Bioimage informatics: a new area of engineering biology (original) (raw)
Abstract
In recent years, the deluge of complicated molecular and cellular microscopic images creates compelling challenges for the image computing community. There has been an increasing focus on developing novel image processing, data mining, database and visualization techniques to extract, compare, search and manage the biological knowledge in these data-intensive problems. This emerging new area of bioinformatics can be called ‘bioimage informatics’. This article reviews the advances of this field from several aspects, including applications, key techniques, available tools and resources. Application examples such as high-throughput/high-content phenotyping and atlas building for model organisms demonstrate the importance of bioimage informatics. The essential techniques to the success of these applications, such as bioimage feature identification, segmentation and tracking, registration, annotation, mining, image data management and visualization, are further summarized, along with a brief overview of the available bioimage databases, analysis tools and other resources.
Contact: pengh@janelia.hhmi.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
In the last several decades, numerous biomedical imaging techniques were developed, ranging from the whole organism level (millimeter resolution) down to the single molecule level (nanometer resolution) (Murphy, 2001; Tsien, 2003). Some of the most widely used biological imaging methods include confocal or two-photon laser scanning microscopy (LSM) (Pawley, 2006), scanning or transmission electron microscopy (EM) (Bozzola and Russell, 1999), etc. Novel imaging techniques such as PALM (Betzig et al., 2006), STORM (Rust et al., 2006), STED (Hell, 2003) that far surpass the resolution of conventional optical microscopes currently can pinpoint the location of individual proteins that are only several nanometers apart. Along with the dramatic advances of many related techniques such as image signal digitization and storage, biological tissue labeling [e.g. green fluorescent proteins (GFP) and enhanced GFP (EGFP) (Heim _et al._, 1995; Shimomura et al., 1962), Dronpa (Ando et al., 2004), Brainbow combinatorial labeling (Livet et al., 2007)], the number of biological images (e.g. cellular and molecular images, as well as medical images) acquired in digital forms is growing rapidly. Large bioimage databases such as Allen Brain Atlas (Lein et al., 2007) and the Cell Centered Database CCDB; (Martone et al., 2002) are becoming available. These image data could involve (1) two-dimensional (2D) or 3D spatial information, (2) multiple colors which may correspond to various molecular reporters, (3) 4D spatio-temporal information for developing tissues or moving cells, (4) various co-localized biological signals such as mRNA expression levels of different genes (Lein et al., 2007; Long et al., 2007b; Peng et al., 2007) or (5) other screening experiments related to RNA interference (RNAi), chemical compounds, etc. (Echeverri and Perrimon, 2006; Moffat et al., 2006; Sepp et al., 2008). Analyzing these images is critical for biologists to seek answers to many biological problems, such as differentiating cancer cell phenotypes (Long et al., 2007a), categorization of neurons (Jefferis et al., 2007), etc.
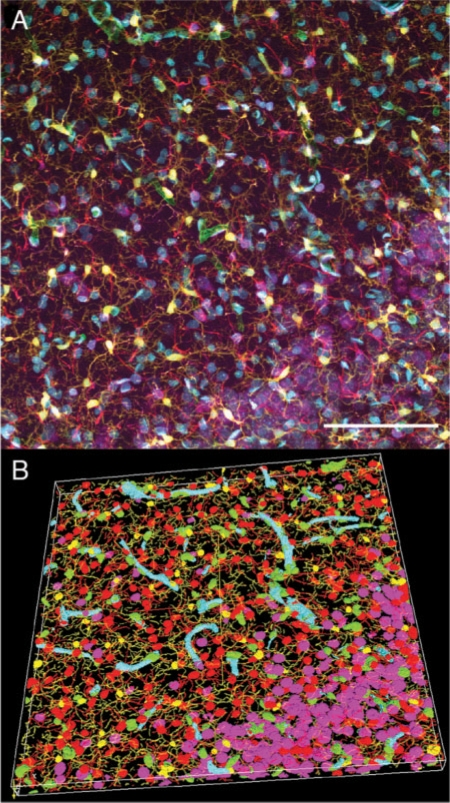
The deluge of complicated biological and biomedical images poses significant challenges for the image computing community. As a natural extension of the existing biomedical image analysis field, an emerging new engineering area is to develop and use various image data analysis and informatics techniques to extract, compare, search and manage the biological knowledge of the respective images. This new field can be called bioimage informatics. However, due to the great complexity and information content in bioimages, such as the very high density of cells (e.g. astrocytes, microglia, neurons) intertwined together (Fig. 1A), or very rapid microtubule growing process in a 4D movie of live cells, it is very challenging to directly apply existing medical image analysis methods to these bioimage informatics problems. Special techniques such as those developed in the FARSIGHT project (Roysam, 2008) will be necessary to analyze these complicated image objects (Fig. 1B). In addition, usually a single biological image stack has a large size (several hundreds of megabytes or even several gigabytes) and several color channels. The objects of interest in such an image, for instance the 3D structures of neurons, could have dramatic variations of morphology and intensity variations from image to image. It is yet not uncommon that thousands of images need to be automatically analyzed in a high-throughput way, in terms of the number of hours or days, but not months or years of manual work. All these difficulties make it necessary to develop novel bioimage informatics algorithms and systems, especially from three aspects: image processing and mining, image database and visualization.
Fig. 1.
(A) Maximum projection of a 5-channel confocal 3D image of a 100 μm thick section of rat hippocampus. Red: GFAP-labeled astrocytes; green: EBA-labeled blood vessels; yellow: Iba1-labeled microglia; cyan: CyQuant-labeled cell nuclei; purple: NeuroTrace-labeled Nissl substance; scale bar=50 μm. (B) 3D rendering (with a similar color scheme) of the segmented and classified cells produced using the FARSIGHT techniques for (A). Image courtesy of Badrinath Roysam (Bjornsson et al., 2008)
Many studies of bioimage informatics are either underway or have been done over the last few years. Several very successful workshops (e.g. bioimageinformatics.org) were organized to discuss the latest developments of this field. The goal of this essay is to briefly review the advance of bioimage informatics from the angles of applications, key techniques, available tools and resources. First, in Section 2 several application studies on the high-through biology, model organisms, etc., are introduced. Further, in Section 3 the desired computational techniques, including bioimage feature identification, segmentation, registration, annotation, mining, indexing, retrieval and visualization, are discussed. In Sections 4 and 5 the available tools and resources are summarized. While in this short article, it is difficult to include all the important work, and to explain the details of the discussed applications and computing methods (such as their biological objectives, challenges and findings), I hope that the presented facts and links can be helpful for both researchers in this field and general audiences who may have interests in learning the basic ideas of bioimage informatics.
2 APPLICATIONS
Just like many other engineering fields, bioimage informatics is application-driven, as one can see from the following non-exclusive instances.
2.1 High-throughput and high-content analysis of cellular phenotypes
Large-scale screening of cellular phenotypes, at whole-cell or sub-cellular levels, is of importance for determination of gene functions, delineating cellular pathways, drug discovery and even cancer diagnosis. The CellProfiler system (Carpenter et al., 2006; Lamprecht et al., 2007) was developed to screen cellular images rapidly and gather information such as number of cells, size and other morphological features of cells, per-cell protein levels, cell cycle distribution, etc. This system has been used to detect various cell phenotypes, such as Drosophila Kc167 cells, whose images are often textured and clumpy, and human HT29 cells, which are smooth and elliptical. Intelligent human–computer interface and content-based image retrieval relevance feedback were also used to enable high-content screening of Drosophila (fruit fly) neurons (Hong, 2006; Lin et al., 2007). Analysis of the morphological signatures of cells was used to study signaling pathways related to cell protrusion, adhesion and tension (Bakal et al., 2007).
For high-resolution intracellular analysis, 3D protein location patterns associated with a number of subcellular organelles and components such as nucleus, nucleolus, mitochondria, cytoskeleton, etc., can be described and classified using fluorescence image features, such as Haralick textures features and Zernike moments (Murphy et al., 2003). Spatial patterns may also be considered in clustering analysis and used for prediction of breast cancers (Long et al., 2007a). More systematic descriptions, such as generative models for subcellular locations of proteins, can provide information for systems biology study (Zhao and Murphy, 2007).
