has become particularly important in recent years as the continued increase in the number of experimentally determined protein structures, together with the explosion of genome sequence information, has made it possible to produce comparative models of a large number of protein structures with wide utility2. Ideally, these models would consistently approach the resolution offered by X-ray crystallography, enabling precise drug design and a deeper understanding of catalysis and binding. Accurate high resolution models can in principle be achieved by searching for the lowest energy structure given the sequence of the protein. However, despite progress3, the large number of degrees of freedom in a protein chain and the ruggedness of the energy landscape produced by strong atomic repulsion at short distances greatly complicates this search for sequences lacking close homologues of known structure.
An important application for predicted structures is to help solve the X-ray crystallographic phase problem.4, 5 Converting X-ray diffraction data into electron density maps of proteins requires the inference of phases associated with each diffraction peak. While phase estimates can be obtained through the preparation of heavy atom derivatives, the problem can be solved without additional experimental information by the technique of molecular replacement4, 5 given a structure model that has high structural similarity (better than 1.5 Å root-mean-squared deviation) to the crystallized protein over a large fraction of the molecule. As an example of the stringency of this condition, models of protein structures derived from nuclear magnetic resonance (NMR) data typically do not give good molecular replacement models for crystallographic data on the same proteins.6 Perhaps the most successful approach to molecular replacement is the use of prior crystal structures of highly sequence-similar (>40%) templates as search models. In cases of lower sequence similarity, structure prediction tools can frequently help build comparative models that give better molecular replacement solutions; however, the success rate drops rapidly as the template sequence identity falls below 30%.4, 5 In cases where structurally similar experimental models are not available, ab initio phasing techniques have had some success for targets with simple folds of high symmetry7, 8 or with new structures that have been rationally designed from first principles9, but ab initio phasing of diffraction data for natural globular proteins remains an unsolved problem.
In this study, we present a new energy-based rebuilding-and-refinement method (Figure 1 and Methods Summary) that consistently improves models derived from NMR, from sequence-distant templates, and from de novo folding methods. The final models include high-resolution features not present in the starting models, including the packing of core side-chains. Bringing together these results from all-atom structure prediction with state-of-the-art algorithms for molecular replacement and automated rebuilding,10-12 we show that distant-template-based and de novo models can reach the accuracy required to solve the X-ray crystallographic phase problem.
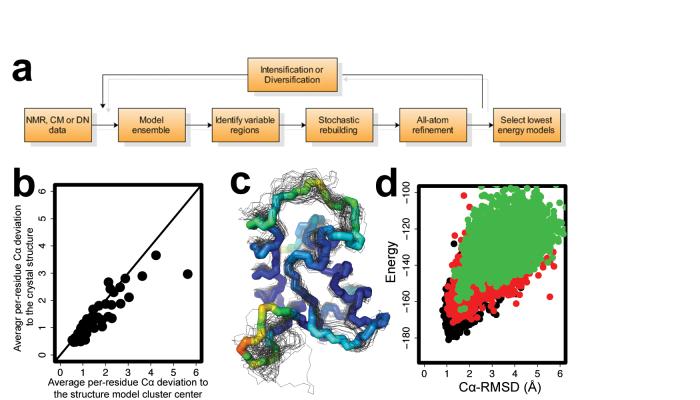
Figure 1.
Overview of rebuilding-and-refinement method. a, Schematic diagram of the rebuilding-and-refinement method applied to structures from nuclear magnetic resonance (NMR), from comparative modeling (CM), and from de novo (DN) modeling approaches. b, Strong correlation between the per-residue backbone conformation variation in model ensemble and the deviation from the native structure for target T0199 from the Sixth Critical Assessment of Structure Prediction (CASP6). c, Superposition of the native structure of CASP6 target T0199 with fifty low energy all-atom refined models. The native structure backbone is shown as a thick line, and the models are shown as thinner lines. Residues of the native backbone structure are colored by the average per residue Cα root-mean-squared deviation (RMSD) to the native from 4.5Å (red) to 0.5 Å (blue). D. Iterative rebuilding and refinement yields low energy native-like models. The energy and the Cα-RMSD of models generated during three iterations of the loop-relax protocol are displayed for iteration 1 (green), iteration 4 (red), and iteration 7 (black). The Rosetta all-atom energy includes the enthalpy plus the solvation contribution to the entropy but not the configurational entropy.
Improving NMR models
As a first test of the new rebuilding-and-refinement method, we sought to improve the accuracy of protein structure models derived from moderate-resolution NMR experiments. NMR is an important method for determining folds of proteins at atomic resolution that has the advantage of not requiring crystals. In some cases, however, NMR models can contain errors due to either insufficient data or ambiguities in interpretation of the input NMR spectra.13 We applied the method outlined in Figure 1a to ten ensembles of NMR models deposited in the Protein Data Bank for which independently determined high-resolution X-ray crystal structures provide tests of model accuracy.14, 15 Regions with high variation in initial all-atom refined ensembles were stochastically rebuilt as well as regions assessed as poorly packed (see Methods) to allow for possible over-convergence of the initial NMR ensemble in regions with incorrect constraints.
In eight of the ten cases, the lowest energy refined model was closer to the crystal structure than any member of the starting NMR ensemble (typically twenty members) in terms of backbone agreement, as assessed by GDT-HA [Geometric Distance Test (High Accuracy)16]. Comparison of the best of five lowest energy refined models to the NMR ensemble indicates improvement in backbone accuracy and core packing in all cases (see Table 1 and Supplementary Figures S3 and S4). In addition, the quality of the refined models was consistently better than the starting NMR models in terms of clash score, number of rotamer outliers, and number of backbone (Ramachandran) outliers, as assessed by the MolProbity server (Supplementary Table S2).17 Four examples of this energy-based structural improvement are shown in Figure 2a-d. It should be noted that no NMR data were included in these rebuilding-and-refinement tests; judicious use of experimental NMR information to focus all-atom refinement (for example, using inferential structure determination15) could yield still better results.
Table 1.
