Subunit Organization of Mcm2-7 and the Unequal Role of Active Sites in ATP Hydrolysis and Viability (original) (raw)
Abstract
The Mcm2-7 (_m_ini_c_hromosome _m_aintenance) complex is a toroidal AAA+ ATPase and the putative eukaryotic replicative helicase. Unlike a typical homohexameric helicase, Mcm2-7 contains six distinct, essential, and evolutionarily conserved subunits. Precedence to other AAA+ proteins suggests that Mcm ATPase active sites are formed combinatorially, with Walker A and B motifs contributed by one subunit and a catalytically essential arginine (arginine finger) contributed by the adjacent subunit. To test this prediction, we used copurification experiments to identify five distinct and stable Mcm dimer combinations as potential active sites; these subunit associations predict the architecture of the Mcm2-7 complex. Through the use of mutant subunits, we establish that at least three sites are active for ATP hydrolysis and have a canonical AAA+ configuration. In isolation, these five active-site dimers have a wide range of ATPase activities. Using Walker B and arginine finger mutations in defined Mcm subunits, we demonstrate that these sites similarly make differential contributions toward viability and ATP hydrolysis within the intact hexamer. Our conclusions predict a structural discontinuity between Mcm2 and Mcm5 and demonstrate that in contrast to other hexameric helicases, the six Mcm2-7 active sites are functionally distinct.
Replicative helicases are essential motor proteins that use nucleoside triphosphate-fueled conformational changes to unwind duplex DNA (reviewed in reference 25). In eukaryotes, a ring-shaped heterohexameric ATPase called the _m_ini_c_hromosome _m_aintenance (Mcm2-7) complex is believed to fulfill this role (reviewed in reference 12). In combination with several other DNA replication factors (Cdc45 [31] and the GINS complex [14]), Mcm2-7 is physically present and functionally required for both the initiation and elongation phases of DNA replication (reviewed in reference 3). These properties are reminiscent of the Escherichia coli DNA replicative helicase DnaB (24) and strongly implicate Mcm2-7, possibly in combination with Cdc45 or the GINS complex, as the eukaryotic replicative helicase (reviewed in reference 2).
In contrast to homohexameric helicases, Mcm2-7 is formed from six different and individually essential subunits (numbered 2 to 7) that are all members of the AAA+ ATPase family (20). This unique situation makes Mcm2-7 an attractive system for studying the coordination between ATP hydrolysis and motor function, since mutant complexes can be engineered with defined alterations at precise locations within the complex for study (e.g., see reference 29). The differences between the six subunits are likely of functional significance, since each Mcm subunit forms a conserved and essential gene family dating back to the earliest evolutionary split between eukaryotes and the archaea (19).
Although Mcm2-7 awaits structural analysis, precedence to other AAA+ members suggests that Mcm ATPase active sites are formed in trans from a conserved Walker A and B motif from one subunit and a catalytically essential “arginine finger” from the other (11, 20). Five unique dimeric combinations likely corresponding to ATPase active sites have been identified (9). For one dimer, composed of Mcm3 and Mcm7 (Mcm3/7), mutational analysis demonstrated that this dimer is a true active site with a typical AAA+ composition (9, 29). However, it is unknown if the remaining dimers have a similar active-site arrangement.
Unlike a typical homohexameric helicase in which the six active sites participate equally (e.g., see references 10 and 13), the ATPase active sites in Mcm2-7 may be functionally nonequivalent. Although historically Mcm2-7 lacks in vitro helicase activity, an Mcm subcomplex specifically containing only Mcm4, -6, and -7 (Mcm467 complex) has a weak helicase activity (18, 22, 23). This observation differentiates between Mcm subunits involved in helicase activity (Mcm4, -6, and -7) and those that lack helicase activity (Mcm2, -3, and -5). In addition, the five putative Mcm active-site dimers have a wide range of ATPase activities (9). However, both observations involve Mcm subcomplexes, and the relevance of these data to the intact Mcm2-7 hexamer is unknown.
To compare the activity of the isolated dimers to their corresponding activity within the Mcm2-7 hexamer, we generated and studied mutants within the six Walker B and arginine finger motifs. In addition to Mcm3/7, at least two additional active sites, Mcm7/4 and Mcm6/2, function in a combinatorial nature similar to that of other AAA+ proteins. Our data support a specific subunit architecture for Mcm2-7 that contains a physical discontinuity between Mcm2 and Mcm5. We also find a good correlation between the varied ATPase activity of isolated dimers and their contribution toward ATPase activity and viability within the Mcm2-7 hexamer. These data demonstrate that unlike the case with other hexameric helicases, active sites within Mcm2-7 contribute unequally toward activity.
MATERIALS AND METHODS
Nucleotides, buffers, and other reagents.
Radiolabeled [32P]ATP was purchased from MP Biomedical or Perkin Elmer. Unlabeled ATP was obtained from GE Healthcare, and its concentration was calculated from the absorbance at 259 nm. Buffer S/0.1 was 25 mM potassium-HEPES (pH 7.3), 100 mM potassium chloride, 5 mM magnesium acetate, 50 μM zinc acetate, 100 μM EDTA, 10% glycerol, 0.02% NP-40, and 1 mM dithiothreitol. Oligonucleotides were obtained from IDT (Coralville, IA). All other reagents were of the highest available purity.
MCM mutagenesis and cloning.
Details of mutagenesis and plasmid construction are available upon request. Briefly, the mcm Walker B and arginine finger alleles of Saccharomyces cerevisiae (Table 1) were generated by megaprimer mutagenesis (27) and verified by complete sequencing of each respective mcm gene. As was done previously (29), these alleles were cloned into three different vectors: a TRP1 ARS/CEN vector for genetic complementation testing, which expresses a hemagglutinin (HA)/His10 C-terminal epitope-tagged version of each allele under the control of the MCM5 promoter; a LEU2 integration vector for dominance testing, which expresses each allele under the inducible GAL1-10 promoter and adds a C-terminal 3× HA, 3× Myc, or FLAG epitope tag as needed; and bacmid vectors to generate recombinant baculovirus for protein expression (Invitrogen Bac-to-Bac system). These C-terminal epitope tags have been previously shown not to interfere either with Mcm complex formation during recombinant protein production or with viability during genetic complementation (29). Yeast strains used in this study are bar1 lys2 derivatives of S. cerevisiae W303 (32).