2.2 Atlas building for model organisms
Bioimage informatics methods were used to study widely used model organisms, such as mouse (Dorr et al., 2008; Lein et al., 2007; Ng et al., 2007), fruit fly (Luengo Hendriks et al., 2006, Luengo Hendriks et al., 2006, Peng and Myers, 2004, Peng and Myers, 2004; H. Peng et al., unpublished data), Caenorhabditiselegans (Liu et al., 2008; Long et al., 2007b), zebrafish (Megason et al., 2007), etc. One very important aspect is to build various digital atlases of these organisms, and further integrate the respective anatomical and ontological knowledge into databases.
Allen Brain Atlas (Lein et al., 2007) integrates the genome-wide RNA in situ hybridization (ISH) gene expression information of 20 000 mouse genes. Besides a manually generated reference atlas, the Anatomic Gene Expression Atlas (AGEA) is an interactive 3D atlas of the adult mouse brain based on ISH gene expression images. AGEA is based on approximately 4000 coronal gene sets, which allows anatomic specification and browsing based on 3D spatial coordinates and expression threshold control. With the pixel resolution at ∼25 μm, Allen Brain Atlas provides very useful information for studies close to the cellular level.
Single-cell analysis for an entire animal is useful for understanding the cell functions, such as the neuronal circuit mapping based on 3D cellular images of a brain. This task is possible if the cells have unique identities, indicated by the stereotypy of their 3D locations, 3D morphology, birth orders (lineages), gene expression patterns or other functional properties. Several systems do have these distinct properties. In C.elegans, each cell has a unique lineage and identity. A recent development is the building of the single-cell atlas for the L1 stage of C.elegans (Long et al., 2007b). It is based on a series of bioimage-processing and mining techniques including C.elegans worm body straightening (Peng et al., 2008a), nuclei segmentation (Long et al., 2007c), annotation and cell identification (Long et al., 2008; Peng et al., 2008b) and atlas modeling. With this atlas, systematic and high-throughput analysis of gene expression at the truly single-cell level, instead of clusters of cells, becomes feasible (Liu et al., 2008). Several other pieces of similar work are underway for different systems, e.g. a fruit fly adult brain (H. Peng et al. unpublished data).
2.3 Understanding the dynamic processes in cells and living organisms
For intracellular processes, the microtubule, one class of the cytoskeleton polymers that is constantly assembled and disassembled, receives much attention in studies of various cell functions, e.g. cell division. By imaging GFP fused to the distal ends of microtubules, it is possible to analyze the different dynamic patterns of microtubules, such as the velocity and acceleration, for mutants or under other conditions. Computationally, the microtubule growing, shortening and other dynamic patterns can be tracked in time-lapse microscopy images, via mixture analysis of hidden Markov models (Altinok et al., 2006; Altinok et al., 2007), minimum shared decomposition of directed graphs derived from the microtubule spots (Swidan et al., 2007), particle filtering (Smal et al., 2008), multiscale tip and body model (Jiang et al., 2005), detecting individual segments and linking (Danuser et al., 2000; Hadjidemetriou et al., 2004; Meijering et al., 2006). Hierarchical, agglomerative clustering analysis of various yeast mutants based on kinetochore microtubule dynamics was also reported (Jaqaman et al., 2007).
For developmental biology, visualizing how genes are expressed in living organisms allows us to gain insight in the interactions of gene products. For developing zebrafish embryos, in toto imaging based on time-lapse, LSM were used to track cells in the four dimensions of space and time (Megason et al., 2008). Image analysis methods were developed to read out quantitative, cell-based protein expression patterns and transcriptional expression patterns in vivo. The in toto imaging analysis approach is suitable for studying animal development from a systems biology perspective. For cases where it is difficult to directly observe how 3D spatial patterns of gene expression change over time, manifold learning can be used to computationally reconstruct the 4D spatio-temporal developmental dynamics of these patterns. For developing fruit fly embryos, spatial registration and comparison of 3D gene expression patterns were developed and conjugated with an approximation algorithm of the Traveling Salesman problem, to reconstruct the developing dynamics of genes such as ftz and snail (Peng et al., 2005a).
2.4 Reconstruction of 3D neuronal structures and the wiring diagram of a brain
For neuroscience, there have been a lot of efforts on tracing and reconstruction of 3D structures of neurons, based on optical and electron microcopy images. Neurolucida (Glaser and Glaser, 1990), a pioneering software package in this sort, permits users to digitally trace neuronal structures in images. Many automated approaches were developed recently. Directional kernels were used to exploratorily search neuronal topology in confocal images (Al-Kofahi et al., 2002, 2003). A repulsive force-based snake model was proposed to segment axons in 2D images and then track them in 3D confocal images of transgenic mice that express fluorescent protein (Cai et al., 2006). A graph cut method was used to segment neuronal structures in electron micrographs (Vu and Manjunath, 2008). The convolutional neural network was used to reconstruct the nanometer scale image objects from scanning electron microscopy (SEM) images (Jain et al., 2007). Several automated 3D reconstruction software packages for optical and EM images were also built (e.g. Maack et al., 2007); Y. Mishchenko, personal communication). The FARSIGHT project (Fig. 1), which targets integrating the automated 3D segmentation and tracing algorithms for astrocytes, microglia, neurons, etc., uses a systematic divide and conquer strategy for associative bioimage analysis (Bjornsson et al., 2008; Roysam et al., 2008). Thousands of reconstructed neurons have also been organized into publicly available databases, such as NeuroMorpho.org (Ascoli, 2006). Along with a number of on-going projects on categorizing the types of neuronal structures, or mapping the neuronal circuits, these resources will provide very valuable information to understand and manipulate neuronal circuits.
One of the most exciting challenges in science is to understand how a brain works. The reverse-engineering approach to tackle this problem needs to reconstruct either the anatomical wiring diagram of the brain of an animal (e.g. a fruit fly's brain with 100 000 or so neurons), or the functional wiring diagram of this brain, or both. The aforementioned 3D neuron tracing techniques, as well as image segmentation and neuron classification methods are needed to identify neurons and study their wirings based on electron, optical or functional imaging (e.g. Ca2+) data.
2.5 Joint analysis using both bioimage informatics and other bioinformatics methods
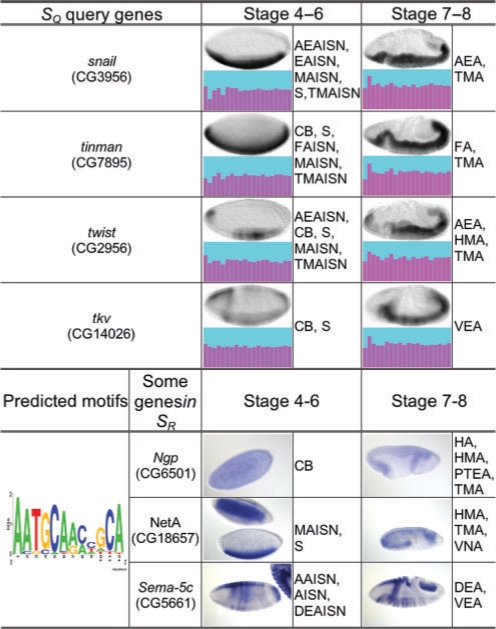
Bioimage informatics techniques can also be paired with conven-tional bioinformatics methods. For example, clustering embryonic gene expression patterns of fruit fly can be conjugated with com-parative genomics approach to predict sequence motifs that may have regulatory functions (Fig. 2) (Peng et al., 2007).
Fig. 2.