Improving the quality of protein structure models and molecular replacement solutions by rebuilding-and-refinement of NMR models, comparative models (CM), and de novo (DN) models
| X-ray structure | Starting Modela | Length (N)b | Sequence identity to best template (%)c | GDT-HAd | TFZe in molecular replacement | Auto-traced residues backbone,side-chainf | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| best template | refined model | best template | refined model | best template | refined model | |||||
| NMR | 1hb6 | 2abd | 86 | N/A | 0.58 | 0.79 | 4.1 | 11.3 | 12,0 | 80,80 |
| 1who | 1bmw | 94 | N/A | 0.59 | 0.68 | 5.7 | 8.3 | 25,12 | 47,44 | |
| 1gnu | 1kot | 119 | N/A | 0.64 | 0.73 | 6.6 | 10.6 | 62,53 | 82,78 | |
| 1a19 | 1ab7 | 89 (2) | N/A | 0.63 | 0.78 | 3.74.5 | 8.812.5 | 31,2014,0 | 48,3744,35 | |
| 1fvk | 1a24 | 189 (2) | N/A | 0.49 | 0.69 | 3.44.3 | 6.912.4 | 66,5055,43 | 97,9185,68 | |
| 1mzl | 1afh | 93 | N/A | 0.60 | 0.66 | 4.6 | 5.1 | 36,29 | 58,44 | |
| 1tvg | 1xpw | 143 | N/A | 0.63 | 0.74 | 4.3 | 6.7 | 15,6 | 103,86 | |
| 2snm | 2sob | 97 | N/A | 0.45 | 0.48 | 3.8 | 4.8 | 17,16 | 43,37 | |
| 1agr | 1ezy | 129 | N/A | 0.49 | 0.76 | N/Ag | N/Ag | |||
| 1abq | 1awo | 56 | N/A | 0.58 | 0.83 | N/Ah | N/Ah | |||
| CM | 2hhz (T0331) | 1ty9A | 149 | 14.5 | 0.49 | 0.58 | 5.4 | 8.8 | 28,24 | 68,63 |
| 2hr2 (T0368) | 2c21C | 158 (6) | 14.8 | 0.57 | 0.67 | 6.0 | 5.4 | 37,37 | 20,14 | |
| 2hq7 (T0380) | 2fhqA | 145 (2) | 25.4 | 0.58 | 0.69 | 4.44.6 | 6.614.2 | 47,2330,17 | 92,8360,59 | |
| 2ib0 (T0385) | 1jgcB | 170 (2) | 7.8 | 0.62 | 0.69 | 5.15.8 | 7.915.5 | 63,3750,2 | 56,5652,52 | |
| 2hi0 (T0329_D2) | 1rqIA | 92 (2) | 8.8 | 0.52 | 0.67 | N/Ag | N/Ag | |||
| 2hcf (T0330_D2) | 1lvhB | 75 | 14.1 | 0.51 | 0.65 | N/Ag | N/Ag | |||
| 2hi6 (T0357)i | 1aco | 132 | 8.4 | 0.45 | 0.52 | N/Ai | N/Ai | |||
| DN | 2hh6 (T0283) | 2b2j | 112 | 3.6 | 0.22 | 0.64 | 5.4 | 9.0 | 26,12 | 112,112 |
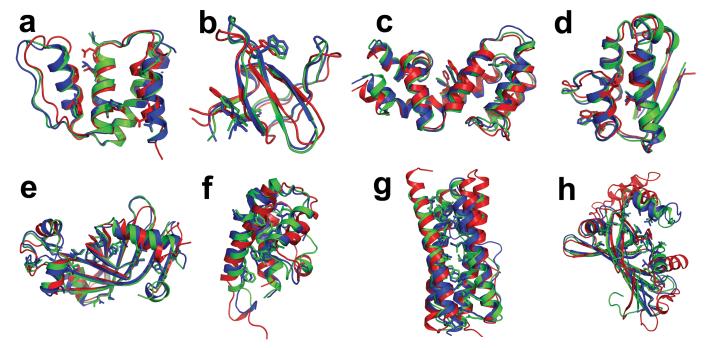
Figure 2.
Improvement in model accuracy produced by rebuilding-and-refinement. a-d are NMR refinement tests, displaying superpositions of the crystal structure (blue), model 1 of NMR ensemble (red), and the all-atom refined model (green) for four NMR refinement test cases (a, acyl CoA binding protein 2abd; b, SH3 domain of ABL tyrosine kinase 1awo; c, guanine nucleotide binding protein 1ezy; and d, barstar 1ab7). e-h are blind predictions produced by comparative modeling, displaying superpositions of the native structure (blue), the best template in the Protein Data Bank (red), and the best of our five submitted models (green) for four CASP7 targets (e, T0380; f, T0385; g, T0330 domain 2; and h, T0331). A subset of the core side-chains are shown in stick representation to illustrate the accuracy of core packing. Figures were prepared in PyMOL (Delano Scientific, Palo Alto, CA).
As noted above, NMR structures often do not give good molecular replacement models for crystallographic data,6 and we hypothesized that the all-atom refined models would yield better solutions. Indeed, we found such improvement in molecular replacement scores for all eight cases in which diffraction data were publicly available (Table 1), using the sensitive and widely used Phaser software18. Further, using phases from the molecular replacement trial with the highest translation function Z-score, electron density maps were generated and in seven of the eight cases the widely used ARP/wARP11 or RESOLVE12 automatic map tracing programs could build the majority of the residues with no human intervention (Table 1). An example of the improvement in density is shown in Figure 3a and 3b. These results suggest that all-atom rebuilding-and-refinement may be a powerful supplement to existing strategies of trial-and-error trimming of NMR ensembles to improve molecular replacement solutions for crystallographic data.6
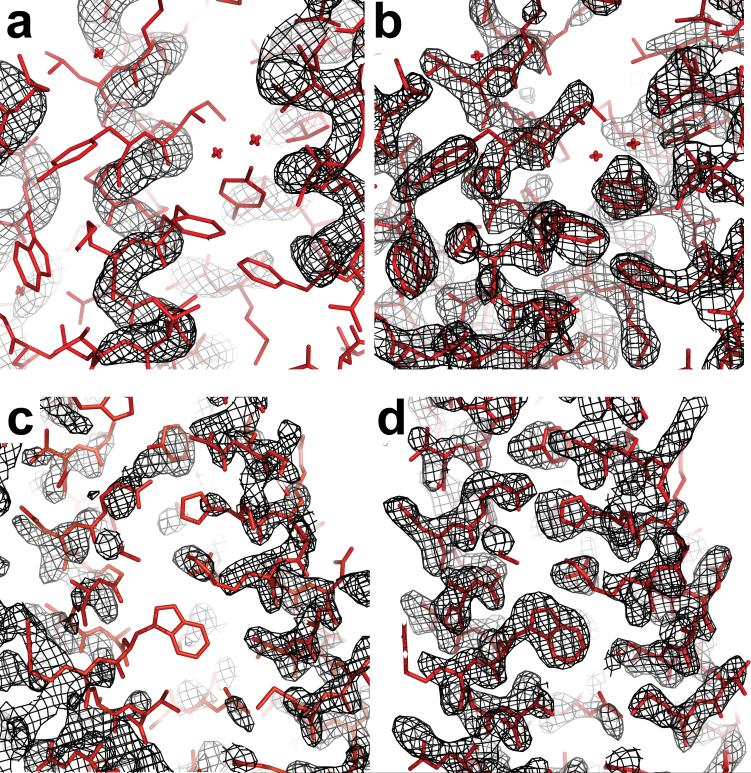
Figure 3.