TABLE 1.
MCM alleles
| Allele | Sequencea |
|---|---|
| Walker B mutant | |
| mcm2DENQ | 604-LIDEFDKMN |
| mcm3DENQ | 470-CIDEFDKMT |
| mcm4DENQ | 629-CIDEFDKMS |
| mcm5DENQ | 477-CIDEFDKMR |
| mcm6DENQ | 636-CIDEFDKMD |
| mcm7DENQ | 521-CIDEFDKMD |
| MCM consensus | IDEFDKM |
| Arg finger mutant | |
| mcm2R676A | 673-LSRFD |
| mcm3R542A | 539-LSRFD |
| mcm4R701A | 698-LSRFD |
| mcm5R549A | 546-LSRFD |
| mcm6R708A | 705-MSRFD |
| mcm7R593A | 590-LSRFD |
| MCM consensus | SRF |
Protein expression and purification.
All S. cerevisiae Mcm2-7 protein preparations were expressed in baculovirus-infected insect cells and contained a C-terminal His10 epitope tag on Mcm4 and -7 with the other subunits being untagged. They were purified through metal affinity, gel filtration, and ion exchange chromatography; the final fractions were pooled and dialyzed against S/0.1, and the protein concentration was quantified from Sypro orange-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels as done previously (29). Mcm monomers and dimers containing C-terminal polyhistidine tags as detailed in the figure legends were similarly expressed and purified.
Protein characterization.
The final Mcm preparations were characterized by several criteria. The relative abundance of Mcm subunits within each preparation was quantified either directly following Sypro orange staining of SDS-PAGE gels or by quantitative Western blot analysis as done previously (4). Subunit oligomerization was analyzed by gel filtration analysis (4). The stability of Mcm subunit interactions was examined by either immunoprecipitation (4) or pull-down experiments targeting the polyhistidine tag on specific Mcm subunits. For the pull-down experiments, small aliquots of chelating Sepharose Fast Flow resin (Amersham Biosciences) were treated with either 0.1 M NiSO4 (charged) or double-distilled water (uncharged) and then equilibrated in binding buffer (1× phosphate-buffered saline [pH 7.5], 10% glycerol). About 0.35 μg of Mcm dimer was incubated with 20 μl of the 50% chelating Sepharose slurry at room temperature for 20 min. The beads were washed with excess binding buffer and the bound proteins eluted by with SDS-PAGE loading buffer. The samples were separated by electrophoresis and visualized by Sypro orange staining. These pull-down experiments routinely retain >70% of the input protein, and mock experiments using uncharged resin retain <1% of input protein. Analysis indicates that the majority of subunits in Mcm2-7 preparations form high-molecular-weight complexes consistent with hexameric size, and identification and quantitation of the least-prevalent Mcm subunit in each preparation (usually Mcm2) indicates that ≥30 to 50% of these complexes contain all six Mcm subunits (see Fig. S1 in the supplemental material). The characterization of Mcm dimers will be presented below.
ATPase assay.
Steady-state ATP hydrolysis assays were performed as described previously (29). In short, known amounts of the Mcm protein (0.2 to 1 μg) were incubated with 5 mM unlabeled ATP spiked with a small amount of [α32P]ATP in ATPase buffer for predetermined periods of time at room temperature. Samples of each time point were stopped by addition of SDS, ATP was separated from ADP by thin-layer chromatography, the ratios of radiolabeled ATP to ADP were quantified using a phosphorimager, and the resulting reaction rate was plotted as a function of time. ATP hydrolysis under these conditions is linear with respect to time and protein concentration (29). Results are reported as the number of molecules of ATP hydrolyzed/Mcm complex/min (turnover) and represent the average for at least two independent sets of assays performed in duplicate.
RESULTS
Isolation of Mcm dimers.
Mcm2-7 forms a toroidal hexamer; if active sites form at dimer interfaces, the complex should contain six specific dimeric active sites. Although a previous study assembled five distinct Mcm dimers from purified subunits (9), we have observed that S. cerevisiae Mcm4 and Mcm5 are poorly soluble in isolation (below), raising the possibility that some potential dimer combinations might have been missed. As an alternative approach, we assayed stable dimer formation following coexpression of all possible subunit combinations within a common cytoplasm. Each of the 15 pairwise combinations of Mcm-encoding baculovirus were individually used to coinfect insect cells. In each case, only one of the two subunits contained a C-terminal polyhistidine (10×) affinity tag, a modification that does not compromise cell growth or viability (29).
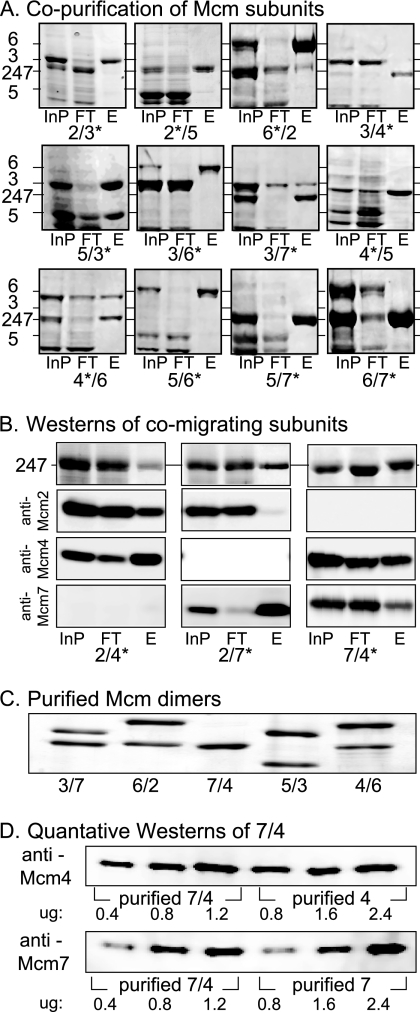
To assay Mcm subunit association, the coexpressed Mcms were subjected to metal affinity chromatography and the retention of both subunits was examined (Fig. 1A). Since Mcm2, -4, and -7 resolve poorly by SDS-PAGE, Western blot analysis was used to assess the subunit composition of these combinations (Fig. 1B). Although Mcm2/4 demonstrated a low level of association upon the initial purification (Fig. 1B, left), this apparent interaction was lost upon additional purification (data not shown) and was excluded from further analysis. In total from among the 15 possible subunit combinations, 5 dimeric associations were recovered for further study: Mcm3/7, -6/2, -7/4, -4/6, and -5/3. Since this experimental procedure enriches for His-tagged subunits irrespective of their physical association to the untagged subunits, these preparations were further purified to homogeneity. Quantification of band intensities from these purified dimeric preparations (Fig. 1C) or by quantitative Western blots of the Mcm7/4 preparation (Fig. 1D) indicates that the two subunits within each final preparation have nearly equal stoichiometry, with an excess of one subunit over the other of ≤30%.