Clustering analysis of embryonic in situ mRNA gene expression patterns of fruit fly genes and its utility in assisting prediction of the regulatory sequence motifs. Based on clustering the eigen-embryo profiles (purple–cyan plot) of representative gene expression patterns, four genes in _S_Q are detected to be co-expressed genes. This prediction is consistent with their known gene regulation relationship for fly mesoderm patterning. Further, _S_Q can be used to predict sequence motifs. The motif example shown is detected using the entire upstream regions of the homologous genes in eight fly species D.melanogaster, D.simulans, D.yakuba, D.erecta, and D.ananassae, D.pseudoobscura, D.virilis, and D.mojavensis, along with three randomly selected example genes in the subsequent genome-wide motif scanning results. BDGP (fruitfly.org) ISH images (in blue) and annotations are also shown, without image cropping or orientation correction. Short terms of annotations: AAISN, amnioserosa anlage in statu nascendi; AISN, anlage in statu nascendi; AEA, anterior endoderm anlage; AEAISN, anterior endoderm anlage in statu nascendi; CB, cellular blastoderm; DEA, dorsal ectoderm anlage; DEAISN, dorsal ectoderm anlage in statu nascendi; EAISN, endoderm anlage in statu nascendi; FA, foregut anlage; FAISN, foregut anlage in statu nascendi; HMA, head mesoderm anlage; HA, hindgut anlage; MAISN, mesoderm anlage in statu nascendi; PTEA, posterior endoderm anlage; S, subset; TMA, trunk mesoderm anlage; TMAISN, trunk mesoderm anlage in statu nascendi; VEA, ventral ectoderm anlage; VNA, ventral neuroderm anlage. Original image source: (Peng et al., 2007)
Besides the above examples, several bioimage informatics applications (e.g. functional genomics) have also been discussed in recent articles such as Megason and Fraser (2007) and Meijering et al., (2006).
3 CRITICAL TECHNIQUES
In order to cope with the complexity in bioimage data, a number of image analyses, machine learning and data mining techniques are needed. Data management and visualization techniques are also required in most bioimage informatics applications. Notably, some particular problems, such as tracking of fibrous microtubule or neuronal structures, may be tackled using different methods, e.g. segmentation versus classification. Therefore, I only review the basic categories of key techniques, but explain very briefly or ignore those more complicated combinations of these basic categories, such as various techniques for modeling. Due to the length limitation, I will also have to skip the signal-processing techniques for biomedical images, such as attenuation correction, deconvolution (Heintzmann, 2007), mixture model estimation, etc., as well as techniques that may be used for general scientific computing but not limited to bioimage informatics, such as supercomputing with particular computer architecture (Rao et al., 2007).
3.1 Feature extraction and selection
Image features are the fundamental description of pixels/voxels and all higher level objects. Useful image features can correspond to statistical, geometrical, morphological properties and frequency of image pixels and regions, as well as the topological relationship of multiple image objects. Almost all bioimage-related studies rely on recognizing certain image features. For instance, points, edges, curves, corners, ridges, textures have been considered in analyzing (e.g. tracking) dynamic fluorescence images (Dorn et al., 2008).
One way to extract features is based on domain knowledge, as seen in the analyses of fruit fly embryogenesis in situ mRNA gene expression patterns. Local features based on Gaussian mixture model decomposition can be utilized to describe and compare gene expression patterns (Peng and Myers, 2004). Global decomposition based on eigen-embryo analysis can be used for clustering these patterns (Peng et al., 2006). Wavelet features that capture both global and local frequency properties of these patterns can be used to recognize these gene expression patterns and thus enable automatic annotation (Peng et al., 2007; Zhou and Peng, 2007). Other useful features, such as those obtained via independent component analysis (Pan et al., 2006) and invariant moments (Gurunathan et al., 2004), were also proposed. Another way for effective features extraction is to consider as many image transformations as possible, and thus generate a rich set of image features. For instance, Murphy et al. (2003) considered many features such as texture and moments to characterize the 3D protein location patterns associated with major subcellular organelles and structures. The WND-CHARM system (Orlov et al., 2008) of multipurpose bioimage classification uses compound image features. Five types of features, including pixel statistics, textures, polynomial decompositions, high contrast features (e.g. object number, spatial distribution, size, shape, etc.), and standard image transforms (Fourier, wavelet, Chebyshev) were produced. These features together were used to classify image patterns. One problem with the rich feature set is that it may contain redundant features, which will degrade the performance of classifiers. The minimum-redundant maximum-relevant (mRMR) feature selection algorithm (Ding and Peng, 2005; Peng et al., 2005b) has been used to determine an optimal set of least redundant features, yielding significantly improved recognition accuracy of gene expression patterns (Zhou and Peng, 2007).
3.2 Segmentation
Image segmentation is one of the most basic processing steps in many bioimage informatics applications. While the goal is simply to segment out the meaningful objects of interest in the respective image, this task is non-trivial in many cases. Very complicated cases also exist due to problems such as a low signal–noise ratio and a big variability of image objects. Remarkably, bioimage segmentation strongly depends on the features used. For example, for chromatin composition, texture features can be used, whereas for nuclear morphology, the concavity features may be considered.
Practically speaking it seems intuitive to categorize image segmentation methods for molecular and cellular images based on the overall shape of an image object. One class of segmentation problems is to segment globular objects such as nuclei/cells in 2D or 3D images of cell-based assay, where nuclear compartment may be fluorescently labeled for localization of molecules. Several widely used methods, e.g. globular-template-based segmentation, watershed segmentation, Gaussian mixture model estimation and active contour/snake methods, which can be further improved by considering different shape or intensity cues of the objects (Cong and Parvin, 1999; Han et al., 2007; Lin et al., 2003, 2005; Long et al., 2007c; Parvin et al., 2002). Gradient information will also provide useful cues in some cases (Li et al., 2007). Model-based merging was considered to reduce the over-segmentation (Lin et al., 2005; Long et al., 2007c). Note that sometimes the globular object segmentation could be very tricky, due to the irregular stains of the objects. For example, for a DAPI-stained nucleus, its nucleolus (or nucleoli) may not be stained. As a result, the nucleus will appear to be hollow. This requires special processing such as hole filling before applying the watershed (Long et al., 2007c). Watershed segmentation has also been used for EM image segmentation where the object morphology is irregular and very complicated (Y. Mishchenko, personal communication).
Non-globular object segmentation is often more complicated. One problem of interest is the tracing of neurons in optical images. Some of the latest developments were discussed earlier in Section 2.4. Generally, local search and fitting methods, such as the directional kernels (Al-Kofahi et al., 2002), 2003) have been found effective. Some of these techniques have been commercialized in neuroanatomical analysis software such as Neurolucida (http://www.mbfbioscience.com). Other available tools include the ImageJ plugin NeuronJ (Meijering et al., 2004), NeuriteTracer (Longair, 2008).
Image object tracking in fluorescent time-lapse images is another well-studied topic that relies on image segmentation. Many pieces of related work were discussed in Section 2.3.