Improvement in electron density using models from rebuilding-and-refinement in molecular replacement searches. Examples are presented for the NMR structure of acyl CoA binding protein 2abd (a and b) and CASP7 comparative modeling target T0385 (c and d). Black mesh represents electron density (2_m_Fo-_D_Fc; 1.5σ contour) using experimental structure factors and phases from molecular replacement with the starting model (a and c) or the refined model (b and d). The coordinates deposited in the Protein Data Bank, determined using experimental phase information, are shown in stick representation. Note that the “refinement” applied to the models refers to the all-atom energy-based protocol (see Figure 2 and text) and not to refinement against the diffraction data. The accurate modeling of side-chains by Rosetta was critical for the illustrated map improvement; molecular replacement trials gave significantly better solutions if the Rosetta-predicted side-chains were retained rather than truncated.
Improved blind predictions based on templates
As a further challenging test, we used the new energy-based rebuilding-and-refinement method to make blind structure predictions for twenty-six proteins with lengths less than 200 residues that had distant homologues (sequence identity lower than 30%) with known structure during the seventh Critical Assessment of Techniques for Protein Structure Prediction (CASP7). Ensembles of starting models based on different alignments to one or more of these distant homologues were generated as described in the Supplementary Information, and the rebuilding-and-refinement protocol was carried out, with several rounds of iteration to more broadly explore conformational space (Figure 1a; see also Supporting Material). Five representative low energy structures from the final population were submitted to the CASP organizers. For eighteen of the twenty-six cases, at least one of these five models was closer to the correct structure than the closest homologous structure in the Protein Data Bank (PDB), as assessed by the GDT-HA score.19 Dramatic improvement was observed in seven cases, with a 10-30% increase in this measure of model quality; see Table 1. This is a particularly notable result because improving on the best template structure has been a long standing challenge for comparative modeling - due to the high dimensionality of conformational space, there are many more ways to degrade a reasonably accurate model than to improve it. Superpositions of the closest homologous structure, the submitted refined models, and the native structure for cases with the greatest improvement are shown in Figures 2e-h. The improvement in the refined structures is evident even in core secondary structural elements.
Out of the seven high resolution predictions, there were four targets for which diffraction data were available and the modeled sequence constituted the entire crystallized construct, enabling tests of molecular replacement. In each of these cases, we found that the best prior templates in the Protein Data Bank failed to produce clear-cut molecular replacement solutions (Phaser Z-scores greater than 7), even after using knowledge of structurally alignable regions and a side-chain truncation approach to trim back the search models to their most accurate atoms.4 Other template-based models submitted to CASP7, based on methods that typically did not employ aggressive all-atom refinement, gave similarly low molecular replacement scores (Table 1). For three of the four cases, however, the refined models that we submitted for CASP7 gave significantly better molecular replacement solutions than the best template (Table 1). For these targets, the maps produced by combining phases from the blindly predicted model with the experimental diffraction amplitudes were of sufficient quality to permit the automatic chain tracing program RESOLVE12 to build a large fraction of each structure with high accuracy (Table 1). An example of the dramatic improvement in electron density upon using the refined models is shown in Figure 3c and 3d.
Ab initio phasing by ab initio modeling
To the best of our knowledge, a de novo structure prediction for a natural protein with an asymmetric, globular fold has never been used successfully for molecular replacement. However, the accuracy of de novo prediction methods has been improving rapidly. In particular, the use of all-atom refinement to follow low resolution modeling by the Rosetta de novo modeling method20 led to several blind predictions in CASP7 for proteins of all-α, all-β, and α+β secondary structure classes that placed the majority of backbone elements and core side-chains with high accuracy (see Figures 4a-c).19 This progress in de novo modeling, along with the successes above with refined NMR and template-based models, encouraged us to attempt molecular replacement with an exceptional prediction for the 112-residue α-helical CASP7 target T0283.
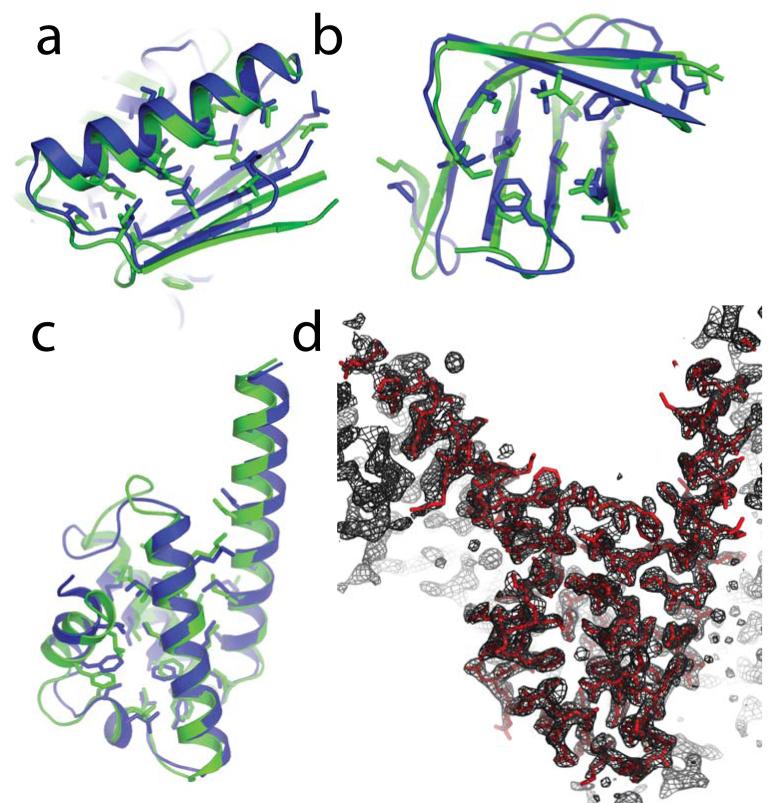
Figure 4.
Ab initio phasing by ab initio modeling. a-c, Superpositions of blind Rosetta de novo structure predictions (green) and the subsequently released crystal structures (blue) for CASP7 targets T0354 (a), domain 3 of T0316 (b), and T0283 (c). Buried side-chains and backbone-aligned residues are displayed. d, Electron density map (2_m_Fo-_D_Fc; 2σ contour) produced by automatic refinement of the molecular replacement solution obtained from the T0283 structure prediction (black mesh; 1σ contour) agrees with the coordinates deposited in the Protein Data Bank (red), solved with experimental phase information. The electron density map immediately after molecular replacement is shown in Supplementary Figure S5.
The best of five models for T0283 blindly predicted without the use of templates matched the subsequently released crystal structure (2hh621) with a Cα-RMSD of 1.4 Å over 90 residues (Figure 4c). The closest previously known fold in the Protein Data Bank, identified from structure superpositions by CASP7 assessors (2b2j22), was significantly different from the T0283 crystal structure, aligning 70 residues with a Cα-RMSD of 3.1 Å (note also the poor GDT-HA score in Table 1).