FIG. 1.
Identification and ATPase activity of stable Mcm dimeric assemblies. (A) SDS-PAGE gel of column fractions following metal chelate chromatography of coexpressed Mcm subunits. The subunit labeled with an asterisk contains the C-terminal polyhistidine tag. InP, input extract; FT, column flowthrough; E, purified eluate fraction. (B) Association of subunits that comigrate in SDS-PAGE. The analysis is the same as that for panel A, except that Western blotting using subunit-specific antibodies was performed. Top row, indicated fractions stained with Coomassie blue; bottom three rows, Western blots with indicated Mcm antibody. (C) Silver-stained SDS-PAGE of purified stable dimeric Mcm subunits. (D) Quantitative Western blots to measure subunit stoichiometry within the purified Mcm7/4 dimer.
Physical characterization of dimeric Mcm preparations.
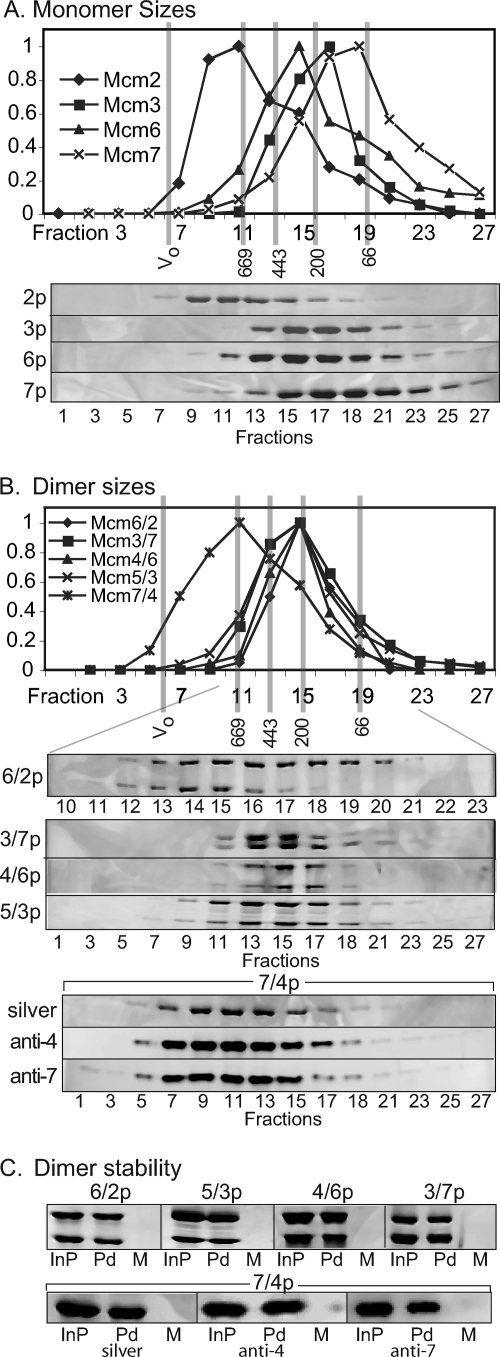
Gel filtration was used to confirm the physical association between Mcm subunits and determine their oligomerization state. Isolated Mcm subunits were first examined to provide a point of reference. The six His-tagged Mcm subunits were individually expressed in our baculovirus system. Preparations of Mcm2, -3, -6, and -7 yielded soluble proteins that were subsequently purified to homogeneity, while Mcm4 and Mcm5 were largely insoluble and their analysis was not further pursued (data not shown). Based upon their estimated molecular masses (which range from 86 kDa [Mcm5] to 113 kDa [Mcm6]), the resulting elution volumes of these single-subunit preparations were consistent with a molecular mass in the monomer/dimer range for Mcm3 and -7 and in the dimer/trimer range for Mcm6, while the elution volume of Mcm2 was consistent with a hexameric oligomerization (Fig. 2A). These results are consistent with the previous observation that isolated Mcm subunits have a variable propensity to oligomerize (9).
FIG. 2.
Oligomerization and stability of Mcm dimers. (A) Gel filtration of the indicated Mcm single-subunit preparations. (B) Gel filtration of final purified dimeric preparations. In both panels A and B, the elution volumes of the indicated molecular weight markers are shown; Sypro orange staining or Western blot analysis of the indicated fractions is shown. (C) Pull-downs of indicated Mcm dimers by metal affinity chromatography. InP, input protein; Pd, pull-down of protein retained on the beads; M, mock, represents analogous pull-down experiment conducted without charging the affinity resin with nickel.
Aliquots of the final dimeric preparations were next analyzed. To claim stable dimerization, both participating subunits must coelute, and additionally, one or both subunits must demonstrate a different elution profile from that of the corresponding single-subunit preparation. By this criterion, physical interaction between Mcm subunits was demonstrated for four of the five dimeric preparations. Mcm3/7, -5/3, and -6/2 eluted as apparent dimers, whereas Mcm7/4 eluted as a hexamer (Fig. 2B). Physical association between subunits in the Mcm4/6 preparation could not be confirmed, since the elution of Mcm4 was not examined, and the resulting Mcm4/6 preparation eluted at the same position as Mcm6 alone. The sizes of our Mcm3/7, -5/3, and -6/2 complexes were smaller than previously observed, possibly reflecting a structural difference between dimers formed by in vivo coexpression (this study) as opposed to in vitro reconstitution (9, 29). In contrast, the hexameric size of Mcm7/4 was significantly larger than previously reported (9), although it should be noted that a significant amount of Mcm7/4 eluted at a hexameric size in this prior study.