3.3 Registration
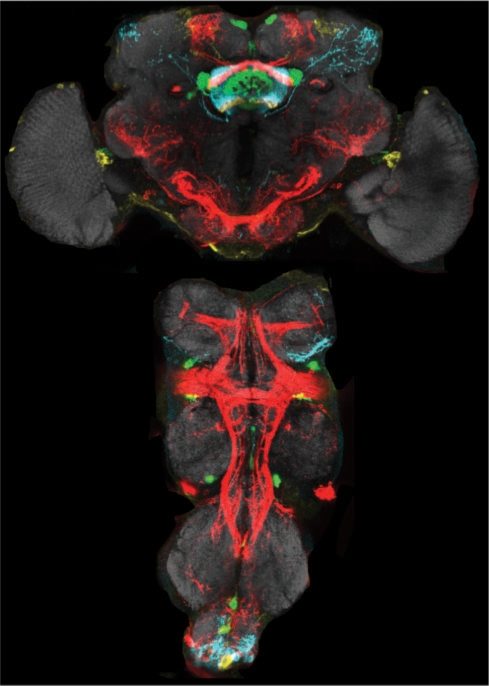
Bioimage registration is essential in many applications that need to compare multiple image subjects of different conditions. Quantitative measurements and visualization of comparing patterns in the registered images can be done directly in a ‘standard’ space. Image registration was used in applications such as building the brain atlases (Carson et al., 2005; Ng et al., 2007; Toga and Thompson, 2001), comparison of neuron morphology and gene expression patterns in fruit fly (Ahammad et al., 2005; Jefferis et al., 2007; H. Peng et al., unpublished data), cardiac imaging of Zebrafish embryos (Liebling et al., 2005), standardization of C.elegans images (Peng et al., 2008a). Figure 3 shows one example of the 3D registered fruit fly nervous system, where different GAL4 neuronal patterns highlighted in different colors are mapped into a ‘standard’ space (H. Peng et al., unpublished data). Many of the 2D and 3D image registration methods proposed for medical image analysis, such as the mutual information registration (Volla and Wells, 1997), spline-based elastic registration (Rohr et al., 2003), invariant moment feature-based registration (Shen and Davatzikos, 2002), congealing registration (Miller, 2006; Zollei et al., 2005), etc., can be extended to align the molecular and cellular images. However, due to the great complexity and variation of patterns, the big volume of images, (e.g. 2048 × 2048 × 300 pixels), and a low signal–noise ratio, 3D bioimage registration remains very challenging in general.
Fig. 3.
Maximum projection of 3D registered and overlaid neuronal patterns of multiple fruit fly central complexes (top) and thoracic ganglia (bottom), each with a different GAL4 line (Peng et al., unpublished data). Red: a205; Green: EB1; Cyan: NP2320; Yellow: NP6510; gray: NC82-labeled neuropil. Raw confocal images were produced by Julie Simpson and Phuong Chung.
Image registration will also help to produce a panoramic scene of the 2D or 3D images that correspond to tiles of tissues. This is often called montaging or tiling. In serial EM, many physical sections are generated for imaging. Each section may also be imaged as many overlapping tiles. Hence, there are two alignment problems: first, stitching all corresponding tiles into a complete single picture, and second, aligning adjacent sections if they have different orientations and deformations (e.g. stretch, shear, compression) introduced during sample preparation stages such as sectioning and fixation/dehydration/embedding. The first alignment problem can be solved via maximizing the cross-correlation of overlapping regions of neighboring tiles. The second alignment problem can be solved via finding a global 2D affine transformation for adjacent sections, followed by slight local non-linear deformation. Many previous tutorials provide the details (Szeliski, 2006).
Sometimes registration needs to be considered in the domain of extracted image objects, besides aforementioned pixel-domain image alignment. For fruit fly blastoderm embryos, each nucleus can be described using a point in the 3D space. Point cloud registration method was used to generate a virtual fruit fly embryo (Fowlkes et al., 2008). The fairly broad expression patterns of the reference markers, such as the transcriptional factor evenskipped which is expressed as seven stripes around an embryo, and the non-trivial variation of the number of nuclei (in the ±10% range), make it difficult to achieve the single nucleus accuracy for the registered point clouds. For C.elegans and the embryonic central nervous system of fruit fly, both the single-cell-level automatic cell recognition technique (Long et al., 2008) and 3D annotation tool WANO (Peng et al., 2008b) have been developed to determine the identities of cells/nuclei and produce digital point-cloud atlases at single-cell/nucleus resolution.
3.4 Clustering, classification and annotation
Many applications such as phenotyping cells and determination of subcellular locations of proteins require the pattern clustering and classification techniques (Arif and Rajpoot, 2007; Chen et al., 2006; Newberg and Murphy, 2008). Multiresolution classification of HeLa cells was proposed (Chebira et al., 2007). Graph-partition-based clustering, such as the minimum-spanning-tree-cut (Peng et al., 2006), was used to group potentially in situ mRNA expression patterns of co-regulated genes and thus to detect sequence motifs (Peng et al., 2007). Pattern classification can also help other processing and analysis tasks, for instance the watershed segmentation and grouping of over-segmented objects (Lin et al., 2005; Long et al., 2007c). Automatic determination of cell identities (Long et al., 2008) is also developed, which uses both the absolute 3D location of cells and their relative location patterns to determine the identities of cells. This technique is essential for both high-throughput measuring gene expression level at the single-cell level and manipulating single cells based on optogenetic methods. Cell identity tracking can also be combined with temporal information, as shown in the work to trace lineage of dividing embryonic cells of C.elegans (Bao et al., 2006).
Annotation of bioimage objects converts the image content information to concrete semantically meaningful information that is usually texts and can be conveniently organized and searched. This task is often accomplished manually, such as the anatomical and ontological annotation of the gene expression patterns collected for about 5000 fruit fly genes in BDGP database (www.fruitfly.org). Automatic annotation of bioimage patterns has begun to be studied (Peng et al., 2007; Zhou and Peng, 2007). Bioimage patterns could correspond to many (e.g. 100 or more) anatomical and ontological annotation terms. Thus this problem can be formulated as pattern classification with hundreds of mutually non-exclusive classes, which falls outside of the framework of conventional multiclass classification that involves a much smaller number (e.g. 10) of mutually exclusive classes. This challenging annotation problem can be solved via parallel classifiers, each performing a bi-classification to indicate if a specific target annotation term should be assigned to the image pattern or not (Zhou and Peng, 2007).
3.5 Indexing and retrieval
Currently there are two ways to access the bioimage data in databases. The prevailing method is to provide and organize the text descriptors. These metadata are indexed and thus searchable. They serve as the proxy to find the real image data. Existing relational database indexing and searching techniques can be used. Comparison of biological image patterns is often complicated due to the lack of standards in nomenclature; therefore, it will be a big advantage if annotations stored in a bioimage database are organized based on the controlled/standard ontological vocabulary. The web-based annotation system for fruit fly gene expression patterns in BDGP (Tomancak et al., 2002) provides a set of controlled ontological words used by the curator to assign to an image displayed. Of note, techniques of biomedical ontology and semantic web techniques (www.semanticweb.org) can be naturally blended into bioimage databases. New ontology systems were introduced, e.g. subcellular anatomy of the nervous system (Larson et al., 2007).
The second way is to enable content-based access of the image data in term of raw and processed data. Comparing image patterns requires aforementioned feature extraction, selection and data clustering and classification methods. Various distance metrics, such as Euclidean distances and the earth mover's distance (EMD) (Peleg et al., 1989), can be considered. Recent work (Ljosa et al., 2006) shows that a multiresolution LB-index approach can be used to index the EMD scores. Lower bounds were derived to compute EMD at various resolutions. This approach led to faster similarity query than conventional methods for a database of fluorescent confocal retina images consisting of microglial cells and blood vessels. Query and retrieval on the probability density functions, which may be modeled by adaptive-piecewise-linear approximations, have been developed (Ljosa and Singh, 2007).
3.6 Visualization
Bioimage visualization is a subfield of the general scientific data visualization. The widely used techniques for both the original and processed bioimages are volume, surface, flow visualization. Tools for interactive processing and visualization of images for protein surfaces, retinal optical coherence tomographic data and gene expression images of early stage fruit fly embryogenesis were recently developed (Staadt et al., 2007). Scalable volume visualization was used to study cell lineage and gene expression of developing C.elegans embryos (Cedilnik et al., 2007). On the other hand, immersive visualization systems, where a user walks into the data volume/model, may enable one to analyze the data like playing a video game. A few systems, such as NCMIR's ATLAS in silico system that utilizes CalIT2’s 100-million-pixel autosterographic display (West, 2007), the ImmersaDeskTM system (Ai et al., 2005), etc., support such immersive visualization, which requires virtual reality methods.
4 AVAILABLE TOOLS
Many tools have been developed for various aspects of the above techniques as well as applications. Some popular tools are summarized below.