After truncating the Rosetta prediction to a consensus core (residues 10 to 88, for which four of the five submitted models coincided to within 2.5 Å Cα-RMSD), molecular replacement by Phaser showed clear features for the omitted N- and C-terminal helices (see Supplementary Figure S5 and caption). Starting from this molecular replacement solution, the ARP/wARP software was able to complete the structure automatically, tracing all 112 residues correctly. The final result (Figure 4d) is in excellent agreement with the structure deposited in the Protein Data Bank, which used phases experimentally derived by selenium single-wavelength anomalous dispersion, with an RMSD of 0.13 Å for all 112 Cα atoms. In contrast, attempts to solve the structure by molecular replacement with the closest existing “template” 2b2j failed to produce a clear-cut phasing solution (Table 1), even when knowledge of the optimal superposition was used to trim this search model back to the seventy residues that aligned best to the actual structure. It will be of great interest to investigate whether this result can be generalized to rapidly phase diffraction data for proteins of new folds.
Improving model accuracy and molecular replacement
The results described here show that an all-atom rebuilding-and-refinement protocol can produce protein structure models of high accuracy. The iterative protocol outlined in Figure 1a brings together the individually quite powerful global optimization ideas underlying Monte Carlo minimization23, tabu search24, and conformational space annealing25 while targeting aggressive sampling to regions most likely to be incorrect. The substantial improvements achieved in prediction quality - in several cases enabling molecular replacement phasing of X-ray diffraction data - suggest that structure prediction has matured considerably. Nevertheless, we emphasize that there is still considerable room for improvement: our high resolution rebuilding-and-refinement protocol does not always improve starting models, and T0283 is the only CASP7 target predicted de novo for which the models were accurate enough for molecular replacement. We look forward to advances in both the energy function, notably the addition of configurational entropy, and in conformational sampling. The significant energy gap between the refined models and the refined crystal structure20 for most of the cases studied here suggests that sampling is still the primary bottleneck for high accuracy all-atom structure prediction.
At the present date, the Protein Structure Initiative lists hundreds of proteins with lengths less than 200 residues that have been crystallized but not yet solved. Publication of diffraction data sets that have not yielded to experimental phasing could catalyze the development of new hybrid prediction/phasing algorithms, much like the blind CASP trials have accelerated progress in the field of structure prediction. With continuing advances in high resolution structure prediction, in molecular replacement tools, and in the interface between these two fields, we expect that in silico phasing will become an increasingly important component of the crystallographer’s toolkit.
In the present study, aggressive all-atom refinement was carried out in the absence of any experimental information. The incorporation of experimental data into the rebuilding-and-refinement protocol could help overcome the current shortcomings in both the energy function and conformational sampling and allow more consistent high resolution structural inference. In practical applications to molecular replacement trials, the diffraction data do not need to be set aside as a stringent post facto test of model accuracy, as was carried out in this study. Diffraction data without phases would be useful in screening large numbers of trial structures for molecular replacement or in complementing the physical energy terms with diffraction data derived likelihood scores29 during rebuilding-and-refinement. Weak phase information, e.g. based on anomalous scattering from intrinsic sulfur atoms30, could also be exploited, for instance after using an initial molecular replacement model to locate the anomalous scatterer sites18. Although not used in the present study, NMR chemical shift, nuclear Overhauser effect, and residual dipolar coupling data can help to pinpoint regions of the models to rebuild and regions to constrain during all-atom refinement. On a larger scale, mass spectrometry techniques coupled with hydrogen/deuterium exchange26, chemical cross-linking27, and radical footprinting28 show great promise for providing high-throughput, residue-level information that may rapidly constrain structure prediction and, in the absence of crystallographic data, help validate models. We anticipate that the combination of high resolution modeling with limited experimental structural data will become an increasingly powerful approach for characterizing the structures of biological macromolecules and complexes in the years to come.
Methods Summary: Targeted rebuilding-and-refinement
We have developed a new approach for refining protein models that combines the targeting of aggressive sampling to regions most likely in error with powerful global optimization techniques. The new protocol is outlined in Figure 1a. The first step of this protocol is the energy-based optimization of an input ensemble of models using the previously described Rosetta all-atom refinement method. This method combines Monte-Carlo minimization with side-chain remodeling to relieve inter-atomic clashes and to optimize side-chain packing and hydrogen bonding, as encoded by an all-atom force field..20, 31 Briefly, in each Monte Carlo move, a random perturbation to the protein backbone torsion angles is followed by discrete optimization of the side-chain conformations, which allows efficient crossing of side-chain torsional barriers. Then, quasi-Newton optimization of the side-chain and backbone torsion angles is carried out prior to the decision on whether to accept the move. Because of the final minimization, each point on the landscape is mapped to the closest local minimum, flattening energy barriers.23 While making it possible to recognize near-native predictions based on their low energies1, 20, this all-atom refinement alone does not consistently produce significant improvements in model quality (Supplementary Figure S1).
The second step in the new protocol is the identification of regions of variation in the ensemble of refined models. We have found a striking correlation between the extent of variation in the coordinates of a residue in the refined structures and the deviation of the coordinates of the residue in the refined models from the native structure. An example is shown in Figures 1b and 1c: positions exhibiting small variance across the models are usually quite close to the correct structure, whereas positions for which the variance is large often deviate considerably from the native structure. This correlation arises from the relatively short range of the force field and the energy gap between the native structure and the models: since the energy of the entire system is roughly equal to the sum of its parts, for most portions of the protein, the correct conformation will be lower in energy than non-native conformations. Regions of the protein that can access the native conformation are likely to converge on this conformation and thus exhibit less variation, while locally incorrect conformations are likely to be spread throughout the landscape and exhibit more variation. We observe this correlation for many different proteins in both the Cartesian coordinates and the internal torsion angles; a related principle has recently been used in the Pcons method for assessing protein models.32
The third step in the new protocol targets aggressive sampling to the regions most likely to be in error. A fragment-based segment rebuilding method (see Supplementary Material) is used to completely rebuild regions of models with relatively high variation in the model population. Because the precise regions that are incorrect cannot be identified unambiguously, we carry out many independent calculations in which different segments in the higher variation regions are randomly selected for complete rebuilding. The partially rebuilt models are then subjected to the Rosetta all-atom refinement protocol described above.20, 31 In the segment rebuilding process, side-chains are initially represented as soft interaction centers and the connectivity of the chain is temporarily broken, thus permitting the traversal of much larger barriers than those crossed by all-atom refinement alone.
As indicated in Fig 1a, if the lowest energy refined structures have not converged, the rebuilding-and-refinement protocol is applied iteratively using a selection process inspired by natural evolution to guide convergence on the global minimum. At each iteration, a subset of models that are low in energy yet structurally diverse is chosen to seed the next round: the regions to be rebuilt are determined based on the backbone variation in the selected population. Bringing together ideas from tabu search24 and conformational space annealing25, the selection process alternates between the propagation of a structurally diverse population into the next round (“diversification”) and focusing in on the lowest energy regions of the energy landscape explored thus far (“intensification”). The lowest energy models after ten iterations are selected as the final predictions. As illustrated in Fig 1d, models with progressively lower energies and more native-like structures can be obtained with increasing number of iterations; results on a number of refinement problems are summarized in Suppl. Fig S2.