To confirm the stability of these subunit associations, pull-down experiments that target the polyhistidine epitope tags on an individual subunit were conducted (Fig. 2C). Quantitation of the two subunits within each preparation either by Sypro staining of SDS-PAGE gels or by quantitative Western blotting (e.g., with Mcm7/4) indicate for all of the preparations except Mcm4/6 approximately the same subunit stoichiometry in the pull-down fraction as in the input (≤30% excess of one subunit over another). Although these experiments confirm subunit association within the Mcm4/6 dimer, the pull-down recovered approximately twice as much Mcm6 as Mcm4, suggesting that this dimer may be relatively unstable. These experiments are in good agreement with our gel filtration results and with the possible except of Mcm4/6 indicate a good physical association between subunits in these preparations.
The involvement of the Walker B motif in the Mcm3/7 active site.
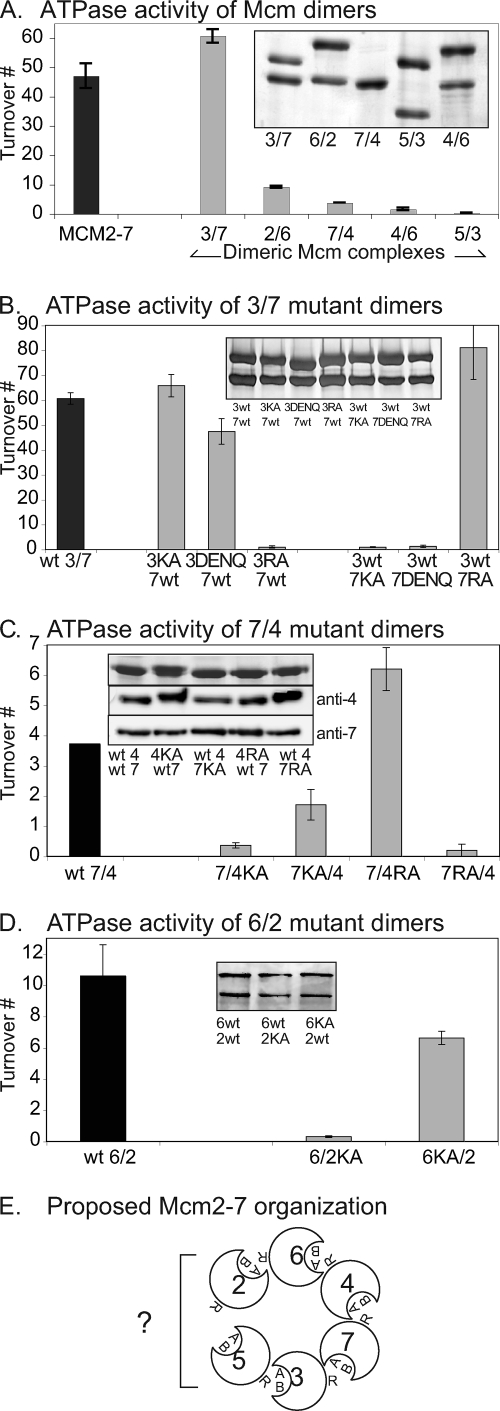
The resulting five dimeric complexes were assayed for ATP hydrolysis (Fig. 3A) and were found to differ greatly in activity: Mcm3/7 demonstrated high ATPase activity, similar in magnitude to that of the intact Mcm2-7 hexamer, Mcm6/2 and Mcm7/4 had moderate activity, and Mcm5/3 and -4/6 nearly lacked activity. Since no stimulation of ATPase activity had been previously observed upon addition of single-stranded DNA (ssDNA) to either the intact Mcm2-7 complex (29) or isolated dimers (9), ssDNA was omitted from these reactions. These levels of ATP hydrolysis in our dimer preparations are similar to those in the previous study (9), confirming the validity of our experimental approach.
FIG. 3.
Contribution of ATPase motifs to active-site function. (A) ATPase activities of stable dimeric preparations. MCM2-7, activity of the wild-type Mcm2-7 heterohexameric preparation. (B to D). ATPase activities of Mcm dimers containing the indicated mutant or wild-type (wt) Mcm subunits. Insets show silver-stained SDS-PAGE gels or Western blot analysis of equivalent amounts of the final preparations. (E) Proposed subunit organization of Mcm2-7.
To study the composition of ATPase motifs in individual active sites, appropriate mcm mutations were generated (see Materials and Methods; Table 1). Mutations in the Mcm Walker A (KA) and arginine finger (RA) motifs have been described previously (9, 29). Since previous attempts to study Mcm Walker B motifs with alleles containing a single aspartate-to-alanine substitution (DEFD→AEFD) proved unsuccessful (29), double mutations were constructed that changed the first two acidic residues into their uncharged amide counterparts (DE→NQ, referred to subsequently as DENQ alleles). In contrast to the previous Walker B alleles (29), the DENQ alleles in five of the six subunits proved lethal (Table 2), consistent with the hypothesis that they functionally ablate the corresponding active site.
TABLE 2.
MCM mutant phenotypes
| Expt | Gene | Result for wild type | Growth of mutant | ||
|---|---|---|---|---|---|
| Walker Ac | Walker B | Arg finger | |||
| Genetic | MCM2 | + | − | ± | − |
| complementationa | MCM3 | + | − | − | − |
| MCM4 | + | − | − | + | |
| MCM5 | + | − | − | − | |
| MCM6 | + | − | + | − | |
| MCM7 | + | − | − | − | |
| Overexpression | MCM2 | − | + | − | +++ |
| lethalityb | MCM3 | − | − | ± | +++ |
| MCM4 | − | +++ | − | − | |
| MCM5 | − | + | ± | + | |
| MCM6 | − | +++ | − | − | |
| MCM7 | − | +++ | +++ | +++ |
Dimeric complexes containing either the Mcm3 DENQ or Mcm7 DENQ mutation were expressed, purified, assayed for ATPase activity (Fig. 3B), and compared to analogous dimers that contain either the KA mutations (29) or the RA mutations (9). As previously shown, Mcm7 contributes the catalytically essential Walker A motif, while Mcm3 contributes the essential arginine finger motif (9, 29). Our results demonstrated that Mcm7 but not Mcm3 also contributes the Walker B motif to the Mcm3/7 active site (Fig. 3B). In addition, these data confirm that our Walker B and arginine finger mutants eliminate catalysis with little or no effect on subunit association, validating them as good candidates to test the role of individual ATPase active sites within the Mcm2-7 hexamer (below).