4.1 Image formats and I/O
Microscope vendors use different file formats to store their raw image data. In addition, users may add customary metadata/tags to the raw or processed images. It is very useful to be able to read, write and convert different file formats. ImageJ (http://rsb.info.nih.gov/ij/, Abramoff et al., 2004), empowered by a number of free codes/plugins contributed by volunteers, has the ability to read and write a number of bioimage file formats, such as the Bio-rad PIC file, Zeiss LSM file and others.
Embedding the reading/writing engines of different bioimage formats in one's own code is also desirable. One useful standalone Java library for importing/exporting various bioimage data is Bio-Formats (http://www.loci.wisc.edu/ome/formats.html). It can be used in ImageJ, Matlab, etc. With the ability to parsing both pixels and metadata for a large number of formats, it finds a range of applications in the bioimage informatics. Sometimes these images, such as the Zeiss LSM files, are variants of the TIFF images, therefore can be handled using the open-source libtiff library (http://www.libtiff.org/).
4.2 Image analysis tools
ImageJ (http://rsb.info.nih.gov/ij/, Abramoff et al., 2004) is a Java-based cross-platform tool for biomedical image processing and measurement. A number of image analysis toolboxes such as fluorophore tracking, filament detection, etc., were developed by various groups (Unser, 2008). ImageJ is not only useful for daily use to view and small-scale analysis of images, but can also be deployed to run large-scale analysis in batch.
ITK (www.itk.org, (Yoo et al., 2002) provides a number of image segmentation and registration functions. In this category similar tools include the Matlab image-processing toolbox and other third party toolbox such as the DIPimage toolbox (www.diplib.org).
More and more sophisticated bioimage analysis tasks need tools to perform heavy duty image tasks such as 3D registration of animals’ brains, 3D automatic neuron tracing, etc. Several projects are currently underway, such as V3D (H. Peng et al., unpublished data), which tries to integrate a suite of convenient 3D image segmentation, registration, standardization and visualization tools to improve the efficiency of the workflow. Several labs have begun to use the alpha test version of V3D to study the fruit fly nervous systems at embryonic, larval and adult developmental stages. ZFIQ (Zebrafish Image Quantitator) (Liu et al., 2008) is another toolkit, which provides a set of image analysis tools for quantitative, reproducible and accurate interpretation of zebrafish imaging data. Cell-ID (Gordon et al., 2007), an open-source cell finding and tracking package, was developed first for yeast cells, can be used for other regularly shaped cells as well. Other useful analysis packages include CellProfiler (Carpenter et al., 2006; Lamprecht et al., 2007), STARRYNITE (Bao et al., 2006), Neuron Image Quantitator (neuroniq.cbi-platform.net) and those listed at the NCMIR site (http://ncmir.ucsd.edu/downloads/software).
4.3 Database and annotation tools
OME (Open Microscopy Environment) (openmicroscopy.org) (Swedlow et al., 2003) is a microscopic image and metadata management system. It is divided into several parts, the OME server, which implements image-based analysis or cellular localization and phenotypes, as well as an OME-XML schema language, and OMERO, which is a suite of java-based tools for data storage, management and annotation.
The UCSB Bisque system (http://dough.ece.ucsb.edu/bisquik/) provides an integrated online environment for users to upload, search, edit and annotate images. It also includes a few analysis and visualization modules.
Several other systems can also build images database and manage tens of thousands of images and associated metadata entries in a scalable way; examples include XNAT (Extensible Neuroimaging Archive Toolkit, www.xnat.org) (Marcus et al., 2007), Biotrue CDMS (www.biotrue.net) and Axiope e-CAT (www.axiope.com).
Annotating segmented image objects in 3D is another interesting topic. One tool available is WANO (Peng et al., 2008b), http://research.janelia.org/peng/proj/wano/index.html), a QT-based cross-platform 3D annotator, which provides a spreadsheet of all segmented 3D-image objects linked to both the 3D view of the raw image and that of the segmentation mask. WANO enables a user to quickly add or edit the annotations such as cell names/properties in images, as well as editing the segmentation results such as adding or removing segmented objects. This tool has been used to build digital atlases of C.elegans and fruit fly (Long et al., 2007b).
4.4 Visualization tools
For visualization of multidimensional multicolor images, such as confocal image stacks, commercially available products include Amira (Mercury), Volocity (Improvision), etc. Free visualization tools include Voxx (www.nephrology.iupui.edu/imaging/voxx/), Chimera (www.cgl.ucsf.edu/chimera/), Volume Rover (cvcweb. ices.utexas.edu/software/), many ImageJ plugins, etc. Blender (http://www.blender.org/) is often considered in rendering models.
For displaying and browsing large 2D/3D image set such as the stitched EM sections, each of which could easily exceed the size 100 000 pixels by 100 000 pixels, some tools such as Zoomify (http://www.zoomify.com/) and HDView (Microsoft) can be used to build the atlas view of a big image, similar to the Google map.
To develop visualization systems, many studies have relied on VTK (www.vtk.org), which provides multilanguage interfaces to a rich set of visualization functions. For heavy-duty visualization tasks such as large volume rendering, people may consider using OpenGL or even GPU programming directly. For building of cross-platform GUI, QT (http://trolltech.com/products/qt) and Java are often considered.
5 OTHER RESOURCES
5.1 Bench test datasets
There are a number of bioimage databases available for various model organisms, including for example: the Allen Brain Atlas database (www.brain-map.org) with genome-wide in situ gene expression patterns for the mouse brain; the interactive and multiresolution database for scanned and annotated images of serial sections of both primate and non-primate brains (Brainmaps.org); the BDGP database (www.fruitfly.org) containing in situ embryogenesis gene expression patterns of about 5000 fruit fly genes; the GFP expression pattern database for C.elegans (gfpworm.org) and the ZFin FishNet (www.fishnet.org.au, Bryson-Richardson, 2007) that is a 3D database of zebrafish development from the early embryo to adult.
For different disciplines, there are also many established databases, such as the 3D neuronal structure database Neuromorpho (neuromorpho.org) (Ascoli, 2006), which arranges the neuronal structures based on animal species, brain regions, neuron types, research labs, etc., and also provides useful neuron structure measuring, comparison and visualization tools. Similarly useful databases include CCDB (ccdb.ucsd.edu), which provides a venue for sharing and mining cellular and subcellular data derived from light and electron microscopy, including correlated imaging. CCDB provides the raw data, reconstructed and segmented data for download and includes 2D images and animations. Another interesting database is PSLID (pslid.cbi.cmu.edu), a database of protein subcellular location images. This database collects 2D through 5D fluorescence microscope images, annotations and derived features in a relational schema. There are also efforts to establish some general bench test datasets. Some authors have contributed data for the OME bench test database currently with about 10 datasets (ome.grc.nia.nih.gov/iicbu2008/). The Biomedical Informatics Research Network (BIRN, www.nbirn.net) is a multisite collaboration to facilitate data sharing of different labs; biomedical images and associated metadata of various animal models are available for downloading.
5.2 Conferences, special issues and books
There is an increasing interest for research meetings in this new area. The 2005 Bioimage Informatics meeting was held at Stanford University (bioimageinformatics.org). The 2008 meeting at UC Santa Barbara attracted about 150 frontier researchers in this field. The upcoming conference in 2009 will be held at Janelia Farm Research Campus, Howard Hughes Medical Institute. Many other events include workshops on Microscopic Image Analysis with Applications in Biology (miaab.org), several workshops related to bioimage analysis in the annual IEEE ISBI conferences (biomedicalimaging.org), NIST workshop on 2D/3D image content representation, analysis and retrieval (www.nist.gov), etc.
There are several special issues of journals and books on the topics of bioimage informatics, molecular and cellular image analysis, etc. BMC Cell Biology published a special issue in 2007 (http://www.biomedcentral.com/1471-2121/8?issue=S1), including nine papers covering new image analysis and mining algorithms, data visualization, biological applications, enabling supercomputing techniques, and computer vision and machine learning methods to solve other biology problems. It also includes a short summary of the bioimage informatics challenges (Auer et al., 2007), including the demand for bioimage informatics techniques, the need of multiscale imaging, collaboration and communication between biologists and engineers, common bioimage informatics problems and bench test datasets and modeling. Other special editions include for example the IEEE Transactions on Image Processing 2005 special issue on Molecular and Cellular Bioimaging (edited by Murphy, R, Meijering, E. and Danuser, G.), etc. Artech Publishing House is going to publish a book on the Microscopic Image Analysis for Life Science Applications in 2008 (edited by Rittscher, J., Machiraju, R. and Wong, S.).