Supplementary Material
Supplementary Material
Acknowledgments
We thank Rosetta@home participants for contributing computing power that made testing of many new ideas possible; the DOE INCITE program for access to Blue Gene/L at Argonne National Lab and the IBM Blue Gene Watson supercomputers; and the NCSA, SDSC and Argonne National Lab supercomputer centers for computer time and help with porting Rosetta to Blue Gene. We thank David Kim and Keith Laidig for developing the computational infrastructure underlying Rosetta@home; Jan Abendroth for help with RESOLVE and ARP/wARP software; Mike Kennedy of NESG for the NMR structure coordinates of protein 1xpw and for help with the molecular replacement calculations; and Chu Wang and Jim Havranek for helpful comments on the manuscript. We also thank the CASP organizers and contributing structural biologists for providing an invaluable test set for new structure refinement methods. This work was funded by the National Institute of General Medical Sciences, National Institutes of Health (to DB), the Wellcome Trust, U.K. (to RJR), the Howard Hughes Medical Institute (DB), a Leukemia and Lymphoma Society Career Development fellowship (to BQ), and a Jane Coffin Childs fellowship (to RD).
APPENDIX
Methods
We present detailed descriptions of six methods discussed in the main text:
- Rebuilding-and-refinement protocol
- Identification of regions to rebuild from the NMR structure ensemble
- Preparation of blind predictions
- Metrics for comparing models with crystal structures
- Screening of models for suitability for molecular replacement
- Assessing model quality with MolProbity
1. Rebuilding-and-refinement protocol
We describe the key steps of the rebuilding-and-refinement protocol - segment rebuilding, all-atom refinement, and iterative evolution - in the next three sub-sections.
Segment-rebuilding protocol
We used a new segment rebuilding protocol to rebuild regions with high structural variation in the model population as these regions are often incorrect (see, e.g., Figure 1b). Because of uncertainties in the precise locations of incorrect regions, the portions of the model to be rebuilt were chosen stochastically from the regions with high variance at the beginning of each simulation. Up to 90% of all the separate regions were rebuilt in a given run - this allows for compensatory changes in interacting segments to occur.
The coordinates in the region to be rebuilt were generated using the Rosetta fragment insertion based de novo folding protocol35. After each fragment insertion, the decision to accept or to reject was made according to the standard Metropolis criterion based on the total energy of the system. To maintain the connectivity of the protein chain, cyclic coordinate descent [CCD 36] was used to close the chain-break at a stochastically selected position of the region rebuilt. The rebuilding process was divided into ten stages. At each successive stage, an increasing chain-break score (a penalty to the deviation of the peptide bond length at the chain-break from the ideal peptide bond length) was applied. In each of the first five stages, the number of fragment insertion trials was ten times the number of residues in the region being rebuilt. In a fragment insertion trial, randomly chosen nine-residue, three-residue, or one-residue fragments were inserted into randomly chosen positions in the region being rebuilt, and the Metropolis Monte-Carlo criterion was used to accept or reject the newly inserted fragment based on the Rosetta low-resolution energy function31. In each of the 5 last stages, in addition to the fragment insertion trials, we also performed cyclic-coordinate-descent-based backbone torsion angle moves (CCD-moves) in which the cyclic coordinate descent solution was calculated and the backbone torsion angles for five randomly picked positions in the region being rebuilt were modified according to the CCD solution.
If after the ten rebuilding stages described above, any chain-break remained larger than 0.2 Å, the region to be rebuilt was expanded by one residue on both sides. The above fragment insertion and chain-break closing process was repeated using a harmonic tether to the starting values of the torsion angles in the newly included regions (which may fall into regions with low variance in the starting population) and another stochastically selected chain-break position. The regions to be rebuilt were allowed to expand by up to five residues upstream and downstream of the original starting and ending positions, until chain closure was achieved. This procedure was usually sufficient to ensure the recovery of a continuous peptide chain. In very rare cases where the chain could not be closed in a rebuilt region, it was merged with an adjacent region to be rebuilt along with the fixed portion of the model between these two regions and the rebuilding process was repeated. With the added flexibility of a larger region being rebuilt, the peptide chain could essentially always be closed. Variable regions at the chain termini were rebuilt using the fragment insertion-based de novo protocol without steps for chain-break closure.
The segment rebuilding protocol is implemented in the “loop_relax” subroutine in the freely available Rosetta source code.
All-atom refinement protocol
The segment rebuilding protocol described above aggressively employs fragment insertion moves to sample a broad range of conformations. The all-atom refinement protocol, described in this section, then searches for local minima in the vicinity of the structures produced by segment rebuilding using a detailed all-atom force-field.
The Rosetta all-atom energy function is largely dominated by short-range interactions9, primarily Lennard-Jones interactions and orientation-dependent hydrogen-bonding, and the Laziridis-Karplus implicit solvation model37. The torsional states of backbone and side-chains are evaluated using knowledge-based potentials derived from amino acid specific Ramachandran maps and the rotamer probabilities and χ angle standard deviations in the backbone-dependent rotamer library developed by Dunbrack and colleagues38.
During all-atom refinement, all the backbone and side-chain atoms in the protein are explicitly represented. The bond lengths and angles are kept fixed at ideal values39 and the polypeptide chain is described in internal coordinates (the backbone and side-chain torsion angles). A single move in the all-atom refinement protocol consists of the following steps: (1) one of the several types of perturbations to the backbone torsion angles described below, (2) greedy optimization of the side-chain rotamer conformations (“rotamer-trials”40) for the new backbone conformation (3) minimization of the energy with respect to either the backbone degrees of freedom only (first half of refinement procedure) or backbone and side-chain degrees of freedom (second half of refinement procedure) using the Davidson-Fletcher-Powell (DFP) algorithm. The convergence criterion for exiting this quasi-Newton minimization was decreased from 10-3 to 10-5 during the course of refinement to enable more complete minimization in the final stages of refinement. (4) the compound move (steps 1-3) is accepted or rejected according to the Metropolis Monte Carlo criterion. These compound moves extend the Monte Carlo Minimization procedure found to be quite powerful in previous studies41 by incorporating discrete optimization of side-chain conformations; this allows energy-directed barrier hopping at the level of the side-chains.
The following backbone perturbations are used at step (1) in the Monte Carlo Minimization move described above and in a previous reference31. The “small” and “shear” moves are small perturbations of the backbone at five to ten randomly chosen positions. In “small” moves, ϕ and Ψ are perturbed randomly by up to 1° in helix or strand regions or 1.5° in loop regions. In “shear” moves, ϕ is perturbed randomly by up to 2° in helix or strand regions or 3° in loop regions and the preceding Ψ is perturbed by the same amount of degrees in the opposite direction to produce a compensatory shear motion in the peptide plane. The “wobble” and “crank” moves involve insertion of fragments and are more aggressively perturbing than the small and shear moves31. For both of these move types, the fragment set35 is filtered to exclude those which cause a mean square deviation in the coordinates of the downstream atoms of more than 60 Å and one of the remaining fragments is chosen randomly for insertion. In “wobble” moves, the torsion angles belonging to the three residues immediately following the site of the one or three residue fragment insertion are varied to minimize the downstream perturbation still further. In “crank” moves, one residue is varied immediately following the insertion site, and three more residues at a site spaced by 6-20 residues from the fragment insertion site; this produces a “crankshaft” like movement of the intervening portion of the chain. “small-wobble” moves involve an initial 10-20° random change in the torsion angles of a single residue, followed by minimization of the perturbation over the three adjacent residues. The minimization of the perturbation in the wobble and crank moves is carried out using the fast gradient based algorithm described previously31. After all five move types, the side-chains are optimized and the energy is minimized as described in the preceding paragraph.