Characterization of the Mcm7/4 and Mcm6/2 active sites and subunit architecture of Mcm2-7.
In contrast to the Mcm5/3 and Mcm4/6 dimers, the Mcm7/4 and Mcm6/2 dimers retained sufficient ATPase activity to allow their functional analysis. To study these sites, a set of mutant Mcm7/4 and Mcm6/2 dimers was expressed and purified. Although neither the Mcm6RA nor Mcm2RA mutations affected the stability of the Mcm2-7 hexamer (see Fig. S1 in the supplemental material), we were unsuccessful in purifying stable dimers containing these alleles (data not shown). The remaining dimers were assayed for ATP hydrolysis (Fig. 3C and D).
In contrast to the case with Mcm3/7, analysis of the Mcm7/4 and Mcm6/2 dimers yielded more-complex results (Fig. 3C and D). The Mcm7RA mutation specifically blocked ATP hydrolysis in Mcm7/4 and thus indicated that Mcm7 contributes the catalytically essential arginine finger residue. However, both Walker A motifs contributed to ATPase activity: the Mcm4KA mutation nearly eliminated ATPase activity, while the Mcm7KA mutation significantly reduced ATP hydrolysis. Mcm6/2 provided similar results; in this case, Mcm2KA nearly eliminated ATP hydrolysis whereas Mcm6KA noticeably reduced this activity. Assuming that the Walker A motif that has the greatest effect on ATPase activity is at the dimer interface, our results support the predicted participation of these motifs (9). Although the low ATPase activity of the Mcm5/3 and Mcm4/6 dimers prevented a similar analysis, as a working hypothesis we will assume that they represent AAA+ ATPase active sites as previously proposed (9). Hereafter, we will name each active site such that the subunit contributing the arginine finger residue comes first, followed by the subunit contributing the Walker motifs.
To summarize our data, we propose a subunit organization for the Mcm2-7 complex (Fig. 3E). Although these data predict a linear structure with Mcm2 or -5 at either end, hexamerization of Mcm2-7 into a toroid likely forces these two subunits into close juxtaposition to form an active site (see Discussion). This model is similar to that previously proposed (9).
Importance of the Walker B and arginine finger motifs to Mcm2-7 in vivo function.
The Walker A box KA mutations of Mcm2-7 have been previously analyzed and cause two effects: not only do they block ATP hydrolysis at the corresponding active site, they also block ATP hydrolysis of the remaining wild-type active sites (29). To separate the catalytic versus regulatory contributions of the Mcm active sites, alleles that uncouple these two activities are required. Studies of other ATPases have found that the Walker B and arginine finger motifs are often needed only for ATP hydrolysis, not for binding (reviewed in reference 17), suggesting that unlike the KA mutations, the latter mutations might be suitable for studying the contributions of individual active sites without blocking ATP hydrolysis of the entire complex.
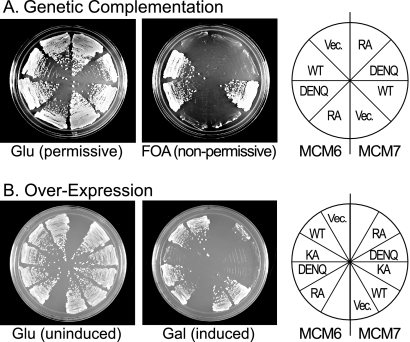
To this end, we tested the Walker B (DENQ) and arginine finger (RA) alleles of all six Mcm genes for genetic complementation in S. cerevisiae (Table 2). As an example, the phenotypes of the mcm6 and mcm7 alleles are shown (Fig. 4). Although all mcm constructs are viable in the presence of an additional plasmid-borne copy of the wild-type gene (Fig. 4A, Glu), most mcm alleles demonstrate little or no growth when the Walker B or arginine finger allele provides the sole copy of that MCM gene (Fig. 4A, FOA) (Table 2 shows the results for all 12 mutations). The lack of viability is not due to the defective expression of these alleles, since they express protein at levels comparable to those of the corresponding wild-type gene (data not shown). Interestingly, several alleles retained viability; mcm2DENQ had a slow-growth phenotype, whereas mutations in the Mcm4/6 active site (mcm4RA and -6DENQ) are apparently associated with normal growth (Table 2). Fluorescence-activated cell sorter analysis of the mcm2DENQ mutant indicates that it accumulates cells in S phase relative to its congenic wild-type strain (E. Tsai, C. Poth, and A. Schwacha, unpublished), suggesting a DNA replication defect. No other defects are evident for these three viable mcm alleles; in an otherwise wild-type strain background, they have normal viability and resistance to the DNA replication inhibitors hydroxyurea and methylmethanesulfonate (R. Elbakri and A. Schwacha, observation). These results suggest that ATP hydrolysis by the Mcm4/6 active site is dispensable for in vivo Mcm2-7 function.
FIG. 4.
Analysis of the Walker B and arginine finger alleles of mcm6 and mcm7. (A) Genetic complementation. Yeast tester strains contained chromosomal deletions of either MCM6 or MCM7 as indicated, and their viability is maintained by a URA3 plasmid expressing a wild-type (WT) copy of the corresponding MCM gene. TRP1 plasmids containing either an expressed copy of the indicated allele (wild type, DENQ, or RA) or an empty TRP1 vector (Vec.) were transformed into these strains. Glu, permissive conditions; FOA, nonpermissive conditions in which viability depends upon the indicated mcm allele. (B) Dominant overexpression of mcm alleles. The indicated mcm alleles were expressed under the inducible GAL1-10 promoter and integrated into the chromosomal LEU2 gene. Glu, uninduced conditions; Gal, induced conditions.