6 DISCUSSIONS AND CONCLUSION
While in the earlier sections, the molecular and cellular images are emphasized, many of the techniques can be used for other biological image or video data. Characterizing the behaviors of living animals in videos relies on a similar set of tracking techniques to phenotyping and tracking microtubule activities. Recent developments include tracking of C.elegans, fruit fly, mouse and fish (Armstrong, 2005; Branson and Belongie, 2005; Fontaine et al., 2007; Fontaine et al., 2008; Fry et al., 2003; Geng et al., 2004); R. Kerr, personal communication; (Roussel et al., 2007; Tsechpenakis et al., 2007). Other examples, include the analysis of gel and microarray images (Angulo and Serra, 2003; Jung and Cho, 2002; White et al., 2005; Young et al., 2004), etc.
Remarkably bioimage computing methods are also demanded to improve the quality and throughput of novel digital imaging techniques, e.g. the super-resolution PALM (Betzig et al., 2006) and correlative microscopy (Grabenbauer et al., 2005; Robinson et al., 2001). It is also possible to adaptively acquire fluorescence microscopic images with consideration of image classification accuracy (Merryman and Kovačević, 2005).
The ultimate evaluation standard of bioimage informatics is how these computational techniques can be used to enhance our understanding of the biological entities and ability to solve the respective problems. With a number of new computing tools and databases that are increasingly shared by different research labs, this new engineering biology field will see a boom in the coming years.
Supplementary Material
[Supplementary Data]
ACKNOWLEDGEMENTS
I thank Fuhui Long, Yuriy Mishchenko, Ting Zhao and the Associate Editor Jonathan Wren for all the suggestions, comments and criticisms that help improve the article significantly, Margaret Jefferies for improvement of the technical writing. I also thank Badrinath Roysam for providing Figure 1, Julie Simpson and Phuong Chung for generating the raw images used for Figure 3, and the anonymous reviewers for suggesting several references.
Conflict of Interest: none declared.
REFERENCES
- Abramoff MD, et al. Image processing with ImageJ. Biophoto. Int. 2004;11:36–42. [Google Scholar]
- Ahammad P, et al. Joint nonparametric alignment for analyzing spatial gene expression patterns in Drosophila imaginal discs. IEEE CVPR 2005. 2005;2:20–25. [Google Scholar]
- Ai Z, et al. Reconstruction and exploration of three-dimensional confocal microscopy data in an immersive virtual environment. Comput. Med. Imaging Graph. 2005;29:313–318. doi: 10.1016/j.compmedimag.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Al-Kofahi K, et al. Rapid automated three-dimensional tracing of neurons from confocal image stacks. IEEE Trans. Inf. Technol. Biomed. 2002;6:171–187. doi: 10.1109/titb.2002.1006304. [DOI] [PubMed] [Google Scholar]
- Al-Kofahi K, et al. Median based robust algorithms for tracing neurons from noisy confocal microscope images. IEEE Trans. Inf. Technol. Biomed. 2003;7:302–317. doi: 10.1109/titb.2003.816564. [DOI] [PubMed] [Google Scholar]
- Altinok A, et al. Activity analysis in microtubule videos by mixture of hidden Markov models. IEEE CVPR. 2006;2:1662–1669. [Google Scholar]
- Altnok A, et al. Model based dynamics analysis in live cell microtubule image. BMC Cell Biol. 2007;8(Suppl. 1):S4. doi: 10.1186/1471-2121-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanda M, et al. Automated microarray image analysis toolbox for MATLAB. Bioinformatics. 2005;21:3578–3579. doi: 10.1093/bioinformatics/bti576. [DOI] [PubMed] [Google Scholar]
- Ando R, et al. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- Angulo J, Serra J. Automatic analysis of DNA microarray images using mathematical morphology. Bioinformatics. 2003;19:553–562. doi: 10.1093/bioinformatics/btg057. [DOI] [PubMed] [Google Scholar]
- Arif M, Rajpoot N. Classification of potential nuclei in prostate histology images using shape manifold learning. IEEE Int. Conf. Machine Vision. 2007:113–118. [Google Scholar]
- Ascoli G. Mobilizing the base of neuroscience data: the case of neuronal morphologies. Nat. Rev. Neurosci. 2006;7:318–324. doi: 10.1038/nrn1885. [DOI] [PubMed] [Google Scholar]
- Auer M, et al. Development of multiscale biological image data analysis: review of 2006 international workshop on multiscale biological imaging, data mining and informatics, Santa Barbara, USA (BII06) BMC Cell Biol. 2007;8(Suppl. 1):S1. doi: 10.1186/1471-2121-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C, et al. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–1756. doi: 10.1126/science.1140324. [DOI] [PubMed] [Google Scholar]
- Bao Z, et al. Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2006;103:2707–2712. doi: 10.1073/pnas.0511111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bjornsson CS, et al. J. Neurosci. Methods. in press; 2008. Associative image analysis: a method for automated quantification of 3D multi-parameter images of brain tissue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzola J, Russell LD. Electron Microscopy. 2nd edn. Jones & Bartlett Publishers; 1999. [Google Scholar]
- Branson K, Belongie S. Tracking multiple mouse contours (without too many samples) Proceedings of the IEEE CVPR 2005. 2005:1039–1046. [Google Scholar]
- Bryson-Richardson R, et al. FishNet: an online database of zebrafish anatomy. BMC Biol. 2007;5:34. doi: 10.1186/1741-7007-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess HA, Granato M. Modulation of locomotor activity in larval zebrafish during light adaptation. J. Exp. Biol. 2007;210:2526–2539. doi: 10.1242/jeb.003939. [DOI] [PubMed] [Google Scholar]
- Cai H, et al. Repulsive force based snake model to segment and track neuronal axons in 3D microscopy image stacks. NeuroImage. 2006;32:1608–1620. doi: 10.1016/j.neuroimage.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JP, et al. A digital atlas to characterize the mouse brain transcriptome. PLoS Comp. Biol. 2005;1:e41. doi: 10.1371/journal.pcbi.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedilnik A, et al. Integration of information and volume visualization for analysis of cell lineage and gene expression during embryogenesis. Proc. SPIE. 2007;6809 [Google Scholar]
- Chebira A, et al. A multiresolution approach to automated classification of protein subcellular location images. BMC Bioinformatics. 2007;8:210. doi: 10.1186/1471-2105-8-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, et al. Automated segmentation, classification, and tracking of cancer cell nuclei in time-lapse microscopy. IEEE Trans. Biomed. Eng. 2006;53:762–766. doi: 10.1109/TBME.2006.870201. [DOI] [PubMed] [Google Scholar]
- Cong G, Parvin B. Model based segmentation of nuclei. IEEE CVPR'99. 1999:23–25. [Google Scholar]
- Danuser G, et al. Tracking differential interference contrast diffraction line images with nanometre sensitivity. J. Microsc. 2000;198:34–53. doi: 10.1046/j.1365-2818.2000.00678.x. [DOI] [PubMed] [Google Scholar]
- Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J. Bioinform. Comput. Biol. 2005;3:185–205. doi: 10.1142/s0219720005001004. [DOI] [PubMed] [Google Scholar]
- Dorn JF, et al. Computational processing and analysis of dynamic fluorescence image data. Methods Cell Biol. 2008;85:497–538. doi: 10.1016/S0091-679X(08)85022-4. [DOI] [PubMed] [Google Scholar]
- Dorr AE, et al. Neuroimage. in press; 2008. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user's guide. Nat. Rev. Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- Fontaine E, et al. Model-based tracking of multiple worms and fish. 2007 In ICCV Workshop on Dynamical Vision. [Google Scholar]
- Fontaine E, et al. Automated visual tracking for studying the ontogeny of zebrafish swimming. J. Exp. Biol. 2008;211:1305–1316. doi: 10.1242/jeb.010272. [DOI] [PubMed] [Google Scholar]