The all-atom refinement protocol is divided into three stages:
- Ramp-up. The ramp-up stage consists of sets of ten small and shear moves preceded by combinatorial optimization of the side-chain rotamer conformations. The weight on the repulsive part of the Lennard-Jones potential is progressively increased from 0.05 to 1.0 over eight such move sets. The gradual ramping up of the repulsive weight facilitates a smooth rearrangement of the side-chains with small perturbations of the backbone and ensures a reasonably well-packed low energy model before the more aggressive second stage.
- Aggressive sampling stage: Alternating “wobble”, “small-wobble” and “crank” compound Monte Carlo minimization moves are carried out; the total number of attempts for each move type is equal to the number of residues in the protein. A full combinatorial search over side-chain rotamer conformations is carried out after every 25 attempts of each type of move. The more aggressive nature of the moves employed at this stage allows the traversal of modest energy barriers. The convergence tolerance for the DFP minimization is set to 10-4.
- Fine optimization stage: Alternating “small” and “shear” moves are carried out, again for a total number of attempts equal to the number of residues in the protein. The more subtle backbone conformation changes brought about by these moves assist convergence on a relatively low energy local minimum. The convergence tolerance for minimization is set to 10-5. Following these three stages, a final minimization with respect to all degrees of freedom is carried out with a convergence tolerance of 10-6.
The refinement protocol described in this section is implemented in the “fullatom_relax” subroutine in Rosetta; the CPU cost is about 20 minutes for a 100 residue protein on an Intel Pentium IV 1.6 GHz processor.
Ensemble evolution by alternate cycles of diversification and intensification
The challenge in refinement is to focus sampling on the lowest energy regions of the energy landscape identified up to that point while maintaining a broad enough search to avoid converging on a local energy minimum. Towards this end, we developed a protocol that balances intensification of the search in low energy regions with diversification to maintain subpopulations exploring alternative energy minima. The approach adopts the idea of explicit controlling of the search intensity from tabu search24, and is a generalization of the conformational space annealing (CSA) technique, which has achieved success in a broad range of optimization problems25.
In both the intensification and diversification steps, an input population of 200 models was clustered using the method described in ref20 to identify distinct populations of structures. The clustering threshold was chosen such that the largest cluster contained 10% of the models. For each cluster, ten models were selected (if there were fewer than 10 models in a cluster, all were selected) and each model was subjected to nine independent segment rebuilding plus all-atom refinement runs initialized with different random number seeds.
In the diversification stages (iterations 1, 3, 5, 7 and 9), the models in the parent population were kept in their original cluster assignment. A newly generated model was assigned to the closest cluster if the root-mean-squared deviation over alpha carbons (Cα-RMSD) between this model and the closest cluster member was less than the current diversity threshold (see below), and the highest energy member of the cluster was thrown away. If the RMSD between a newly generated model and its closest cluster member was higher than the current diversity threshold, then the model with the highest energy in the current parent population was thrown away, and the newly generated model formed a cluster of its own. This is analogous to speciation in natural evolution. As a model is discarded for each new model added, the population size stayed unchanged. The diversification step favors a broad exploration of the conformational space by maintaining the distinct populations of clusters: there is competition for low energy within but not between clusters. Combined with the initial clustering step, it ensures that the new population will not be dominated by overly closely related structures which could result in premature convergence away from the global minimum.
In the intensification stages (iterations 2, 4, 6, 8 and 10), all but the lowest energy 10% of the entire population (parents plus offspring) is discarded to bring the population back to a size of 200. The remaining models from the parent population keep their original cluster assignment. A newly generated model was assigned to the closest remaining cluster if the RMSD between this model and the closest cluster member was lower than the current diversity threshold; otherwise it formed a new cluster of its own. This stage differs from the diversification stage in that the energy based selection is carried out across all clusters and hence higher energy clusters can be eliminated completely. This stage allows more thorough exploration of the most promising (lowest energy) regions of the energy landscape explored thus far.
The diversity threshold used to maintain distinct populations and to guide the spawning of new populations was reduced at each iteration to allow gradual convergence on the global energy minimum. The starting value was the clustering threshold in the original population, and this was reduced by 0.1Å at each iteration. This annealing of the diversity threshold was introduced in the CSA strategy.25
The new parent population generated by the diversification or intensification procedures was used to seed the next generation, and nine independent segment rebuilding plus all-atom refinement calculations were again carried out for each parent. After ten iterations, the low energy models were clustered and the lowest energy models in the largest 5 clusters were selected as the final predictions. The overall iterative procedure took approximately 2,000 CPU hours per target. For molecular replacement efforts, this computational effort would likely be significantly reduced if phasing trials with diffraction data are used to screen models.
2. Identification of regions to rebuild from the NMR structure ensemble
The test cases for NMR refinement were chosen to be proteins representing different fold topologies for which an NMR structure and a high-resolution crystal structure (with structure factors deposited in PDB) existed. These were chosen from the datasets used by Garbuzynskiy et al42 and Grishaev et al14.
For investigations of refinement of NMR structures, we rebuilt two sets of regions. The first are regions that vary within the NMR ensemble. As in the comparative modeling case, we have observed that regions that vary within the NMR ensemble are likely to be the regions that are most different from a high-resolution crystal structure. These are most likely loops that are either inherently dynamic in the NMR structure or loops that are held in place with insufficient restraints. [Applying all-atom refinement to the NMR ensembles gave essentially the same list of variable regions (data not shown).]
The second set of regions are segments that are internally consistent within the NMR ensemble but systematically under-packed. To estimate packing we used a recently developed packing metric (W. Sheffler, personal communication) based on the relative accessible surface areas of groups of atoms. For each buried atom, we compute the largest sphere tangent to that atom which can fit into empty space within the protein. A group composed of all-atoms within 5 Å of the center is defined for each sphere. For each group of atoms, accessible surface (SASA) to small and large spherical probes (radii 0.9 Å and 2 Å) is computed; given that a ball of atoms has a certain area accessible to a large sphere, less accessible area to a small sphere indicates better packing. A summary percentile score is computed based on a reference set of crystal structures, approximating the fraction of native proteins which are better packed than the scored structure.