We also tested the DENQ and RA alleles for dominant lethality upon overexpression, a phenotype previously associated with other Mcm alleles (Table 2) (15, 29). Toward this end, the mcm Walker B and arginine finger alleles were integrated into an otherwise wild-type strain and expressed by the strongly inducible GAL promoter. As an example, analysis of the mcm6 and -7 alleles is shown (Fig. 4B). All mcm7 alleles (7KA, 7DENQ, and 7RA) demonstrate a strong dominant-lethal phenotype upon exposure to galactose, whereas among alleles of mcm6, only the mcm6KA allele demonstrates dominant lethality (Fig. 3). Table 2 shows the results with the other alleles. Dominant-negative effects were generally weak with the DENQ alleles. Only mcm2DENQ and mcm3DENQ demonstrated a slight growth defect. The RA alleles in general demonstrated a stronger dominant effect; mcm2RA and mcm3RA caused lethality, whereas mcm5RA resulted in slow growth. Note that MCM7 is unique, since it is the only subunit that demonstrates a strong dominant-lethal phenotype for all three mutations (KA, DENQ, and RA), suggesting a special role for Mcm7 within the complex (see below). With the exception of MCM7, however, our findings suggest that dominant effects are associated with particular classes of mutants rather than particular active sites.
Mcm active sites contribute differentially to ATP hydrolysis.
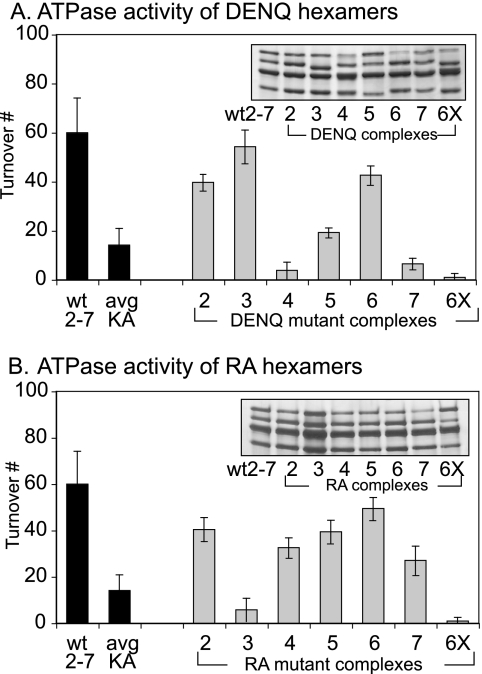
If Mcm active sites contribute equally to ATP hydrolysis, Mcm2-7 complexes containing any of the Walker B or arginine finger mutants should have similar defects in ATP hydrolysis. To directly test this hypothesis, we expressed and purified Mcm2-7 complexes that contained the specified mutant subunit in the company of five other wild-type subunits (see Materials and Methods). The resulting mutant Mcm2-7 preparations formed complexes of hexameric size that contained all six Mcm subunits (Fig. 5A; also see Fig. S1 in the supplemental material).
FIG. 5.
ATPase activity of Mcm2-7 heterohexamers containing subunits with either Walker B or arginine finger mutations. Panels A and B show the ATPase activities of Mcm2-7 complexes (2 to 7, Mcm2 to -7; 6X, all six complexes) containing the indicated mutant Walker B (A) or arginine finger allele (B) subunit in the presence of the five other wild-type (wt) subunits. Each inset shows a silver-stained gel of equivalent amounts of the indicated Mcm complexes. Avg KA, average ATPase activity of Mcm2-7 containing the Walker A mutations in any of the six individual subunits (29).
We assayed the mutant Mcm2-7 complexes for ATPase activity. To verify that the DENQ and RA alleles in fact eliminate ATP hydrolysis, we generated and assayed complexes simultaneously containing either the DENQ or RA mutations in all six subunits; these complexes are completely devoid of ATP hydrolysis (Fig. 5A and B). The single-mutant Mcm2-7 preparations were next assayed. Although most of these alleles are lethal, many of the corresponding mutant Mcm2-7 complexes demonstrate only relatively minor defects in ATP hydrolysis (Fig. 5A and B), suggesting that unlike the Walker A mutations (29), the Walker B and arginine finger mutations appear to block ATP hydrolysis only at the affected active site, rather than blocking ATP hydrolysis of the entire Mcm2-7 complex.
Five Mcm2-7 mutant preparations showed a significant loss of ATP hydrolysis. The Mcm4DENQ, Mcm7DENQ, and Mcm3RA mutant complexes were the most severely affected, whereas preparations containing either the Mcm5DENQ or Mcm7RA mutation decreased ATP hydrolysis to the same levels observed for an Mcm2-7 complex with a single Walker A mutant subunit (29). Four of these five mutations target the Mcm3/7 and -7/4 active sites, demonstrating that the bulk of in vitro ATP hydrolysis of Mcm2-7 comes from these two sites. The exception to this is the Mcm5DENQ mutation. However, unlike the other four mutant complexes, the ATPase activity of a complex containing the corresponding arginine finger allele (Mcm2RA) demonstrated only a small defect in ATP hydrolysis. This may suggest that the Mcm5DENQ mutation causes an allele-specific loss of ATP binding to this site, thus mimicking the effect of the Mcm5KA allele in blocking ATP hydrolysis from the entire Mcm2-7 complex. These results directly demonstrate that the active sites within Mcm2-7 contribute unequally to Mcm function, with the relative contribution of each active site in the hexamer toward ATP hydrolysis and in vivo function roughly corresponding to relative ATPase activities of the isolated active-site dimers (i.e., Mcm3/7 contributed more than either Mcm4/7 or Mcm6/2, which in turn contributed more than Mcm5/3 or Mcm4/6).
DISCUSSION
Our analysis of the Mcm2-7 active sites, both as isolated dimers and within the context of the Mcm2-7 heterohexamer, strongly indicates that they contribute unequally to ATP hydrolysis and viability. This suggests that in contrast to various models of helicase function (10, 13), equal involvement of the six active sites is not mechanistically essential for Mcm2-7 function.
Mcm subunit architecture.
Our dimer association experiments support a previously predicted subunit architecture and active-site configuration for Mcm2-7 (Fig. 3E) (9). Although we explicitly demonstrate canonical AAA+ ATPase active-site composition only for Mcm3/7 and Mcm7/4, these associations severely constrain the possible composition of the remaining active sites. Importantly, these dimer pairs can be arranged in a linear order, with Mcm2 and Mcm5 at either end. However, since Mcm2-7 forms a toroidal structure (1, 4, 6, 25, 28), the association of Mcm2 and Mcm5 with the remaining Mcm subunits likely drives circularization and formation of the Mcm2/5 active site. Although neither this study nor a previous study (9) provides direct evidence for the Mcm2/5 site, denaturing immunoprecipitation of Mcm subunits from Drosophila (8) and humans (35) both have identified a specific cross-linking-dependent physical interaction between Mcm2 and Mcm5 that is highly labile in the absence of cross-linking (26).