- Fowlkes C, et al. A quantitative spatiotemporal atlas of gene expression in the drosophila blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- Fry S, et al. The aerodynamics of free-flight maneuvers in Drosophila. Science. 2003;300:495–498. doi: 10.1126/science.1081944. [DOI] [PubMed] [Google Scholar]
- Gelasca ED, et al. Proceedings of the IEEE ICIP 2008. San Diego, CA: 2008. Evaluation and benchmark for biological image segmentation. [Google Scholar]
- Geng W, et al. Automatic tracking, feature extraction and classification of C. elegans phenotypes. IEEE Trans. Biomed. Eng. 2004;51:1811–1820. doi: 10.1109/TBME.2004.831532. [DOI] [PubMed] [Google Scholar]
- Giuliano K, et al. Advances in high content screening for drug discovery. Assay Drug Dev. Technol. 2003;1:565–577. doi: 10.1089/154065803322302826. [DOI] [PubMed] [Google Scholar]
- Glaser JR, Glaser EM. Neuron imaging with Neurolucida – a PC-based system for image combining microscopy. Comput. Med. Imaging Graph. 1990;14:307–317. doi: 10.1016/0895-6111(90)90105-k. [DOI] [PubMed] [Google Scholar]
- Glory E, Murphy RF. Automated subcellular location determination and high throughput microscopy. Dev. Cell. 2007;12:7–16. doi: 10.1016/j.devcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Gordon A, et al. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat. Methods. 2007 doi: 10.1038/nmeth1008. [DOI] [PubMed] [Google Scholar]
- Grabenbauer M, et al. Correlative microscopy and electron tomography of GFP through photooxidation. Nat. Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- Gurunathan R, et al. Identifying spatially similar gene expression patterns in early stage fruit fly embryo images: binary feature versus invariant moment digital representations. BMC Bioinformatics. 2004;5:202. doi: 10.1186/1471-2105-5-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjidemetriou S, et al. Automatic quantification of microtubule dynamics. Proceedings of the IEEE ISBI 2004. 2004 [Google Scholar]
- Han J, et al. Segmentation of mammosphere structures from volumetric data. IEEE ISBI 2007. 2007:524–527. [Google Scholar]
- Heim R, et al. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Heintzmann R. Estimating missing information by maximum likelihood deconvolution. Micron. 2007;38:136–144. doi: 10.1016/j.micron.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Hell SW. Toward fluorescence nanoscopy. Nat. Biotechnol. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- Heward JA, et al. Proceedings of Measuring Behavior 2005, 5th International Conference on Methods and Techniques in Behavioral Research. The Netherlands: Wageningen; 2005. flyTracker: real-time analysis of insect courtship; pp. 409–410. August 30–September 2 2005. [Google Scholar]
- Hong P. CA, USA: Santa Barbara; 2006. Interactive analysis of high-content cellular images via relevant feedback. [Google Scholar]
- Jain V, et al. Supervised learning of image restoration with convolutional networks. ICCV 2007. 2007 [Google Scholar]
- Jaqaman K, et al. Phenotypic clustering of yeast mutants based on kinetochore microtubule dynamics. Bioinformatics. 2007;23:1666–1673. doi: 10.1093/bioinformatics/btm230. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, et al. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, et al. Automated extraction of microtubules and their plus-ends. IEEE Workshop on Applications of Computer Vision. 2005:336–341. [Google Scholar]
- Jung H.-Y, Cho H.-G. An automatic block and spot indexing with k-nearest neighbors graph for microarray image analysis. Bioinformatics. 2002;18:S141–S151. doi: 10.1093/bioinformatics/18.suppl_2.s141. [DOI] [PubMed] [Google Scholar]
- Lamprecht MR, et al. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Larson SD, et al. A formal ontology of subcellular neuroanatomy. Front. Neuroinform. 2007;1:3. doi: 10.3389/neuro.11.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li G, et al. 3D cell nuclei segmentation based on gradient flow tracking. BMC Cell Biol. 2007;8:40. doi: 10.1186/1471-2121-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebling M, et al. Four-dimensional cardiac imaging in living embryos via postacquisition synchronization of nongated slice sequences. J. Biomed. Opt. 2005;10 doi: 10.1117/1.2061567. eid 054001. [DOI] [PubMed] [Google Scholar]
- Lin C, et al. Boston, MA, USA: 2007. Intelligent interfaces for mining large-scale rnai-hcs image databases. In IEEE 7th International Conference on Bioinformatics and Biomedical Engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, et al. A hybrid 3-d watershed algorithm incorporating gradient cues & object models for automatic segmentation of nuclei in confocal image stacks. Cytometry. 2003;56A:23–36. doi: 10.1002/cyto.a.10079. [DOI] [PubMed] [Google Scholar]
- Lin G, et al. Hierarchical, model-based merging of multiple fragments for improved 3-D segmentation of nuclei. Cytometry. 2005;63A:20–33. doi: 10.1002/cyto.a.20099. [DOI] [PubMed] [Google Scholar]
- Liu T, et al. ZFIQ: a software package for zebrafish biology. Bioinformatics. 2008;24:438–439. doi: 10.1093/bioinformatics/btm615. [DOI] [PubMed] [Google Scholar]
- Liu X, et al. Molecular signatures and gene expression at the single cell level in C. elegans. Stanford University Technical Report. 2008 [Google Scholar]
- Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Ljosa V, Singh AK. APLA: indexing arbitrary probability distributions. Proceedings of the 23rd International Conference on Data Engineering (ICDE). 2007 [Google Scholar]
- Ljosa V, et al. Indexing spatially sensitive distance measures using multi-resolution lower bounds. Proceedings of the 10th International Conference on Extending Database Technology. 2006:865–883. [Google Scholar]
- Long F, et al. Phenotype clustering of breast epithelial cells in confocal images based on nuclear protein distribution analysis. BMC Cell Biol. 2007a;8(Supp.1):S3. doi: 10.1186/1471-2121-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, et al. A 3D digital cell atlas for the first larval stage of C. elegans hermaphrodite. HHMI JFRC Technical Report. 2007b [Google Scholar]
- Long F, et al. Automatic segmentation of nuclei in 3D microscopy images of C. elegans. Proceedings of the IEEE ISBI 2007. 2007c:536–539. [Google Scholar]
- Long F, et al. Lecture Notes in Computer Science: Research in Computational Molecular Biology. Berlin, Heidelberg: Springer; 2008. Automatic recognition of cells (ARC) for 3D images of C. elegans; pp. 128–139. [Google Scholar]
- Longair M. 2008. [Google Scholar]
- Luengo Hendriks CL, et al. 3D morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biol. 2006;7:R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maack N, et al. 3D reconstruction of neural circuits from serial EM images. 31st Göttingen Neurobiology Conf. 2007;31:1195. [Google Scholar]
- Marcus DS, et al. The extensible neuroimaging archive toolkit (XNAT): an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Martone ME, et al. A cell centered database for electron tomographic data. J. Struct. Biol. 2002;138:145–155. doi: 10.1016/s1047-8477(02)00006-0. [DOI] [PubMed] [Google Scholar]
- Megason S, Fraser S. Imaging in systems biology. Cell. 2007;130:784–795. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- Megason SG, et al. The digital fish project – in toto imaging and fliptraps for digitizing development. FASEB J. 2008;22:253.3. [Google Scholar]
- Meijering E, et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry. 2004;58A:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Meijering E, et al. Tracking in molecular bioimaging. IEEE Signal Proc. Mag. 2006:46–53. [Google Scholar]