3. Preparation of blind predictions
Template selection and alignment ensemble for comparative modeling
The initial set of template-based models was obtained from the 3D-Jury server43 and subjected to all-atom refinement using Rosetta all-atom energy function. Up to ten templates from which the very lowest energy models were derived were used as the candidate templates. Alignment ensembles between the candidate templates and the target sequence were parametrically generated using the K*Sync alignment method44. The alignment ensemble was turned into a model ensemble by placing the sequence of the query onto the backbone of the parent based on each alignment. Missing densities from the insertion and deletion regions of the alignment was modeled using the segment modeling protocol described in the “segment-rebuilding protocol” section above. The full-chain models were then subjected to the all-atom refinement procedure as described in the “all-atom refinement” section, constrained by a set of Cα-Cα distance constraints, described next.
The Cα-Cα distance constraints were generated from the 3D-Jury43 template-based models with the lowest Rosetta all-atom energies after all-atom refinement. A Cα-Cα pair was used to derive constraints only when the associated distance was less than 8 Å in more than 80% of the selected constraint-generating models. Upper and lower bounds for each of these pairs were determined by padding the highest and lowest of these distances by one standard deviation of the Cα-Cα distance distribution function, as described in ref45. For computational efficiency, we further trimmed down the number of constraint pairs by eliminating neighboring pairs separated by 1 or 2 residues. During all-atom refinement, a penalty score is applied when the Cα-Cα distances in the model exceed the upper or lower limit of the corresponding constraints. If a distance exceeds the upper or lower constraint limit by d (in Å), then the penalty score Ec is _d_2 when d < 0.5 Å, and (d - 0.25 Å) when d ≥ 0.5 Å. The resulting ensemble of low-energy comparative models became the inputs to further rounds of rebuilding-and-refinement (Fig. 1a).
De novo modeling
For targets without clear templates identified by the 3D-Jury server43, the full chain was fully modeled by fragment assembly starting from an extended chain, followed by the all-atom refinement procedure described above. The convergence of the Rosetta de novo prediction protocol can differ significantly for different sequence representatives of a given fold.20, 46 For T0283, one of seven tested sequence homologues gave exceptionally well converged low energy models that, after sequence mapping, allowed structure prediction for the target sequence with the rebuilding-and-refinement protocol.19, 20
Computational cost
About 100,000 all-atom refined models were generated for each modeling target, requiring approximately 100,000 CPU-hours. As noted above, for molecular replacement efforts, this computational effort would likely be significantly reduced if phasing trials with diffraction data are used to screen models; as the predicted models used in this manuscript were prepared as blind predictions for CASP7, such diffraction data were not available at the time of modeling.
4. Metrics for comparing models with crystal structures
As has been discussed previously, no metric for comparing structure models with the crystal structures is perfect47. In this work, we used three different structural metrics for model quality assessment. The Cα-RMSD is a widely used metric for structure comparison, but it can be distorted by large deviations in a small number of residues, especially at the termini or in long surface loops. The Geometric Distance Test (High Accuracy; GDT-HA) score is the average percentage of Cα’s in the model within 0.5, 1.0, 2.0, and 4.0 Å of the corresponding Cα coordinates in the crystal structure; we used TMalign48 to align the structures. This metric is less sensitive than the full chain RMSD to deviations in poorly ordered termini and long loops, and was used in the CASP7 template based modeling assessment.
The core residue all-atom RMSD describes the accuracy of both the backbone and side-chain conformation prediction. We used this metric in the evaluation of NMR refinement since it can be applied to both the starting (NMR ensemble) and ending (Rosetta refined) models. In template based modeling, this metric is not practical since the template usually does not have the same amino acid sequence as the target to be modeled.
In addition, successful molecular replacement using the predicted structure can be regarded as a stringent test for model quality assessment, as suggested in ref 49.
5. Screening of models for suitability for molecular replacement
Searching for molecular replacement solutions involves applying rigid-body transformations along the six rotational and translational degrees of freedom. We carried out this search with the Phaser software, which is described in 18 and references therein. For completeness, the algorithms are briefly summarized here. Phaser uses likelihood functions to judge how well molecular replacement models agree with the measured diffraction data after they have been first rotated and then also translated. Brute-force likelihood calculations over grids of orientations and positions are computationally expensive, so fast-fourier-transform-based approximations are used to compute sets of possible solutions, which are rescored with the full likelihood targets. By using a tree-search-with-pruning strategy, almost all solutions that would be found with a full 6-dimensional search are found, but with a much lower computational cost. As well, this strategy allows effective searches for multiple copies, in crystals with more than one molecule in the asymmetric unit. For each molecule to be placed, a rotation search is first carried out. A translation search is then carried out for each plausible orientation. All plausible rotation/translation solutions are checked for packing in the lattice, and solutions that pack successfully are subjected to rigid body refinement. If more than one copy is present, all plausible partial solutions are fixed in turn while carrying out rotation and translation searches for subsequent copies. In molecular replacement trials with Phaser, the clearest indication of success comes from high values of the Z-score (number of standard deviations above the mean), computed by comparing the log-likelihood-gain (LLG) for the peak with LLG scores for a random sample of search points.
For molecular replacement in each of the NMR modeling cases, we evaluated the combined NMR ensemble as a potential search model and compared these results to trials with the twenty-five lowest energy Rosetta models from rebuilding-and-refinement (Table 1 in main text). Further, we have carried out molecular replacement trials with each of the members of the deposited NMR ensemble individually, with results given in Supplementary Table S1. Finally, for an actual search for a good molecular replacement solution, a larger set of models from rebuilding-and-refinement can be screened rapidly. We thus extended the search to the 1000 lowest energy models from Rosetta rebuilding-and-refinement and the results, notably improved, are presented in Table S1.
For molecular replacement in comparative modeling cases, we prepared search models from the best existing templates and from our comparative modeling predictions. For the best templates, we followed the “mixed model” protocol described in 4 for optimizing molecular replacement. Further, based on the 3DPAIR50 structure alignment between the native structure and the best template structure, the template structure was trimmed to contain only the structurally alignable regions. Then the native sequence was threaded onto the backbone of the corresponding template structure, while retaining the side-chain coordinates of the identical residues between the template and native sequences. Non-identical side-chains longer than serine were mutated to serine, followed by Rosetta side-chain packing protocol 51 to model the mutated serine and the shorter non-identical side-chains, while keeping the identical side-chain conformation fixed. To prepare search models for these predictions, we superimposed one hundred low energy models from the final round of refinement, and defined the model that has the lowest average RMSD to the rest of the models as the reference model. Then we calculated the average per atom distance Da between each of the superimposed models and the reference model. The Rosetta temperature factor is calculated as Te = 8_π_2 _Da_2/3 for each atom and inserted to the B-factor column of the refined model files. The Rosetta temperature factor is intended to represent the uncertainty in the final refined models after extensive refinement in the Rosetta all-atom force field. As suggested earlier25, by using the B-factor effectively to smear each atom over its possible positions, the correlation of the modeled electron density with the true electron density can be maximized.
For the de novo modeling case, target T0283, search models for molecular replacement were trimmed according to residues for which there was consensus among submitted models. Supplementary Figure S5 gives a more detailed description and illustration of the molecular replacement solution.