Possible coordination among Mcm ATPase active sites.
Within the Mcm7/4 and Mcm6/2 dimers, the Walker A motifs in both subunits appear to contribute to ATP hydrolysis. Since the Walker A motif typically serves to bind nucleotide (reviewed in reference 17), this result suggests that binding of ATP to the noncatalytic site of these dimers positively regulates activity at the adjoining catalytically competent site. Such a feature could be used to coordinate ATP hydrolysis among the Mcm active sites; binding and hydrolysis of ATP at one site would stimulate activity of the adjacent site. Owing to the circular nature of the Mcm complex, this would lead to a defined hydrolysis order around Mcm2-7 (discussed more below). Alternatively, in the case of Mcm7/4, which oligomerizes into a hexamer (Fig. 2B), a possible noncanonical active site might be formed between the Mcm7 Walker A motif and the Mcm4 arginine finger motif; inclusion of the Mcm7KA allele into this complex might simply block ATP hydrolysis from this putative site. Note that ATP hydrolysis at Mcm3/7 is independent of the noncatalytic Walker A motif, a fact that may be relevant to the unique functional importance of this site (below).
The unequal involvement of Mcm active sites.
We identified a correlation between the ATPase activity of isolated Mcm dimers and their functional importance within the intact Mcm2-7 complex. The Mcm3/7 and Mcm7/4 active sites demonstrate good to moderate ATP hydrolysis as isolated dimers, and analysis of the ATPase activity of Mcm2-7 complexes containing mutants that affect ATP hydrolysis at these active sites indicated that they are especially critical to hydrolysis within the complete Mcm2-7 hexamer. In contrast, Mcm4/6 lacks ATPase activity in isolation, and mutations within this site cause negligible ATPase defects within isolated Mcm2-7 complexes. This correlation between the activity of isolated dimers and Mcm2-7 mutations is not absolute, since Mcm6/2 demonstrates ATP hydrolysis as an isolated dimer but appears to contribute little to ATP hydrolysis within the Mcm2-7 hexamer. This observation suggests that the activities of individual active sites are likely constrained within the context of the hexamer. It is interesting to note that for several active sites the corresponding Walker B and arginine finger mutations have similar phenotypes, e.g., both the 3RA and 7DENQ mutant hexamers are almost completely defective for ATP hydrolysis, whereas both the 4RA and 6DENQ mutants demonstrate normal viability. This strong correlation between mutations proposed to target the same active site for both in vivo and in vitro activities provides additional support for the proposed active-site composition. Correlating our results with additional data suggests particular functions for specific Mcm active sites.
Mcm3/7- and Mcm7/4-regulated core of the Mcm motor.
Mutations within these two active sites uniquely inhibit steady-state ATP hydrolysis of Mcm2-7. Furthermore, Walker A mutations in either Mcm7 or Mcm4 uniquely inhibit the ability of Mcm2-7 to bind ssDNA in an ATP-dependent manner (4). Since both ATP hydrolysis and ssDNA interactions are directly required for DNA unwinding, our results strongly implicate these active sites in coupling of ATP hydrolysis to helicase activity. Mcm7 appears to be particularly important: it is a common subunit in both active sites, and unlike the case with any other Mcm subunit, overexpression of any of the three mcm7 ATPase mutants (mcm7KA, -DENQ, or -RA) causes lethality. The functional prominence of Mcm7 would make it a good regulatory target for modulation of the ATPase activity of the entire Mcm2-7 complex. It is interesting to note that various putative negative regulators of Mcm2-7 (DNA damage checkpoint proteins Rad17 [33], ATRIP [7, and the retinoblastoma protein [30]) specifically bind to the C terminus of Mcm7.
Mcm4/6: a structural element?
Among our ATPase alleles, only the Mcm6DENQ and the Mcm4RA alleles demonstrated a completely normal viability and growth rate (Fig. 3A; Table 2). In Schizosaccharomyces pombe, the mcm6 Walker B mutations are also viable (15). This is in sharp contrast to the mcm6KA allele, which is lethal and blocks ATPase activity of Mcm2-7 (29). Curiously, however, the Mcm6DENQ mutant blocks helicase activity from both the murine Mcm467 complex (34) and the S. cerevisiae complex (see Fig. S2 in the supplemental material). These data suggest that the Mcm6DENQ mutation (and probably the Mcm4RA mutation) does indeed block ATP hydrolysis at the Mcm4/6 site. Since the Walker B and arginine finger motifs are often involved with ATP hydrolysis, while the Walker A motif often makes larger contributions to nucleotide binding (reviewed in reference 17), these results suggest that the Mcm4/6 site may need to only bind, not hydrolyze, ATP for in vivo function. There is considerable precedence for ATP binding sites serving structural rather than catalytic roles: three of the six ATPase sites in the F1 ATPase need to bind but not hydrolyze ATP (5), and the “stator” subunit of the DNA replication processivity clamp loader retains a nonfunctional ATP active site (16, 21).
Mcm2/5: an ATP-dependent gate?
Additional data from our lab support a unique involvement of the Mcm2/5 active site in both the association rate of Mcm2-7 with ssDNA (4) and its ability to bind circular ssDNA (4a). In combination with the putative labile nature of the Mcm2/5 active site, these results are consistent with an ATP-dependent topological closure of the Mcm2-7 ring at the Mcm2/5 active site (“gate”). The functional significance of this putative gate is under investigation.
Function of other Mcm active sites.
Molecular motors couple ATP binding and hydrolysis to produce work that likely necessitates a fixed ATP hydrolysis cycle in which the participating active sites fire once in a defined order during each cycle. Our analysis indicates that Mcm6/2 and Mcm5/3 sites contribute little to in vitro ATP hydrolysis within Mcm2-7, suggesting that they are not part of a fixed ATP hydrolysis cycle that unwinds DNA. However, they are needed for in vivo function, suggesting that perhaps they have a function that is not directly related to DNA unwinding, e.g., to load the Mcms onto DNA during initiation or alternately assist unloading of the complex during termination.