- Merryman TE, Kovačević J. An adaptive multirate algorithm for acquisition of fluorescence microscopy data sets. IEEE Trans. Image Proc. 2005;14:1246–1253. doi: 10.1109/tip.2005.855861. [DOI] [PubMed] [Google Scholar]
- Miller E. Data driven image models through continuous joint alignment. IEEE Trans. Pattern Anal. Mach. Intell. 2006;28:236–250. doi: 10.1109/TPAMI.2006.34. [DOI] [PubMed] [Google Scholar]
- Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Murphy DB. Fundamentals of Light Microscopy and Electronic Imaging. Wiley–Liss Inc; 2001. [Google Scholar]
- Murphy RF, et al. Robust numerical features for description and classification of subcellular location patterns in fluorescence microscope images. J. VLSI Sig. Proc. 2003;35:311–321. [Google Scholar]
- Newberg J, Murphy RF. A framework for the automated analysis of subcellular patterns in human protein atlas images. J. Proteome Res. 2008;9 doi: 10.1021/pr7007626. [DOI] [PubMed] [Google Scholar]
- Ng L, et al. Neuroinformatics for genome-wide 3-d gene expression mapping in the mouse brain. IEEE/ACM Trans. Comput. Biol.Bioinform. 2007;4:382–393. doi: 10.1109/tcbb.2007.1035. [DOI] [PubMed] [Google Scholar]
- Pan JY, et al. Automatic mining of fruit fly embryo images. Proceedings of the 12th ACM SIGKDD 2006. 2006 [Google Scholar]
- Parvin B, et al. BioSig: an imaging bioinformatic system for studying phenomics. Computer. 2002;35:65–71. [Google Scholar]
- Parvin B, et al. Iterative voting for inference of structural saliency and localization of subcellular structures. IEEE Trans. on Image Process. 2007;16:615–623. doi: 10.1109/tip.2007.891154. [DOI] [PubMed] [Google Scholar]
- Pawley JB. Handbook of Biological Confocal Microscopy. 3rd edn. Berlin: Springer; 2006. [Google Scholar]
- Peleg S, et al. A unified approach to the change of resolution: space and gray-level. IEEE Trans. Pattern Anal. Mach. Intell. 1989;11:739–742. [Google Scholar]
- Peng H, Myers EW. Proceedings of the RECOMB 2004. USA: San Diego; 2004. Comparing in situ mRNA expression patterns of Drosophila embryos; pp. 157–166. [Google Scholar]
- Peng H, et al. 2005 Drosophlia Meeting. CA: San Diego; 2005a. Reconstructing a developmental time series of 3D gene expression patterns in Drosophila embryos. Available at http://research.janelia.org/peng/papersall/docpdf/2005_FlyMeeting_poster.pdf. [Google Scholar]
- Peng H, et al. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005b;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- Peng H, et al. Clustering gene expression patterns of fly embryos. Proceedings of the IEEE ISBI 2006. 2006:1144–1147. [Google Scholar]
- Peng H, et al. Automatic image analysis for gene expression patterns of fly embryos. BMC Cell Biol. 2007;8(Supp. 1):S7. doi: 10.1186/1471-2121-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, et al. Straightening Caenorhabditis elegans images. Bioinformatics. 2008a;24:234–242. doi: 10.1093/bioinformatics/btm569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, et al. WANO: a 3D bioimage annotation system. HHMI JFRC Technical Report. 2008b [Google Scholar]
- Robinson JM, et al. Correlative fluorescence and electron microscopy on ultrathin cryosections: bridging the resolution gap. J. Histochem. Cytochem. 2001;49:803–808. doi: 10.1177/002215540104900701. [DOI] [PubMed] [Google Scholar]
- Rohr K, et al. Spline-based elastic image registration, integration of landmark errors and orientation attributes. Comput. Vis. Image Underst. 2003;90:153–168. [Google Scholar]
- Roussel N, et al. A computational model for C. elegans locomotory behavior: application to multi-worm tracking. IEEE Trans. Biomed. Eng. 2007;54:1786–1797. doi: 10.1109/TBME.2007.894981. [DOI] [PubMed] [Google Scholar]
- Roysam B, et al. The FARSIGHT project: associative multi-dimensional image analysis methods for optical microscopy. In: Rittscher J, et al., editors. Microscopic Image Analysis for Life Science Applications. Artech Publishing House; 2008. [Google Scholar]
- Rust M, et al. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp K, et al. From flies to mice: identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet. 2008 doi: 10.1371/journal.pgen.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: heirarchical attribute matching mechanism for elastic registration. IEEE Trans. Med. Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Shi J, Malik J. Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2000;22:888–905. [Google Scholar]
- Shimomura O, et al. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell Comp. Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- Smal I, et al. Particle filtering for multiple object tracking in dynamic fluorescence microscopy images: application to microtubule growth analysis. IEEE Trans. Med. Imaging. 2008 doi: 10.1109/TMI.2008.916964. [DOI] [PubMed] [Google Scholar]
- Swedlow JR, et al. Informatics and quantitative analysis in biological imaging. Science. 2003;300:100–102. doi: 10.1126/science.1082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidan F, et al. MAD: minimum shared decomposition of DAGs for multitarget tracking. HHMI JFRC Technical Report. 2007 [Google Scholar]
- Szeliski R. Image alignment and stitching: a tutorial. 2006;2:1–104. [Google Scholar]
- Toga AW, Thompson PM. The role of image registration in brain mapping. Image Vis. Comput. 2001;19:3–24. doi: 10.1016/S0262-8856(00)00055-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. research0088.1-0088.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsechpenakis T, et al. Tracking C. elegans populations in fluid environments for the study of different locomotory behaviors. In. Proceedings of the MIAAB 2007. 2007 [Google Scholar]
- Tsien RY. Imagining imaging's future. Nat. Rev. Mol. Cell Biol. 2003;4:SS16–SS21. [PubMed] [Google Scholar]
- Unser M. Advanced image processing and analysis using ImageJ. In: Davos GR, editor. 8th European Light Microscopy Initiative Meeting. Switzerland: 2008. May 27–30, 2008. [Google Scholar]
- Viola P, Wells WM. Alignment by maximization of mutual information. Int. J. Comput. Vis. 1997;24:137–154. [Google Scholar]
- Vu N, Manjunath BS. Proceedings of the IEEE ICIP 2008. San Diego, CA.: 2008. Graph cut segmentation of neuronal structures from transmission electron micrographs. October 2008. [Google Scholar]
- West RG. ACM SIGGRAPH 2007 Art Gallery. Vol. 225. California: San Diego; 2007. ATLAS in silico. August 05–09, 2007. [Google Scholar]
- Yang Q, Parvin B. Harmonic cuts and regualrized centroid transform for localization of subcellular structures. IEEE Trans. Bioeng. 2003;50:469–476. doi: 10.1109/TBME.2003.809493. [DOI] [PubMed] [Google Scholar]
- Yoo TS, et al. Engineering and algorithm design for an image processing API: a technical report on ITK – the insight toolkit. In: Westwood J, editor. Proceedings of the Medicine Meets Virtual Reality. Amsterdam: IOS Press; 2002. pp. 586–592. [PubMed] [Google Scholar]
- Young N, et al. GelScape: a web-based server for interactively annotating, manipulating, comparing and archiving 1D and 2D gel images. Bioinformatics. 2004;20:976–978. doi: 10.1093/bioinformatics/bth033. [DOI] [PubMed] [Google Scholar]
- Zhao T, Murphy RF. Automated learning of generative models for subcellular location: building blocks for systems biology. Cytometry. 2007;71A:978–990. doi: 10.1002/cyto.a.20487. [DOI] [PubMed] [Google Scholar]
- Zhou J, Peng H. Automatic recognition and annotation of gene expression patterns of fly embryos. Bioinformatics. 2007;23:589–596. doi: 10.1093/bioinformatics/btl680. [DOI] [PubMed] [Google Scholar]
- Zollei L, et al. Efficient population registration of 3D data. 2005 In ICCV Workshop on Computer Vision for Biomedical Image Applications: Current Techniques and Future Trends. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]