6. Assessing model quality with MolProbity
For the investigations of refinement of NMR models, we used the MolProbity software17 to investigate the quality of the refined models versus that of the starting NMR ensemble. For purposes of comparison, we chose the lowest energy refined model and the first member of the deposited NMR structure. Supplementary Table S2 shows the clash score, number of rotamer outliers and number of Ramachandran outliers of the NMR and refined models. The refined models consistently have better model quality than the starting NMR structure based on these metrics.
References
- 1.Misura KM, Baker D. Progress and challenges in high-resolution refinement of protein structure models. Proteins. 2005;59:15–29. doi: 10.1002/prot.20376. [DOI] [PubMed] [Google Scholar]
- 2.Pieper U, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–5. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15:285–9. doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Schwarzenbacher R, Godzik A, Grzechnik SK, Jaroszewski L. The importance of alignment accuracy for molecular replacement. Acta Crystallogr D Biol Crystallogr. 2004;60:1229–36. doi: 10.1107/S0907444904010145. [DOI] [PubMed] [Google Scholar]
- 5.Giorgetti A, Raimondo D, Miele AE, Tramontano A. Evaluating the usefulness of protein structure models for molecular replacement. Bioinformatics. 2005;21(Suppl 2):ii72–6. doi: 10.1093/bioinformatics/bti1112. [DOI] [PubMed] [Google Scholar]
- 6.Chen YW, Dodson EJ, Kleywegt GJ. Does NMR mean “not for molecular replacement”? Using NMR-based search models to solve protein crystal structures. Structure. 2000;8:R213–20. doi: 10.1016/s0969-2126(00)00524-4. [DOI] [PubMed] [Google Scholar]
- 7.Strop P, Brzustowicz MR, Brunger AT. Ab initio molecular-replacement phasing for symmetric helical membrane proteins. Acta Crystallogr D Biol Crystallogr. 2007;63:188–96. doi: 10.1107/S0907444906045793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossmann MG. Ab initio phase determination and phase extension using non-crystallographic symmetry. Curr Opin Struct Biol. 1995;5:650–5. doi: 10.1016/0959-440x(95)80058-1. [DOI] [PubMed] [Google Scholar]
- 9.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–8. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 10.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 11.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–63. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 12.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doreleijers JF, Rullmann JA, Kaptein R. Quality assessment of NMR structures: a statistical survey. J Mol Biol. 1998;281:149–64. doi: 10.1006/jmbi.1998.1808. [DOI] [PubMed] [Google Scholar]
- 14.Grishaev A, Bax A. An empirical backbone-backbone hydrogen-bonding potential in proteins and its applications to NMR structure refinement and validation. J Am Chem Soc. 2004;126:7281–92. doi: 10.1021/ja0319994. [DOI] [PubMed] [Google Scholar]
- 15.Rieping W, Habeck M, Nilges M. Inferential structure determination. Science. 2005;309:303–6. doi: 10.1126/science.1110428. [DOI] [PubMed] [Google Scholar]
- 16.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–4. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovell SC, et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 18.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das R, et al. CASP7 Structure Prediction with Rosetta@home. Proteins. 2007 in press. [Google Scholar]
- 20.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–71. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 21.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–3. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrade SL, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc Natl Acad Sci U S A. 2005;102:14994–9. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wales DJ, Scheraga HA. Global optimization of clusters, crystals, and biomolecules. Science. 1999;285:1368–72. doi: 10.1126/science.285.5432.1368. [DOI] [PubMed] [Google Scholar]
- 24.Glover F, Laguna M. Tabu Search. Springer. 1997. [Google Scholar]
- 25.Lee J, Liwo A, Scheraga HA. Energy-based de novo protein folding by conformational space annealing and an off-lattice united-residue force field: application to the 10-55 fragment of staphylococcal protein A and to apo calbindin D9K. Proc Natl Acad Sci U S A. 1999;96:2025–30. doi: 10.1073/pnas.96.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Englander JJ, et al. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc Natl Acad Sci U S A. 2003;100:7057–62. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young MM, et al. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci U S A. 2000;97:5802–6. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu Rev Biophys Biomol Struct. 2006;35:251–76. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 29.Pannu NS, Read RJ. Improved structure refinement through maximum likelihood. Acta Cryst. A. 1996;52 doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- 30.Dauter Z. New approaches to high-throughput phasing. Curr Opin Struct Biol. 2002;12:674–8. doi: 10.1016/s0959-440x(02)00372-x. [DOI] [PubMed] [Google Scholar]
- 31.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 32.Wallner B, Elofsson A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Sci. 2006;15:900–13. doi: 10.1110/ps.051799606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–21. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller T, Oehlenschlager F, Buehner M. Human interleukin-4 and variant R88Q: phasing X-ray diffraction data by molecular replacement using X-ray and nuclear magnetic resonance models. J Mol Biol. 1995;247:360–72. doi: 10.1006/jmbi.1994.0144. [DOI] [PubMed] [Google Scholar]
- 35.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–25. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 36.Canutescu AA, Dunbrack RL., Jr. Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci. 2003;12:963–72. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–52. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 38.Dunbrack RL, Jr., Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–81. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engh RA, Huber R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta. Cryst. 1991;A47:392–400. [Google Scholar]
- 40.Wang C, Schueler-Furman O, Baker D. Improved side-chain modeling for protein-protein docking. Protein Sci. 2005;14:1328–39. doi: 10.1110/ps.041222905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Scheraga HA. Monte Carlo-minimization approach to the multiple-minima problem in protein folding. Proc Natl Acad Sci U S A. 1987;84:6611–5. doi: 10.1073/pnas.84.19.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garbuzynskiy SO, Melnik BS, Lobanov MY, Finkelstein AV, Galzitskaya OV. Comparison of X-ray and NMR structures: is there a systematic difference in residue contacts between X-ray- and NMR-resolved protein structures? Proteins. 2005;60:139–47. doi: 10.1002/prot.20491. [DOI] [PubMed] [Google Scholar]
- 43.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–8. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 44.Chivian D, Baker D. Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res. 2006;34:e112. doi: 10.1093/nar/gkl480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 46.Bonneau R, Strauss CE, Baker D. Improving the performance of Rosetta using multiple sequence alignment information and global measures of hydrophobic core formation. Proteins. 2001;43:1–11. doi: 10.1002/1097-0134(20010401)43:1<1::aid-prot1012>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 47.Moult J, Fidelis K, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)--round 6. Proteins. 2005;61(Suppl 7):3–7. doi: 10.1002/prot.20716. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–9. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petsko GA. The grail problem. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-comment002. COMMENT002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plewczynski D, Pas J, Von Grotthuss M, Rychlewski L. Comparison of proteins based on segments structural similarity. Acta Biochim Pol. 2004;51:161–72. [PubMed] [Google Scholar]
- 51.Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc Natl Acad Sci U S A. 2000;97:10383–8. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material