Relationship between Mcm ATPase active sites and helicase activity.
Although our analysis of the Mcm Walker B and arginine finger mutants indicates the unequal involvement of these sites in Mcm2-7 function, definitive verification will require being able to study the DNA binding and helicase activities of these mutant complexes. Despite the historic inability to demonstrate helicase activity from purified Mcm2-7 (4, 18, 22, 23), a recent report demonstrating in vitro helicase activity from this complex in the absence of either the GINS complex or CDC45 (4a) offers the promise of being able to finally study the mechanistic contribution of the Mcm ATP active sites toward DNA unwinding.
Supplementary Material
[Supplemental material]
Acknowledgments
This work was supported by a postdoctoral fellowship from the Leukemia Society and a Research Scholar grant, RSG-05-113-01-CCG, from the American Cancer Society to A.S. and by the Howard Hughes Medical Institute and a grant from the NIH (GM58701) to S.P.B. S.P.B. is an employee of the Howard Hughes Medical Institute.
Footnotes
▿
Published ahead of print on 28 July 2008.
REFERENCES
- 1.Adachi, Y., J. Usukura, and M. Yanagida. 1997. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 2467-479. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio, T., A. Ibarra, and J. Mendez. 2006. Cdc45-MCM-GINS, a new power player for DNA replication. Cell Div. 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71333-374. [DOI] [PubMed] [Google Scholar]
- 4.Bochman, M. L., and A. Schwacha. 2007. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J. Biol. Chem. 28233795-33804. [DOI] [PubMed] [Google Scholar]
- 4a.Bochman, M., and A. Schwacha. 2008. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell 31287-293. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, P. D. 1993. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim. Biophys. Acta 1140215-250. [DOI] [PubMed] [Google Scholar]
- 6.Chong, J. P., M. K. Hayashi, M. N. Simon, R. M. Xu, and B. Stillman. 2000. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 971530-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortez, D., G. Glick, and S. J. Elledge. 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA 10110078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crevel, G., A. Ivetic, K. Ohno, M. Yamaguchi, and S. Cotterill. 2001. Nearest neighbour analysis of MCM protein complexes in Drosophila melanogaster. Nucleic Acids Res. 294834-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey, M. J., C. Indiani, and M. O'Donnell. 2003. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J. Biol. Chem. 278:4491-4499. [DOI] [PubMed] [Google Scholar]
- 10.Enemark, E. J., and L. Joshua-Tor. 2006. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442270-275. [DOI] [PubMed] [Google Scholar]
- 11.Erzberger, J. P., and J. M. Berger. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 3593-114. [DOI] [PubMed] [Google Scholar]
- 12.Forsburg, S. L. 2004. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 68109-131, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gai, D., R. Zhao, D. Li, C. V. Finkielstein, and X. S. Chen. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 11947-60. [DOI] [PubMed] [Google Scholar]
- 14.Gambus, A., R. C. Jones, A. Sanchez-Diaz, M. Kanemaki, F. van Deursen, R. D. Edmondson, and K. Labib. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8358-366. [DOI] [PubMed] [Google Scholar]
- 15.Gomez, E. B., M. G. Catlett, and S. L. Forsburg. 2002. Different phenotypes in vivo are associated with ATPase motif mutations in Schizosaccharomyces pombe minichromosome maintenance proteins. Genetics 1601305-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenther, B., R. Onrust, A. Sali, M. O'Donnell, and J. Kuriyan. 1997. Crystal structure of the delta′ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell 91335-345. [DOI] [PubMed] [Google Scholar]
- 17.Hanson, P. I., and S. W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6519-529. [DOI] [PubMed] [Google Scholar]
- 18.Ishimi, Y. 1997. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 27224508-24513. (Erratum, 273:23616, 1998.) [DOI] [PubMed] [Google Scholar]
- 19.Iyer, L. M., and L. Aravind. 2006. The evolutionary history of proteins involved in pre-replication complex assembly, p. 751-757. In M. L. DePamphilis (ed.), DNA replication and human disease. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Iyer, L. M., D. D. Leipe, E. V. Koonin, and L. Aravind. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 14611-31. [DOI] [PubMed] [Google Scholar]
- 21.Jeruzalmi, D., M. O'Donnell, and J. Kuriyan. 2001. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell 106429-441. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan, D. L., M. J. Davey, and M. O'Donnell. 2003. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 27849171-49182. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J. K., and J. Hurwitz. 2000. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem. 27518871-18878. [DOI] [PubMed] [Google Scholar]
- 24.Messer, W. 2002. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26355-374. [DOI] [PubMed] [Google Scholar]
- 25.Patel, S. S., and K. M. Picha. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69651-697. [DOI] [PubMed] [Google Scholar]
- 26.Sakwe, A. M., T. Nguyen, V. Athanasopoulos, K. Shire, and L. Frappier. 2007. Identification and characterization of a novel component of the human minichromosome maintenance complex. Mol. Cell. Biol. 273044-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8404-407. [PubMed] [Google Scholar]
- 28.Sato, M., T. Gotow, Z. You, Y. Komamura-Kohno, Y. Uchiyama, N. Yabuta, H. Nojima, and Y. Ishimi. 2000. Electron microscopic observation and single-stranded DNA binding activity of the MCM4,6,7 complex. J. Mol. Biol. 300421-431. [DOI] [PubMed] [Google Scholar]
- 29.Schwacha, A., and S. P. Bell. 2001. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell 81093-1104. [DOI] [PubMed] [Google Scholar]
- 30.Sterner, J. M., S. Dew-Knight, C. Musahl, S. Kornbluth, and J. M. Horowitz. 1998. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol. 182748-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tercero, J. A., K. Labib, and J. F. Diffley. 2000. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 192082-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56619-630. [DOI] [PubMed] [Google Scholar]
- 33.Tsao, C. C., C. Geisen, and R. T. Abraham. 2004. Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling. EMBO J. 234660-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.You, Z., Y. Komamura, and Y. Ishimi. 1999. Biochemical analysis of the intrinsic Mcm4-Mcm6-Mcm7 DNA helicase activity. Mol. Cell. Biol. 198003-8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, Z., D. Feng, and C. Liang. 2004. Pairwise interactions of the six human MCM protein subunits. J. Mol. Biol. 3401197-1206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]